Abstract
The cellular response to glucocorticoid receptor (GR) activation involves a highly orchestrated series of regulatory actions influenced at multiple levels by a variety of mechanisms including the action of transcription factors and chromatin modifiers. Because the majority of GR binding sites (glucocorticoid-responsive elements (GREs)) are distant from promoters, it is likely that interactions at a distance play an important role in GR action. To determine whether long range chromosomal associations play a role in transcription regulation by GR, we utilized a chromosome conformation capture-based technique (associated chromosome trap) to identify unknown, remote sequences that interact with the GR-induced Lipocalin2 (Lcn2) gene. Our screen revealed that the Lcn2 GRE interacts with the Ciz1 gene, nearly 30 kb upstream. Ciz1 was subsequently found to be a novel GR-responsive gene. The GRE proximal to the Lcn2 promoter apparently functions to regulate both the Lcn2 gene and the distal Ciz1 gene. Using quantitative chromosome conformation capture, we find that a loop structure is organized between these two genes. This structure is hormone-independent and present only in cell types where the genes are active. The strong correlation between gene expression and loop structure in different cell lines suggests that high order interactions play a role in determining tissue-specific gene regulation.
The glucocorticoid receptor (GR)2 is a member of the nuclear hormone receptor superfamily. Hormone-activated GR binds to glucocorticoid-responsive elements (GREs) in the vicinity of target genes (1, 2) inducing both positive and negative regulation of transcription (2). The transcriptional activity of GR depends on its interaction with coactivators and corepressors in multiprotein complexes that modulate chromatin structure and interact with the basal transcriptional machinery (3).
An additional level of complexity in transcription regulation in eukaryotes resides in the action of distant genomic elements such as enhancers (4, 5) and locus control regions (6). The recently developed chromosome conformation capture (3C) technique (7) has been valuable in constructing a picture of the regulatory architecture, stressing the spatial organization of gene enhancer interactions and other interactions.
Although 3C has proved to be invaluable in deciphering complex interacting partners located on one chromosome (in cis) or on different chromosomes (in trans) (8), this method is limited by the need for prior identification of candidate partners. Modifications of the 3C technique, such as associated chromosome trap (ACT), enable identification of previously unknown interacting partners of a specific region of interest (9).
Glucocorticoid receptor and estrogen receptor binding sites across the genome have been recently shown to be located at great distances from the transcription start site of regulated genes (2, 10–14), suggesting that GREs and EREs loop to their target genes to participate in transcription regulation.
Using the ACT approach to find potential interacting targets for the GR-regulated gene, Lipocalin2 (Lcn2), we find that the Lcn2 GRE is associated with the upstream region of Cip1-interacting zinc finger protein (Ciz1). Ciz1 was subsequently found to be a GR-regulated gene. To characterize this interaction, we carried out a detailed 3C analysis and found that the loop structure between Lcn2 and Ciz1 is hormone-independent, suggesting that GR binding itself is not the determinative event in formation of the long range interaction. Moreover, loop formation is evident only in cell types where the two loci are active. Our data suggest that active GREs loop to their target sites prior to hormone induction. It is possible that such an organization allows a rapid transcriptional response to hormone stimulation at responsive loci.
EXPERIMENTAL PROCEDURES
Cell Culture—Mouse mammary epithelial adenocarcinoma cells (3134) (15) pituitary (AtT-20) (16) and hepatocyte (Hepa1C1C7) cell lines (17) were maintained as per standard protocols. Culture conditions are described in detail in the supplemental materials.
Associated Chromosome Trap—ACT was performed after treating the cells with 100 nm dexamethasone (Dex) for 1 h as described previously (9). Lcn2-specific primers and a detailed procedure are provided in the supplemental materials.
ChIP, Reverse Transcription-PCR, and Real-time Quantitative-PCR—Experiments were carried out after treating the cells with either vehicle or 100 nm Dex for the indicated time as described (14). A detailed procedure is provided in the supplemental materials.
Chromosome Conformation Capture—The 3C assay was performed as described previously (8, 18) with minor modifications. A detailed procedure is provided in the supplemental materials.
ChIP Loop Assay—The ChIP loop assay was carried out as described previously (19, 20) with minor modifications described in the supplemental materials.
RESULTS
ACT Screen Uncovers an Lcn2-associated Partner—To determine whether long range chromosomal associations (or chromosome looping) play a role in transcriptional regulation by GR, we performed an ACT assay to identify unknown, remote sequences that interact with the Lcn2 promoter. ACT is a variation of the 3C technique. Unlike the 3C technique, which requires prior identification of both elements of the interacting DNA segments, the ACT assay permits a search for putative interacting DNA elements using a known sequence as bait.
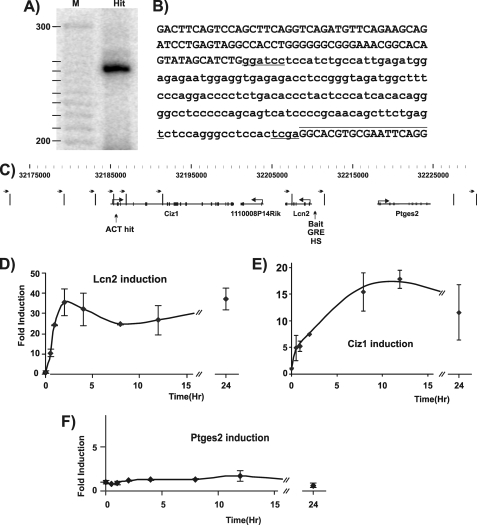
Lipocalin2 (Lcn2) was selected to serve as a bait for the ACT screen for several reasons; Lcn2 transcription increases rapidly in response to Dex hormone treatment in mammary cells (3134 cell line), peaks at 2 h after induction, and then plateaus up to 24 h from induction (Fig. 1D). The Lcn2 promoter was carefully characterized and found to harbor a GRE by tiled ChIP-on-chip as well as by conventional ChIP and was found to become hypersensitive to DNase I in response to Dex treatment (14). Using Lcn2 as bait, we identified several non-continuous, interacting DNA sequences throughout the genome. One of these events represented a putative interaction between the Lcn2 promoter and a region located 300 bp downstream of Ciz1 ATG, which is located 24,881 bp from the Lcn2 bait (Fig. 1, A–C). This interaction was chosen for future characterization.
FIGURE 1.
The ACT assay reveals Ciz1 as an Lcn2 associated partner under hormone regulation. A, PCR reactions products were run on 5% polyacrylamide electrophoresis gels and analyzed with PhosphorImager, showing a 260-bp DNA product. B, the PCR product was sequenced and revealed Ciz1 (lowercase letters), flanked by Lcn2 (uppercase letters) and a BamHI restriction site (lowercase underline letters) on one side, and the linker (uppercase letters, overline) together with a TaqI restriction site (lowercase underline letters) on the other side. C, graphical representation of the Lcn2-Ciz1 locus. Gene structures from transcription start site to end are shown with introns (lines) and exons (boxes) and 3C and ChIP loop assays primers (black arrows on top), HindIII restriction sites (vertical black lines), GRE, and the Dex-induced DNase I hypersensitivity site (HS). D–F, time course analysis of Lcn2, Ciz1, and Ptges2 transcript levels following hormone treatment. Samples were collected before DEX treatment (0) and at 0.5, 1, 2, 4, 8, 12, and 24 h after Dex induction. Lcn2 (D) and Ciz1 (E) are up-regulated in response to Dex treatment, whereas Ptges2 transcription (F) is unaffected. Results shown are the average of two independent experiments, and S.D. values are indicated (error bars).
Ciz1 Is a GR-regulated Gene—We asked whether Ciz1, a putative Lcn2-associated partner, is also regulated by hormone (Dex). Analysis of Ciz1 transcription showed that Ciz1 is up-regulated following Dex stimulation. Moreover, the kinetics of Ciz1 induction is similar to that of Lcn2, with a rapid induction followed by persistently elevated transcript production (Fig. 1E). Because there could be local GR-mediated expression of multiple genes in the Lcn2 neighborhood, we asked whether Ptges2 expression, which is proximal to Lcn2 in the opposite direction from Ciz1 (Fig. 1C), is also up-regulated by Dex and found that it is not a glucocorticoid-responsive gene (Fig. 1F). Interestingly, Ciz1 was not previously known to be a hormone-responsive gene and was not identified as such by a screen using genomic tools (14, 21). In summary, we took an unbiased approach to search for loci interacting with Lcn2 and identified Ciz1, which was then observed to be a GR-regulated gene.
Lcn2-Ciz1 Interact in Vivo—Loci along a chromatin fiber might be in physical proximity despite the linear distance between them, and they will randomly interact as the result of the intrinsic flexibility of chromatin (7). This trait is usually reflected in the 3C assay and in the 3C-based screens (22–24), resulting in amplification of a 3C DNA product, which may not indicate a functional loop between the bait and the nearby sequences. Nevertheless it is essential to test whether there is a loop between Lcn2 and Ciz1 by using quantitative 3C. We applied 3C using one primer proximal to the HindIII restriction site within the Lcn2 promoter together with HindIII sites across the locus, comparing the non-induced and the hormone-induced states. Our 3C results show that there is a decrease followed by elevation in the interaction frequency 30–35 kb from the Lcn2 GRE, indicating that there is a loop between the Lcn2 GRE and the Ciz1 upstream region. In the absence of a loop, the interaction frequency would be expected to stay low after the drop in the interaction frequency at 26 kb from Lcn2 GRE. As all the primer pairs amplify the Bac control template with similar efficiencies, we confirmed the validity of the sharp drop-off and rise in the interaction frequency with alternative primers and found that the pattern remains (supplemental Fig. 1). Interestingly, the interaction frequency between the two loci is not dependent on the presence of hormone (Fig. 2A). The interaction frequency in the Ciz1 direction decreased 26 kb away from Lcn2 GRE, whereas in the Ptges2 direction, it dropped after 19 kb, which may suggest that there is an interaction between the lcn2 GRE and elements within the 26-kb domain.
FIGURE 2.
Long range interactions between Lcn2 GRE and Ciz1. A, 3C analysis shows high interaction frequencies between Lcn2 GRE and Ciz1 upstream region in the presence (diamond points) or absence (square points) of hormone. The Lcn2 GRE primer was used as an anchor and paired with primers across and surrounding the Ciz1 gene. The graph presents relative cross-linking frequency on the vertical axis. The x axis indicates the position in kb relative to the Lcn2 GRE in the middle of the HindIII fragment. S.E. values are indicated. Results shown are the average of two independent biological experiments with PCR reactions from each experiment performed in duplicates for 6–8 times. Each signal was normalized to control templates to correct for primer efficiency and also to the cross-linking frequency for two close fragments of the Gapd locus as an internal control to correct for differences in 3C preparations in different conditions and repeats. B, Lcn2 GRE interacts with Ciz1 upstream region and GR protein in cells under Dex stimulation as shown by quantitative PCR analysis of GR ChIP loop DNA. Each signal was normalized to the GR ChIP loop signal of non-induced samples. Data shown are the average of three independent experiments. S.D. values are indicated. C, time course GR ChIP loop assay between Lcn2 GRE and the Ciz1 upstream region (32 kb apart) indicates a stable interaction frequency. Cells were harvested at 1, 4, and 8 h after Dex treatment. Data were normalized to GR ChIP loop signal with non-induced samples and to GR occupancy at the Lcn2 GRE. Results shown are the average of two independent experiments. S.D. values are indicated.
To confirm the presence of an Lcn2-Ciz1 loop and to ask whether GR is present in this loop, we performed a ChIP loop assay (19, 20). In the ChIP loop assay, immunoprecipitation following cross-linking and restriction enzyme processing enriches the sample for fragments bound to a specific protein complex. Following ligation, the chimeric DNA fragments are then subjected to quantitative PCR, determining the interaction frequency. We took advantage of the fact that GR is bound to the promoter region of Lcn2 in the presence of hormone (14) and carried out a ChIP loop assay with GR antibodies. We found that Lcn2 and Ciz1 form a loop and that GR interacts with the loop (Fig. 2B). The peak of interaction with Lcn2 over the Ciz1 locus is 30 kb apart, in agreement with the 3C results (Fig. 2A). We could not detect interactions between Lcn2 and loci at the Ptges2 locus ∼20 kb from Lcn2 in the opposite direction to Ciz1 and also at regions surrounding the peak (Fig. 2B), suggesting that the interaction detected between Lcn2 and Ciz1 upstream regions represents a loop rather than merely a trait of the chromatin structure, allowing interactions within proximal promoter regions.
A time course ChIP analysis shows that equilibrium GR binding at the Lcn2 GRE is unchanged over a 24-h period, as long as Dex is continuously applied to the cells.3 We then determined the frequency of GR binding to the loop using a time course ChIP loop assay. We found that the frequency of GR binding in the loop is stable up to 8 h after induction (Fig. 2C). In contrast to the robust binding of GR at the Lcn2 promoter, ChIP experiments with GR antibodies did not reveal any significant enrichment for GR over a 12-kb region spanning the Ciz1 transcription start site, suggesting that Lcn2 and Ciz1 might both be regulated by a common GR site located proximal to Lcn2 (supplemental Fig. 2).
Cell Type-specific Loop Structure Is Associated with Transcription—If the loop structure plays a role in transcription regulation in a tissue-specific manner, we would expect to find a correlation between the two. To explore this possibility, we first characterized the hormone-dependent response of Ciz1 and Lcn2 in different cell lines. We found that both Lcn2 and Ciz1 are induced by Dex in the hepatocyte cell line, Hepa1C1C7 (17), but not in the AtT20 pituitary cell (16) (Fig. 3, C–F). Furthermore, 3C analysis across the Lcn2-Ciz1 locus with one fixed primer at the Lcn2 GRE together with primers spanning the entire Lcn2-Ciz1 region shows that the loop topology in Hepa1C1C7 cells is similar to 3134 cells (Fig. 3B). However, the loop structure is completely absent in AtT20 cells where Lcn2 and Ciz1 are not expressed (Fig. 3A). The correlation between expression and loop structure in different cell lines suggests that loop formation plays a role in transcriptional regulation of both genes (Fig. 4).
FIGURE 3.
Cell-specific expression of Lcn2 and Ciz1 is correlated with loop formation. A and B, 3C analysis over the Lcn2-Ciz1 locus shows interaction frequencies between the Lcn2 GRE and the Ciz1 upstream region in the presence (diamond) or absence (square) of hormone in AtT20 cells (A) and in Hepa1 cells (B). Controls and symbols are described in the legend for Fig. 2. C–F, mRNA analysis shows that Lcn2 and Ciz1 transcription is strongly up-regulated in response to Dex treatment in Hepa1 liver cells but not in AtT20 pituitary cells. Results shown are the average of three independent experiments. S.D. values are indicated.
FIGURE 4.
Loop formation in the Lcn2-Ciz1 domain. A long range interaction is detected between the immediate upstream regions of the Lcn2 and Ciz1 promoters, both in the presence and in the absence of hormone (Dex). This interaction is likely to result from dynamic and transient pairing between the regions occurring on a rapid time scale. The glucocorticoid receptor, although not a critical component of the loop interaction, stimulates transcriptional activity of both promoters by mechanisms related to the loop-induced proximity of the promoters. Other long range interactions between the Lcn2 GRE and intermediate regions of the Ciz1 domain are possible (– – –).
DISCUSSION
The observation that distant regulatory elements can associate with, and activate, core promoter elements has been well established in several genetic systems. In other cases, long range interactions have been associated with transcriptional repression (25, 26). The majority of nuclear receptor binding sites are located distant (greater than 10 kb) from transcription start sites, as exemplified by EREs in human breast cancer (10, 11) and mouse liver cells (12) and by GREs in human lung cells (2). This suggests that GREs and EREs loop to their target genes. In this respect, we wanted to probe the nuclear environment of the GRE of the strongly hormone-induced gene, Lcn2. We took advantage of the newly developed ACT assay (9) and performed an unbiased search for physical association of genome regions with the Lcn2 GRE. Our screen revealed Ciz1 as an associated partner of Lcn2. Quantitative 3C and ChIP loop assays demonstrated an interaction between the Lcn2 GRE and the upstream region of Ciz1, with GR participating in this preformed 30-kb loop after hormone induction. Ciz1 transcription analysis revealed that Ciz1, like Lcn2, is up-regulated by GR; moreover, the two genes share the same time-dependent expression kinetics.
The frequency of interaction between the Lcn2 GRE and Ciz1 upstream regions is not affected by hormone or the transcriptional status of the two genes. Our 3C studies extend the understanding of GREs and gene-looping phenomenon in higher eukaryotes and show that two elements can be organized in a higher order structure prior to hormone induction. This preorganized environment may facilitate a rapid transcriptional response to hormone stimulus.
Different cell types vary in their response to GR. We hypothesized that if loop structure plays a role in transcription, we would expect a correlation between the two. We observed that Lcn2 and Ciz1 are co-regulated by GR in mammary cell lines and liver cell lines. Furthermore, 3C analysis of the locus showed that the two loci form long range interactions in both cell types. In contrast, in pituitary AtT20 cells, neither of these genes are GR-regulated, and neither are found in the 30-kb loop structure. These findings indicate that specific long range interactions play a role in cell type-specific GR function.
The kinetics of GR-stimulated transcriptional output is quite complex, often with alternate activation and repression phases (27). The common transcription profile of Lcn2 and Ciz1 suggests that the two genes are regulated by a common mechanism. It is possible that genes sharing the same kinetic behavior are also co-localized in the nucleus in specialized microenvironments and that these microenvironments contain a set of factors governing a common transcriptional response (22, 24, 28–30). An examination of multiple sets of GR-regulated genes will be required to explore the validity of this concept.
The molecular basis of looping is not yet clear, although interactions between structural proteins and transcription factors are all candidates (25, 26, 31, 32). To understand the role of the GR protein in the loop, we performed a GR ChIP loop assay and found that GR participates in the loop between Lcn2 and Ciz1. The fact that GR participates in the loop may suggest that GR binding sites would be found at the two ends of the loop by ChIP. However, a detailed GR ChIP across a 12-kb region upstream of the Ciz1 gene did not reveal any direct GR binding events. Moreover a high throughput sequencing of GR binding sites (ChIP-seq) shows that the Lcn2 GRE is the only one within 200 kb around the locus,3 suggesting that the Ciz1 response is mediated through the Lcn2 GRE. GR binding to a given GRE has been uniformly associated with chromatin transitions, as reflected by increased hypersensitivity to DNase I (DHS) (14). In agreement with the ChIP data, the only induced DHS site found in the Lcn2-Ciz1 locus is at the Lcn2 GRE (14), further suggesting that the Lcn2 GRE is the only GRE present in this large chromatin domain. These findings support a hypothesis by which a single GRE at the Lcn2 gene regulates the expression of both genes.
Given the dynamic nature of transcription factor movement in living cells (33), it seems possible that the long range interactions are transient in nature. The interactions detected by biochemical analysis between the Ciz1 upstream region and the Lcn2 proximal region likely reflect an average in the cell population that is fixed by formaldehyde (34). Therefore, one conclusion from our work is that the interaction frequency between Lcn2 GRE and GR is larger than the interaction frequency between the Lcn2 GRE to the Ciz1 locus (Fig. 4). The fact that the interaction frequency is reduced only 26 kb away from Lcn2 GRE may suggest that the structure is more complex than a simple loop: i.e. there may also be a long range interaction between Lcn2 GRE and elements within the 26 kb. A general finding from this study is that nuclear receptor elements on the genome interact with nuclear receptors more frequently than they loop to their targets. Several studies (2, 10–14) show that most nuclear receptor elements are distant from their promoter target sites. However, these binding elements are rarely found cross-linked to transcription start sites, consistent with the observations described here for Ciz1. In addition, imaging studies show that GR foci are not co-localized with RNA polymerase II foci (35), suggesting that the two proteins are bound distant from each other on the chromatin fiber on most of the sites across the genome.
Although the reaction to GR is fast, it is possible that within this time, the responsive genes are rearranged in the nucleus, forming new interaction networks. To gain a wider view of the nuclear organization in response to GR induction, it will be necessary to look at the Lcn2 environment more extensively by additional approaches, such as the novel 3C-on-chip (4C) methodology (22, 24).
Supplementary Material
This work was supported, in whole or in part, by a National Institutes of Health grant from the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, two supplemental figures, and four supplemental tables.
Footnotes
The abbreviations used are: GR, glucocorticoid receptor; GRE, glucocorticoid-responsive element; ERE, estrogen-responsive element; 3C, chromosome conformation capture; ACT, associated chromosome trap; Dex, dexamethasone; ChIP, chromatin immunoprecipitation.
S. John et al., unpublished results.
References
- 1.Schoneveld, O. J., Gaemers, I. C., and Lamers, W. H. (2004) Biochim. Biophys. Acta 1680 114–128 [DOI] [PubMed] [Google Scholar]
- 2.So, A. Y., Chaivorapol, C., Bolton, E. C., Li, H., and Yamamoto, K. R. (2007) PLoS. Genet. 3 e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hager, G. L., Nagaich, A. K., Johnson, T. A., Walker, D. A., and John, S. (2004) Biochim. Biophys. Acta 1677 46–51 [DOI] [PubMed] [Google Scholar]
- 4.Banerji, J., Rusconi, S., and Schaffner, W. (1981) Cell 27 299–308 [DOI] [PubMed] [Google Scholar]
- 5.Wasylyk, B., Wasylyk, C., Augereau, P., and Chambon, P. (1983) Cell 32 503–514 [DOI] [PubMed] [Google Scholar]
- 6.Grosveld, F., van Assendelft, G. B., Greaves, D. R., and Kollias, G. (1987) Cell 51 975–985 [DOI] [PubMed] [Google Scholar]
- 7.Dekker, J., Rippe, K., Dekker, M., and Kleckner, N. (2002) Science 295 1306–1311 [DOI] [PubMed] [Google Scholar]
- 8.Spilianakis, C. G., Lalioti, M. D., Town, T., Lee, G. R., and Flavell, R. A. (2005) Nature 435 637–645 [DOI] [PubMed] [Google Scholar]
- 9.Ling, J. Q., Li, T., Hu, J. F., Vu, T. H., Chen, H. L., Qiu, X. W., Cherry, A. M., and Hoffman, A. R. (2006) Science 312 269–272 [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., Liu, X. S., Brodsky, A. S., Li, W., Meyer, C. A., Szary, A. J., Eeckhoute, J., Shao, W., Hestermann, E. V., Geistlinger, T. R., Fox, E. A., Silver, P. A., and Brown, M. (2005) Cell 122 33–43 [DOI] [PubMed] [Google Scholar]
- 11.Carroll, J. S., Meyer, C. A., Song, J., Li, W., Geistlinger, T. R., Eeckhoute, J., Brodsky, A. S., Keeton, E. K., Fertuck, K. C., Hall, G. F., Wang, Q., Bekiranov, S., Sementchenko, V., Fox, E. A., Silver, P. A., Gingeras, T. R., Liu, X. S., and Brown, M. (2006) Nat. Genet. 38 1289–1297 [DOI] [PubMed] [Google Scholar]
- 12.Gao, H., Falt, S., Sandelin, A., Gustafsson, J. A., and Dahlman-Wright, K. (2007) Mol. Endocrinol. 22 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magee, J. A., Chang, L. W., Stormo, G. D., and Milbrandt, J. (2006) Endocrinology 147 590–598 [DOI] [PubMed] [Google Scholar]
- 14.John, S., Sabo, P. J., Johnson, T. A., Sung, M. H., Biddie, S. C., Lightman, S. L., Voss, T. C., Davis, S. R., Meltzer, P. S., Stamatoyannopoulos, J. A., and Hager, G. L. (2008) Mol. Cell 29 611–624 [DOI] [PubMed] [Google Scholar]
- 15.Walker, D., Htun, H., and Hager, G. L. (1999) Methods (Amst.) 19 386–393 [DOI] [PubMed] [Google Scholar]
- 16.Richardson, U. I., and Schonbrunn, A. (1981) Endocrinology 108 281–290 [DOI] [PubMed] [Google Scholar]
- 17.Hankinson, O. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F., and de, L. W. (2002) Mol. Cell 10 1453–1465 [DOI] [PubMed] [Google Scholar]
- 19.Horike, S., Cai, S., Miyano, M., Cheng, J. F., and Kohwi-Shigematsu, T. (2005) Nat. Genet. 37 31–40 [DOI] [PubMed] [Google Scholar]
- 20.Cai, S., Lee, C. C., and Kohwi-Shigematsu, T. (2006) Nat. Genet. 38 1278–1288 [DOI] [PubMed] [Google Scholar]
- 21.Johnson, T. A., Elbi, C., Parekh, B. S., Hager, G. L., and John, S. (2008) Mol. Biol. Cell 19 3308–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonis, M., Klous, P., Splinter, E., Moshkin, Y., Willemsen, R., de, W. E., van, S. B., and de, L. W. (2006) Nat. Genet. 38 1348–1354 [DOI] [PubMed] [Google Scholar]
- 23.Wurtele, H., and Chartrand, P. (2006) Chromosome Res. 14 477–495 [DOI] [PubMed] [Google Scholar]
- 24.Zhao, Z., Tavoosidana, G., Sjolinder, M., Gondor, A., Mariano, P., Wang, S., Kanduri, C., Lezcano, M., Sandhu, K. S., Singh, U., Pant, V., Tiwari, V., Kurukuti, S., and Ohlsson, R. (2006) Nat. Genet. 38 1341–1347 [DOI] [PubMed] [Google Scholar]
- 25.Fraser, P. (2006) Curr. Opin. Genet. Dev. 16 490–495 [DOI] [PubMed] [Google Scholar]
- 26.Ling, J. Q., and Hoffman, A. R. Pediatr. Res. (2007) 61 11R–16R [DOI] [PubMed] [Google Scholar]
- 27.John, S., Johnson, T. A., Sung, M. H., Biddie, S. C., Trump, s., koch-Paiz, C. A., Davis, S. R., Walker, R., Meltzer, P. S., and Hager, G. L. (2009) Endocrinology, in press [DOI] [PMC free article] [PubMed]
- 28.Xu, M., and Cook, P. R. (2008) J. Cell Biol. 181 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser, P., and Bickmore, W. (2007) Nature 447 413–417 [DOI] [PubMed] [Google Scholar]
- 30.Misteli, T. (2007) Cell 128 787–800 [DOI] [PubMed] [Google Scholar]
- 31.de, L. W., and Grosveld, F. (2007) Curr. Opin. Genet. Dev. 17 456–464 [DOI] [PubMed] [Google Scholar]
- 32.de, L. W. (2007) Curr. Opin. Cell Biol. 19 317–320 [DOI] [PubMed] [Google Scholar]
- 33.McNally, J. G., Muller, W. G., Walker, D., Wolford, R., and Hager, G. L. (2000) Science 287 1262–1265 [DOI] [PubMed] [Google Scholar]
- 34.Voss, T. C., John, S., and Hager, G. L. (2006) Mol. Endocrinol. 20 2641–2655 [DOI] [PubMed] [Google Scholar]
- 35.Muller, W. G., Rieder, D., Karpova, T. S., John, S., Trajanoski, Z., and McNally, J. G. (2007) J. Cell Biol. 177 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.