Abstract
The orphan nuclear receptor chicken ovalbumin upstream promoter transcription factor I (COUP-TFI) plays key roles in development and homeostasis. A tandem affinity purification procedure revealed that COUP-TFI associated with a number of transcriptional regulatory proteins in HeLa S3 cells, including the nuclear receptor corepressor (NCoR), TIF1β/KAP-1, HDAC1, and the SWI/SNF family member Brahma. The proapoptotic protein DBC1 was also identified in COUP-TFI complexes. In vitro experiments revealed that COUP-TFI interacted directly with NCoR but in a manner different from that of other nuclear receptors. DBC1 stabilized the interaction between COUP-TFI and NCoR by interacting directly with both proteins. The gene encoding the anti-apoptotic protein TNFAIP8 (tumor necrosis factor α (TNFα)-induced protein 8) was identified as being repressed by COUP-TFI in a manner that required several of the component proteins of the COUP-TFI complex. Finally, our studies highlight a central role for COUP-TFI in the induction of the TNFAIP8 promoter by TNFα. Together, these studies identify a novel COUP-TFI complex that functions as a repressor of transcription and may play a role in the TNFα signaling pathways.
The eukaryotic genome is highly compacted into chromatin, an organized mélange of nucleic acid and histone proteins (1). Chromatin serves several important cellular functions. First, it solves a packing problem by extensively compacting the DNA into a bundle small enough to fit into the eukaryotic nucleus. Second, chromatin modifications underlie the basis of epigenetic regulation of gene expression, providing scalable control of gene expression that is critical for the maintenance of cellular homeostasis, differentiation, and proliferation (2, 3).
Nuclear receptors belonging to the steroid/thyroid hormone receptor superfamily are ligand-dependent transcription factors that regulate critical processes in growth, development, and homeostasis (4, 5). Some nuclear receptors, such as thyroid hormone receptors and retinoic acid receptors, bind to regulatory elements in the target gene promoters in the absence of their cognate ligands. These aporeceptors function as repressors of transcription by interactively recruiting nuclear receptor corepressor (NCoR)2- or SMRT-containing corepressor complexes, which harbor multiple histone deacetylases, to the promoter template (6, 7). Agonist binding to nuclear receptors appears to stabilize a receptor conformation that is not permissive for corepressor binding but instead favors the cyclical recruitment of a series of multiprotein coactivator complexes, many of which contain multiple histone-modifying enzymes, to the responsive promoter (7). The dynamic agonist-driven exchange of transcriptional coregulatory proteins on nuclear receptor-bound promoter templates likely underlies the molecular basis of regulation of gene expression by this family of transcription factors (8).
Three mammalian genes encoding chicken ovalbumin upstream promoter transcription factor (COUP-TF) proteins, which are orphan members of the steroid/thyroid hormone receptor superfamily, have been cloned (9, 10): COUP-TFI (also known as Nr2f1 or EAR3), COUP-TFII (Nr2f2 or ARP1), and COUP-TFIII (Nr2f6 or EAR2). COUP proteins play unique roles in fetal development, including neurogenesis and angiogenesis, and possibly in metabolic homeostasis in adult organisms (11). In the mouse, deletion of COUP-TFI or COUP-TFII results in perinatal and embryonic lethality, respectively, possibly due to disrupted neuronal development (12–14) and aberrant angiogenesis, skeletal muscle development, and cardiac development (15, 16). COUP-TFIII–/– mice exhibit disruption of noradrenergic homeostasis in the locus coeruleus and rostral cerebral cortex, along with enhanced nociception and dysregulation of period1 and period2, clock genes that are important for proper circadian timing (17). These findings highlight the absolute essentiality of the COUP-TF family of orphan nuclear receptors in developmental processes and indicate that these proteins do not function redundantly in vivo.
A number of COUP-interacting proteins have been described, and the majority of these were identified and/or characterized by yeast two-hybrid systems: transcription factor IIB (18), a p56lck ligand (19), NCoR and SMRT (20), Sp1 (18, 21), Alien (22), CTIP1 and CTIP2 (23), Tat (24), FOG-2 (25), CIP-1 (26), CIP-2 (27), and Ubc9 (an E2 conjugating enzyme of the small ubiquitin-like modifier (SUMO)-1 family) (28, 29). To our knowledge, none of these COUP-interacting proteins have been demonstrated to be recruited to the promoter of target genes in a COUP-dependent manner, and a systematic study of COUP-TFI complexes in mammalian cells has not been conducted.
In these studies, we identified a number of COUP-TFI-interacting proteins using a tandem affinity purification strategy, and we found that the orphan receptor and several of these proteins co-occupied the promoter of a COUP-TFI target gene that was first identified herein, TNFAIP8 (tumor necrosis factor α-induced protein 8). Our data strongly suggest that the induction of TNFAIP8 by tumor necrosis factor α (TNFα) involves relief of COUP-TFI-mediated repression of the corresponding promoter. It is believed that TNFAIP8, via direct inhibition of caspase activity, serves to dampen the apoptotic response of cells that are stimulated by TNFα (30). Thus, our findings implicate COUP-TFI in the TNFα signaling pathways in mammalian cells.
MATERIALS AND METHODS
Cell Culture—HeLa S3 and 293T cells were grown at 37 °C in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (Atlas Biologicals) and 1% penicillin/streptomycin (Invitrogen). SK-N-MC cells were grown under the same conditions except that 1% sodium pyruvate (Invitrogen) was added to the medium.
Constructs and Generation of Stable Cell Lines—The plasmid pOZ-COUP was prepared by PCR amplification of human COUP-TFI with primers containing appropriate restriction sites for insertion into the pOZ-N vector (a bicistronic retroviral expression vector, which was a kind gift from Pat Nakatani, Dana-Farber Cancer Institute) (31–33). The pOZ-N vector contains a tandem epitope tag (hemagglutinin (HA) and FLAG) upstream of the multiple cloning site and an internal ribosomal entry site downstream of the cloning site. The internal ribosomal entry site is followed by the coding sequence of the extracellular domain of CD25 (interleukin-2 receptor α). Thus, the pOZ-COUP vector, which was used to prepare pOZ-COUP cells (see below), encodes FLAG-HA-COUP-TFI (referred to as FH-COUP) and CD25, the latter of which facilitated cell sorting of infected cells (see below). The NCoR expression vector and associated deletion mutants were described previously (34). Plasmids encoding glutathione S-transferase (GST) fusions of COUP-TFI and NCoR fragments were prepared by inserting amplicons into pGEX2T (GE Healthcare) using standard methodology. All DBC1 constructs were kind gifts from Eileen White (Rutgers University) (35). The expression vector for FLAG-HA-tagged full-length COUP-TFI in pcDNA3.1(+) was constructed by PCR amplification and insertion into pcDNA3.1(+). The COUP-TFI-responsive reporter construct harboring a tetramerized COUP-TFI-binding site, (DR1)4-tk-CAT, was constructed in pBL2-CAT. All constructs were sequenced to confirm authenticity.
Recombinant retroviruses were produced using the BD Retro-X universal packaging system (BD Biosciences) and used to infect HeLa S3 cells growing in suspension. Following retroviral infection, the proportion of cells expressing CD25 varied from 0.5 to 15% as determined by flow cytometry on an EPICS XL flow cytometer (Beckman Coulter) using a phycoerythrin-conjugated anti-CD25 antibody (Miltenyi Biotec). The transduced HeLa S3 cultures were enriched for CD25+ cells by magnetic sorting either using an autoMACS system or manually using magnetic beads (Miltenyi Biotec). The enriched cells were cultured for several days to increase cell number and again subjected to the magnetic enrichment procedure. After three to five rounds of enrichment, cultures reached 96–98% purity as determined by flow cytometry and were then transferred to a suspension culture for large-scale production. Purified cell populations have maintained 96–98% purity after >4 months in continuous culture as determined by weekly flow cytometric analyses. HeLa S3 cells infected with recombinant retrovirus derived from the empty vector (pOZ-N cells) were similarly purified and analyzed and used as a control in the tandem affinity purification procedure.
Transfections—293T cells (2 × 106 cells) were plated onto a 10-cm plate and transfected 24 h later using the calcium phosphate method. Each transfection consisted of varying amounts of the (DR1)4-tk-CAT reporter gene, FH-COUP, and an empty expression vector to standardize the total amount of DNA transfected. The medium was changed, and the cells were harvested 24 and 48 h after transfection, respectively.
Antibodies—The anti-FLAG antibody was purchased from Sigma. The anti-HA antibody was purchased from Roche Applied Science. Antibodies against TIF1β, DBC1, HDAC1, and TBLR1 were obtained from Bethyl Laboratories (Montgomery, TX). Anti-COUP-TFI (T19), anti-BAF170, and anti-SIN3A antibodies as well as control IgGs (mouse, rabbit, and goat) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-HSP70 antibody was purchased from StressGen, and the anti-NCoR antibody was a generous gift from Dr. Geoff Rosenfeld (University of California, San Diego).
Tandem Affinity Purification—Approximately 3 × 109 purified HeLa S3 cells stably expressing FH-COUP or control pOZ-N cells were harvested during log phase (∼106 cells/ml). Nuclear extracts were prepared essentially as described by Shapiro et al. (36) with minor modifications. Briefly, nuclear extract (∼150 mg) was incubated with 400 μl of anti-FLAG-agarose (Sigma) for 5 h with constant rotation at 4 °C. This resin was then washed extensively with ice-cold phosphate-buffered saline containing 0.05% Nonidet P-40, and protein complexes were eluted with a peptide corresponding to a dimerized FLAG epitope (Sigma). The eluted material was then loaded directly onto 200 μl of anti-HA-agarose resin (Roche Applied Science), which was then incubated, washed, and eluted with the HA peptide (Sigma) as described above.
Protein Identification by Tandem Mass Spectrometry—Twice-purified material was electrophoretically separated on a 4–12% SDS-polyacrylamide gradient gel, and individual proteins were visualized by a mass spectrometry (MS)-friendly Coomassie stain (Bio-Rad), excised, and subjected to an in-gel tryptic digest as described previously (37). Following digestion, samples were desalted using a micro-C18 tip (Millipore) and dried. Samples were then resuspended in 5 μl of 0.1% formic and analyzed by liquid chromatography/electrospray ionization tandem mass spectrometry (MS/MS) with a Nano2D LC (Eksigent) coupled to an LTQ FT mass spectrometer (Thermo Electron) using an instrument configuration as described (38). Data were collected in a data-dependent mode in which a high mass resolution/high mass accuracy MS scan (in the FT part of the instrument) was followed by MS/MS scans of the five most abundant ions from the preceding MS scan. The five selected ions for MS/MS were placed on an exclusion list and not selected for subsequent MS/MS for 1.5 min, allowing less intense ions to be interrogated for MS/MS. Proteins were identified from MS data using a modified version of the open source X!Tandem (Beavis Informatics) automated protein data base search algorithm. The score function of native X!Tandem was replaced with a dot product-based score algorithm that is compatible with PeptideProphet (39). Search results were considered correct if at least two peptides had raw scores >200 for +1 ions, >300 for +2 ions, and >300 for +3 ions, percent ions of >15%, and PeptideProphet scores >0.9 and if the identification did not appear in a blank portion of the gel.
Co-immunoprecipitation and Immunoblotting—Co-immunoprecipitations and immunoblotting were conducted as described previously (40).
Size-exclusion Chromatography—Size-exclusion chromatography was conducted using 50 mg of nuclear extract from stably transduced HeLa S3 or control cells and a Superose 6 column as described previously (40, 41).
GST Pulldown Analyses—GST pulldown analyses were conducted as described previously using bacterially expressed GST fusion proteins as baits and [35S]methionine-labeled proteins as prey (42).
Reporter Gene Assays—Chloramphenicol acetyltransferase (CAT) reporter gene assays were conducted and quantified as described previously (41).
Chromatin Immunoprecipitation Studies—Both chromatin immunoprecipitation (ChIP) and re-ChIP studies were conducted essentially as described previously (40). The following primers were used for amplification reactions: TNFAIP8, 5′-TCCTCCTTCCCTGCACGCT-3′ (forward primer) and 5′-CCAGGAGCCACTTACTCGGA-3′ (reverse primer; amplification product, 278 bp); and IGF2, 5′-GATCATCGTCCAGGCAGTTT-3′ (forward primer) and 5′-CTTCCCCTCCTTCAGAAACC-3′ (reverse primer; amplification product, 227 bp).
Small Interfering RNA Transfections—HeLa S3 cells were transfected with 30 nmol of negative control small interfering RNA (siRNA) or specific siRNA against COUP-TFI, NCoR, or TIF1β using siPORT NeoFX transfection reagent (Ambion). The medium was replaced with fresh growth medium after 24 h, and cells were harvested for RNA extraction 48 h after transfection.
Quantitative Reverse Transcription-PCR—RNA was prepared using the RNeasy minikit (Qiagen), and 1 μg of total RNA was reverse-transcribed using reverse transcriptase and oligo(dT) (Invitrogen). The resulting cDNA (1 μl) was then used for the following amplification reactions: TNFAIP8, 5′-TGAGCTAGCATTGATGGAGA-3′ (forward primer) and 5′-TCCAACATTTTGTTGATACCA-3′ (reverse primer); and hypoxanthine-guanine phosphoribosyltransferase, 5′-ATTGTAATGACCAGTCAACAGGG-3′ (forward primer) and 5′-GCATTGTTTTGCCAGTGTCAA-3′ (reverse primer). These primer sets generate amplicons of 300 and 117 bp, respectively. An Applied Biosystems 7500 real-time PCR instrument and SYBR Green technology (Qiagen) were used for all quantitative PCR analyses.
Compound Treatments and Apoptosis Assay—HeLa S3 cells were grown on coverslips to 70% confluency, followed by 8 h of TNFα (5 μg/ml) and cycloheximide (CHX; 30 μg/ml) treatments. After drug treatment, cells were fixed with paraformaldehyde, and the nuclei were stained with 4′,6-diamidino-2-phenylindole. Non-apoptotic cells with intact nuclei were counted using a fluorescence microscope, and the percentage of non-apoptotic cells after TNFα/CHX or mock treatment was calculated.
RESULTS
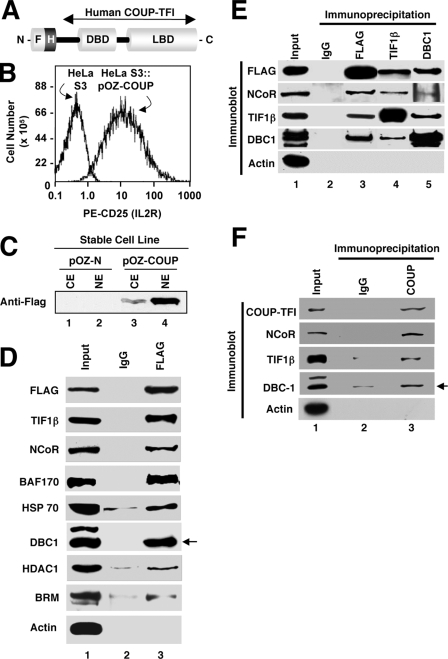
Purification and Identification of Component Proteins of COUP-TFI Complexes—Stable cell lines were established in the HeLa S3 background using a recombinant retrovirus directing expression of COUP-TFI harboring a tandem epitope tag (FLAG-HA) at its N terminus (FH-COUP) (Fig. 1A), which allowed immunoprecipitation and immunodetection of COUP-TFI with high efficiency and specificity. The bicistronic retroviral vector used to create the cell line (pOZ-COUP) stably expressing FH-COUP also encoded the extracellular domain of the interleukin-1 receptor (CD25), which facilitated cell sorting to enrich infected cells to near homogeneity (Fig. 1B). As a control, HeLa S3 cells were infected with a recombinant retrovirus prepared using the empty pOZ-N vector (pOZ-N cells). Both pOZ-COUP and pOZ-N cells were purified to >95% homogeneity by multiple rounds of magnetic-based cell sorting (data not shown) and then expanded in suspension culture. Stable expression of FH-COUP was validated by immunoblotting (Fig. 1C), and subcellular distribution analyses demonstrated that the tagged protein was localized mostly in the nucleus, consistent with previous studies of COUP-TFI (29, 43, 44).
FIGURE 1.
Validation of component proteins of FH-COUP complexes. A, schematic diagram of tandem epitope-tagged COUP-TFI. The sequences encoding FLAG and HA tags are located upstream of the human COUP-TFI cDNA in a retroviral vector. LBD, ligand-binding domain. B, flow cytometric analysis of uninfected HeLa S3 cells and HeLa/FH-COUP cells after multiple rounds of magnetic cell purifications indicating >95% purity of the latter. PE, phycoerythrin; IL2R, interleukin-2 receptor. C, immunodetection of FH-COUP in cytoplasmic (CE) or nuclear (NE) extracts demonstrating that cells infected with FH-COUP-programmed but not control retrovirus express FH-COUP and that the majority of the fusion protein localizes in the nucleus. D, co-IP studies in pOZ-COUP cells. Nuclear lysates were immunoprecipitated with the anti-FLAG antibody or nonspecific IgG, and these immune complexes were analyzed after gel electrophoresis by immunoblotting using the antibodies indicated on the left. Input represents 10% of the material used for immunoprecipitation. E, co-IP analyses of FH-COUP, NCoR, TIF1β, and DBC1 using nuclear extract from pOZ-COUP cells. Immunoprecipitation and immunoblotting were conducted using the indicated antibodies. Input corresponds to 5% of the nuclear extract used for immunoprecipitation. F, NCoR, TIF1β, and DBC1 co-IP with endogenous COUP-TFI from nuclear extracts prepared from untransfected SK-N-MC neuroblastoma cells. All studies shown in D–F are representative of three to eight independent experiments.
Protein complexes containing FH-COUP were isolated from nuclear extracts of pOZ-COUP cells by tandem affinity purification (supplemental Fig. S1A) essentially as described by Nakatani and co-workers (32, 33). Purified protein complexes were resolved by SDS-PAGE and visualized by Coomassie staining (supplemental Fig. S1B), which revealed that immunopurified FH-COUP co-purified with ∼25 other polypeptides, most of which were not present in twice-immunopurified material from pOZ-N cells (supplemental Fig. S1B, compare lanes 1 and 2).
MS analysis of FH-COUP complexes after sequential immunoaffinity chromatography revealed the co-purification of several components of transcriptional repressive complexes along with FH-COUP. These included NCoR, HDAC1, TBLR1, TIF1β/KAP-1, and MPC-3 (a chromodomain-containing polycomb protein) (Table 1 and supplemental Fig. S1B). Brahma (BRM), an ATP-dependent chromatin-remodeling protein (45), and its associated factors BAF155 and BAF170 were also identified in FH-COUP complexes (Table 1 and supplemental Fig. S1B). The Brahma complex appears to be a bifunctional SWI/SNF family member, having been implicated in both transcriptional activation (1) and repression (46). Proteins not previously implicated in transcriptional regulation were also found to co-purify with FH-COUP, including the DNA repair protein DDB1, DBC1 (a pro-apoptotic protein that is deleted in breast cancer), HSP70, HSP90, and HYD1 (a ubiquitin ligase). Finally, several components of the spliceosome assembly were identified (SFR1, SF3A1, and SF3B1). Collectively, our tandem affinity purification strategy revealed that FH-COUP may associate with a number of transcriptional regulatory proteins in HeLa S3 cells as well as other classes of proteins that have not been previously implicated in the regulation of gene expression.
TABLE 1.
Components of the COUP-TFI complexes
| Protein symbol | GenBank™ codes | Unique peptides | Function |
|---|---|---|---|
| COUP-TFI | NP_005645 | 12 | Transcription factor |
| TIF1β | NP_005753 | 7 | Transcriptional corepressor |
| NCoR | NP_006302 | 8 | Transcriptional corepressor |
| HDAC1 | NP_004955 | 7 | Transcriptional repressor |
| TBLR1 | NP_078941 | 8 | Transcriptional repression |
| MPC-3 | NP_065700 | 2 | Transcriptional repression |
| TRIM33 | NP_056990 | 4 | Transcriptional corepressor |
| FIR | NP_055096 | 4 | Transcriptional repression |
| BAF170 | NP_620706 | 2 | ATP-dependent chromatin-remodeling protein |
| BAF155 | NP_003065 | 2 | ATP-dependent chromatin-remodeling protein |
| BRM | NP_620614 | 4 | ATP-dependent chromatin-remodeling protein |
| THOC4 | NP_005773 | 4 | Transcriptional coactivator |
| TFIIIC | NP_036336 | 2 | Transcriptional activator |
| TAF9 | NP_003178 | 2 | Transcriptional coactivator |
| DBC1 | NP_066997 | 27 | Apoptosis |
| BAT3 | NP_542434 | 7 | Apoptosis |
| DDB1 | CAA05770 | 10 | DNA repair |
| HSP70 | NP_005338 | 11 | Protein chaperone |
| HSP90 | NP_031381 | 5 | Protein chaperone |
| Human HYD1 | U95000_1 | 49 | Ubiquitin ligase |
| EF1α | NP_001393 | 6 | Elongation factor |
| SFR1 | NP_008855 | 6 | Splicing factor |
| SF3A1 | NP_005868 | 3 | Splicing factor |
Characterization of COUP-TFI-associated Proteins—We undertook a multipartite approach to verify association of the identified proteins with FH-COUP complexes in mammalian cells. First, we conducted co-immunoprecipitation (co-IP) studies in nuclear extracts from HeLa S3 cells stably expressing FH-COUP. Endogenous TIF1β, NCoR, BAF170, BRM, HSP70, DBC1, and HDAC1 were efficiently co-immunoprecipitated with FH-COUP using the anti-FLAG antibody (Fig. 1D).
Second, we performed a series of reciprocal immunoprecipitation experiments in nuclear extracts prepared from pOZ-COUP cells. These studies utilized antibodies directed against both FH-COUP and representative endogenous COUP-interacting proteins. The results of these studies confirmed that endogenous NCoR, TIF1β, and DBC1 were specifically immunoprecipitated with FH-COUP by the anti-FLAG antibody (Fig. 1E). The reciprocal immunoprecipitations also indicated that FH-COUP was immunoprecipitated with antibodies directed against TIF1β and DBC1 (Fig. 1E). Notably, TIF1β, NCoR, and DBC1 all appeared to co-immunoprecipitate with each other (Fig. 1E, lanes 4 and 5). These results strongly suggest that COUP-TFI stably associates with endogenous NCoR, TIF1β, and DBC1 within the same or a highly related protein complex in HeLa S3 cells.
Third, we conducted co-IP experiments using nuclear extracts prepared from untransfected SK-N-MC neuroblastoma cells, which express relatively high levels of COUP-TFI endogenously. The results of these co-IP experiments revealed the specific association of endogenous COUP-TFI with NCoR, TIF1β, BRM, and DBC1 in SK-N-MC cells (Fig. 1F).
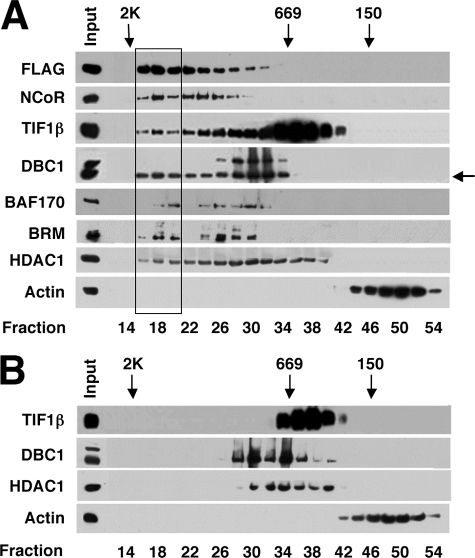
Fourth, we verified that putative component proteins of FH-COUP complexes co-chromatographed with FH-COUP on a Superose 6 size-exclusion column. We found that the majority of FH-COUP immunoreactivity present in nuclear extracts of pOZ-COUP cells eluted from the Superose 6 column with an apparent mass between 1 and 2 MDa (Fig. 2A). This peak of FH-COUP immunoreactivity was not symmetrical but rather eluted with a tailing shoulder extending nearly to the 669-kDa marker. All of the putative FH-COUP complex proteins examined co-eluted with FH-COUP in the high molecular mass fractions (fractions 16–20) to varying degrees, but the chromatographic behavior of these proteins differed dramatically. This is perhaps best exemplified by TIF1β, the majority of which eluted independently of FH-COUP as a fairly symmetrical peak centered around 669 kDa (Fig. 2A, fractions 32–38). However, the leading shoulder of the TIF1β peak extended up into the MDa range of the profile, where it appeared to co-elute with FH-COUP as well as with other putative proteins of the COUP-TF complex (Fig. 2A). Other proteins of the putative COUP-TFI complex (DBC1 and HDAC1) exhibited similar chromatographic behavior, which prompted us to examine if these anomalous elution patterns might result from overexpression of COUP-TFI. To test this possibility, we examined the chromatographic properties of these proteins in control pOZ-N cells (prepared with the empty pOZ-N vector). In the absence of FH-COUP overexpression, TIF1β, DBC1, and HDAC1 all appeared to elute from the Superose 6 column as fairly symmetrical peaks corresponding to the lower mass species observed in pOZ-COUP cells (Fig. 2B; see also Fig. 2A). This finding suggests that stable overexpression of FH-COUP in HeLa S3 cells alters the chromatographic behavior of these proteins, consistent with the possibility that these proteins may be authentic components of FH-COUP complexes. Considered together, these data validate the identified proteins as components of COUP-TFI complexes in mammalian cells.
FIGURE 2.
Gel filtration analysis of FH-COUP complexes. Nuclear extracts from pOZ-COUP (A) and pOZ-N (B) cells were subjected to size-exclusion chromatography using a Superose 6 column. Fractions were collected and analyzed by immunoblotting using the antibodies indicated on the left. The elution peaks of the molecular mass standards are indicated at the top: blue dextran, 2000 kDa (2K); thyroglobulin, 669 kDa; and alcohol dehydrogenase, 150 kDa. Fraction numbers are indicated at the bottom.
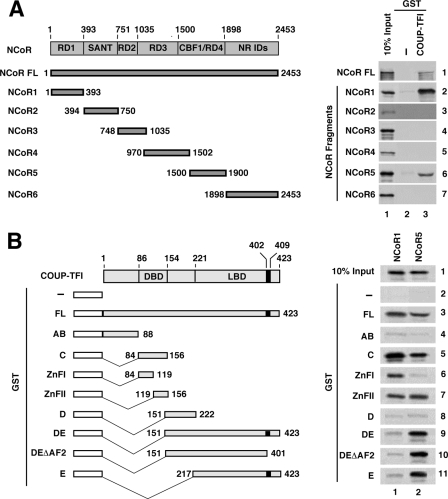
COUP-TFI Interacts Directly with Two Different Regions in NCoR—Because NCoR has been identified as a corepressor for several hormone nuclear receptors (8, 47), we hypothesized that NCoR may interact directly with COUP-TFI, nucleate the FH-COUP complex, and serve as a scaffold for other protein/protein interactions within the FH-COUP complex. Moreover, Shibata et al. (20) previously reported a direct interaction between COUP-TFI and a large fragment of NCoR in yeast and in vitro, but neither defined the COUP-TFI interaction domain of NCoR precisely nor directly demonstrated a role for NCoR in COUP-mediated repression. GST pulldown experiments were performed to investigate the former in detail, and these studies revealed that GST-COUP-TFI interacted with full-length NCoR (Fig. 3A, panel 1) as well as with NCoR fragments containing repression domain (RD) 1 (corresponding to amino acids 1–393, NCoR1) (panel 2) and RD4 (also known as the CBF1/Su[H] domain, corresponding to amino acids 1500–1900, NCoR5) (panel 6). Other isolated regions of NCoR, including the canonical nuclear receptor interaction domain (NCoR6) (Fig. 3A, panel 7), did not appear to interact with GST-COUP-TFI. In reciprocal pulldown experiments, we confirmed that COUP-TFI was specifically pulled down by both GST-NCoR1 and GST-NCoR5 (supplemental Fig. S2).
FIGURE 3.
NCoR interacts directly with COUP-TFI in an atypical manner. A, diagram of NCoR and deletion mutants (left) used for GST pulldown experiments (right). NCoR fragments were translated in vitro as [35S]Met-labeled proteins, and the presence of NCoR fragments in the pulldown was detected by autoradiography. Lane 1 in all cases was loaded with 10% of the input of individual NCoR fragments. B, diagram of FH-COUP and deletion mutants (left) used for GST pulldown experiments (right) using various GST-COUP-TFI fragments and in vitro translated NCoR1 (lane 1) or NCoR5 (lane 2). The putative AF2 of COUP-TFI (amino acids 402–409) is denoted by a black box. Representative experiments are shown. NR IDs, nuclear receptor interaction domains; FL, full-length; LBD, ligand-binding domain; ZnFI and ZnFII, zinc fingers I and II, respectively.
The NCoR interaction domains of COUP-TFI were similarly mapped in reciprocal pulldown experiments. These studies revealed that the isolated RD1 and RD4 interacted directly with full-length COUP-TFI (Fig. 3B, panel 3). However, the two NCoR fragments interacted differentially with isolated regions of COUP-TFI. For example, NCoR1, which contains RD1, interacted strongly with any deletion mutant containing the COUP-TFI DNA-binding domain (DBD) (Fig. 3B, panels 5–7, lane 1). NCoR1 appeared to interact with zinc fingers I and II with equivalent efficiency (compare lane 1 in Fig. 3B, panels 6 and panel 7), neither of which was as efficient as the two fingers together (panel 5, lane 1). In contrast, NCoR5, which harbors RD4, interacted directly with the isolated COUP DBD (Fig. 3B, panel 5, lane 2) and any mutant containing the ligand-binding domain (Fig. 3B, panels 9–11, lane 2). The interaction of NCoR5 with the COUP-TFI DBD differed from that of NCoR1 because NCoR5 appeared to interact with zinc finger II but not with zinc finger I and did so as efficiently as with the full-length DBD (compare lane 2 in Fig. 3B, panels 5–7). Previous work has identified the AF2 region near the C terminus of COUP-TFI as an important region for COUP-TFI-mediated repression (20). However, the AF2 domain of COUP-TFI was dispensable for the interaction of the orphan receptor with NCoR5 (Fig. 3B, panel 10, lane 2), and neither the intact COUP ligand-binding domain (panel 9, lane 1) nor derivative mutants (panels 8, 10, and 11, lane 1) interacted with NCoR1. We conclude that two distinct domains of COUP-TFI interact differentially with two regions of NCoR: the COUP-TFI DBD was found to interact with fragments harboring both NCoR RD1 and CBF1/RD4, whereas the COUP-TFI ligand-binding domain interacted only with the NCoR fragment harboring the CBF1/RD4 region. Thus, NCoR appears to play a crucial role in nucleating the large FH-COUP-containing complex that we observed in HeLa S3 cells, but the manner by which COUP-TFI interacts with NCoR appears to be different from that of other members of the nuclear receptor superfamily.
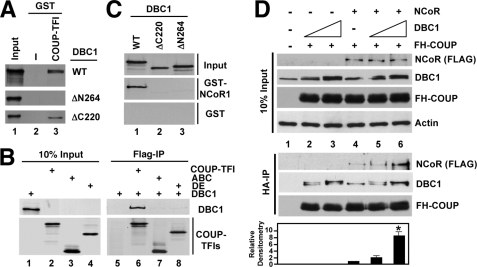
DBC1 Interacts Directly with Both COUP-TFI and NCoR—We found that COUP-TFI interacted directly with DBC1 in GST pulldown studies in a manner that required the N but not the C terminus of DBC1 (Fig. 4A, lane 3). Whereas full-length FH-COUP interacted with wild-type DBC1 (Fig. 4B, lane 3), we could not define the DBC1 interaction domain(s) of COUP-TFI more precisely by truncation mutagenesis, as neither the isolated N- or C-terminal regions of COUP interacted with DBC1 (Fig. 4B, lanes 4 and 5). This suggests that a higher degree of structural complexity of COUP-TFI is required for its interaction with DBC1.
FIGURE 4.
DBC1 interacts directly with both COUP-TFI and NCoR. A, in vitro pulldown of DBC1 by GST or GST-COUP-TFI. Lane 1 was loaded with 10% of the input used in pulldown reactions. DBC1 protein (full-length or truncation mutants) was translated in the presence of [35S]Met and detected by autoradiography. WT, wild-type. B, in vitro pulldown of DBC1 by full-length FH-COUP or FH-COUP fragments by immunoprecipitation with the anti-FLAG antibody. The upper panels correspond to the signal for full-length DBC1 ([35S]Met-labeled protein), and the lower panels demonstrate that equal amounts of FLAG-COUP proteins were immunoprecipitated. Lanes 1–4 contain 10% of the input used in the immunoprecipitation reactions shown in lanes 5–8. ABC and DE refer to regions of COUP-TFI, as defined in Fig. 3B. C, in vitro pulldown of DBC1 and deletion mutant proteins by GST-NCoR1 and GST. DBC1 and its deletion mutants were detected as described for A. D, co-IP of transfected FH-COUP, DBC1, and NCoR in 293T cell extracts. Cell extracts were immunoprecipitated using the anti-HA antibody to pull down FH-COUP, and immune complexes were analyzed by immunoblotting with the antibodies indicated on the right. Note that the lower band in the DBC1 blot (middle panel) corresponds to endogenous protein, whereas the signal for the upper band (*) is derived from transfected DBC1-V5His. The NCoR expression vector used in this experiment harbors a C-terminal FLAG epitope, and transfected NCoR protein was detected using the anti-FLAG antibody. The lower panel depicts a densitometric analysis of the NCoR (FLAG) blot, indicating the amount of NCoR (FLAG) pulled down in each immunoprecipitation. This experiment is representative of two additional studies.
DBC1 also interacted with GST-NCoR1 (Fig. 4C, lane 1), which contains RD1 (see above). However, unlike FH-COUP, DBC1 did not interact with NCoR5, the CBF1/RD4 region of the corepressor (supplemental Fig. S3, panel e, lane 4). DBC1 also did not interact with other NCoR fragments tested (data not shown). Both the C- and N-terminal regions of DBC1 appeared to be required for the interaction with NCoR (Fig. 4C, lanes 1–3).
The results described above suggest that DBC1 may function as an adaptor protein between COUP-TFI and NCoR, and this was tested in co-IP studies in human embryonic kidney cells that had been transiently transfected with combinations of FH-COUP, DBC1, and NCoR. As illustrated in Fig. 4D (lanes 3–5), transfection of increasing amounts of DBC1 significantly increased the amount of NCoR that co-immunoprecipitated with FH-COUP in 293T cells. Collectively, our data revealed direct interactions between COUP-TFI, DBC1, and NCoR, suggesting that DBC1 may function as adaptor protein that stabilizes the interaction of COUP-TFI and NCoR and possibly maintains the integrity of the FH-COUP complex.
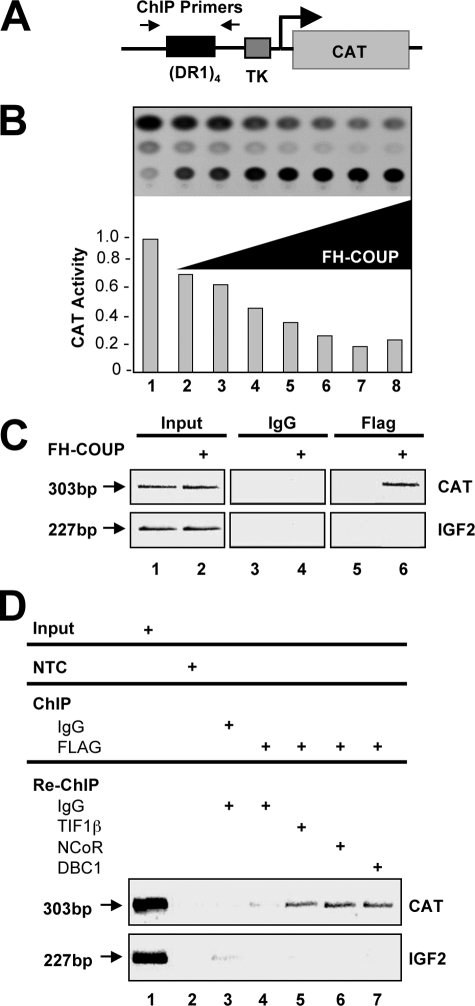
NCoR, TIF1β, and DBC1 Co-occupy the Promoter of an FH-COUP-responsive Promoter—To investigate the association of FH-COUP and its identified interaction proteins on a COUP-responsive promoter, we transiently cotransfected 293T cells with an expression vector encoding FH-COUP and a reporter construct harboring a multimerized DR1 motif in the context of the thymidine kinase promoter upstream of the CAT reporter (Fig. 5A). Transfection of increasing amounts of FH-COUP expression vector resulted in repression of this reporter, which reached a maximum of ∼5-fold (Fig. 5B). We next determined whether NCoR, TIF1β, and DBC1 co-occupied the COUP-responsive promoter of the CAT reporter together with FH-COUP. As expected, FH-COUP was found on this template in cells transfected with the FH-COUP expression vector but not in cells transfected with the empty vector (Fig. 5C). NCoR, TIF1β, and DBC1 were all found to co-occupy the FH-COUP-responsive promoter with FH-COUP in transiently transfected cells (Fig. 5D, upper and lower panels, respectively, lanes 5–8). These data demonstrate that a COUP-TFI complex minimally containing FH-COUP, NCoR, TIF1β, and DBC1 is present on the promoter template of a COUP-TFI-responsive reporter gene.
FIGURE 5.
Co-occupation of a COUP-responsive promoter by FH-COUP, NCoR, TIF1β, and DBC1. A, schematic diagram of the (DR1)4-tk-CAT reporter construct. ChIP primers were designed to amplify the region encompassing the multimerized DR1 element as indicated by the arrows. TK, thymidine kinase. B, repression of the basal CAT repression by COUP-TFI in 293T cells. Relative CAT activities are showed in the lower panel. C, ChIP analyses demonstrating that COUP-TFI associates with the promoter region of the CAT reporter in transiently transfected cells. A region of the IGF2 promoter (–1198 to –1424 bp upstream of the transcriptional start site, 227-bp amplicon) was used as a negative control to demonstrate the specificity of FH-COUP binding to the (DR1)4-tk-CAT promoter. D, re-ChIP analyses demonstrate that FH-COUP, NCoR, TIF1β, and DBC1 co-occupy the (DR1)4-tk-CAT promoter. NTC, no template control.
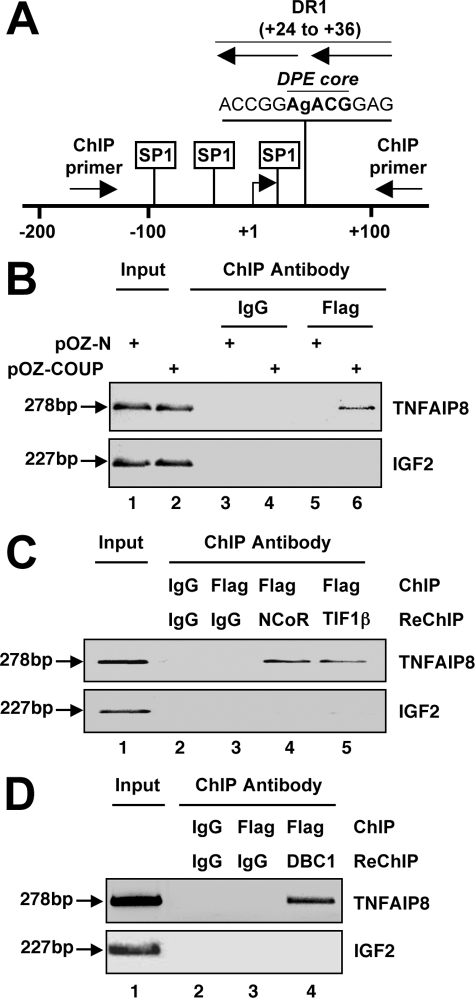
The COUP-TFI Complex Regulates the Promoter of TNFAIP8—To extend the above results to natural chromatinized promoters, we first identified putative COUP-TFI target genes using a genome-wide ChIP-chip approach.3 From this screen, we identified the anti-apoptotic gene TNFAIP8 as a putative target of COUP-TFI. The promoter region of TNFAIP8 has not been experimentally characterized, but computational examination of the predicted promoter identified a degenerate DR1-like response element located ∼30 bp downstream of the transcriptional start site. This DR1-like element is found within the putative downstream core promoter element (sequence RGWYVT) (Fig. 6A), which has been characterized by Kadonaga and co-workers (48). Further analyses also revealed that the TNFAIP8 promoter lacks a conventional TATA box but harbors multiple Sp1-binding sites within 100 bp of the predicted start site (Fig. 6A). ChIP analyses conducted in pOZ-COUP cells validated the presence of FH-COUP on the TNFAIP8 promoter (Fig. 6B). Moreover, re-ChIP analyses demonstrated that FH-COUP, NCoR, TIF1β, and DBC1 all co-occupied the same fragment of the TNFAIP8 promoter (Fig. 6, C and D).
FIGURE 6.
Identification of an endogenous target of the COUP-TFI·NCoR complex in HeLa S3 cells. A, schematic diagram of the TATA-less promoter region of the TNFAIP8 gene highlighting the locations of the GC box (Sp1-binding site), the downstream core promoter element (DPE), and the putative COUP-TFI response element (DR1). The locations of ChIP primers, designed to amplify the region covering the putative COUP-TFI-binding site, are indicated by arrows. B, ChIP analysis demonstrating FH-COUP on the TNFAIP8 promoter in pOZ-COUP but not pOZ-N cells. The primers used for the amplification reaction are shown schematically in A. C, and D, re-ChIP analyses demonstrating co-occupancy of the TNFAIP8 promoter by FH-COUP, NCoR, TIF1β, and DBC1.
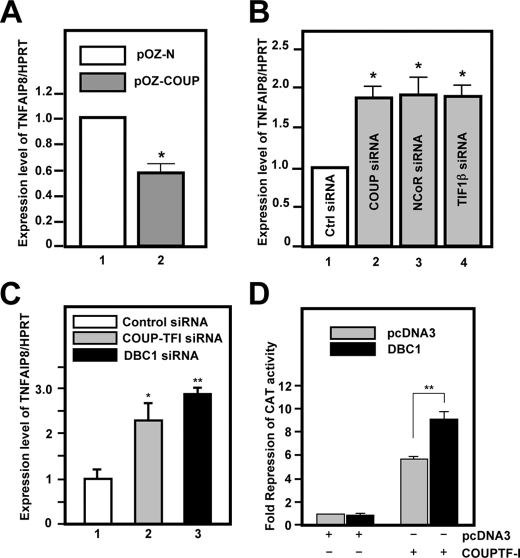
Roles of NCoR, TIF1β, and DBC1 in COUP-TFI-mediated Repression—To determine the transcriptional outcome of the interaction of the FH-COUP complex with the TNFAIP8 promoter, we first examined the effect of COUP-TFI overexpression on TNFAIP8 transcript levels in HeLa S3 cells. Quantitative reverse transcription-PCR (RT-qPCR) analyses revealed that TNFAIP8 mRNA levels were down-regulated (∼50%) in pOZ-COUP cells relative to pOZ-N cells (Fig. 7A). This was confirmed by knocking down endogenous COUP-TFI expression, which resulted in an ∼2-fold increase in TNFAIP8 expression (Fig. 7B). Similarly, knockdown of NCoR and TIF1β resulted in derepression of the TNFAIP8 promoter to a level that was indistinguishable from that of COUP-TFI knockdown (Fig. 7B). The siRNA-mediated knockdown of COUP-TFI, NCoR, and TIF1β expression was verified by immunoblot analyses (supplemental Fig. S4A). Collectively, our results indicate that the COUP-TFI·NCoR complex represses expression of TNFAIP8 expression in untransfected HeLa S3 cells.
FIGURE 7.
NCoR, TIF1β, and DBC1 are required for COUP-mediated transcriptional repression. A, repression of TNFAIP8 expression by stable overexpression of FH-COUP in pOZ-COUP cells as revealed by RT-qPCR analyses. The relative -fold change in TNFAIP8 mRNA expression was calculated relative to the -fold change in the expression of a housekeeping gene, hypoxanthineguanine phosphoribosyltransferase (HPRT). Bars represent mean expression levels ± S.E. (n = 3) of TNFAIP8 relative to hypoxanthine-guanine phosphoribosyltransferase from independent determinations. Statistical significance was determined using Student's t test (*, p = 0.03). B, HeLa S3 cells transfected with negative control (Ctrl) siRNA or siRNA targeting COUP-TFI, NCoR, or TIF1β and subjected to RT-qPCR analyses. The expression level of TNFAIP8 was determined by RT-qPCR as described above. C, siRNA-mediated COUP-TFI and DBC1 knockdown results in derepression of TNFAIP8 expression in 293T cells. Transfection conditions and analyses were as described above. D, DBC1 stimulates COUP-TFI-dependent gene repression in 293T cells. Cells were transfected with the (DR1)4-tk-CAT reporter, FH-COUP, and DBC1-V5His DNA constructs as indicated. CAT activities were quantified, and the level of -fold repression was calculated relative to the basal reporter activity. Addition of DBC1 significantly stimulated the -fold repression of CAT activity mediated by COUP-TFI (**, p < 0.01).
We do not know if DBC1 plays a role in COUP-TFI-mediated repression of the TNFAIP8 promoter in HeLa S3 cells because we could not achieve appreciable knockdown of DBC1 expression in these cells (data not shown). However, DBC1 does appear to play a role in COUP-TFI-mediated repression in HEK293T cells, in which we could knock down DBC1 expression using an siRNA approach (supplemental Fig. S4B). First, similar to HeLa S3 cells, TNFAIP8 expression was also induced upon knockdown of endogenous COUP-TFI in 293T cells, suggesting that TNFAIP8 is a bona fide target of the COUP-TFI transcriptional repressor complex in both HeLa and HEK293 cells. Notably, FH-COUP-mediated repression of the endogenous TNFAIP8 promoter was reversed by knockdown of DBC1 (Fig. 7C). In addition, cotransfection of DBC1 stimulated significantly the FH-COUP-dependent repression of the (DR1)4-tk-CAT reporter in 293T cells (Fig. 7D and supplemental Fig. S4C). Thus, we conclude that NCoR, TIF1β, and DBC1 all play key roles in COUP-TFI-mediated transcriptional repression.
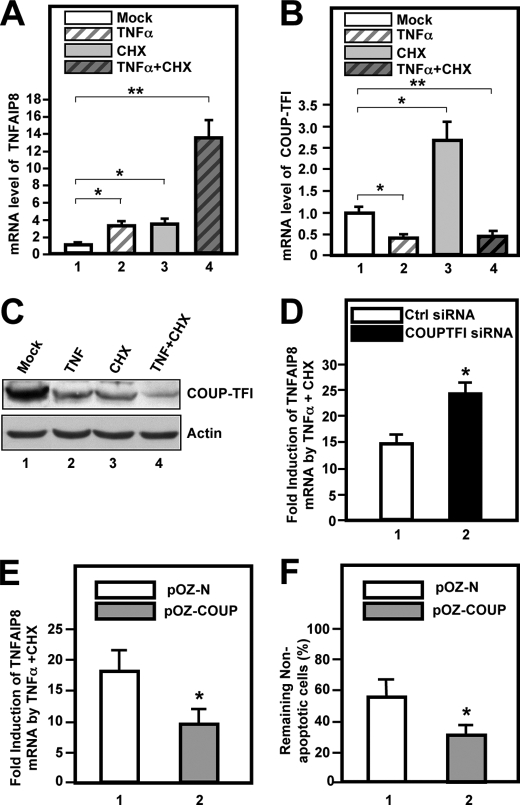
Role of COUP-TFI during TNFα-induced Apoptosis—TNFAIP8 is up-regulated by TNFα treatment (49), and the TNFAIP8 protein functions as a caspase inhibitor, which appears to limit the extent of apoptosis induced by TNFα (50). The results presented above also identified TNFAIP8 as a target of COUP-TFI-mediated repression in HeLa S3 and human embryonic kidney cells. Therefore, we investigated the possibility that COUP-TFI may play a direct or indirect role in the induction of TNFAIP8 by TNFα as well as in TNFα-induced apoptosis in HeLa S3 cells. We confirmed that TNFAIP8 was induced by ∼4-fold by TNFα in HeLa S3 cells (Fig. 8A, bar 2). We also found that expression of TNFAIP8 was induced by the protein synthesis inhibitor CHX (Fig. 8A, bar 3) and that the combination of TNFα and CHX acted synergistically to induce TNFAIP8 in HeLa S3 cells (∼15-fold induction) (Fig. 8A). It is well established that both pro-apoptotic and anti-apoptotic pathways are activated by TNFα (51) and that CHX may inhibit the translation of anti-apoptotic mRNAs induced by TNFα, thus potentiating TNFα-induced apoptosis (51). In contrast, COUP-TFI expression was found to be divergently regulated by TNFα and CHX: TNFα down-regulated COUP-TFI expression by ∼65% (Fig. 8B, bar 2), whereas CHX treatment increased COUP-TFI mRNA levels by nearly 3-fold (bar 3). The latter finding may suggest that a labile repressor dictates basal COUP-TFI expression levels in HeLa S3 cells. Strikingly, concomitant treatment with TNFα and CHX completely reversed the stimulatory effect of CHX on COUP-TFI mRNA levels (Fig. 8B, bar 4). TNFα, alone or in combination with CHX, also down-regulated COUP-TFI protein levels (Fig. 8C). However, the stimulatory effect of CHX on COUP-TFI mRNA levels did not generalize to the protein level, which is likely due to the inhibitory effect of CHX on protein synthesis.
FIGURE 8.
Role of COUP-TFI in TNFα/CHX-induced apoptosis. A, treatment of HeLa S3 cells with TNFα and protein synthesis inhibitor CHX synergistically induced TNFAIP8 expression as determined by RT-qPCR. Cells were treated with vehicle (Me2SO, indicated as Mock), TNFα (5 μg/ml), CHX (30 μg/ml), or TNFα/CHX for 4 h. TNFAIP8 transcript levels were analyzed by RT-qPCR, and statistical significance was determined using Student's t test (*, p < 0.05; **, p < 0.01). B, treatment with TNFα but not CHX down-regulated COUP-TFI transcripts in HeLa S3 cells. COUP-TFI transcript levels were determined by RT-qPCR and expressed relative to those of hypoxanthine-guanine phosphoribosyltransferase as shown. C, TNFα and CHX down-regulated COUP-TFI protein levels. HeLa S3 cells were treated as indicated for 4 h, and cells were lysed immediately and subjected to immunoblot analyses using antibody against endogenous COUP-TFI or actin as a loading control. D, knockdown of COUP-TFI stimulated induction of TNFAIP8 by TNFα/CHX treatment. HeLa S3 cells were transfected with COUP-TFI siRNA for 40 h prior to TNFα/CHX treatment. Induction of TNFAIP8 was measured by RT-qPCR as shown. E, TNFAIP8 induction by TNFα/CHX was attenuated in pOZ-COUP cells relative to pOZ-N cells. F, overexpression of COUP-TFI sensitized HeLa S3 cells to apoptosis initiated by TNFα/CHX. pOZ-COUP or pOZ-N cells were grown on coverslips and treated with TNFα/CHX for 8 h. The percentage of the remaining non-apoptotic cells after this treatment was determined by 4′,6-diamidino-2-phenylindole staining. * and **, statistical significance at the p < 0.05 and p < 0.01 levels, respectively, when comparing treatments with mock treatment (A and B), COUP-TFI-specific siRNA with control siRNA (D), and pOZ-COUP cells with pOZ-N cells (E and F).
The above results prompted us to determine whether TNFα/CHX-induced down-regulation of COUP-TFI protein levels played a role in the induction of TNFAIP8 mRNA by TNFα/CHX, and we addressed this using two approaches. First, we examined the effect of COUP-TFI knockdown on induction of TNFAIP8 mRNA by TNFα/CHX. We found that knockdown of COUP-TFI potentiated TNFα/CHX-mediated induction of the TNFAIP8 promoter by ∼2-fold (Fig. 8D). Second, we compared induction of TNFAIP8 by TNFα/CHX in cells overexpressing COUP-TFI (pOZ-COUP cells) and control cells (pOZ-N cells). We found that overexpression of COUP-TFI impaired the induction of TNFAIP8 by TNFα/CHX relative to control cells (Fig. 8E), and as a functional correlate of this, cells overexpressing COUP-TFI were also more sensitive to TNFα/CHX-induced apoptosis (Fig. 8F). Considered together, these results suggest that COUP-TFI plays a direct or indirect role in the induction of the TNFAIP8 gene by TNFα with or without inhibition of protein synthesis by CHX.
DISCUSSION
The orphan nuclear receptor COUP-TFI is a transcription factor that plays essential roles in the regulation of key biological processes, presumably in the context of a multiprotein complex. To our knowledge, this is the first study to identify component proteins of cellular complexes containing COUP-TFI using a proteomic approach, which revealed that COUP is associated with a large number of proteins in HeLa S3 cells.
As with many other nuclear receptor complexes, NCoR appears to serve as a scaffold protein that couples COUP-TFI to histone deacetylases, which may underlie the mechanistic basis for COUP-TFI-mediated transcriptional repression. However, the molecular basis for the interaction of COUP-TFI with NCoR appeared to differ from that of other previously studied nuclear receptors in that COUP-TFI did not interact with the canonical nuclear receptor interactions domains located in the C terminus of NCoR (8, 52). Rather, the DNA- and putative ligand-binding domains of COUP-TFI interacted with RD1 and RD4 of NCoR, respectively. RD4 of NCoR also interacted with the COUP-TFI DBD but did so in a manner that differed from that of NCoR RD1. However, it remains unclear if the relatively small COUP-TFI DBD can interact with one or two domains of NCoR while simultaneously binding DNA in a sequence-specific manner. This seems unlikely without some type of facilitation, and we propose that the adaptor protein DBC1 functions to stabilize COUP-TFI/NCoR interaction on the promoter of target genes such as TNFAIP8. Indeed, DBC1 was found to co-occupy the TNFAIP8 promoter with COUP-TFI, NCoR, and TIF1β, and all of these proteins were crucial for the repression of the TNFAIP8 promoter by COUP-TFI. Further studies are clearly needed to map the protein/protein interaction network within the COUP-TFI complex in greater detail and to define the role of other complex proteins such as BRM and TIF1β in the transcriptional regulatory activity of COUP-TFI.
Previous studies by Torchia and co-workers (53) revealed the existence of at least two chromatographically distinct NCoR complexes: NCoR1, containing NCoR, HDAC3, TIF1β, BAF155, BAF170, and SF3A/3B; and NCoR2, containing NCoR, SIN3A, HDAC1, and HDAC1/2. Subsequently, Roeder and co-workers (54) described a SIN3A-less NCoR complex containing NCoR, HDAC3, TBL, TBLR1, and GPS2, which may be the same as or highly related to that described by Wong and co-workers (55). These findings support the idea that NCoR can interact with a variety of proteins in an extremely flexible manner. The FH-COUP·NCoR complex described herein appears to be most similar to the SIN3A-less NCoR1 complex described by Torchia and co-workers (53) in that the complex that we identified clearly contains TIF1β, the BRM-associated factors, and SF3A/3B. However, unlike NCoR1 or the NCoR complex described by the Roeder (54) and Wong (55) laboratories, we found no evidence for the presence of HDAC3 in the FH-COUP·NCoR complex. Moreover, HDAC1, which is not present in NCoR1, was identified by peptide sequencing to be a component of FH-COUP complexes, and this was verified by co-IP analyses. Considered together, our findings suggest the existence of a novel NCoR complex that harbors FH-COUP, as well as other proteins such as DBC1, and that has no appreciated role in transcriptional regulation. Nonetheless, as observed previously in the case of other nuclear receptor complexes, NCoR appears to play the role of a scaffold protein in FH-COUP complexes that couples the orphan receptor to the transcriptional repression machinery (i.e. histone deacetylases).
DBC1 is a pro-apoptotic protein that was originally cloned from human chromosome 8p21, a region that is homozygously deleted in some breast cancer cells (56). DBC1 is localized exclusively in the nucleus of healthy cells (35) and has been reported to interact directly with and inhibit the catalytic activity of the class III histone deacetylase SIRT1 (57). Upon apoptotic stimulation, DBC1 undergoes caspase-dependent processing with rapid degradation of the N terminus, and the C-terminal fragment translocates from the nucleus to the mitochondria, promoting apoptosis by sensitizing cells to apoptotic signals (35). As reported herein, DBC1 also appears to associate with the COUP-TFI complex on the promoter of COUP-TFI target genes and functions to stabilize interaction between NCoR and COUP-TFI, thus contributing to COUP-TFI-mediated transcriptional repression. In this study, we found that the DBC1 N terminus is required for the interaction with COUP-TFI. Therefore, as a result of caspase cleavage, DBC1 may no longer be able to interact with COUP-TFI during TNFα-induced apoptosis. Thus, it may be of interest to study other dynamic changes taking place within the COUP-TFI complex during TNFα-induced apoptosis.
TIF1β, which is also known as KAP-1 (KRAB-associated protein 1) and TRIM28 (tripartite motif containing 28) in humans and mice, is a transcriptional corepressor of the KRAB domain-containing zinc finger protein family (58). TIF1β harbors multiple protein/protein interaction and functional domains, including an N-terminal region with homology to RBCC (ring finger/B-box/coiled-coil) proteins, a HP1 (heterochromatin protein 1)-binding domain, an NCoR homology domain, and a C-terminal region known as the PB region (which contains a plant homeodomain (PHD) finger and a bromodomain in tandem) (64). The PHD of TIF1β interacts strongly with the SUMO E2 transfer protein Ubc9 and functions as an intramolecular E3 SUMO ligase that directs Ubc9 to sumoylate specific lysine residues in the bromodomain of TIF1β (64). The sumoylated bromodomain of TIF1β recruits the nucleosome-remodeling and d–eacetylation NuRD complex, which deacetylates template-associated histones, including Lys9 of histone H3. In addition, the sumoylated bromodomain of KAP-1 appears to recruit the lysine-specific methyltransferase SETDB1, which methylates this residue, forming a binding site for HP1 on the template, thus promoting the silenced state of the promoter. TIF1β/KAP-1 is clearly an integral component of the COUP-TFI complex and is required for COUP-TFI-mediated transcriptional repression of the TNFAIP8 promoter. However, it remains to be determined if the intramolecular E3 SUMO ligase activity of TIF1β/KAP-1 is required for the transcriptional repressive properties of the COUP-TFI complex on target gene promoters.
The SWI/SNF complex has been shown to disrupt nucleosome structure and to increase DNA accessibility for transcriptional activators (1). Although it is tempting to speculate that the Brahma-containing COUP-TFI complex may be involved in COUP-TFI-mediated transcriptional activation, for example, of ovalbumin (59), cholesterol 7α-hydroxylase (60), and aldosterone synthase (29) promoters, we cannot rule out the possibility that Brahma and associated factors may be part of the COUP-TFI·NCoR complex that represses transcription. Indeed, recent studies have found that the SWI/SNF remodeling activity of Brahma is required for transcriptional repression as well as transcriptional activation (46). Brahma appears to interact with the methyl-CpG-binding protein MeCP2 to form a repressive complex that also contains SIN3A and HDAC1 (46). Moreover, a number of studies have demonstrated that Brahma is associated with HDAC1 and HDAC2 (61, 62) as well as NCoR and TIF1β (53). Further studies are required to define the specific role(s) of the SWI/SNF complex in the transcriptional regulatory activity of COUP-TFI.
TNFAIP8, which is also known as SCC-S2 or NDED, is a novel oncogene that plays a role in tumor progression. The N terminus of TNFAIP8 harbors a sequence that is highly homologous to death effector domain II of FLICE inhibitory proteins (FLIP), which has been shown to block death receptor-mediated apoptosis by preventing caspase-8 activation (50) TNFAIP8 expression is up-regulated by TNFα stimulation and by activation of NF-κB in tumor cell lines (49, 50). Overexpression of TNFAIP8 leads to enhanced survival and inhibition of apoptosis through inhibition of the apoptotic proteins caspase-3 and caspase-8 (30).
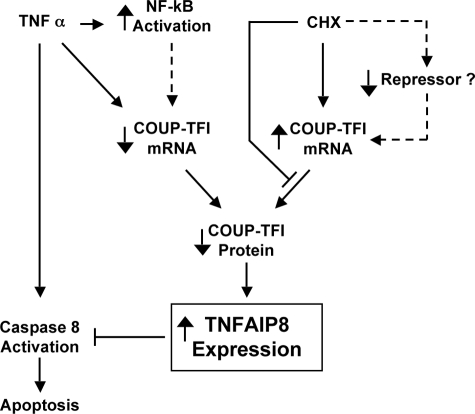
We identified a novel anti-apoptotic pathway that was initiated by TNFα and involved down-regulation of COUP-TFI expression, derepression of the newly identified COUP-TFI target gene TNFAIP8, and subsequent inhibition of apoptosis (Fig. 9). Thus, down-regulation of COUP-TFI expression, which resulted in less COUP-TFI on the TNFAIP8 promoter (supplemental Fig. S5), would appear to be an important component of the signaling pathway leading to induction of TNFAIP8 expression by TNFα. At present, we do not know the mechanistic connection between TNFα signaling and repression of COUP-TFI expression. However, the proximal region of the COUP-TFI promoter harbors seven putative, but highly conserved, NF-κB-binding sites, and it is conceivable that NF-κB may work through one or more of these binding sites to repress COUP-TFI expression (Fig. 9) in a manner similar to that by which NF-κB represses the Bmp4 promoter (63).
FIGURE 9.
Role of COUP-TFI during TNFα-induced apoptosis. Solid and dashed lines represent known and hypothesized events, respectively. Note that treatment with either TNFα or CHX reduces endogenous COUP-TFI protein levels, which relieves repression of the TNFAIP8 promoter, resulting in inhibition of caspase-8 activation and reduced apoptosis. Conversely, ectopically overexpressed COUP-TFI represses TNFAIP8 expression in a manner that is insensitive TNFα, resulting in loss of TNFAIP8 protein and increased sensitivity to TNFα-induced apoptosis. Thus, COUP-TFI may play a central role in the sensitivity of a cell to TNFα-induced apoptosis.
Finally, it is of note that most of the FH-COUP-associated proteins identified herein appear to be involved in transcriptional repression, even though COUP-TFI has been identified as a transcriptional activator of a number of genes (29, 59, 60). The basis for our findings is unclear, but it is conceivable that our proteomic studies were conducted under conditions in which an activating COUP-TFI “signal” was not present. Such an activating signal could include small molecules and/or post-translational modification of the protein; for example, phosphorylation (64, 65). As described previously for many other nuclear receptors, activating COUP-TFI signals may influence the dynamics of coregulator exchange at the level of the promoter-bound orphan receptor, thus influencing the rate of transcription of a subset of COUP-TFI target genes (8). Studies aimed at identifying these putative signals will provide further molecular insight into the regulation of gene expression by COUP-TFI.
Supplementary Material
Acknowledgments
We thank Valerie Peterson for outstanding technical assistance, Dr. Siva Kolluri for very helpful suggestions, and Drs. Wayne Kradjan and Gary DeLander for continuous support and encouragement.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60852 (to M. L.). This work was also supported by the Oregon State University College of Pharmacy. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: NCoR, nuclear receptor corepressor; COUP-TF, chicken ovalbumin upstream promoter transcription factor; SUMO, small ubiquitin-like modifier; TNFα, tumor necrosis factor α; HA, hemagglutinin; GST, glutathione S-transferase; MS, mass spectrometry; MS/MS, tandem mass spectrometry; CAT, chloramphenicol acetyltransferase; ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; CHX, cycloheximide; co-IP, co-immunoprecipitation; RD, repression domain; DBD, DNA-binding domain; RT-qPCR, quantitative reverse transcription-PCR.
L. Zhang and M. Leid, unpublished data.
References
- 1.Narlikar, G. J., Fan, H. Y., and Kingston, R. E. (2002) Cell 108 475–487 [DOI] [PubMed] [Google Scholar]
- 2.Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007) Cell 128 635–638 [DOI] [PubMed] [Google Scholar]
- 3.Kadonaga, J. T. (1998) Cell 92 307–313 [DOI] [PubMed] [Google Scholar]
- 4.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., and Evans, R. M. (1995) Cell 83 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson-Rechavi, M., Escriva Garcia, H., and Laudet, V. (2003) J. Cell Sci. 116 585–586 [DOI] [PubMed] [Google Scholar]
- 6.Zamir, I., Harding, H. P., Atkins, G. B., Horlein, A., Glass, C. K., Rosenfeld, M. G., and Lazar, M. A. (1996) Mol. Cell. Biol. 16 5458–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perissi, V., Aggarwal, A., Glass, C. K., Rose, D. W., and Rosenfeld, M. G. (2004) Cell 116 511–526 [DOI] [PubMed] [Google Scholar]
- 8.Glass, C. K., and Rosenfeld, M. G. (2000) Genes Dev. 14 121–141 [PubMed] [Google Scholar]
- 9.Avram, D., Ishmael, J. E., Nevrivy, D. J., Peterson, V. J., Lee, S. H., Dowell, P., and Leid, M. (1999) J. Biol. Chem. 274 14331–14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toulouse, A., Rochefort, D., Roussel, J., Joober, R., and Rouleau, G. A. (2003) Genomics 82 162–171 [DOI] [PubMed] [Google Scholar]
- 11.Pereira, F. A., Tsai, M. J., and Tsai, S. Y. (2000) CMLS Cell. Mol. Life Sci. 57 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu, Y., Pereira, F. A., DeMayo, F. J., Lydon, J. P., Tsai, S. Y., and Tsai, M. J. (1997) Genes Dev. 11 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Studer, M., Filosa, A., and Rubenstein, J. L. (2005) Brain Res. Bull. 66 394–401 [DOI] [PubMed] [Google Scholar]
- 14.Zhou, C., Qiu, Y., Pereira, F. A., Crair, M. C., Tsai, S. Y., and Tsai, M. J. (1999) Neuron 24 847–859 [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. T., Li, L., Takamoto, N., Martin, J. F., Demayo, F. J., Tsai, M. J., and Tsai, S. Y. (2004) Mol. Cell. Biol. 24 10835–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira, F. A., Qiu, Y., Zhou, G., Tsai, M. J., and Tsai, S. Y. (1999) Genes Dev. 13 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warnecke, M., Oster, H., Revelli, J. P., Alvarez-Bolado, G., and Eichele, G. (2005) Genes Dev. 19 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, Y., and Dufau, M. L. (2003) Mol. Cell. Biol. 23 6958–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus, S. L., Winrow, C. J., Capone, J. P., and Rachubinski, R. A. (1996) J. Biol. Chem. 271 27197–27200 [DOI] [PubMed] [Google Scholar]
- 20.Shibata, H., Nawaz, Z., Tsai, S. Y., O'Malley, B. W., and Tsai, M. J. (1997) Mol. Endocrinol. 11 714–724 [DOI] [PubMed] [Google Scholar]
- 21.Rohr, O., Aunis, D., and Schaeffer, E. (1997) J. Biol. Chem. 272 31149–31155 [DOI] [PubMed] [Google Scholar]
- 22.Dressel, U., Thormeyer, D., Altincicek, B., Paululat, A., Eggert, M., Schneider, S., Tenbaum, S. P., Renkawitz, R., and Baniahmad, A. (1999) Mol. Cell. Biol. 19 3383–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avram, D., Fields, A., Pretty On Top, K., Nevrivy, D. J., Ishmael, J. E., and Leid, M. (2000) J. Biol. Chem. 275 10315–10322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohr, O., Schwartz, C., Hery, C., Aunis, D., Tardieu, M., and Schaeffer, E. (2000) J. Biol. Chem. 275 2654–2660 [DOI] [PubMed] [Google Scholar]
- 25.Huggins, G. S., Bacani, C. J., Boltax, J., Aikawa, R., and Leiden, J. M. (2001) J. Biol. Chem. 276 28029–28036 [DOI] [PubMed] [Google Scholar]
- 26.Shibata, H., Kobayashi, S., Kurihara, I., Saito, I., and Saruta, T. (2003) Horm. Res. 59 Suppl. 1, 85–93 [DOI] [PubMed] [Google Scholar]
- 27.Kurihara, I., Shibata, H., Kobayashi, S., Saito, I., and Saruta, T. (2002) Endocr. Res. 28 581. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, S., Shibata, H., Kurihara, I., Yokota, K., Suda, N., Saito, I., and Saruta, T. (2004) J. Mol. Endocrinol. 32 69–86 [DOI] [PubMed] [Google Scholar]
- 29.Kurihara, I., Shibata, H., Kobayashi, S., Suda, N., Ikeda, Y., Yokota, K., Murai, A., Saito, I., Rainey, W. E., and Saruta, T. (2005) J. Biol. Chem. 280 6721–6730 [DOI] [PubMed] [Google Scholar]
- 30.Kumar, D., Gokhale, P., Broustas, C., Chakravarty, D., Ahmad, I., and Kasid, U. (2004) Oncogene 23 612–616 [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, H., Ishiguro, K., Gaubatz, S., Livingston, D. M., and Nakatani, Y. (2002) Science 296 1132–1136 [DOI] [PubMed] [Google Scholar]
- 32.Shi, Y., Sawada, J., Sui, G., Affar el, B., Whetstine, J. R., Lan, F., Ogawa, H., Luke, M. P., and Nakatani, Y. (2003) Nature 422 735–738 [DOI] [PubMed] [Google Scholar]
- 33.Nakatani, Y., and Ogryzko, V. (2003) Methods Enzymol. 370 430–444 [DOI] [PubMed] [Google Scholar]
- 34.Picard, F., Kurtev, M., Chung, N., Topark-Ngarm, A., Senawong, T., Machado De Oliveira, R., Leid, M., McBurney, M. W., and Guarente, L. (2004) Nature 429 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundararajan, R., Chen, G., Mukherjee, C., and White, E. (2005) Oncogene 24 4908–4920 [DOI] [PubMed] [Google Scholar]
- 36.Shapiro, D. J., Sharp, P. A., Wahli, W. W., and Keller, M. J. (1988) DNA (N. Y.) 7 47–55 [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850–858 [DOI] [PubMed] [Google Scholar]
- 38.Yi, E. C., Lee, H., Aebersold, R., and Goodlett, D. R. (2003) Rapid Commun. Mass Spectrom. 17 2093–2098 [DOI] [PubMed] [Google Scholar]
- 39.Keller, A., Eng, J., Zhang, N., Li, X., and Aebersold, R. (2005) Mol. Syst. Biol. 1 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topark-Ngarm, A., Golonzhka, O., Peterson, V. J., Barrett, B., Jr., Martinez, B., Crofoot, K., Filtz, T. M., and Leid, M. (2006) J. Biol. Chem. 281 32272–32283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senawong, T., Peterson, V. J., Avram, D., Shepherd, D. M., Frye, R. A., Minucci, S., and Leid, M. (2003) J. Biol. Chem. 278 43041–43050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dowell, P., Ishmael, J. E., Avram, D., Peterson, V. J., Nevrivy, D. J., and Leid, M. (1997) J. Biol. Chem. 272 33435–33443 [DOI] [PubMed] [Google Scholar]
- 43.Wang, L. H., Tsai, S. Y., Cook, R. G., Beattie, W. G., Tsai, M. J., and O'Malley, B. W. (1989) Nature 340 163–166 [DOI] [PubMed] [Google Scholar]
- 44.Wang, L. H., Tsai, S. Y., Sagami, I., Tsai, M. J., and O'Malley, B. W. (1987) J. Biol. Chem. 262 16080–16086 [PubMed] [Google Scholar]
- 45.Wang, W., Xue, Y., Zhou, S., Kuo, A., Cairns, B. R., and Crabtree, G. R. (1996) Genes Dev. 10 2117–2130 [DOI] [PubMed] [Google Scholar]
- 46.Harikrishnan, K. N., Chow, M. Z., Baker, E. K., Pal, S., Bassal, S., Brasacchio, D., Wang, L., Craig, J. M., Jones, P. L., Sif, S., and El-Osta, A. (2005) Nat. Genet. 37 254–264 [DOI] [PubMed] [Google Scholar]
- 47.Horlein, A. J., Naar, A. M., Heinzel, T., Torchia, J., Gloss, B., Kurokawa, R., Ryan, A., Kamei, Y., Soderstrom, M., Glass, C. K., and Rosenfeld, M. G. (1995) Nature 377 397–404 [DOI] [PubMed] [Google Scholar]
- 48.Juven-Gershon, T., Hsu, J. Y., Theisen, J. W., and Kadonaga, J. T. (2008) Curr. Opin. Cell Biol. 20 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar, D., Whiteside, T. L., and Kasid, U. (2000) J. Biol. Chem. 275 2973–2978 [DOI] [PubMed] [Google Scholar]
- 50.You, Z., Ouyang, H., Lopatin, D., Polver, P. J., and Wang, C. Y. (2001) J. Biol. Chem. 276 26398–26404 [DOI] [PubMed] [Google Scholar]
- 51.Gaur, U., and Aggarwal, B. B. (2003) Biochem. Pharmacol. 66 1403–1408 [DOI] [PubMed] [Google Scholar]
- 52.Seol, W., Mahon, M. J., Lee, Y. K., and Moore, D. D. (1996) Mol. Endocrinol. 10 1646–1655 [DOI] [PubMed] [Google Scholar]
- 53.Underhill, C., Qutob, M. S., Yee, S. P., and Torchia, J. (2000) J. Biol. Chem. 275 40463–40470 [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J., Kalkum, M., Chait, B. T., and Roeder, R. G. (2002) Mol. Cell 9 611–623 [DOI] [PubMed] [Google Scholar]
- 55.Yoon, H. G., Chan, D. W., Huang, Z. Q., Li, J., Fondell, J. D., Qin, J., and Wong, J. (2003) EMBO J. 22 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamaguchi, M., Meth, J. L., von Klitzing, C., Wei, W., Esposito, D., Rodgers, L., Walsh, T., Welcsh, P., King, M. C., and Wigler, M. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, W., Kruse, J. P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008) Nature 451 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanov, A. V., Peng, H., Yurchenko, V., Yap, K. L., Negorev, D. G., Schultz, D. C., Psulkowski, E., Fredericks, W. J., White, D. E., Maul, G. G., Sadofsky, M. J., Zhou, M. M., and Rauscher, F. J., III (2007) Mol. Cell 28 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sagami, I., Tsai, S. Y., Wang, H., Tsai, M. J., and O'Malley, B. W. (1986) Mol. Cell. Biol. 6 4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroup, D., Crestani, M., and Chiang, J. Y. (1997) J. Biol. Chem. 272 9833–9839 [DOI] [PubMed] [Google Scholar]
- 61.Pal, S., Yun, R., Datta, A., Lacomis, L., Erdjument-Bromage, H., Kumar, J., Tempst, P., and Sif, S. (2003) Mol. Cell. Biol. 23 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sif, S., Saurin, A. J., Imbalzano, A. N., and Kingston, R. E. (2001) Genes Dev. 15 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, N. L., Li, C., Huang, H. H., Sebald, M., Londhe, V. A., Heisterkamp, N., Warburton, D., Bellusci, S., and Minoo, P. (2007) Gene (Amst.) 393 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kane, C. D., and Means, A. R. (2000) EMBO J. 19 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gay, F., Barath, P., Desbois-Le Peron, C., Metivier, R., Le Guevel, R., Birse, D., and Salbert, G. (2002) Mol. Endocrinol. 16 1332–1351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.