Abstract
Variant histone H2AZ-containing nucleosomes are involved in the regulation of gene expression. In Saccharomyces cerevisiae, chromatin deposition of histone H2AZ is mediated by the fourteen-subunit SWR1 complex, which catalyzes ATP-dependent exchange of nucleosomal histone H2A for H2AZ. Previous work defined the role of seven SWR1 subunits (Swr1 ATPase, Swc2, Swc3, Arp6, Swc5, Yaf9, and Swc6) in maintaining complex integrity and H2AZ histone replacement activity. Here we examined the function of three additional SWR1 subunits, bromodomain containing Bdf1, actin-related protein Arp4 and Swc7, by analyzing affinity-purified mutant SWR1 complexes. We observed that depletion of Arp4 (arp4-td) substantially impaired the association of Bdf1, Yaf9, and Swc4. In contrast, loss of either Bdf1 or Swc7 had minimal effects on overall complex integrity. Furthermore, the basic H2AZ histone replacement activity of SWR1 in vitro required Arp4, but not Bdf1 or Swc7. Thus, three out of fourteen SWR1 subunits, Bdf1, Swc7, and previously noted Swc3, appear to have roles auxiliary to the basic histone replacement activity. The N-terminal region of the Swr1 ATPase subunit is necessary and sufficient to direct association of Bdf1 and Swc7, as well as Arp4, Act1, Yaf9 and Swc4. This same region contains an additional H2AZ-H2B specific binding site, distinct from the previously identified Swc2 subunit. These findings suggest that one SWR1 enzyme might be capable of binding two H2AZ-H2B dimers, and provide further insight on the hierarchy and interdependency of molecular interactions within the SWR1 complex.
The eukaryotic genome is packaged into chromatin that can regulate genome metabolism and gene expression (1). The fundamental unit of chromatin is the nucleosome in which ∼146 bp of DNA is wrapped around a histone octamer (2). Epigenetic regulation at the level of nucleosome is mediated by specific multiprotein histone modification or ATP-dependent chromatin remodeling enzymes (3, 4). These enzymes act by covalently modifying histones, repositioning, or displacing nucleosomes or exchanging histone variants.
The histone variant H2AZ constitutes a minor fraction of chromatin as compared with that of conventional histone H2A (5). It has a role in the regulation of gene expression, the maintenance of silent chromatin boundaries and the higher order folding of the chromatin fiber, as well as embryonic development in higher eukaryotes (6–8). Post-translational modification of chromatin-bound H2AZ by acetylation is important for its function, and there is also evidence that H2AZ acetylation facilitates its incorporation into chromatin (9–11). Localization of H2AZ in yeast, Drosophila or human by conventional chromatin immunoprecipitation (ChIP),4 high resolution tiling array (12–14) or nucleosomal DNA sequencing (15–17) in chromosome-wide or genome-wide studies reveals that H2AZ maps mostly at intergenic regions, as one or two nucleosomes near transcription start site of gene promoters.
Genetic and biochemical studies have revealed that this site-specific incorporation of H2AZ in yeast is mediated by the highly conserved Swi2/Snf2-related SWR1 ATPase complex (18–20). The purified SWR1 complex catalyzes displacement of histone H2A from conventional nucleosome arrays and its replacement with histone H2AZ (18). Enzymes with similar components and histone replacement function have also been identified in higher eukaryotes (21–24). As such, studies of the yeast SWR1 enzyme could provide insight into the functions of the related higher eukaryotic counterparts. The yeast SWR1 enzyme contains fourteen subunits: the Swr1 ATPase, Swc2, Bdf1, Swc3, Arp6, Swc5, Yaf9, Swc6, and Swc7 subunits are encoded by genes not essential for cell viability; Rvb1, Rvb2, Arp4, Swc4 (also known as Eaf2), and Act1 are encoded by essential genes (18–20). Some subunits are not unique to the SWR1 complex and thus have functions apart from SWR1. For example, Rvb1, Rvb2, Act1, and Arp4 are shared components with another ATP-dependent chromatin remodeling complex INO80 (25, 26). Act1 and Arp4, along with Swc4 and Yaf9, are also shared with the histone acetyltransferase complex NuA4 (27, 28). Bdf1 interacts with TFIID at TATA-less promoters during RNA polymerase II transcription initiation (29, 30). Given the complex nature of SWR1 and its subunits, the study of their specific roles in the SWR1 complex would benefit from biochemical analysis.
Deletion analysis of a number of nonessential SWR1 subunits has revealed that chromatin deposition of H2AZ in vivo is dependent on Swc2, Arp6, Swc6, and Yaf9 as well as the Swr1 ATPase (19, 31). Further studies have begun to address how individual subunits are involved in H2AZ deposition (32). Because the SWR1-mediated H2AZ replacement process is a stepwise reaction consisting of assembly of the SWR1 complex, binding to substrates, followed by H2A-H2B eviction, and H2AZ-H2B deposition, the individual subunits of the SWR1 complex may be important during any step of the reaction. By examining their role in SWR1 complex assembly or integrity, and binding and transfer of H2AZ in vitro, we have previously uncovered Swc2 as an essential subunit required for direct binding and transfer of H2AZ into chromatin. Swc6 and Arp6 are necessary for the association of Swc2 to the ATPase domain of Swr1. Other subunits, Swc5 and Yaf9, are required for H2AZ transfer but are involved in steps separate from H2AZ binding, while the Swr1 ATPase subunit also provides a structural framework for assembly of the entire complex (32). However, the role of several other SWR1 subunits including Bdf1, Swc7 and Arp4 in the assembly of the SWR1 complex and the H2AZ replacement process remained to be explored.
In addition to binding to TFIID, Bdf1 has two bromodomains that bind to acetylated histone H3 and H4 tails (33, 34). Bromodomains occur frequently in ATP-dependent chromatin remodeling complexes and their presence, for example, in the RSC chromatin remodeling complex, enhance the binding of RSC to acetylated nucleosomes (35, 36). Genome-wide studies have showed that strains lacking Bdf1 display a reduction in site-specific incorporation of H2AZ at many promoters (12, 30). Besides its interaction with acetylated histone tails, whether Bdf1, like Swc2, Swc6, or Arp6, is required for the basic H2AZ histone replacement activity of SWR1, is unknown. Moreover, although Arp4 has a characterized role in the NuA4 and INO80 complexes (37–39), its activity in the SWR1 complex has not been examined. Finally, little is known about the smallest SWR1 subunit, Swc7. In this study, we characterized the role of Bdf1, Swc7, and Arp4 in complex assembly and H2AZ histone replacement activity. In addition, we sought to further define the molecular interactions on the N-terminal region of the Swr1 ATPase, which contains the helicase-SANT-associated (HSA) domain (40), critical for protein association, including association of actin and actin-related proteins (41, 42). We also uncovered a second H2AZ binding site located on the N-terminal region of the Swr1 ATPase subunit itself.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions—Strains used in this study are listed in supplemental Table S1. All deletion and tagged strains were confirmed by colony PCR amplification of the targeted locus. Strains YCW424 to YCW427 were generated in a previous study (32). The swc7Δ strain was generated using the hphMX dominant drug-resistance cassette transformed into parental strain MBY121 essentially as previously described (32). Three copies of the Flag epitope tag were fused to the C terminus of SWR1 in AL246 to produce strain YCW850. The temperature-sensitive lethal degron allele arp4-td was generated essentially as described in Ref. 43, with the degron cassette integrated at the start of the ARP4 open reading frame. A 3Flag-hphMX cassette5 was integrated at the C terminus of either SWR1 or HTZ1 before the stop codon to generate YCW830 or YCW831. YCW870 was created by insertion of a 3Flag-KanMX cassette and stop codon after nucleotide A2043 of SWR1.
YP glucose (YPD), YP raffinose (YPR), and YP galactose (YPG) media were prepared according to standard recipes. Growth conditions for the arp4-td strain were essentially as described (43). The arp4-td strains were grown at 24 °C in YPD medium until A600 0.8. Cells were harvested and transferred to YPR medium plus 0.5 μg/ml of doxycycline and incubated at 24 °C for 6 h, followed by incubation in YPG medium (0.5 μg/ml doxycycline) at 24 °C for 1 h. Degradation of Arp4 was induced by transferring cells to 37 °C pre-warmed YPG (20 μg/ml doxycycline) medium. Samples were taken at 0, 0.5, 1, 2, 3, 4, and 5 h and analyzed by Western blotting to determine the time when Arp4 was maximally degraded. Samples taken at 4 h were used for purification of the SWR1arp4-td complex or H2AZ chromatin immunoprecipitation (ChIP).
SWR1 Protein Complex Purification—Purification of the SWR1 complex from the arp4-td strain used 2-liter (A600 1–1.5) cells harvested after 4-h growth at 37 °C in YPG (20 μg/ml doxycycline) medium. Affinity purification of wild-type and mutant SWR1 protein complexes were as described (32). Protein complexes were purified using the standard 0.5 m KCl wash condition unless stated otherwise in the text or figure legends.
Antibody Preparation and Western Blotting—Recombinant Swr1, Bdf1, Arp4, Rvb1, Rvb2, Swc5, Yaf9, and Swc7 full-length or truncated proteins were expressed from pET28c (Novagen). The His6-tagged proteins were purified using Ni-NTA HisBind resin (Novagen). The purified proteins were used as immunogen for the preparation of polyclonal antibodies by Pocono Rabbit Farm and Laboratory.
For the analysis of subunit association of SWR1 or H2AZ association with the SWR1 complex, we loaded equivalent amounts of the Swr1 subunit, as detected by Western blotting using horseradish peroxidase-conjugated anti-Flag or anti-Swr1. Subunits in wild-type and mutant SWR1 complexes were resolved by 4–20% SDS-PAGE followed by Western blotting analysis. The PVDF membrane strips with SWR1 subunits were probed with the corresponding antibodies. Antibody against SWR1 subunits were generated and used for Western blotting at the following dilutions: anti-Bdf1, 1:10,000; anti-Arp4, 1:4,000; anti-Rvb1, 1:3,000; anti-Rvb2, 1:5,000; anti-Swc5, 1:2,000; anti-Yaf9, 1:6,000; anti-actin, 1:4000; anti-Swc7, 1: 5,000; anti-Swr1, 1:4,000. Polyclonal anti-Swc4, anti-Swc6, and anti-Arp6 were used at 1:4,000, 1:3,000, 1:4,000, respectively (32). Rabbit polyclonal anti-H2AZ was used at 1:5,000 (32). Horseradish peroxidase-conjugated anti-Flag M2 (Sigma) was used at 1:2,000 dilution. Prior to the generation of a Bdf1 antibody by the Wu laboratory, Western blotting of the Bdf1 subunit in wild-type and mutant SWR1 complexes in Fig. 1A was examined by A. Ladurner with antibodies generated in his laboratory.
FIGURE 1.
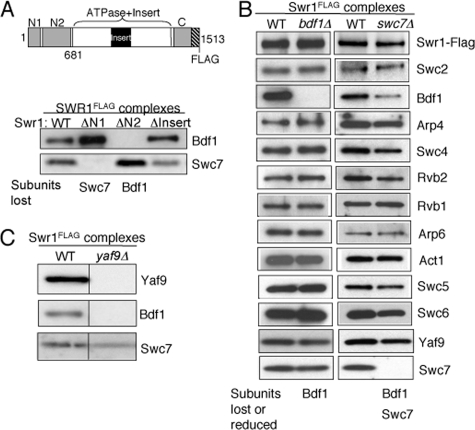
Association of Bdf1 and Swc7 is dependent on the N terminus of Swr1. A, SDS-PAGE and Western blotting analysis of Bdf1 and Swc7 in partial SWR1 complexes purified from wild-type and mutant swr1 strains as indicated. PVDF membrane strips corresponding to subunit(s) were probed with antibodies against Bdf1 and Swc7. The schematic diagram of Swr1 indicates subunit interaction domains of Swr1 and fragments used in this study: N1 (residues 1–210), N2 (residues 278–681), Insert (residues 1002–1221), Swr1 (residues 1–681). SWR1(ΔN1), SWR1(ΔN2), and SWR1(ΔInsert) are mutant SWR1 complexes isolated from cells lacking the N1, N2, and Insert regions of the Swr1 ATPase subunit, respectively (32). B, SDS-PAGE and Western blotting analysis of Swr1, Bdf1, Arp4, Swc4, Yaf9, and Swc7 in SWR1 complexes isolated from wild-type, bdf1Δ and swc7Δ strains. PVDF membranes were probed with anti-Flag (to reveal Swr1), and antibodies against individual SWR1 subunits as indicated. C, SDS-PAGE (4–20% gel) and Western blotting analysis of Yaf9, Bdf1, and Swc7 in SWR1 complexes isolated from wild-type and yaf9Δ cells. PVDF membranes were probed with antibodies against Yaf9, Bdf1, and Swc7, respectively.
H2AZ Histone Replacement Assay—The H2AZ histone replacement assay was carried out as previously described (18). Quantitative Western blotting of Flag-tagged Swr1 protein was used to normalize the amount of wild-type or mutant SWR1 complex used in the histone replacement assay.
Recombinant Protein Expression and Protein Binding Assay—Swr1 (residues 1–681) was cloned into pET100 (Invitrogen), while Swr1 (residues 371–681) and Swc7 were cloned into pET28c. Swr1 (residues 1–681) was expressed and purified from the BL-21 codon plus strain of E. coli (Stratagene). Swr1 (residues 371–681) and Swc7 were expressed and purified from the BL-21 DE3 pLysS strain of E. coli (Stratagene). The His6-tagged proteins were purified using HisPur™ cobalt resin (Thermo Scientific). Proteins were retained on the beads and stored in buffer B (25 mm HEPES-KOH, pH7.6, 1 mm EDTA, 2 mm MgCl2, 10% glycerol, 0.01% Nonidet P-40, 0.5 mm dithiothreitol, 0.1 m KCl, protease inhibitors) at 4 °C. Each protein binding reaction contained 3 μg of bead-bound His6-tagged protein and 30 ng of H2AZ-H2B or H2A-H2B dimers reconstituted from bacterially expressed proteins. The binding assay was performed as described (32). Rabbit polyclonal anti-H2B (44) was used at 1:8,000 dilution.
RESULTS
The Association of Bdf1 and Swc7 Depends on the N-terminal Region of Swr1—Previous truncation analysis of the Swr1 ATPase subunit has shown that it provides a framework for the assembly of many SWR1 subunits (32). To determine which region of Swr1 is required for the association of Bdf1 or Swc7, we purified mutant SWR1 complexes using Flag-epitope tagged Swr1 from strains containing deletions of Swr1 regions “N1,” “N2,” and “Insert,” and measured the association of Bdf1 and Swc7 in the complex (Fig. 1A). We observed little or no Bdf1 in the SWR1 complex isolated from the swr1ΔN2 cells, indicating that the association of Bdf1 requires the N2 region (residues 278–681) of Swr1. Loss of Swc7 in the SWR1(ΔN1) complex indicates that the N1 region (residues 1–210) of Swr1 is required for Swc7 association. Deletion of the Insert region did not result in loss of either Bdf1 or Swc7 (Fig. 1A), as did deletion of the entire ATPase domain (see below).
To determine the role of Bdf1 and Swc7 in the association of other SWR1 subunits, we examined the subunit composition of affinity-purified SWR1 complexes from bdf1Δ and swc7Δ strains by Western blotting. Inspection of the protein level of each SWR1 subunit in wild-type and mutant (normalized to the level of the Swr1-Flag component) revealed that Bdf1 was the only subunit absent from the SWR1 complex purified from the bdf1Δ mutant (Fig. 1B). SWR1 complexes purified from the swc7Δ mutant lacked Swc7, as expected, and retained all other examined subunits, with the exception of diminished Bdf1 (Fig. 1B). This indicates that the full association of Bdf1 with the SWR1 complex requires Swc7. A clue to how Swc7 modulates Bdf1 association is provided by examination of the SWR1(ΔN1) complex. Deletion of the N1 region of the Swr1 subunit resulted in the loss of Swc7 but not a concurrent loss of Bdf1, suggesting that the N1 region may have a negative effect on Bdf1 association that is counteracted by Swc7 (Fig. 1A (ΔN1 lane)). In summary, neither Bdf1 nor Swc7 are obligatory for the overall integrity of the SWR1 complex.
Previous analyses have shown that the N-terminal region of Swr1 that includes N1 and N2 is required for the association of Yaf9 with the SWR1 complex and that association of Swc4 requires Yaf9 through a direct interaction (32, 45). To determine whether association of Bdf1 and Swc7 with the N-terminal region of Swr1 is dependent on Yaf9 (and/or Swc4), we measured Bdf1 and Swc7 protein association in SWR1 complexes purified from the yaf9Δ strain. We observed a substantial reduction in Bdf1 level, indicating that association of Bdf1, but not Swc7 is dependent on Yaf9 (Fig. 1C).
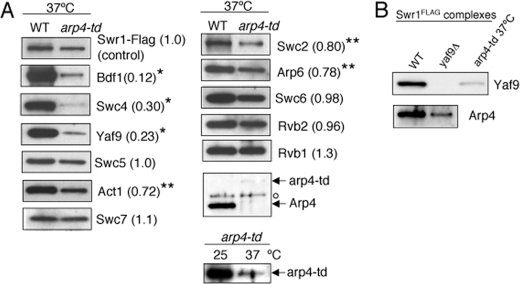
Arp4 Is Required for Association of Bdf1, Yaf9, and Swc4 with the SWR1 Complex—To better understand the hierarchical association of SWR1 subunits and the role of Arp4, which is encoded by an essential gene, we generated a conditional mutant for Arp4 (arp4-td) by tagging it with a heat-inducible degron cassette (46). The mutant arp4-td protein level was substantially reduced 30 min after shifting to the restrictive temperature (37 °C) and was undetectable after 2 h (supplemental Fig. S1A). This heat-induced protein degradation was specific to Arp4, as we did not observe general protein loss in whole cell extracts under the same growth condition (supplemental Fig. S1A). We observed that growth of the arp4-td strain was substantially impaired at the non-permissive temperature.
We next examined subunit association in the SWR1 complex purified from the arp4-td strain incubated at 37 °C. Based on the normalized ratio of the protein level of each subunit in the SWR1 complex purified from the arp4-td strain relative to wild type (see Fig. 2 legend), we divided the subunits into two groups. The first group contained subunits of which association were either unaffected or affected slightly. Association of Swc5, Swc6, Swc7, Rvb1, or Rvb2 was unaffected, while Swc2, Arp6, or Act1 was present at 70–80% of the level in the wild-type SWR1 complex, indicating that the effect of arp4-td depletion on association of these subunits is minor. The second group, Bdf1, Swc4, and Yaf9, was present at only 10 to 20% of the level observed in the wild-type SWR1 complex, indicating that these subunits are substantially dependent on Arp4 for their association (Fig. 2A). Furthermore, the association of Arp4 and Yaf9 are interdependent as shown in Fig. 2B. The decreased level of subunit association in the SWR1 complex purified from the arp4-td strain is not due to changes in expression of the corresponding subunit genes, as their mRNA levels were equivalent if not higher in arp4-td cells (supplemental Fig. S1B).
FIGURE 2.
Arp4 is required for association of several components of the SWR1 complex. A, SDS-PAGE and Western blotting analysis of SWR1 subunits in SWR1 complexes purified from wild-type and arp4-td strains as indicated. Numbers in parentheses indicate subunit abundance in arp4-td strains relative to WT, normalized to the levels of the Swr1-Flag subunit. One asterisk indicates a low level of 10–30% abundance. Two asterisks indicate a moderate level of 70–80% abundance. The open circle indicates a nonspecific signal by anti-Arp4. Level of arp4-td was also compared at 25 and 37 °C in the arp4-td strain using anti-Arp4. A trace amount of arp4-td was detectable at 37 °C when overexposed. B, SDS-PAGE and Western blotting analysis of Yaf9 and Arp4 in SWR1 complexes purified from wild type, yaf9Δ, and arp4-td strains as indicated.
Arp4 but Not Bdf1 or Swc7 Is Required for the Basic H2AZ Replacement Activity of SWR1—While the SWR1 complex is the ATP-dependent chromatin remodeling enzyme responsible for H2AZ site-specific incorporation, a number of regulatory factors including acetylated histone H3 and H4 (12, 14, 30, 47), and gene-specific transcription factors and cofactors (13, 48, 49) have been shown to contribute to efficient H2AZ incorporation in vivo. Conceivably, the purpose of these factors is to regulate global or site-specific recruitment of SWR1 and may not be required for the core or basic H2AZ replacement activity of SWR1. This latter H2AZ replacement activity is represented here as that measured by an in vitro H2AZ histone replacement assay using unmodified nucleosome arrays (18).
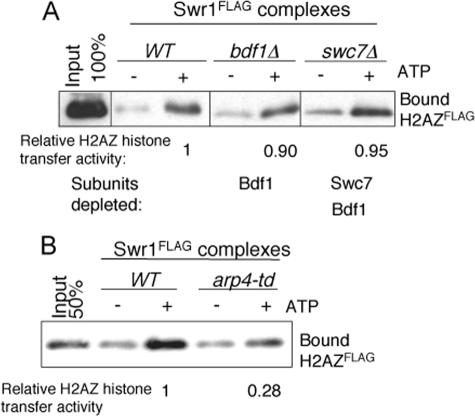
Potentially, SWR1 subunits like Bdf1, Arp4, and Swc7 could be involved as part of the basic H2AZ replacement machinery of the SWR1 complex, or they could act as interaction surfaces to regulate SWR1 recruitment and thus incorporate H2AZ at specific sites in vivo. Indeed, Bdf1 has been shown to be required for efficient deposition of H2AZ into chromatin in vivo, likely through its bromodomain binding to acetylated histone H4 tails (12, 14, 30, 47, 50). However, whether Bdf1 is necessary for the basic H2AZ replacement activity of SWR1, and whether Arp4 and Swc7 are required for H2AZ incorporation both in vivo and in vitro were not clear. To answer these questions, we first examined the in vitro H2AZ histone replacement activity of mutant SWR1 complexes purified from bdf1Δ, swc7Δ and arp4-td strains. In the assay using reconstituted unmodified nucleosome arrays, we failed to observe substantial differences in H2AZ replacement activity between wild-type SWR1 and SWR1bdf1Δ or SWR1swc7Δ enzymes, indicating that the basic H2AZ replacement activity of SWR1 is neither dependent on Bdf1 nor Swc7 (Fig. 3A). In contrast, the basic H2AZ replacement activity is dependent on Arp4, as the mutant SWR1arp4-td enzyme displayed a reduced histone H2AZ replacement activity 28% of wild type (Fig. 3B). This result suggests that Arp4 could have a direct role in H2AZ deposition, or that the Arp4 requirement is indirect, as Arp4 is needed for the association of Yaf9/Swc4, and to a minor degree, Swc2 and Arp6, subunits that have previously been shown to be required for H2AZ deposition in vivo and in vitro (19, 32). Both possibilities are not mutually exclusive.
FIGURE 3.
Arp4 but not Bdf1 or Swc7 is required for basic H2AZ replacement in vitro. SDS-PAGE and Western blotting showing H2AZ-Flag transfer from solution to immobilized, conventional nucleosome arrays (18). Equivalent amounts of SWR1 complexes measured by the Swr1 subunit were used in the reaction. The relative H2AZ histone replacement activity is determined by dividing the ratio of H2AZ signal from ATP (+) lane to the signal from ATP (–) lane from mutant SWR1 complexes by the same ratio obtained from wild-type SWR1 complexes. A, Bdf1 and Swc7 are not essential for functional H2AZ replacement in vitro. The relative H2AZ replacement activity is an average of three independent experiments. B, Arp4 is required for functional H2AZ replacement.
Consistent with its effect on basic H2AZ replacement activity of SWR1, depletion of Arp4 also affected H2AZ deposition in vivo. We observed that H2AZ deposition at regions known to be enriched for H2AZ, measured by H2AZ-Flag ChIP, was substantially reduced in the arp4-td strain grown at the non-permissive temperature (37 °C) (supplemental Fig. S2). As stated above, this effect on H2AZ deposition in chromatin could be either direct or indirect, through the loss of other subunits. Moreover, given that Arp4 is also a stable subunit of the histone acetyltransferase (HAT) complex NuA4 and NuA4 HAT activity is required for efficient chromatin H2AZ deposition (12, 14, 27), the diminished in vivo H2AZ ChIP observed in the arp4-td mutant could also be due to the combined loss of Arp4 function in SWR1 and NuA4 enzyme complexes.
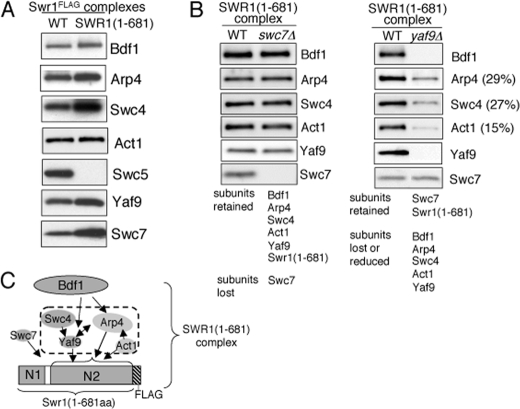
Two Major Regions of Swr1 Direct Subunit Association—Deletion analysis showed that the N-terminal region of Swr1 was necessary for association of Bdf1, Swc7 (Fig. 1A) and Arp4 (32). To determine whether it is sufficient for the association of Bdf1, Swc7, and Arp4, we purified partial SWR1 complexes from strains that contain either a chromosomal Flag-tagged Swr1 fragment corresponding to residues 1–681 of Swr1 (designated SWR1(1–681)). As a control, we purified partial SWR1 complexes from the swr1ΔN2 truncation which contains residues 1–210 fused to residues 682–1513 (designated SWR1(ΔN2)) (32). We then analyzed the subunit composition of the corresponding SWR1(1–681) or SWR1(ΔN2) complexes by Western blotting using antibodies to SWR1 subunits. We observed that Bdf1, Arp4, Swc4, Act1, Yaf9, and Swc7 but not Swc5 are associated with Swr1(1–681) (Fig. 4A), which indicates that the N-terminal region of Swr1 is indeed sufficient for association of those subunits. Western blotting analysis of SWR1(ΔN2) components confirmed previous results showing that Swc2, Rvb1, Rvb2, Arp6, and Swc6 associate with SWR1(ΔN2) (32) (supplemental Fig. S3). Taken together, these results suggest that SWR1 can be divided into two major domains, each with its repertoire of associating components (supplemental Fig. S4). The absence of the Swc5 subunit from both the SWR1(1–681) and SWR1(ΔN2) complexes suggests that, unlike the other examined components, association of Swc5 is dependent on residues within or at the junction of both Swr1 segments.
FIGURE 4.
Subunit association in the SWR1(1–681) complex. Complexes were normalized to the Flag-tagged Swr1 subunit by Western blotting. A, N-terminal region of Swr1 (Swr1(1–681)) is sufficient for association of Bdf1, Arp4, Swc4, Act1, Yaf9, and Swc7 but not Swc5. SDS-PAGE and Western blotting analysis of Bdf1, Arp4, Swc4, Act1, Swc5, Yaf9, and Swc7 abundance in the wild-type SWR1 and SWR1(1–681) complexes. Association of Swc5 was also examined in SWR1(1–681) complex purified using a less stringent condition (0.3 m KCl). Little or no Swc5 was detected (data not shown). B, subunit composition of SWR1(1–681) complexes purified from wild-type, swc7Δ, and yaf9Δ strains. The numbers (%) are determined by measuring protein level of each subunit in mutant relative to the level in wild type. C, association map of SWR1(1–681) components.
We next characterized the interdependency of subunit association in the SWR1(1–681) complex by purifying the complex from strains lacking the nonessential subunits Swc7 or Yaf9. As anticipated, while loss of Swc7 had minimal effects on subunit association in the SWR1(1–681) complex, loss of Yaf9 resulted in a reduction of Swc4, Yaf9, and Act1 as well as loss of Bdf1 (Fig. 4B). These results are consistent with the effect of Arp4 depletion on association of Yaf9, Swc4, and Act1, and suggest that these subunits may constitute a module on the N-terminal region of Swr1 (Fig. 4C). (We note that loss of Swc7 did not result in the reduction of Bdf1 in the SWR1(1–681) complex as it did in the complex carrying full-length Swr1. Association of Bdf1 may be sensitive to conformational changes of Swr1 induced by protein truncation.)
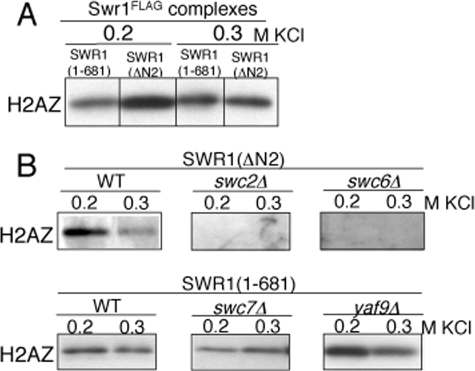
The N-terminal Region of Swr1 Also Binds to H2AZ—We examined H2AZ binding to the SWR1(1–681) and SWR1(ΔN2) complexes and found that both partial complexes were able to bind to H2AZ in pull-down assays (Fig. 5A). Given that the SWR1 complex has been previously shown to bind to H2AZ through the Swc2 subunit (32), this raised the possibility that an additional site for H2AZ binding could occur on other components of the SWR1 complex. To uncover the responsible subunit(s), we pulled down SWR1(1–681) and SWR1(ΔN2) complexes from strains lacking nonessential SWR1 subunits, and examined H2AZ binding by Western blotting analysis (Fig. 5B). As anticipated, deletion of Swc2 or Swc6 resulted in the loss of Swc2 from the SWR1(ΔN2) complex (supplemental Fig. S4) and the loss of H2AZ binding to SWR1(ΔN2) (Fig. 5B). These results are consistent with previous in vivo and in vitro findings that Swc2 binds to H2AZ (32).
FIGURE 5.
H2AZ binding to the SWR1(1–681) or SWR1(ΔN2) complexes. Complexes were normalized to the Swr1 subunit by Western blotting using anti-Swr1. The presence of H2AZ was detected using anti-H2AZ. A, H2AZ binding of SWR1(1–681) and SWR1(ΔN2) complexes. SDS-PAGE (14% gel) and Western blotting analysis of H2AZ pull-down by SWR1(1–681) or SWR1(ΔN2) complexes at the 0.2 or 0.3 m KCl condition. B, subunits required for H2AZ binding in partial SWR1(1–681) and SWR1(ΔN2) complexes. SDS-PAGE (14% gel) and Western blotting analysis of H2AZ pull-down by SWR1(1–681) and SWR1(ΔN2) complexes isolated from wild-type and subunit deletion strains at the 0.2 or 0.3 m KCl condition.
In contrast, H2AZ binding was retained on the SWR1(1–681) complexes purified from the swc7Δ or yaf9Δ strains (Fig. 5B), indicating that neither Swc7, Yaf9, nor the subunits that associate with Swc7 or Yaf9 are required for H2AZ binding. Deletion of Yaf9 generated a minimal SWR1(1–681) complex that contains Swr1(1–681), Swc7, and substantially reduced levels of Bdf1, Arp4, Swc4, and Act1 (Fig. 4B); yet this complex was still able to bind to H2AZ. Given that Swc7 is not required for H2AZ binding, the N-terminal region of Swr1 itself is likely responsible for direct binding of H2AZ to the SWR1 complex.
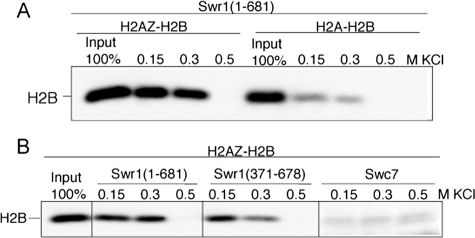
To demonstrate that the N-terminal region of Swr1 binds to H2AZ directly, we analyzed the binding of bacterially expressed Swr1 fragment (residues 1–681) to purified H2AZ-H2B or H2A-H2B dimers that were reconstituted from bacterially expressed proteins. We observed that Swr1(1–681) binds to H2AZ-H2B preferentially over H2A-H2B at the 0.15 m and 0.3 m KCl salt conditions, as detected by Western blotting of the common H2B histone (Fig. 6A). We further expressed a fragment of Swr1 (residues 371–678) in bacteria, and found that Swr1(371–681) exhibited weaker H2AZ-H2B binding than Swr1(1–681), indicating that full H2AZ-H2B binding requires additional Swr1 regions outside residues 371–681. In contrast, bacterially expressed Swc7 did not bind H2AZ-H2B dimers at the same conditions. Taken together, these results indicate that the N-terminal region of Swr1 can bind to H2AZ-H2B directly and preferentially over H2A-H2B, and introduce another histone H2AZ-H2B binding component besides Swc2 and Chz1 (32, 44).
FIGURE 6.
Bacterially expressed Swr1(1–681) binds to H2AZ-H2B in vitro. A, binding of recombinant H2AZ-H2B or H2A-H2B dimers to immobilized, His6-tagged Swr1(1–681) was assayed by Western blotting using anti-H2B. B, binding of recombinant H2AZ-H2B dimers to immobilized, bacterially expressed Swr1(1–681), Swr1(371–678), and Swc7 was determined by Western blotting using anti-H2B.
DISCUSSION
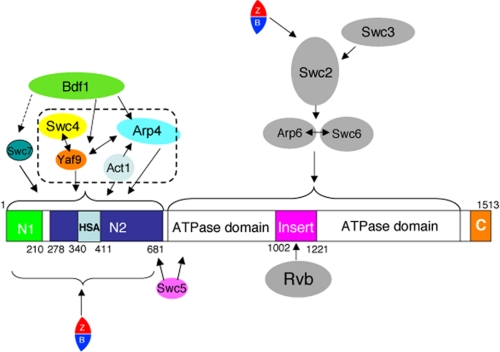
In this study we have further characterized the hierarchy and interdependency of subunit associations within the SWR1 complex and identified an additional H2AZ binding site in the N-terminal region of the Swr1 ATPase (Fig. 7). We observed that both Bdf1 and Swc7 associate with the N terminus of Swr1 and that the association of Bdf1 requires Swc7, Yaf9, and Arp4. In addition, we observed that Arp4 plays a key structural role in SWR1 complex integrity. Arp4 is required for the association of several subunits, including Bdf1, Yaf9, Swc4, and to a minor extent, Act1, Arp6, and Swc2. The association of Yaf9 and/or Swc4 and Arp4 are interdependent. We found that the basic H2AZ replacement activity of SWR1 does not require Bdf1 and Swc7. In contrast, Arp4 is required for functional H2AZ incorporation both in vitro and in vivo, and is therefore important part of histone replacement machinery of the SWR1 complex. Thus, among the fourteen subunits of the SWR1 complex, only three: Swc3 (32), Bdf1, and Swc7 appear to have roles auxiliary to the basic histone replacement activity. Finally, we identified that the N-terminal region of the Swr1 subunit (residues 1–681) contains a H2AZ binding site. This H2AZ binding site is distinct from the previously characterized H2AZ interaction with Swc2, suggesting that a single SWR1 complex might interact with two H2AZ-H2B dimers.
FIGURE 7.
SWR1 subunit association map. One-headed arrows indicate that the association of one subunit with the complex requires the subunit to which the arrow points. Two-headed arrows indicate that the associations are interdependent. Note: Arp4 and Act1 have been shown to bind to the HSA domain (residues 340–411) of Swr1 (42). In the NuA4 complex, with which SWR1 shares the Swc4, Yaf9, Arp4, and Act1 subunits, the association of Yaf9 requires the C-terminal region of Swc4 (51).
Efficient H2AZ deposition by SWR1 has been linked to histone H3 and H4 acetylation by Gcn5, SAS or NuA4 (12, 14, 30, 45, 47, 50, 51), in addition to the presence of gene-specific transcription factor and cofactors (13, 48, 49). It has been suggested that two bromodomains in Bdf1 may play a role in targeting SWR1 to promoters by binding to acetylated histone H3 and/or H4 tails (12, 14, 30). This view is supported by an in vitro study using Drosophila dTip60 (the Domino/p400/Tip60 supercomplex containing both H2AZ replacement and histone acetyltransferase activities), which demonstrated that H2AZ is incorporated more efficiently when nucleosome arrays are acetylated (21). Conservation of a similar supercomplex, the NuA4/Tip60 or TRRAP/Tip60 complex in human (23, 51, 52) further highlights the connection between histone acetylation and H2AZ exchange. Our finding that Bdf1 is not required for the basic histone H2AZ replacement activity of the SWR1 complex is consistent with these findings. (However, it cannot be excluded that Bdf2, the Bdf1 paralog, may substitute the function of Bdf1 by binding to the same interaction surface within the SWR1 complex, as Bdf1 and Bdf2 are partially redundant in promoting H2AZ chromatin incorporation in vivo (14, 30)).
Recent studies have shown that an N-terminal subdomain (residues 340–411) of Swr1, the HSA domain, is sufficient to pull-down Arp4 and Act1 (42), and thus can be considered as a binding platform for Arp4 and actin. Moreover, there is evidence that Swc4 interacts with Yaf9 directly (45, 51). Collectively, these results along with our studies of SWR1 complexes isolated from the arp4-td, yaf9Δ, and swr1(1–681) strains suggest that Yaf9, Swc4(Eaf2), Arp4, and Act1 assemble on the N-terminal region of the Swr1 subunit forming a functional module. Given that these four proteins are the only subunits shared by the SWR1 and NuA4 complexes, their association may serve a similar function in the two enzyme complexes. Because Arp4 is known to interact with histone or nucleosome directly (27, 39) and Swc4/Eaf2 contains a SANT domain implicated in interaction with histone and/or DNA (53), this `module' may be involved in binding to the conventional nucleosome. We note that previous studies observed that the entire NuA4 complex is disrupted in arp4 temperature-sensitive mutants (27). Our results derived from using the arp4-td strain showed that while the association of Bdf1, Yaf9 and Swc4 is diminished, other SWR1 subunits maintained their association with the Swr1 ATPase. It was also feasible to isolate a partial SWR1(ΔN2) complex containing six other subunits. The different results could be related to differences between degron-tagged and temperature-sensitive arp4 mutants, to purification procedures, or to additional functions of Arp4 in maintaining the global integrity of NuA4.
We conducted extensive efforts to delineate the H2AZ binding region within Swr1 (residues 1–681) by subdividing the region into a number of segments for expression in bacteria. However, these efforts were not fruitful as no viable clones expressing the corresponding regions were obtained. Nonetheless, one construct expressing Swr1 (residues 371–678) displayed some H2AZ binding activity, albeit lower than Swr1(1–681). Hence, the H2AZ binding region may require additional residues outside the 371–678 fragment of Swr1. We note that little or no H2AZ binding to SWR1 complex was detected in our previous analysis of SWR1 complex isolated from mutant strains deficient for Swc2 as a common missing subunit (32). This may be attributed to a more stringent wash protocol and nonlinear detection of H2AZ by antibody.
During the course of this work, it was reported that the N-terminal region of Arabidopsis Swr1 binds to H2AZ using a yeast two-hybrid assay (22). Our present results corroborate and extend that finding by showing direct interactions between the N-terminal region of Swr1 and H2AZ in vitro using purified proteins. Of the three H2AZ direct and preferred binding components identified to date, Swc2 and Swr1 are subunits of the SWR1 complex, while Chz1 is a H2AZ nuclear chaperone (32, 44). Interestingly, H2AZ binding to Swr1(1–681) appeared to be weaker than binding to Swc2, as the former was sensitive to washing by 0.5 m KCl. The observation of two H2AZ binding components in the SWR1 complex suggests that one SWR1 enzyme could bind to two H2AZ-H2B dimers. It is tempting to speculate that the two H2AZ-H2B dimer-bound SWR1 complex promotes a more efficient transfer of H2AZ to conventional nucleosomes.
Supplementary Material
Acknowledgments
We thank David Drubin for the actin antibody, Paul Badenhorst for critical reading of the manuscript, Jeremy Daniel, Patrick Grant, Joe Landry, Michael Lichten, Xuetong Shen, Toshi Tsukiyama, and Jan Wisniewski for helpful discussions, Megan Eanes and Kyoko Tomita for technical assistance, the NIH Fellows Editorial Board for editorial assistance.
This work was supported, in whole or in part, by the intramural research program of the NCI, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
Footnotes
The abbreviations used are: ChIP, chromatin immunoprecipitation assay; HSA, helicase-SANT-associated; PVDF, polyvinylidene difluoride.
G. Mizuguchi and C. Wu, unpublished data.
References
- 1.Kornberg, R. D., and Lorch, Y. (1999) Cell 98 285–294 [DOI] [PubMed] [Google Scholar]
- 2.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997) Nature 389 251–260 [DOI] [PubMed] [Google Scholar]
- 3.Fischle, W., Wang, Y., and Allis, C. D. (2003) Curr. Opin. Cell Biol. 15 172–183 [DOI] [PubMed] [Google Scholar]
- 4.Gangaraju, V. K., and Bartholomew, B. (2007) Mutat. Res. 618 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redon, C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W. (2002) Curr. Opin. Genet. Dev. 12 162–169 [DOI] [PubMed] [Google Scholar]
- 6.Eirin-Lopez, J., and Ausio, J. (2007) Curr. Genomics 8 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusch, T., and Workman, J. L. (2007) Subcell Biochem. 41 91–109 [PubMed] [Google Scholar]
- 8.Guillemette, B., and Gaudreau, L. (2006) Biochem. Cell Biol. 84 528–535 [DOI] [PubMed] [Google Scholar]
- 9.Millar, C. B., Xu, F., Zhang, K., and Grunstein, M. (2006) Genes Dev. 20 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keogh, M. C., Mennella, T. A., Sawa, C., Berthelet, S., Krogan, N. J., Wolek, A., Podolny, V., Carpenter, L. R., Greenblatt, J. F., Baetz, K., and Buratowski, S. (2006) Genes Dev. 20 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babiarz, J. E., Halley, J. E., and Rine, J. (2006) Genes Dev. 20 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, H., Roberts, D. N., and Cairns, B. R. (2005) Cell 123 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillemette, B., Bataille, A. R., Gevry, N., Adam, M., Blanchette, M., Robert, F., and Gaudreau, L. (2005) PLoS Biol. 3 e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raisner, R. M., Hartley, P. D., Meneghini, M. D., Bao, M. Z., Liu, C. L., Schreiber, S. L., Rando, O. J., and Madhani, H. D. (2005) Cell 123 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavrich, T. N., Jiang, C., Ioshikhes, I. P., Li, X., Venters, B. J., Zanton, S. J., Tomsho, L. P., Qi, J., Glaser, R. L., Schuster, S. C., Gilmour, D. S., Albert, I., and Pugh, B. F. (2008) Nature 453 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z., Wei, G., Chepelev, I., and Zhao, K. (2007) Cell 129 823–837 [DOI] [PubMed] [Google Scholar]
- 17.Albert, I., Mavrich, T. N., Tomsho, L. P., Qi, J., Zanton, S. J., Schuster, S. C., and Pugh, B. F. (2007) Nature 446 572–576 [DOI] [PubMed] [Google Scholar]
- 18.Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004) Science 303 343–348 [DOI] [PubMed] [Google Scholar]
- 19.Krogan, N. J., Keogh, M. C., Datta, N., Sawa, C., Ryan, O. W., Ding, H., Haw, R. A., Pootoolal, J., Tong, A., Canadien, V., Richards, D. P., Wu, X., Emili, A., Hughes, T. R., Buratowski, S., and Greenblatt, J. F. (2003) Mol. Cell 12 1565–1576 [DOI] [PubMed] [Google Scholar]
- 20.Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., Madhani, H. D., and Rine, J. (2004) PLoS Biol 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusch, T., Florens, L., Macdonald, W. H., Swanson, S. K., Glaser, R. L., Yates, J. R., 3rd, Abmayr, S. M., Washburn, M. P., and Workman, J. L. (2004) Science 306 2084–2087 [DOI] [PubMed] [Google Scholar]
- 22.Choi, K., Park, C., Lee, J., Oh, M., Noh, B., and Lee, I. (2007) Development 134 1931–1941 [DOI] [PubMed] [Google Scholar]
- 23.Cai, Y., Jin, J., Florens, L., Swanson, S. K., Kusch, T., Li, B., Workman, J. L., Washburn, M. P., Conaway, R. C., and Conaway, J. W. (2005) J. Biol. Chem. 280 13665–13670 [DOI] [PubMed] [Google Scholar]
- 24.Wong, M. M., Cox, L. K., and Chrivia, J. C. (2007) J. Biol. Chem. 282 26132–26139 [DOI] [PubMed] [Google Scholar]
- 25.Shen, X. (2004) Methods Enzymol. 377 401–412 [DOI] [PubMed] [Google Scholar]
- 26.Shen, X., Mizuguchi, G., Hamiche, A., and Wu, C. (2000) Nature 406 541–544 [DOI] [PubMed] [Google Scholar]
- 27.Galarneau, L., Nourani, A., Boudreault, A. A., Zhang, Y., Heliot, L., Allard, S., Savard, J., Lane, W. S., Stillman, D. J., and Cote, J. (2000) Mol. Cell 5 927–937 [DOI] [PubMed] [Google Scholar]
- 28.Doyon, Y., and Cote, J. (2004) Curr. Opin. Genet. Dev. 14 147–154 [DOI] [PubMed] [Google Scholar]
- 29.Matangkasombut, O., Buratowski, R. M., Swilling, N. W., and Buratowski, S. (2000) Genes Dev. 14 951–962 [PMC free article] [PubMed] [Google Scholar]
- 30.Durant, M., and Pugh, B. F. (2007) Mol. Cell. Biol. 27 5327–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, H., Richardson, D. O., Roberts, D. N., Utley, R., Erdjument-Bromage, H., Tempst, P., Cote, J., and Cairns, B. R. (2004) Mol. Cell. Biol. 24 9424–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, W. H., Alami, S., Luk, E., Wu, C. H., Sen, S., Mizuguchi, G., Wei, D., and Wu, C. (2005) Nat. Struct. Mol. Biol. 12 1064–1071 [DOI] [PubMed] [Google Scholar]
- 33.Matangkasombut, O., and Buratowski, S. (2003) Mol. Cell 11 353–363 [DOI] [PubMed] [Google Scholar]
- 34.Ladurner, A. G., Inouye, C., Jain, R., and Tjian, R. (2003) Mol. Cell 11 365–376 [DOI] [PubMed] [Google Scholar]
- 35.Carey, M., Li, B., and Workman, J. L. (2006) Mol. Cell 24 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasten, M., Szerlong, H., Erdjument-Bromage, H., Tempst, P., Werner, M., and Cairns, B. R. (2004) EMBO J. 23 1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, X., Ranallo, R., Choi, E., and Wu, C. (2003) Mol. Cell 12 147–155 [DOI] [PubMed] [Google Scholar]
- 38.Downs, J. A., Allard, S., Jobin-Robitaille, O., Javaheri, A., Auger, A., Bouchard, N., Kron, S. J., Jackson, S. P., and Cote, J. (2004) Mol. Cell 16 979–990 [DOI] [PubMed] [Google Scholar]
- 39.Sunada, R., Gorzer, I., Oma, Y., Yoshida, T., Suka, N., Wintersberger, U., and Harata, M. (2005) Yeast 22 753–768 [DOI] [PubMed] [Google Scholar]
- 40.Doerks, T., Copley, R. R., Schultz, J., Ponting, C. P., and Bork, P. (2002) Genome Res. 12 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotter, K. W., Fan, H. Y., Ivey, M. L., Kingston, R. E., and Archer, T. K. (2008) Mol. Cell. Biol. 28 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szerlong, H., Hinata, K., Viswanathan, R., Erdjument-Bromage, H., Tempst, P., and Cairns, B. R. (2008) Nat. Struct. Mol. Biol. 15 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M. M., and Wu, C. (2007) Cell 129 1153–1164 [DOI] [PubMed] [Google Scholar]
- 44.Luk, E., Vu, N. D., Patteson, K., Mizuguchi, G., Wu, W. H., Ranjan, A., Backus, J., Sen, S., Lewis, M., Bai, Y., and Wu, C. (2007) Mol. Cell 25 357–368 [DOI] [PubMed] [Google Scholar]
- 45.Bittner, C. B., Zeisig, D. T., Zeisig, B. B., and Slany, R. K. (2004) Eukaryot. Cell 3 976–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dohmen, R. J., and Varshavsky, A. (2005) Methods Enzymol. 399 799–822 [DOI] [PubMed] [Google Scholar]
- 47.Shia, W. J., Li, B., and Workman, J. L. (2006) Genes Dev. 20 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gligoris, T., Thireos, G., and Tzamarias, D. (2007) Mol. Cell. Biol. 27 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gevry, N., Chan, H. M., Laflamme, L., Livingston, D. M., and Gaudreau, L. (2007) Genes Dev. 21 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogan, N. J., Baetz, K., Keogh, M. C., Datta, N., Sawa, C., Kwok, T. C., Thompson, N. J., Davey, M. G., Pootoolal, J., Hughes, T. R., Emili, A., Buratowski, S., Hieter, P., and Greenblatt, J. F. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auger, A., Galarneau, L., Altaf, M., Nourani, A., Doyon, Y., Utley, R. T., Cronier, D., Allard, S., and Cote, J. (2008) Mol. Cell. Biol. 28 2257–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J., and Nakatani, Y. (2000) Cell 102 463–473 [DOI] [PubMed] [Google Scholar]
- 53.Boyer, L. A., Latek, R. R., and Peterson, C. L. (2004) Nat. Rev. Mol. Cell. Biol. 5 158–163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.