Abstract
Resistin antagonizes insulin action in mouse, making it a potential therapeutic target for treating metabolic diseases such as diabetes. To better understand how mouse resistin gene (Retn) expression is restricted to fat tissue, we identified an adipocyte-specific enhancer located ∼8.8-kb upstream of the transcription start site. This region contains a binding site for the master adipogenic regulator peroxisome proliferator-activated receptor γ (PPARγ), and binds endogenous PPARγ together with its partner retinoid-X receptor α (RXRα). It also contains three binding sites for CCAAT/enhancer-binding protein (C/EBP), and is bound by endogenous C/EBPα and C/EBPβ in adipocytes. Exogenous expression of PPARγ/RXRα and C/EBPα in non-adipocyte cells synergistically drives robust expression from the enhancer. Although PPARγ ligands repress Retn transcription in adipocytes, rosiglitazone paradoxically stimulates the enhancer activity, suggesting that the enhancer is not directly involved in negative regulation. Unlike expression of Retn in mouse, human resistin (RETN) is expressed primarily in macrophages. Interestingly, the region homologous to the mouse Retn enhancer in the human gene contains all three C/EBP elements, but is not conserved for the sequence bound by PPARγ. Furthermore, it displays little or no binding by PPARγ in vitro. Taken together, the data suggest that a composite enhancer binding both PPARγ and C/EBP factors confers adipocyte-specific expression to Retn in mouse, and its absence from the human gene may explain the lack of adipocyte expression in humans.
Obese individuals have increased risk of insulin resistance, which leads to type 2 diabetes, hypertension, and cardiovascular disease (1). Thus, adipose tissue has been intensively studied to better understand the molecular mechanism linking obesity and insulin resistance. For a long time, adipose tissue was considered as a passive fuel depot. However, after the discovery of leptin, adipose tissue has been widely recognized as an endocrine organ that actively controls whole body metabolism by secreting bioactive molecules including non-esterified fatty acid, adiponectin, and tumor necrosis factor-α (2, 3).
Resistin was identified as a peptide hormone secreted from adipocytes (4). It belongs to a novel class of cysteine-rich proteins termed Resistin-like Molecules (RELMs)/Found in Inflammatory Zone (FIZZ) (4–6), and circulates in blood as forms of hexamers or trimers (7). Plasma resistin levels are increased in obese rodents (4). Various mouse models, including infusion of recombinant resistin, overexpression of resistin in liver, or deletion of Retn, have consistently shown that resistin antagonizes insulin action (8–10). Although these reports clearly indicate that resistin plays major roles in normal glucose homeostasis as well as the pathophysiology of insulin resistance in rodents, it is controversial whether these conclusions translate to the human situation (11). In humans, resistin is minimally expressed in adipose tissue, but predominantly expressed in mononuclear cells including macrophages (7, 12, 13). Some studies showed that plasma resistin levels and single-nucleotide polymorphism were associated with obesity, insulin resistance, and type 2 diabetes (14–22), while other studies did not show these associations (23–26).
The expression of mouse Retn is highly restricted to adipocytes (4, 10, 27), but the mechanism of fat-specific expression remains unclear. Differentiation of adipocytes is controlled by a cascade of transcription factors: notably, basic-helix-loop-helix transcription factors, CCAAT/enhancer-binding proteins (C/EBPs)3 and a nuclear hormone receptor, peroxisome proliferator-activated receptor γ (PPARγ) (28, 29). C/EBPβ and C/EBPδ are rapidly induced in the early stage of adipogenesis (30, 31). These transcription factors subsequently drive C/EBPα and PPARγ expression (32–34). C/EBPα and PPARγ can induce each other in a positive feedback loop and directly activate many of the genes for terminal adipocyte differentiation (28, 29, 35). PPARγ can induce adipogenesis in C/EBPα-deficient MEFs (35), whereas C/EBPα is incapable of driving the adipogenic program in the absence of PPARγ (36). Thereby, PPARγ is considered as a master regulator of adipogenesis (36). C/EBPα and PPARγ have been shown to regulate many of adipocyte-specific genes including adipocyte P2 (aP2) (37–39) and leptin (40, 41). Based on these reports, we thought C/EBPs and PPARγ might be possible candidates that explain adipocyte-specific expression of Retn.
We have previously reported a functional C/EBPα-binding site at the proximal promoter of Retn, and that a proximal 264-bp fragment of Retn was sufficient for expression in adipocytes (42). However, the expression level of the mouse Retn promoter was quite modest in comparison with the robust expression of endogenous Retn in adipocytes. Moreover, unlike Retn, expression of C/EBPα is not restricted to adipose tissue (30, 43), and thus C/EBPα alone is not sufficient to explain the adipocyte-specific expression of Retn. A functional Sp1 binding site has also been reported in the proximal promoter of the Retn gene, but Sp1 is a ubiquitously expressed transcription factor (44). We have also reported that Retn is dramatically induced by retroviral infection of constitutively active PPARγ (fusion of viral transcriptional activator VP16 and PPARγ) in 3T3-L1 preadipocytes (45), though PPARγ failed to activate a reporter gene containing –6.2 kb of the Retn 5′-flanking sequence in our previous study (42). Based on these observations, we hypothesized that additional cis-regulatory elements, presumably PPARγ-binding sites, might exist outside the region we have previously analyzed to dictate and enhance adipocyte-specific expression of Retn.
Here, we have combined analysis of epigenetic marks and transcription factor binding by chromatin immunoprecipitation to identify a novel adipocyte-specific enhancer element at ∼8.8-kb upstream to the transcription start site (TSS) of Retn. It contains a binding site for PPARγ and three C/EBP-binding sites, and endogenous PPARγ, C/EBPα, and C/EBPβ bound to this region in adipocytes. Simultaneous expression of C/EBPα and PPARγ synergistically and robustly activated the Retn enhancer in non-adipocytic cells. Interestingly, the human resistin gene (RETN) contains a sequence homologous to the mouse enhancer, but lacks the functional PPARγ-binding site. Thus, murine-specific cooperation between C/EBPα and PPARγ dictates adipocyte specific expression of mouse Retn.
MATERIALS AND METHODS
Chromatin Immunoprecipitation Followed by Genomic Chip (ChIP-chip) Analysis—ChIP-chip was performed as described previously (46). In short, input and chromatin IP DNA were amplified linearly using in vitro transcription, and hybridized to a custom, densely tiled DNA microarray. ChIP signals (IP/input) were normalized to total histone H3 occupancy, and plotted as a moving average of 19 contiguous probes spanning 510 bp.
Plasmids—pGL3-basic and pGL4.27 were purchased from Promega (Madison, WI). Various lengths of the 5′-flanking region of mouse Retn were inserted into MluI and XhoI sites of pGL3-basic using bacterial artificial chromosome (BAC) recombination methods as described previously (47, 48). Sequence information was obtained from the UCSC genome browser. The mouse Retn enhancer element (–8872 to –8676 and –8843 to –8695 bp) and human genomic DNA fragment corresponding to the mouse Retn enhancer (–13262 to –13110 bp) were amplified by PCR and subcloned into BglII and XhoI sites of the pGL4.27 vector.
Cell Culture—3T3-L1, HEK 293T, and HepG2 cells were obtained from ATCC. HEK 293T and HepG2 cells were cultured in Dulbecco's-modified Eagle's Medium containing 10% fetal bovine serum and antibiotics in 5% CO2 incubator at 37 °C. 3T3-L1 cells were cultured and differentiated as previously described (49).
Transfection and Luciferase Assay—3T3-L1 adipocytes and preadipocytes were transfected by electroporation (Nucleofector II, AMAXA). Briefly, either preadipocytes or mature adipocytes (days 8–10 after differentiation) were detached from culture dishes with 0.25% trypsin, washed twice with phosphate-buffered saline, and resuspended in electroporation buffer (solution V, AMAXA). Approximately 1 × 106 cells were electroporated (electroporation program T-020) with equimolar amount of reporter vector (0.25∼1.0 μg) and β-galactosidase expression vector (0.3 μg), and seeded into triplicate wells of a 24-well plate. HEK 293T and HepG2 cells were transfected using Lipofectamine 2000 (Invitrogen). Medium was replaced with fresh culture medium after 16 h of transfection. The cells were harvested, and luciferase activities were measured by the luciferase assay system (Promega) after 48 h. β-Galactosidase activity was measured as described elsewhere (50). Light units were normalized to β-galactosidase activity. Fold activations relative to control were calculated, and means and standard errors from triplicate samples were plotted.
Chromatin Immunoprecipitation—ChIP-QPCR for the transcription factors PPARγ, C/EBPα, and C/EBPβ was performed as previously described (51). Briefly, lysates from mature adipocytes (Day 10 post-hormonal induction) were used for ChIP with the following antibodies: anti-PPARγ (81B8, Cell Signaling Technology), anti-C/EBPα (sc-61, Santa Cruz Biotechnology), and anti-C/EBPβ (sc-150, Santa Cruz Biotechnology). Isolated DNA was used for QPCR with Power SYBR Green PCR Master Mix (Applied Biosystems) and the PRISM 7500 instrument (Applied Biosystems). Relative enrichment at target sites was obtained by normalization to a negative control region in the Arbp gene where none of the transcription factors binds. The following primers were used for QPCR: F-retn-50bp: 5′-TCCCTCCTCTGGGACCTCTA-3′; R-retn-50bp: 5′-CCCATCCTGCCTTGGATAAT-3′; F-retn-8.8kb: 5′-GTAAGGGGGTGGCCTGATAG-3′; R-retn-8.8kb: 5′-ATTCCCTTCTCCCACCAAGT-3′; F-36b4: 5′-CTGGGACGATGAATGAGGAT-3′; R-36b4: 5′-AGCAGCTGGCACCTAAACAG-3′; F-ins1: 5′-CTTCAGCCCAGTTGACCAAT-3′; and R-ins1: 5′-AGGGAGGAGGAAAGCAGAAC-3′.
Genome-wide ChIP-chip analysis for PPARγ- and C/EBPα-binding sites in adipocytes was described elsewhere (51).
Preparation of Nuclear Extract—Nuclear extract from 3T3-L1 preadipocytes and adipocytes were prepared as described previously (52) with slight modifications. In brief, ∼2 × 107 cells were washed twice with ice-cold phosphate-buffered saline, scraped in 3 ml of phosphate-buffered saline, and spun briefly at 3,000 × g. They were resuspended in 10–20-packed cell volumes of cold Buffer A (10 mm HEPES-KOH pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiothreitol (DTT), 0.2 mm phenylmethanesulfonyl fluoride (PMSF), and Complete protease inhibitor mixture (Roche Applied Science)), and allowed to swell on ice for 10 min. Then, the cells were subjected to a tissue homogenizer with a B pestle, monitoring lysis of the cytosol fraction with a microscope. The nuclei were pelleted down at 3,000 × g at 4 °C for 5 min. The nuclei from adipocytes were further washed twice with the same volume of Buffer A to get rid of residual lipid. The pellet was resuspended with 3–4 packed nuclear volumes of cold Buffer C (20 mm HEPES-KOH pH 7.9, 25% (vol/vol) glycerol, 1.5 mm MgCl2, 420 mm KCl, 0.5 mm DTT, 0.2 mm PMSF, and 1× Complete), rocked at 4 °C for 30 min, and spun at 15,000 × g for 2 min at 4 °C. The supernatant fraction containing nuclear protein was collected and subjected to dialysis in dialysis buffer containing 20 mm HEPES-KOH pH 7.9, 25% (vol/vol) glycerol, 1.5 mm MgCl2, 50 mm KCl, 0.5 mm DTT, 0.2 mm PMSF, and 1× Complete to reduce the concentration of KCl.
Electrophoretic Mobility Shift Assay (EMSA)—EMSA was slightly modified from the method previously reported (53). Binding reactions were performed in 20 μl, containing 10 mm HEPES-KOH pH 7.9, 50 mm KCl, 1 mm EDTA, 5 mm DTT, 0.5 mm PMSF, 10% (vol/vol) glycerol, 0.25 μg/μl bovine serum albumin, and 0.1 μg/μl poly(dI-dC). DNA probes were prepared by end-labeling double-stranded oligonucleotides with T4 polynucleotide kinase and [γ-32P]deoxy-ATP (6,000 Ci/mmol). Oligonucleotide probes used in this study were as follows: A: 5′-CCCGCCATTCAATTCTTTATCTAGAACATTTTTGTCTTGTGTGAAATTGCTCAAAT-3′; B: 5′-GTCTTGTGTGAAATTGCTCAAATCTGAAAACAAGTTGACCAATAGCTCCTTACTG-3′; C: 5′-TGACCAATAGCTCCTTACTGCCAAGAACATTGTGAAAATGTGTCTGGGGTAAGGG-3′; D: 5′-AAATGTGTCTGGGGTAAGGGGGTGGCCTGATAGAAAGGGAGTGCTTTCTGGGGCA-3′; E: 5′-AGGGAGTGCTTTCTGGGGCAAGGATGACAGCACCAGGGCTCAGCGTTAGCAACAC-3′; mouse Retn PPARγ-binding site (mouse): 5′-CTTTCTGGGGCAAGGATGACAGCAC-3′, and the human sequence corresponding to the mouse Retn PPARγ-binding site (human): 5′-CTGGTGGGATCAGAGATGAGGTGTC-3′.
2 μg of nuclear extract or 2 μl of in vitro translated mouse PPARγ and RXRα, synthesized using the TnT T7-coupled transcription/translation system (Promega), were used for the binding reaction. The supershift assay was performed by adding 0.1 μg of rabbit polyclonal IgG specific for C/EBPα, C/EBPβ, C/EBPδ, PPARγ, or control IgG (sc-61, 746, 636, 7196, and 2027, Santa Cruz Biotechnology) to each reaction. All the reactions were carried out at room temperature, and each reaction was preincubated 10 min before the addition of probe (10,000 cpm). To resolve the complexes, the reactions were applied to 4 or 5% native acrylamide gels (acrylamide-bisacrylamide, 29:1) in 0.5× TBE buffer (44.5 mm Tris, 44.5 mm boric acid, 1 mm EDTA) and 5% (vol/vol) glycerol.
RNA Analysis—The effect of rosiglitazone on Retn mRNA stability was examined in mature 3T3-L1 adipocytes. Adipocytes were pretreated with either DMSO or 1 μm rosiglitazone for 48 h, and 5 μg/ml actinomycin D (Sigma-Aldrich) were added. The cells were harvested for RNA at several time points, and total RNA was isolated using the RNeasy Mini Kit (Qiagen). Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed using the PRISM 7500 instrument and Taqman Universal PCR Master Mix (Applied Biosystems). Retn mRNA levels were normalized to Arbp mRNA. Primers and probes used were: F-Retn: 5′-AAGAAGGAGCTGTGGGACAGG-3′, R-Retn: 5′-CAGCAGTTCAGGGACAAGGAA-3′, Retn-probe: 5′-TGTGTCCTGCTAAGTCCTCTGCCACG-3′, F-36B4: 5′-TCATCCAGCAGGTGTTTGACA-3′, R-36B4: 5′-GGCACCGAGGCAACAGTT-3′, and 36B4-probe: 5′-AGAGCAGGCCCTGCACTCTCG-3′.
Retn precursor mRNA (pre-mRNA) was analyzed as follows. 1 μg of total RNA was pretreated with DNase I (Invitrogen) and reverse-transcribed using random primers. Primers encoding exon-1 and intron-1 of Retn were used to amplify pre-mRNA and quantified using Power SYBR Green PCR Master Mix and PRISM 7500. The values were normalized to Arbp mRNA. Primers used were: F-Retn-ex-1: 5′-CCTCTGCCACGGTAAGTGAT-3′ and R-Retn-int-1: 5′-TTAGTTCCTGAGGGCACACAT-3′.
RESULTS
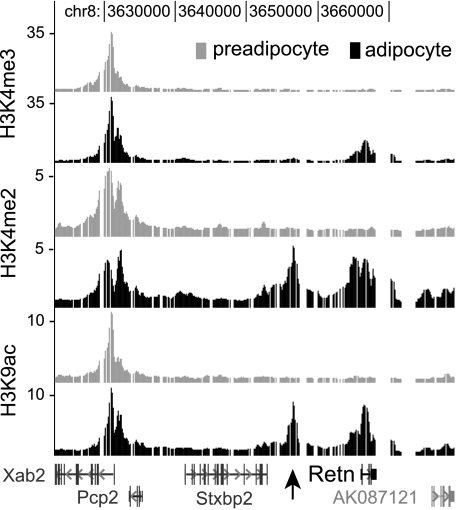
Histone H3 Lysine Acetylation and Methylation Profiles Identify a Peak of Intergenic Enrichment 9-kb Upstream of the Retn Transcription Start Site in Mouse Adipocytes—To identify potential cis-acting regulatory sequences that may reside far from the Retn proximal promoter, we examined histone modifications along the region of mouse chromosome 8 containing Retn and several of its neighboring genes. This strategy was based on recent observations from genome-wide mapping studies indicating that the chromatin of transcriptionally active genes is enriched for specific post-translational histone modifications. Although the combinatorial patterns of histone modifications at active genes can be quite complex (54), it is often the case that regions with high histone H3 lysine 4 trimethylation (H3K4me3) tend to be TSSs, whereas DNA elements containing high levels of H3 acetylation and H3K4me2, but low H3K4me3, tend to be enhancers (55–57). Thus, active enhancers can be enriched for certain chromatin marks, raising the possibility that they can be identified from histone modification profiles.
We used ChIP-chip to map H3K4me3, H3K4me2, and H3K9 acetylation (H3K9ac) at Retn and its 5′- and 3′-flanking regions extending 50 kb from the TSS. ChIP was performed in an established tissue culture model of adipogenesis, whereby 3T3-L1 preadipocyte cells can be induced to differentiate into adipocytes (57) to generate chromatin profiles of Retn in its transcriptionally inactive (preadipocyte) and active (adipocyte) states. Fig. 1 shows that H3K4me3 levels at Retn are similar to background in preadipocytes, while a peak of enrichment is present in adipocytes. Retn profiles for H3K4me2 and H3K9ac are similar such that both display background modification levels in preadipocytes and two distinct peaks of enrichment in adipocytes. As expected, one of the peaks is at the Retn TSS. The other is located 9-kb upstream of the TSS, and it has the characteristics of a tissue-specific enhancer given its low level of H3K4me3 and high levels of H3K4me2 and H3K9ac in adipocytes but not preadipocytes, and its location far from the proximal promoter. The evidence for cell type specificity of the enriched regions at or near Retn is strong considering the histone profiles at the neighboring gene Xab2, a pre-mRNA splicing factor. Expression of Xab2 is not adipocyte-specific, and consistent with this each histone mark is enriched similarly at the 5′-end of Xab2 in both cell types. Taken together, the data suggest that cis-acting regulatory sequences enhancing Retn transcription may reside ∼9-kb upstream of the TSS.
FIGURE 1.
Profiles of histone H3 modifications at the mouse Retn locus on chromosome 8. ChIP-chip mapping of H3K4me3, H3K4me2 and H3K9ac was performed in undifferentiated (preadipocyte) and differentiated (adipocyte) 3T3-L1 cells. Fold-enrichment values are plotted on a linear scale along a region of mouse chromosome 8 containing Retn and neighboring genes. Gaps indicate the presence of repetitive sequence. Retn and Stxbp2 transcription proceeds from left to right, while Pcp2 and Xab2 proceeds from right to left. The arrow indicates the enrichment peak for H3K4me2 and H3K9ac that is positioned 9-kb upstream of Retn.
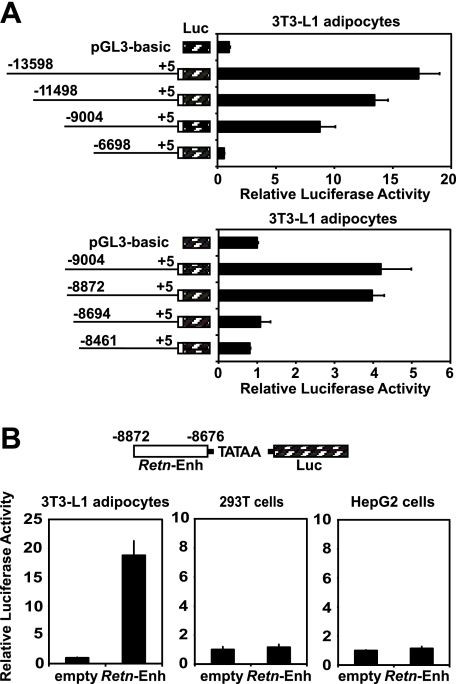
Adipocyte-specific Enhancer Element Is Located between –8872 and –8676 of the Mouse Retn Gene—To determine whether the region at –9 kb regulates Retn transcription and to potentially identify additional cis-acting sequences, we fused 5′-fragments of the Retn promoter upstream of a luciferase reporter and transfected them into 3T3-L1 adipocytes. Truncations between –13,598 and –9004 bp led to minor incremental decreases in reporter activity, while truncation from –9004 to –6698 bp decreased activity 8-fold (Fig. 2A). Promoter studies with a finer series of truncations further defined the region between –8872 and –8694 bp to be important for enhanced reporter activity (Fig. 2A). This small region, ∼200-bp long, was subsequently fused to a luciferase reporter containing a minimal promoter and transfected into several different cell lines. Compared with the control vector, it increased reporter activity 19-fold in 3T3-L1 adipocytes, but not in HEK 293T and HepG2 cells (Fig. 2B). These results demonstrate that the region of Retn between –8872 and –8676 bp is sufficient to activate transcription from a heterologous promoter in a cell type-specific manner, and together with the ChIP-chip analysis, strongly suggest that it functions as an adipocyte-specific enhancer for Retn expression.
FIGURE 2.
Region of mouse Retn between –8872 and –8676 functions as an adipocyte-specific enhancer. A, luciferase activity of Retn reporter constructs transfected into 3T3-L1 adipocytes. B, luciferase activity from 3T3-L1 adipocytes, HEK 293T, and HepG2 cells transfected with a plasmid whereby the mouse Retn enhancer (–8872 to –8676 bp) drives reporter gene expression. Data presented here and in all subsequent transfections are expressed as mean ± S.E. (n = 3 per condition).
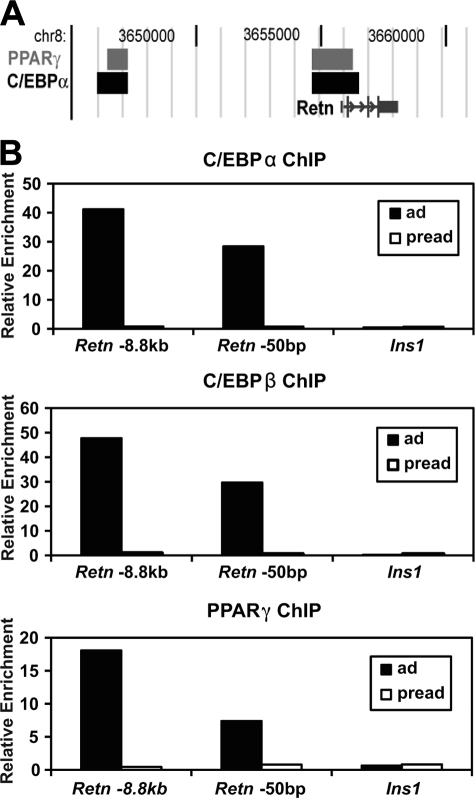
C/EBPα, C/EBPβ, and PPARγ Bind to the Retn Adipocyte Enhancer—We have recently reported genome-wide binding locations for PPARγ and C/EBPα based on ChIP-chip in 3T3-L1 adipocytes (51). For Retn, PPARγ and C/EBPα were found to occupy a region near the TSS and another overlapping with the region identified as the adipocyte enhancer at –8.8 kb (Fig. 3A). To confirm binding to these sites, we used quantitative PCR to analyze ChIP DNA isolated from PPARγ, C/EBPα, or C/EBPβ immunoprecipitations. Compared with insulin, which does not contain C/EBP- and PPARγ-binding sites, the enhancer and promoter showed substantial enrichment in C/EBPα, C/EBPβ, and PPARγ immunoprecipitations from adipocytes, but not preadipocytes, indicating that all three factors bind to these regions in adipocyte cells (Fig. 3B).
FIGURE 3.
C/EBPα, C/EBPβ, and PPARγ bind to the Retn enhancer in 3T3-L1 adipocytes. A, ChIP-chip data showing binding regions for PPARγ and C/EBPα at the mouse Retn locus. B, ChIP-QPCR analysis for C/EBPα, C/EBPβ and PPARγ at the enhancer (Retn, –8.8 kb) and proximal promoter (Retn, –50 bp) of Retn in adipocytes (ad) and preadipocytes (pread). The insulin gene (Ins1) is a negative control.
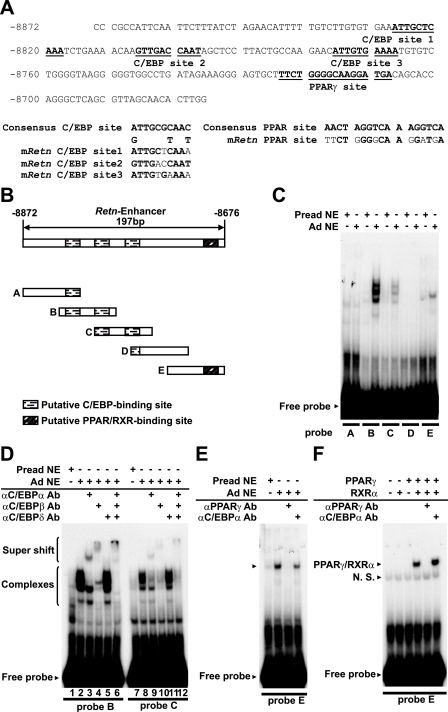
Sequence analysis of the Retn enhancer identified three putative binding sites for C/EBP factors, and a region containing moderate homology to the direct repeat-1 (DR-1) PPARγ-binding site (Fig. 4A). To determine if C/EBPs and PPARγ can bind these sites and if other trans-acting factors target the Retn enhancer, we performed EMSA. The enhancer was divided into five fragments of ∼55 bp, and these were incubated with extract prepared from 3T3-L1 preadipocytes and adipocytes. As shown in Fig. 4C, binding activities abundant in adipocytes, but not preadipocytes, were able to shift probes B, C, and E, but not A and D.
FIGURE 4.
The Retn enhancer contains three C/EBP-binding sites and one PPARγ-binding site. A, sequences with similarity to C/EBP- and PPARγ-binding motifs are underlined in the mouse Retn enhancer region. Comparison of the putative binding sites to the consensus C/EBP and PPARγ motifs is shown. B, schematic representation of the oligonucleotide probes used for this study, with putative C/EBP- and PPARγ-binding sites indicated. C–E, EMSA of activities within 3T3-L1 preadipocyte (Pread) or adipocyte (Ad) nuclear extract (NE) and the indicated probes. F, EMSA with in vitro translated PPARγ/RXRα. For the indicated reactions, antibody (Ab) was added to the nuclear extract or in vitro translated protein ∼10 min before the addition of probe DNA.
As probes B, C, and E contain the putative binding sites for C/EBP and PPARγ, we examined whether their gel-shift complexes contain C/EBP factors and/or PPARγ by performing EMSA in the presence of antibodies recognizing these proteins. It is often the case that antibody binding to a sequence-specific transcription factor will reduce the abundance of its gel-shift complex either by blocking DNA binding or by supershifting the complex to a higher position in the gel. Indeed, addition of C/EBPα antibody reduced a part of gel-shift complexes with probe B and caused a new, slowly migrating complex to appear that likely consists of C/EBPα, probe B, and C/EBPα antibody (Fig. 4D, compare lanes 2 and 3). Addition of C/EBPβ antibody reduced a part of the gel-shift complexes for probe B, and produced a supershift distinct from that generated by the C/EBPα antibody (Fig. 4D, lane 4). These data suggest that protein-DNA complexes for probe B contain C/EBPα and C/EBPβ. In contrast, addition of C/EBPδ antibody had little effect on the abundance or mobility of the gel-shift complexes (Fig. 4D, lane 5). When all three C/EBP antibodies were included in the binding reaction, all of the original gel-shift complexes disappeared, with some of the signal appearing as supershifted complexes (Fig. 4D, lane 6). Similar antibody effects were observed for probe C (Fig. 4D, lanes 7–12), indicating that most, if not all, of the protein-DNA complexes observed with probes B and C are associated with C/EBPα and C/EBPβ. Furthermore, the presence of C/EBPα and C/EBPβ in multiple complexes suggests that they may interact with these sites as both homo- and heterodimers. These data agree with earlier in vitro studies demonstrating that C/EBP isoforms are capable of binding to the same site as homo- and heterodimers in all intrafamilial combinations and that the majority of the binding activity in 3T3-L1 adipocytes is associated with C/EBPα and C/EBPβ (59, 60).
Consistent with the conclusion that C/EBPα and C/EBPβ are the main adipocyte nuclear proteins bound to probes B and C, addition of PPARγ antibody did not alter the gel-shift complexes (data not shown). However, the gel-shift complex generated with probe E was blocked by antibody against PPARγ, but not C/EBPα (Fig. 4E). Probe E contains a putative PPARγ-binding site, and Fig. 4F reveals that in vitro translated PPARγ can bind to it as a heterodimer with RXRα. As before, antibody specific to PPARγ blocked binding. As a whole, the EMSA and ChIP studies make a compelling case that C/EBPα, C/EBPβ, and PPARγ bind to the Retn enhancer in 3T3-L1 adipocytes.
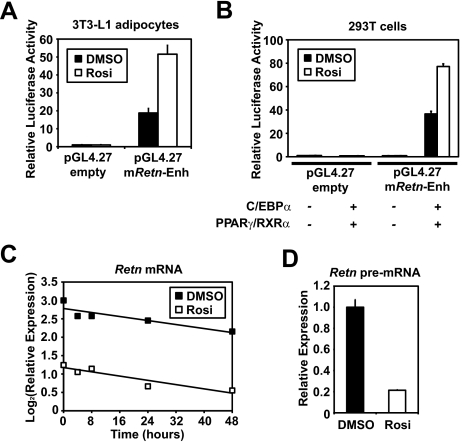
C/EBPα and PPARγ/RXRα Function Synergistically to Transactivate Transcription Driven by the Retn Enhancer—To dissect roles for C/EBPα and PPARγ in the Retn enhancer function, we transfected 293T cells with the reporter construct from Fig. 2B, whereby the Retn enhancer drives reporter transcription from a heterologous promoter, and with expression plasmids for C/EBPα and PPARγ. In the absence of exogenously expressed protein, the enhancer construct displayed similar transcriptional activity to the control carrying only the heterologous promoter and reporter gene, indicating that 293T cells are devoid of activities that function at the enhancer (Fig. 5). Cotransfection of either C/EBPα or PPARγ/RXRα stimulated the Retn enhancer activity 7- and 2-fold, respectively, indicating that the exogenously expressed factors can function at the Retn enhancer in the context of the reporter construct. Of note, simultaneous expression of both C/EBPα and PPARγ/RXRα stimulated the Retn enhancer activity by 22-fold. These results demonstrate that ectopic expression of C/EBPα and PPARγ/RXRα is sufficient for the Retn enhancer activity in non-adipocyte cells, and that both factors working together produce more activity than the sum of activities of either on its own.
FIGURE 5.
PPARγ and C/EBPα function synergistically at the mouse Retn enhancer. Luciferase activity of empty or Retn enhancer (–8872 to –8676 bp) reporter constructs cotransfected with C/EBPα or PPARγ/RXRα expression vectors into HEK 293T cells.
The Retn Enhancer Is Not Directly Involved in the Suppression of Retn mRNA by PPARγ Ligand—PPARγ ligands inhibit Retn expression (4, 42), but the molecular mechanism remains to be elucidated. We examined the effect of rosiglitazone, an anti-diabetic thiazolidinedione (TZD) drug that functions as a PPARγ agonist on the adipocyte-specific Retn enhancer. Rosiglitazone actually stimulated enhancer activity by ∼3-fold in adipocytes as well as in 293T cells cotransfected with PPARγ/RXRα and C/EBPα (Fig. 6, A and B). The Retn-13-kb luciferase reporter was also stimulated by rosiglitazone (data not shown).
FIGURE 6.
The adipocyte-specific Retn enhancer is not directly involved in the suppression of Retn mRNA by synthetic ligand for PPARγ. A, luciferase activity of empty or Retn enhancer (–8872 to –8676 bp) reporter constructs treated with or without 1 μm rosiglitazone (rosi) for 48 h in 3T3-L1 adipocytes. B, luciferase activity of empty or Retn enhancer (–8872 to –8676 bp) reporter constructs cotransfected with C/EBPα and PPARγ/RXRα expression vectors into HEK 293T cells with or without 1 μm rosi for 24 h. C, 3T3-L1 adipocytes were treated with DMSO or 1 μm rosi for 48 h, and 5 μg/ml of actionomycin D were added. RNA was extracted at the indicated time points. Retn mRNA levels were corrected by Arbp expression and plotted as log2. D, 3T3-L1 adipocytes were treated with DMSO or 1 μm rosi for 48 h, and the Retn pre-mRNA levels were measured as described under “Materials and Methods.” Each value represents the mean ± S.E. (n = 3 per condition).
Because the suppression of Retn mRNA could be mediated both transcriptionally and post-transcriptionally, we assessed whether rosiglitazone affected the stability of Retn mRNA. Adipocytes were pretreated with DMSO or rosiglitazone (1 μm) 48 h, then exposed to 5 μg/ml actinomycin D to block transcription. Endogenous Retn mRNA was suppressed by rosiglitazone, as previously reported. Importantly, the treatment with rosiglitazone did not alter Retn mRNA stability (Fig. 6C). Similar effects were seen after simultaneous treatment with rosiglitazone and actinomycin D (data not shown). This suggests that the reduction of Retn mRNA by TZD occurs at the level of transcription. Indeed, Retn precursor mRNA (pre-mRNA) was also suppressed by rosiglitazone (Fig. 6D). These data strongly suggest that Retn mRNA is transcriptionally suppressed by TZDs. However, the mechanism does not directly involve the adipocyte-specific enhancer.
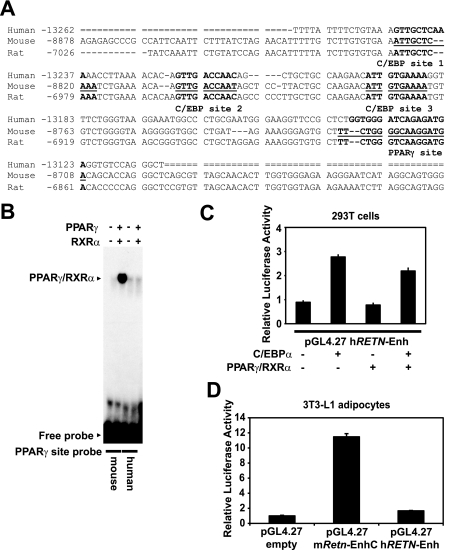
Human RETN Lacks a Functional PPARγ-binding Site—The molecular explanation for why resistin expression is restricted to adipocytes in rodents and macrophages in humans is unknown. One possibility is that the cis-acting regulatory elements differ for the various resistin orthologs. To explore this idea, we investigated whether the mouse Retn enhancer is conserved in humans. Comparative genomic analysis from the UCSC genome website shows that ∼150 bp of the mouse Retn enhancer are conserved in the rat at approximately –7.1 kb, and in the human at approximately –13.3 kb, relative to the TSS (Fig. 7A). Overall similarity was roughly 66% between human and mouse and 88% between rat and mouse. The three C/EBP-binding sites from the mouse gene were very well conserved for human and rat. Consistent with this, C/EBP factors from 3T3-L1 extract bound to the human and mouse sequences with comparable affinities (data not shown). On the other hand, the PPARγ-binding site is well conserved in rat, but not human. Indeed, the human sequence displayed little or no binding of in vitro translated PPARγ and RXRα by EMSA (Fig. 7B). As expected from the binding assays, exogenous expression of C/EBPα significantly activated the human sequence in 293T cells, but PPARγ/RXRα did not, either alone or in combination with C/EBPα and PPARγ/RXRα (Fig. 7C). Furthermore, the human sequence conferred minimal transcriptional activity in 3T3-L1 adipocytes (Fig. 7D). Thus, human RETN lacks the PPARγ-binding site that is critical for enhancer function in mouse, which likely accounts for its low expression in adipocytes.
FIGURE 7.
Human RETN lacks a functional PPARγ-binding site. A, sequence alignment of the mouse, rat, and human resistin genes based on comparative genomic analysis. Sequence coordinates are relative to the resistin TSS for each species. C/EBP and PPARγ motifs from the mouse adipocyte enhancer are underlined. B, EMSA examining binding of in vitro translated PPARγ/RXRα to the mouse PPARγ-binding site (mouse) or the corresponding region of human RETN (human). C, luciferase activity of transfected plasmids in 293T cells. The human sequence corresponding to the mouse enhancer (–13262 to –13110 bp, hRETN-Enh) is fused to a luciferase reporter vector and cotransfected with or without C/EBPα and PPARγ/RXRα expression vectors. D, luciferase activity of transfected plasmids in 3T3-L1 adipocytes. Reporter gene expression is driven by the mouse enhancer (–8843 to –8695 bp, mRetn-EnhC) or the corresponding sequence of human RETN (hRETN-Enh).
DISCUSSION
In this work, we report a novel adipocyte-specific enhancer at ∼8.8-kb upstream of the mouse Retn gene, where PPARγ and C/EBP factors synergistically enhance adipocyte-specific expression of mouse Retn. We discovered the enhancer by ChIP-chip mapping of chromatin marks, and it was independently confirmed by functional dissection of the 5′-flanking sequence by reporter assay, and genome-wide ChIP-chip analysis for PPARγ- and C/EBPα-binding sites. PPARγ and C/EBPs are key transcription factors for adipogenesis, and have been shown to regulate a number of adipocyte genes including adipocyte P2 (aP2) (37–39), phosphoenolpyruvate carboxykinase (PEPCK) (61, 62), and leptin (40, 41). In mouse Retn, PPARγ- and C/EBP-binding sites reside very closely within the 200-bp enhancer sequence. This enhancer region alone revealed strong transcriptional activity in adipocytes, but not in other cell types. In a heterologous reporter assay system, PPARγ/RXRα or C/EBPα alone only partially stimulated the transcriptional activity of the Retn enhancer in non-adipocytic cells. In contrast, simultaneous expression of PPARγ/RXRα and C/EBPα synergistically augmented the robust transcriptional activity of the enhancer. Recent genome-wide ChIP-chip analysis determined that C/EBPα binds nearby at ∼60% of PPARγ-binding sites in 3T3-L1 adipocytes (51), and a simultaneous report of ChIP with deep genomic sequencing also noted C/EBP-binding sites at a high percentage of PPARγ binding locations (63). These data strongly suggest that cooperation between PPARγ and C/EBP factors might be important for other adipocyte genes beyond Retn. At this point, the specific mechanism of the synergism between PPARγ and C/EBPα is not clear, and further study is required.
Unlike rodent Retn, human RETN is only minimally expressed in fat, but predominantly expressed in mononuclear cells including macrophages (7, 12, 13). The mechanism defining the different expression profile was unclear. Although the proximal promoter sequence of human RETN is quite divergent from that of mouse, it seems to conserve binding sites for C/EBP factors, Sp1, and adipocyte determination- and differentiation-dependent factor (ADD1)/sterol regulatory element-binding protein 1c (SREBP1c) as in the mouse promoter (44, 64). In this study, we found that human RETN possesses a sequence that has high homology to the fat-specific enhancer of mouse Retn at –13.3-kb upstream of the TSS. It conserves three C/EBP-binding sites, but lacks the functional PPARγ-binding site, and revealed little transcriptional activity in 3T3-L1 adipocytes. This low level expression of the human homologous sequence in mouse adipocytes is presumably due to the lack of the PPARγ-binding site, though it is also possible that interspecies differences in epigenetic machinery, cellular environment, or transcription factors affect the different expression pattern. A recent report formally demonstrated that the genetic sequence is largely responsible for directing transcriptional programs in homologous tissues (65). This supports our conclusion that the lack of the functional PPARγ-binding site in human RETN could explain the minimal expression of human RETN in fat.
Retn was originally identified as a gene that is suppressed by anti-diabetic drugs, TZDs (4). Rosiglitazone reduced Retn expression with an ED50 similar to its Kd for binding to PPARγ, suggesting that this effect is mediated through PPARγ (42). Here we demonstrated that rosiglitazone suppresses Retn mRNA transcriptionally by demonstrating that it does not destabilize Retn mRNA and suppresses Retn mRNA at pre-mRNA level. However, rosiglitazone stimulated the mouse Retn enhancer activity in 3T3-L1 adipocytes as well as in 293T cells cotransfected with C/EBPα and PPARγ/RXRα. Retn-13kb luciferase reporter activity was also stimulated by rosiglitazone. These observations suggest that the PPARγ-binding site of mouse Retn functions as a positive response element, and the negative regulation of Retn transcription is not directly mediated by cis-regulatory elements present in the ∼13-kb upstream region of Retn that is sufficient for robust adipocyte-specific expression.
Human RETN, in which the PPARγ-binding site is not conserved, has also been reported to be repressed by TZDs (7, 66–68), further suggesting that the PPARγ-binding site in the mouse enhancer is not necessary for the down-regulation of Retn mRNA. Because, so far, no additional PPARγ-binding sites have been found around the Retn locus by our genome-wide ChIP-chip analysis, the repression effect might be mediated by indirect binding of PPARγ, or inducing transcriptional repressors, or modifying transcriptional activity of other transcription factors.
Another interesting feature of mouse Retn is that it is selectively expressed in white adipose tissue (WAT), but not in brown adipose tissue (BAT) (4, 10, 58). BAT is characterized by densely packed mitochondria, and serves to dissipate energy to produce heat in response to cold in contrast to WAT. PPARγ and C/EBPs cannot explain the WAT-specific expression of Retn, since these transcription factors appear to play roles in the differentiation of brown adipocytes as well (69). Recently, Spiegelman and co-workers have discovered PRDM16 (PRD1-BF1-RIZ1 homologous domain containing 16) as a master regulator of brown adipogenesis, which can potentially explain the low expression of Retn in brown fat. PRDM16 is enriched in BAT compared with WAT. Ectopic expression of PRDM16 in white fat cell progenitor could induce brown specific genes and suppress white specific genes including Retn. Conversely, depletion of PRDM16 in brown fat cells causes loss of brown fat characteristics (58). PRDM16 suppresses WAT-specific genes, including Retn, through binding to C-terminal-binding protein-1 (CtBP-1) and CtBP-2 (48). CtBPs are corepressors, and associate with multiple histone modifying enzymes including histone deacetylaces (HDAC1 and HDAC2), histone methyltransferases (G9a and GLP) and a histone methyl transferase (LSD1) (70). Indeed, transient expression of PRDM16 suppressed the Retn –13kb luciferase reporter gene in 3T3-L1 adipocytes (48). However, the molecular mechanism by which PRDM16 represses the expression of Retn in BAT is still unclear, especially which transcription factor is targeted by PRDM16. PRDM16 has two distinct DNA binding domains that recognize distinct binding motifs (71). Although PRDM16 R998Q mutant, which completely lacks DNA binding ability, could repress the resistin expression (58), it is still possible that PRDM16 directly binds to cis-elements through another DNA binding domain. More recently, Spiegelman and co-workers demonstrated that brown, but not white, fat cells arise from common progenitor cells to muscle by lineage-tracing experiment, and PRDM16 stimulates brown adipogenesis by directly binding to PPARγ and enhancing its transcriptional activity (72). Further studies are required to elucidate the mechanism of white adipocyte specific expression of Retn.
In summary, this study reports a novel adipocyte-specific enhancer at ∼8.8-kb upstream of mouse Retn. PPARγ and C/EBP factors synergistically activate fat-specific expression of mouse Retn from the enhancer. The lack of the functional PPARγ site in human RETN may explain its low expression level in fat. Further studies are necessary to elucidate the mechanism of WAT-specific expression and down-regulation by TZDs.
Acknowledgments
We thank Dr. N. Copeland (The National Cancer Institute at Frederick, Frederick, MD) for providing BAC recombination materials.
This work was supported, in whole or in part, by National Institutes of Health Grant DK P01 DK49210 (to M. A. L.). This work was also supported by the Picower Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: C/EBP, CCAAT/enhancer-binding protein; PPARγ, peroxisome proliferator-activated receptor γ; TSS, transcription start site; EMSA, electrophoretic mobility shift assay; TZD, thiazolidinedione; WAT, white adipose tissue; BAT, brown adipose tissue; ChIP, chromatin immunoprecipitation assay; DTT, dithiothreitol; PMSF, phenylmethylsulfonyl fluoride.
References
- 1.Reaven, G., Abbasi, F., and McLaughlin, T. (2004) Recent Prog. Horm. Res. 59 207–223 [DOI] [PubMed] [Google Scholar]
- 2.Ahima, R. S., and Flier, J. S. (2000) Trends Endocrinol. Metab. 11 327–332 [DOI] [PubMed] [Google Scholar]
- 3.Rosen, E. D., and Spiegelman, B. M. (2006) Nature 444 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steppan, C. M., Bailey, S. T., Bhat, S., Brown, E. J., Banerjee, R. R., Wright, C. M., Patel, H. R., Ahima, R. S., and Lazar, M. A. (2001) Nature 409 307–312 [DOI] [PubMed] [Google Scholar]
- 5.Steppan, C. M., Brown, E. J., Wright, C. M., Bhat, S., Banerjee, R. R., Dai, C. Y., Enders, G. H., Silberg, D. G., Wen, X., Wu, G. D., and Lazar, M. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holcomb, I. N., Kabakoff, R. C., Chan, B., Baker, T. W., Gurney, A., Henzel, W., Nelson, C., Lowman, H. B., Wright, B. D., Skelton, N. J., Frantz, G. D., Tumas, D. B., Peale, F. V., Jr., Shelton, D. L., and Hebert, C. C. (2000) EMBO J. 19 4046–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel, L., Buckels, A. C., Kinghorn, I. J., Murdock, P. R., Holbrook, J. D., Plumpton, C., Macphee, C. H., and Smith, S. A. (2003) Biochem. Biophys. Res. Commun. 300 472–476 [DOI] [PubMed] [Google Scholar]
- 8.Rajala, M. W., Obici, S., Scherer, P. E., and Rossetti, L. (2003) J. Clin. Investig. 111 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangwala, S. M., Rich, A. S., Rhoades, B., Shapiro, J. S., Obici, S., Rossetti, L., and Lazar, M. A. (2004) Diabetes 53 1937–1941 [DOI] [PubMed] [Google Scholar]
- 10.Banerjee, R. R., Rangwala, S. M., Shapiro, J. S., Rich, A. S., Rhoades, B., Qi, Y., Wang, J., Rajala, M. W., Pocai, A., Scherer, P. E., Steppan, C. M., Ahima, R. S., Obici, S., Rossetti, L., and Lazar, M. A. (2004) Science 303 1195–1198 [DOI] [PubMed] [Google Scholar]
- 11.Lazar, M. A. (2007) Horm. Metab. Res. 39 710–716 [DOI] [PubMed] [Google Scholar]
- 12.Savage, D. B., Sewter, C. P., Klenk, E. S., Segal, D. G., Vidal-Puig, A., Considine, R. V., and O'Rahilly, S. (2001) Diabetes 50 2199–2202 [DOI] [PubMed] [Google Scholar]
- 13.Yang, R. Z., Huang, Q., Xu, A., McLenithan, J. C., Eisen, J. A., Shuldiner, A. R., Alkan, S., and Gong, D. W. (2003) Biochem. Biophys. Res. Commun. 310 927–935 [DOI] [PubMed] [Google Scholar]
- 14.Osawa, H., Onuma, H., Ochi, M., Murakami, A., Yamauchi, J., Takasuka, T., Tanabe, F., Shimizu, I., Kato, K., Nishida, W., Yamada, K., Tabara, Y., Yasukawa, M., Fujii, Y., Ohashi, J., Miki, T., and Makino, H. (2005) Biochem. Biophys. Res. Commun. 335 596–602 [DOI] [PubMed] [Google Scholar]
- 15.Vidal-Puig, A., and O'Rahilly, S. (2001) Clin. Endocrinol. (Oxf) 55 437–438 [DOI] [PubMed] [Google Scholar]
- 16.McTernan, C. L., McTernan, P. G., Harte, A. L., Levick, P. L., Barnett, A. H., and Kumar, S. (2002) Lancet 359 46–47 [DOI] [PubMed] [Google Scholar]
- 17.McTernan, P. G., McTernan, C. L., Chetty, R., Jenner, K., Fisher, F. M., Lauer, M. N., Crocker, J., Barnett, A. H., and Kumar, S. (2002) J. Clin. Endocrinol. Metab. 87 2407. [DOI] [PubMed] [Google Scholar]
- 18.Wang, H., Chu, W. S., Hemphill, C., and Elbein, S. C. (2002) J. Clin. Endocrinol. Metab. 87 2520–2524 [DOI] [PubMed] [Google Scholar]
- 19.Smith, S. R., Bai, F., Charbonneau, C., Janderova, L., and Argyropoulos, G. (2003) Diabetes 52 1611–1618 [DOI] [PubMed] [Google Scholar]
- 20.Osawa, H., Ochi, M., Tabara, Y., Kato, K., Yamauchi, J., Takata, Y., Nishida, W., Onuma, H., Shimizu, I., Fujii, Y., Miki, T., Ohashi, J., and Makino, H. (2008) Clin. Endocrinol. (Oxf) 69 74–80 [DOI] [PubMed] [Google Scholar]
- 21.Osawa, H., Tabara, Y., Kawamoto, R., Ohashi, J., Ochi, M., Onuma, H., Nishida, W., Yamada, K., Nakura, J., Kohara, K., Miki, T., and Makino, H. (2007) Diabetes Care 30 1501–1506 [DOI] [PubMed] [Google Scholar]
- 22.Osawa, H., Yamada, K., Onuma, H., Murakami, A., Ochi, M., Kawata, H., Nishimiya, T., Niiya, T., Shimizu, I., Nishida, W., Hashiramoto, M., Kanatsuka, A., Fujii, Y., Ohashi, J., and Makino, H. (2004) Am. J. Hum. Genet. 75 678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. H., Chan, J. L., Yiannakouris, N., Kontogianni, M., Estrada, E., Seip, R., Orlova, C., and Mantzoros, C. S. (2003) J. Clin. Endocrinol. Metab. 88 4848–4856 [DOI] [PubMed] [Google Scholar]
- 24.Janke, J., Engeli, S., Gorzelniak, K., Luft, F. C., and Sharma, A. M. (2002) Obes. Res. 10 1–5 [DOI] [PubMed] [Google Scholar]
- 25.Kielstein, J. T., Becker, B., Graf, S., Brabant, G., Haller, H., and Fliser, D. (2003) Am. J. Kidney Dis. 42 62–66 [DOI] [PubMed] [Google Scholar]
- 26.Conneely, K. N., Silander, K., Scott, L. J., Mohlke, K. L., Lazaridis, K. N., Valle, T. T., Tuomilehto, J., Bergman, R. N., Watanabe, R. M., Buchanan, T. A., Collins, F. S., and Boehnke, M. (2004) Diabetologia 47 1782–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajala, M. W., Lin, Y., Ranalletta, M., Yang, X. M., Qian, H., Gingerich, R., Barzilai, N., and Scherer, P. E. (2002) Mol. Endocrinol. 16 1920–1930 [DOI] [PubMed] [Google Scholar]
- 28.Farmer, S. R. (2006) Cell Metab. 4 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosen, E. D., and MacDougald, O. A. (2006) Nat. Rev. Mol. Cell. Biol. 7 885–896 [DOI] [PubMed] [Google Scholar]
- 30.Cao, Z., Umek, R. M., and McKnight, S. L. (1991) Genes Dev. 5 1538–1552 [DOI] [PubMed] [Google Scholar]
- 31.Yeh, W. C., Cao, Z., Classon, M., and McKnight, S. L. (1995) Genes Dev. 9 168–181 [DOI] [PubMed] [Google Scholar]
- 32.Christy, R. J., Kaestner, K. H., Geiman, D. E., and Lane, M. D. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, Z., Xie, Y., Bucher, N. L., and Farmer, S. R. (1995) Genes Dev. 9 2350–2363 [DOI] [PubMed] [Google Scholar]
- 34.Clarke, S. L., Robinson, C. E., and Gimble, J. M. (1997) Biochem. Biophys. Res. Commun. 240 99–103 [DOI] [PubMed] [Google Scholar]
- 35.Wu, Z., Rosen, E. D., Brun, R., Hauser, S., Adelmant, G., Troy, A. E., McKeon, C., Darlington, G. J., and Spiegelman, B. M. (1999) Mol. Cell 3 151–158 [DOI] [PubMed] [Google Scholar]
- 36.Rosen, E. D., Hsu, C. H., Wang, X., Sakai, S., Freeman, M. W., Gonzalez, F. J., and Spiegelman, B. M. (2002) Genes Dev. 16 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christy, R. J., Yang, V. W., Ntambi, J. M., Geiman, D. E., Landschulz, W. H., Friedman, A. D., Nakabeppu, Y., Kelly, T. J., and Lane, M. D. (1989) Genes Dev. 3 1323–1335 [DOI] [PubMed] [Google Scholar]
- 38.Tontonoz, P., Hu, E., Graves, R. A., Budavari, A. I., and Spiegelman, B. M. (1994) Genes Dev. 8 1224–1234 [DOI] [PubMed] [Google Scholar]
- 39.Tontonoz, P., Graves, R. A., Budavari, A. I., Erdjument-Bromage, H., Lui, M., Hu, E., Tempst, P., and Spiegelman, B. M. (1994) Nucleic Acids Res. 22 5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollenberg, A. N., Susulic, V. S., Madura, J. P., Zhang, B., Moller, D. E., Tontonoz, P., Sarraf, P., Spiegelman, B. M., and Lowell, B. B. (1997) J. Biol. Chem. 272 5283–5290 [DOI] [PubMed] [Google Scholar]
- 41.Hwang, C. S., Mandrup, S., MacDougald, O. A., Geiman, D. E., and Lane, M. D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman, H. B., Hu, X., Tyler, K. X., Dalal, C. K., and Lazar, M. A. (2002) J. Biol. Chem. 277 19754–19761 [DOI] [PubMed] [Google Scholar]
- 43.Williams, S. C., Cantwell, C. A., and Johnson, P. F. (1991) Genes Dev. 5 1553–1567 [DOI] [PubMed] [Google Scholar]
- 44.Chung, S. S., Choi, H. H., Cho, Y. M., Lee, H. K., and Park, K. S. (2006) Biochem. Biophys. Res. Commun. 348 253–258 [DOI] [PubMed] [Google Scholar]
- 45.Li, Y., and Lazar, M. A. (2002) Mol. Endocrinol. 16 1040–1048 [DOI] [PubMed] [Google Scholar]
- 46.Steger, D. J., Lefterova, M. I., Ying, L., Stonestrom, A. J., Schupp, M., Zhuo, D., Vakoc, A. L., Kim, J. E., Chen, J., Lazar, M. A., Blobel, G. A., and Vakoc, C. R. (2008) Mol. Cell. Biol. 28 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, P., Jenkins, N. A., and Copeland, N. G. (2003) Genome Res. 13 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kajimura, S., Seale, P., Tomaru, T., Erdjument-Bromage, H., Cooper, M. P., Ruas, J. L., Chin, S., Tempst, P., Lazar, M. A., and Spiegelman, B. M. (2008) Genes Dev. 22 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chawla, A., and Lazar, M. A. (1993) J. Biol. Chem. 268 16265–16269 [PubMed] [Google Scholar]
- 50.Delegeane, A. M., Ferland, L. H., and Mellon, P. L. (1987) Mol. Cell. Biol. 7 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lefterova, M. I., Zhang, Y., Steger, D. J., Schupp, M., Schug, J., Cristancho, A., Feng, D., Zhuo, D., Stoeckert, Jr., C. J., Liu, X. S., and Lazar, M. A. (2008) Genes Dev. 22 2932–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawadogo, M., Van Dyke, M. W., Gregor, P. D., and Roeder, R. G. (1988) J. Biol. Chem. 263 11985–11993 [PubMed] [Google Scholar]
- 54.Wang, Z., Zang, C., Rosenfeld, J. A., Schones, D. E., Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Peng, W., Zhang, M. Q., and Zhao, K. (2008) Nat. Genet. 40 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernstein, B. E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D. K., Huebert, D. J., McMahon, S., Karlsson, E. K., Kulbokas, E. J., 3rd, Gingeras, T. R., Schreiber, S. L., and Lander, E. S. (2005) Cell 120 169–181 [DOI] [PubMed] [Google Scholar]
- 56.Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., Barrera, L. O., Van Calcar, S., Qu, C., Ching, K. A., Wang, W., Weng, Z., Green, R. D., Crawford, G. E., and Ren, B. (2007) Nat. Genet. 39 311–318 [DOI] [PubMed] [Google Scholar]
- 57.Roh, T. Y., Wei, G., Farrell, C. M., and Zhao, K. (2007) Genome Res. 17 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seale, P., Kajimura, S., Yang, W., Chin, S., Rohas, L. M., Uldry, M., Tavernier, G., Langin, D., and Spiegelman, B. M. (2007) Cell Metab. 6 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacDougald, O. A., Cornelius, P., Lin, F. T., Chen, S. S., and Lane, M. D. (1994) J. Biol. Chem. 269 19041–19047 [PubMed] [Google Scholar]
- 60.MacDougald, O. A., Cornelius, P., Liu, R., and Lane, M. D. (1995) J. Biol. Chem. 270 647–654 [DOI] [PubMed] [Google Scholar]
- 61.Tontonoz, P., Hu, E., Devine, J., Beale, E. G., and Spiegelman, B. M. (1995) Mol. Cell. Biol. 15 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park, E. A., Roesler, W. J., Liu, J., Klemm, D. J., Gurney, A. L., Thatcher, J. D., Shuman, J., Friedman, A., and Hanson, R. W. (1990) Mol. Cell. Biol. 10 6264–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nielsen, R., Pedersen, T. A., Hagenbeek, D., Moulos, P., Siersbæk, R., Megens, E., Denissov, S., Børgesen, M., Francoijs, K.-J., Mandrup, S., and Stunnenberg, H. G. (2008) Genes Dev. 22 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo, J. B., Noh, M. J., Yoo, E. J., Park, S. Y., Park, J., Lee, I. K., Park, S. D., and Kim, J. B. (2003) Mol. Endocrinol. 17 1522–1533 [DOI] [PubMed] [Google Scholar]
- 65.Wilson, M. D., Barbosa-Morais, N. L., Schmidt, D., Conboy, C. M., Vanes, L., Tybulewicz, V. L., Fisher, E. M., Tavare, S., and Odom, D. T. (2008) Science 322 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamin, D., Hadigan, C., Lehrke, M., Mazza, S., Lazar, M. A., and Grinspoon, S. (2005) J. Clin. Endocrinol. Metab. 90 3423–3426 [DOI] [PubMed] [Google Scholar]
- 67.Lehrke, M., Reilly, M. P., Millington, S. C., Iqbal, N., Rader, D. J., and Lazar, M. A. (2004) PLoS Med. 1 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung, H. S., Youn, B. S., Cho, Y. M., Yu, K. Y., Park, H. J., Shin, C. S., Kim, S. Y., Lee, H. K., and Park, K. S. (2005) Metabolism 54 314–320 [DOI] [PubMed] [Google Scholar]
- 69.Farmer, S. R. (2008) Genes Dev. 22 1269–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chinnadurai, G. (2007) Int. J. Biochem. Cell Biol. 39 1593–1607 [DOI] [PubMed] [Google Scholar]
- 71.Nishikata, I., Sasaki, H., Iga, M., Tateno, Y., Imayoshi, S., Asou, N., Nakamura, T., and Morishita, K. (2003) Blood 102 3323–3332 [DOI] [PubMed] [Google Scholar]
- 72.Seale, P., Bjork, B., Yang, W., Kajimura, S., Chin, S., Kuang, S., Scime, A., Devarakonda, S., Conroe, H. M., Erdjument-Bromage, H., Tempst, P., Rudnicki, M. A., Beier, D. R., and Spiegelman, B. M. (2008) Nature 454 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]