Abstract
The Helicobacter pylori ArsS-ArsR two-component signal transduction system, comprised of a sensor histidine kinase (ArsS) and a response regulator (ArsR), allows the bacteria to regulate gene expression in response to acidic pH. We expressed and purified the full-length ArsR protein and the DNA-binding domain of ArsR (ArsR-DBD), and we analyzed the tertiary structure of the ArsR-DBD using solution nuclear magnetic resonance (NMR) methods. Both the full-length ArsR and the ArsR-DBD behaved as monomers in size exclusion chromatography experiments. The structure of ArsR-DBD consists of an N-terminal four-stranded β-sheet, a helical core, and a C-terminal β-hairpin. The overall tertiary fold of the ArsR-DBD is most closely related to DBD structures of the OmpR/PhoB subfamily of bacterial response regulators. However, the orientation of the N-terminal β-sheet with respect to the rest of the DNA-binding domain is substantially different in ArsR compared with the orientation in related response regulators. Molecular modeling of an ArsR-DBD-DNA complex permits identification of protein elements that are predicted to bind target DNA sequences and thereby regulate gene transcription in H. pylori.
Helicobacter pylori is a spiral-shaped Gram-negative organism that persistently colonizes the stomach in about half of the human population (1). H. pylori infection is a risk factor for the development of gastric cancer, a leading cause of cancer-related deaths in many parts of the world. Furthermore, H. pylori has been identified as a major risk factor for peptic ulcer disease and gastric lymphoma (1-5).
Within the human stomach, H. pylori encounters a range of acidic pH conditions (6-10). Upon entry of H. pylori into the stomach, the bacterium encounters gastric luminal pH values that range from 1 while fasting to 5 after a meal. After the initial transit of H. pylori through the gastric lumen, the bacterium thrives in the gastric mucus layer, where the pH is thought to be 4.5-6.5 (11). Within the mucus layer, the bacterium is still subject to considerable pH fluctuations as a consequence of the changing luminal pH. Therefore, H. pylori has evolved mechanisms to survive severe acid shock and grow under moderately acidic conditions. One of the important mechanisms by which H. pylori responds to acidic pH involves regulation of gene expression through a two-component signal transduction system (12-20).
Two-component systems (TCS)2 are stimulus-response coupling mechanisms that are used primarily by prokaryotic organisms to regulate cellular functions in response to changing environmental conditions (21). TCSs are composed of a sensor histidine kinase protein and a response regulator (RR) protein. In a typical TCS, the histidine kinase detects and monitors an environmental condition and transmits information via a phosphotransfer event to the cognate RR. Most RRs consist of two domains, an N-terminal regulatory domain and a C-terminal effector domain. Phosphorylation of the RR induces a conformational change in the regulatory domain, which results in activation of the effector domain. The majority of RRs are transcription factors with DNA binding effector domains (DBDs) (22). In contrast to the Escherichia coli genome, which contains 62 open reading frames (ORFs) encoding TCS component proteins (23), the H. pylori genome contains only 10 ORFs encoding TCS-component proteins (24). This paucity of regulatory genes in the H. pylori genome could reflect the pathogen's tight adaptation to its restricted ecological niche and also the lack of competition from other microorganisms in the acidic gastric environment (12, 25).
Several recent studies reported that an H. pylori TCS, ArsS-ArsR (acid-responsive signaling sensor/response regulator), responds to acidic conditions (12-20). This TCS consists of the histidine kinase protein, ArsS, and the RR protein, ArsR. Evidence supporting a cognate relationship between ArsS and ArsR includes immediate proximity of the two genes in the H. pylori chromosome and the demonstration that the two purified proteins can participate in a phosphotransfer reaction in vitro (12). H. pylori ArsS null mutants are viable, but such mutants are impaired in the ability to grow in low pH in vitro, and these mutant strains are unable to colonize mice (14, 15). Attempts to generate ArsR null mutants have been unsuccessful, which suggests that ArsR is essential for H. pylori viability (12).
One approach for identifying members of the ArsRS regulon has been to isolate DNA sequences that bind to ArsR. This approach resulted in the identification of two operons (designated HP1408-1412 and HP427-423 in H. pylori strain 26695) and a family of paralogous genes (exemplified by HP0119) that are regulated by the ArsRS system (13). These genes encode proteins of unknown function. As another approach to identify members of the ArsRS regulon, whole-genome transcriptional profiles of wild-type H. pylori and ArsS mutant strains have been compared. Such studies have been done following growth of the bacteria at either neutral pH or acidic pH. Whole-genome transcriptional profiling of H. pylori strains cultured in low pH conditions identified more than 100 genes that were differentially expressed in the wild-type strain compared with an ArsS-deficient mutant (17). Transcriptional profiling of H. pylori cultured in neutral pH conditions identified a smaller number of genes that were differentially expressed in the wild-type and ArsS null mutant strains (26). Acid-responsive H. pylori genes that are differentially expressed in wild-type and arsS mutant strains include amidases (amiE, amiF) and members of the urease gene cluster (ureA, ureB, ureE, ureF, ureG, ureH, and ureI) (9, 10, 16-18). Gel-shift and DNA-footprinting analyses have shown that ArsR binds directly to the promoter regions of these genes (17, 18). Thus, the ArsRS TCS has an important role in allowing H. pylori to regulate gene expression in response to changes in pH and also has an important role in allowing H. pylori to colonize the gastric mucosa (15).
It has been proposed that the periplasmic domain of ArsS detects low pH conditions (16, 27), triggering an ATP-dependent autophosphorylation reaction at a conserved histidine residue in the protein cytoplasmic domain. This model proposes that the signal from ArsS is then transduced to the cognate RR protein, ArsR, via a phosphotransfer reaction from the phosphohistidine kinase to a conserved aspartate residue (Asp-52) in ArsR, which leads to modification of the ArsR regulatory functions (for review, see Refs. 22 and 27). A derivative of ArsR with a D52N mutation is not phosphorylated in vitro by ArsS. In contrast to an arsR null mutant (which is non-viable) (28), a mutant H. pylori strain expressing the D52N form of ArsR is viable. These findings suggest that this mutant form of ArsR has a function that is sufficient for cell viability. Based on these data, it seems likely that there are two sets of target genes for ArsR. One set of genes in the ArsR regulon is presumed to be essential for cell viability, and regulation of these genes can be accomplished by a non-phosphorylated form of ArsR (exemplified by the D52N mutant protein) (12, 28). A second set of genes in the ArsR regulon is not required for cell viability, and the regulation of these genes occurs by a pathway involving a phosphorylated form of ArsR (i.e. requiring the cognate histidine kinase, ArsS) (13, 17, 18, 20, 26, 29). The products of various genes in the latter group contribute to acid acclimation, and thus host colonization, by H. pylori. It should be noted that genes belonging to the former set (required for cell viability) have not yet been identified.
Although several lines of evidence indicate that the ArsS-ArsR TCS has an important role in H. pylori, many features of this TCS are still not understood. Specifically, very little is known about how the structure and function of ArsR are altered in response to phosphorylation, little is known about how this TCS contributes to acid adaptation, and virtually nothing is known about what functions of ArsR are required for H. pylori viability. Herein, we report the structure of the DNA binding effector domain of ArsR as determined by solution NMR.
EXPERIMENTAL PROCEDURES
Plasmid Construction—The ArsR protein encoded by H. pylori strain J99 comprises 225 amino acids (GenBank™ accession number Q9ZMR6). The DNA-binding domain of this protein, mapped based on comparison to other known response regulators such as PhoB and OmpR, comprises 103 amino acids, beginning at Glu-123 and ending at Tyr-225. The full-length arsR gene and a fragment encoding the DNA-binding domain were amplified by PCR from H. pylori strain J99 (gene JHP0152) using genomic DNA as template and pairs of primers including 5′ BamHI and 3′ KpnI restriction endonuclease sites (5′-CGGGATCCATGATAGAAGTTTTAATGATAGAAGATGATATAG and 5′-CGGGTACCTCAGTATTCTAATTTATAACCAATCCCTCT for full-length ArsR and 5′-GCGGGATCCGAAGAGGTGAGTGAGCCAGGC and 5′-GCGGGTACCTCAGTATTCTAATTTATAACCAATCCCTCT for ArsR-DBD). Purified PCR products were digested with KpnI and BamHI restriction enzymes (Promega, Madison, WI) and ligated into linearized pET-BNK, a modified pET vector (Novagen, Madison, WI) developed specifically for expressing NMR protein targets. The vector contains a 5′-coding sequence for an N-terminal purification tag MRGSHHHHHHGS in-frame with the insert coding for the desired proteins.
Expression and Purification of ArsR and ArsR-DBD—For preparation of NMR samples, transformed E. coli BL21 (DE3) cells were grown in 4.2 liters of LB media containing glucose (2 g/liter) and ampicillin (50 mg/liter). When the culture reached an A600 of 1.0, protein expression was induced for 4 h with 0.8 mm isopropyl-β-d-thiogalactopyranoside. Cells were collected by 10 min of centrifugation at 12,000 × g, suspended in 0.02 m Na2HPO4, 0.5 m NaCl, pH 7.6, and then lysed by sonication 6 times for 30 s on ice. Both 1 mm phenylmethylsulfonyl fluoride and 5 mm Tris(2-carboxyethyl)phosphine hydrochloride were added before sonication. After sonication, the preparation was centrifuged at 31,000 × g for 20 min. The supernatant (∼100 ml) was collected and applied to a 30-ml metal affinity chromatography column (His-Bind, Novagen) charged with Ni2+. The column was washed with a 0-80 mm imidazole gradient to remove proteins bound non-specifically to the column, and the protein of interest was eluted with a 0.08-1.0 M imidazole gradient over 45 ml. 5-ml fractions corresponding to peaks of interest were collected, pooled, and concentrated to a volume of 1 ml in Amicon Ultra-15 centrifugal filters (10-kDa molecular weight cut-off (MWCO) membrane for the full-length protein and a 5-kDa MWCO membrane for the DBD). The purity of the proteins was assessed by Tricine SDS-PAGE on 10% gels. Protein samples estimated to be >95% pure were analyzed by NMR. 15N- and 13C,15N-labeled samples were produced by a process similar to that described above, but cells were grown in M9 medium with the addition of 13C-labeled glucose and 15N-labeled ammonium chloride (CIL, Andover, MA).
Analytical Size Exclusion Chromatography—The proteins were analyzed by gel filtration on a Sephacryl S-100 fast protein liquid chromatography column (GE Healthcare) run at 4 °C in 0.02 m Na2HPO4, 0.5 m NaCl, 1 mm Tris(2-carboxyethyl)phosphine hydrochloride (pH 7.6) and a flow rate of 0.4 ml/min, with ultraviolet absorption measured at 214 nm. ArsR and ArsR-DBD molecular weights (Mr) were determined by calculating their partition coefficient (Kav) values and using a calibration curve, plotting logarithmic values of Mr against calculated Kav values for a set of standard globular monomeric proteins (bovine serum albumin (66 kDa), ovalbumin (45 kDa), and myoglobin (16.7 kDa)), run under identical conditions. Kav values were calculated using the formula Kav = (Ve - Vo)/(Vt - Vo), where Ve is elution volume, Vo is column void volume, and Vt is total column volume.
NMR Structure Calculation—NMR spectra were acquired for ArsR and ArsR-DBD on an Avance 600 Bruker (Billerica, MA) spectrometer with a triple resonance gradient probe and a cryoprobe. Sample protein concentrations were ∼0.2 mm (full-length protein) and ∼0.5 mm (DBD) in 0.5 m NaCl, 0.02 m Na2HPO4, 1 mm Tris(2-carboxyethyl)phosphine hydrochloride (pH 7.6). Spectra were collected at 25 °C for unlabeled (natural abundance), 15N-labeled, and 13C,15N-labeled protein samples. The data were processed using XWINNMR and TOPSPIN software (Bruker) and analyzed with the Sparky suite (Goddard TD and Kneller DG, Sparky 3, University of California, San Francisco). The assignment of backbone resonances was completed using data from two-dimensional 1H,15N HSQC, two-dimensional NOESY, and three-dimensional CBCANH and CBCA(CO)NH experiments. The side chain resonance assignments were completed using data from three-dimensional HCC total correlation spectroscopy (TOCSY), HHC-TOCSY, H(CC)(CO)NH, HCC(CO)NH, HBHA(CO)NH, 15N-edited NOESY, and 13C-edited NOESY experiments (for references, see Ref. 30). The chemical shifts of Hα, Cα, Cβ, and C′ were analyzed with chemical shift index software (31) to produce a prediction of secondary structure elements. J-coupling constants calculated from HNHA experiments were used to determine ϕ angle constraints for structure calculations. The chemical shifts of Hα, Cα, Cβ, C′, and N were also analyzed to calculate angle constraints to be used for structure calculations using TALOS software (32). The structures were calculated using the CYANA Version 2.1 software package (33). Automatic calibration was used to convert the NOE peak intensities into distance constraints. The final calculations were performed for 1000 structures with 40,000 annealing steps for each. The 50 structures with the lowest target functions (≤0.6) were minimized with AMBER (Version 9) (34). 20 structures with the lowest energy were visualized with InsightII (Accelrys, San Diego, CA), Chimera (35), and MOLMOL (36). Electrostatic potentials calculated using Delphi program (37, 38) were used to generate a surface potential map for ArsR-DBD in Chimera. The stereochemistry of the structures was analyzed with PROCHECK-NMR (39).
NMR Analysis of ArsR-DBD in Complex with the Promoter Region of a Target Gene—Two-dimensional 1H,15N HSQC spectra were collected for purified 15N-labeled ArsR-DBD alone (0.1 mm) and ArsR-DBD combined with a 13-bp dsDNA fragment (5′-CGCATCATTAACC) (0.1 mm) from the promoter region of a well characterized ArsR target gene, hp1408 (13). This DNA fragment corresponds to the 5′ half of a DNA binding region identified by footprinting analysis (13).
Sequence Comparison of ArsR and Related Proteins—Sequence alignment of ArsR and several structurally characterized response regulators (OmpR (GenPept accession number AAC76430), PhoB (GenPept accession number AAC73502), DrrD (GenPept accession number 1KGS_A)) was carried out using ClustalW (40). The sequence of ArsR from H. pylori strain J99 was also compared with ArsR from other H. pylori strains (26695 and HPAG1) and orthologs from Helicobacter acinonychis (Sheeba), Helicobacter hepaticus (ATCC 51449), Wolinella succinogenes (strain DSM 1740), Campylobacter lari (RM 2100), Campylobacter curvus (525.92), and Campylobacter concisus (13826).
RESULTS
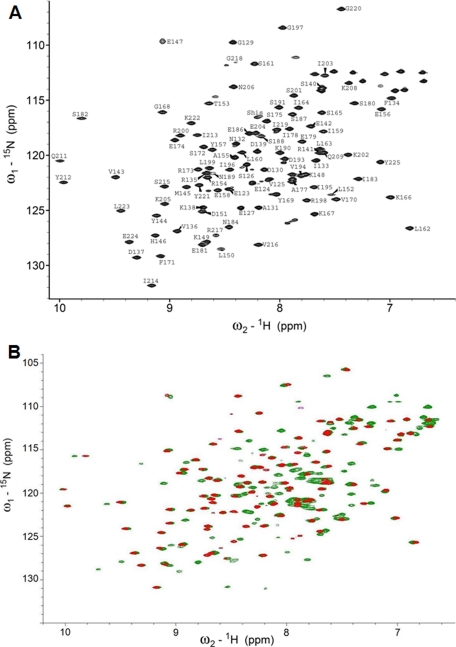
ArsR and ArsR-DBD Biochemical Characterization—The full-length His6-tagged ArsR protein (237 residues) and a His6-tagged ArsR fragment corresponding to the DBD (115 residues) were overexpressed in E. coli, recovered from soluble fractions of the cell lysate, and purified by metal affinity chromatography. Gel filtration experiments conducted on ArsR and ArsR-DBD demonstrated that the proteins migrated with molecular masses of ∼32 kDa (expected monomer size, ∼27 kDa) and ∼19 kDa (expected monomer size, ∼13 kDa), respectively (data not shown). The larger than expected apparent molecular sizes were most likely caused by the 17-residue disordered N termini of proteins. These data suggest that both ArsR and ArsR-DBD are monomeric in solution at concentrations below 0.1 mm. ArsR and ArsR-DBD protein samples were concentrated to ∼0.2 and ∼0.5 mm, respectively, and analyzed by NMR. The directly detected 31P NMR experiments conducted on purified full-length ArsR (in Tris buffer), which contains a putative phosphate-receiving aspartate residue, did not reveal any detectable phosphate signal, suggesting that the majority of the purified protein is in the unphosphorylated form (data not shown). One-dimensional 1H NMR spectra of ArsR showed well dispersed peaks in regions characteristic of amide protons (6-10 ppm) and aliphatic protons (0-4 ppm), indicative of folded protein. ArsR-DBD was produced in natural abundance and in 15N- and 13C,15N-enriched forms. Fig. 1A shows that the 1H,15N correlation NMR spectrum of ArsR-DBD exhibited good chemical shift dispersion. A comparison of 1H,15N HSQCs of full-length ArsR and ArsR-DBD showed that ArsR-DBD amide proton peaks aligned with ArsR amide peaks, indicating that the structure of the isolated ArsR-DBD was very similar to that domain in the full-length protein (Fig. 1B).
FIGURE 1.
NMR spectra of ArsR and ArsR-DBD. A, 1H,15N HSQC spectrum of ArsR-DBD. Peaks are labeled with the one-letter amino acid code and residue numbers. B, comparison of 1H,15N HSQC spectra of ArsR (green) and ArsR-DBD (red).
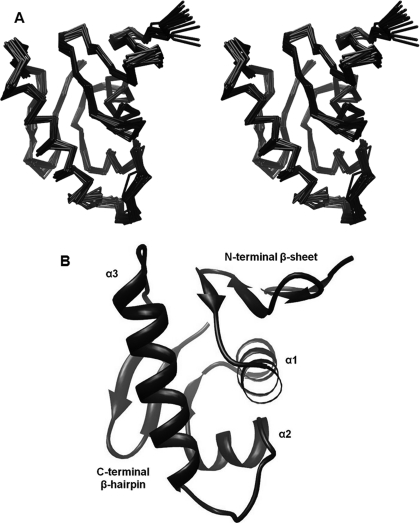
ArsR-DBD Structural Determination—Backbone NH resonance assignments were obtained (Fig. 1A) for all residues of the ArsR-DBD, except for the first 11 residues of the 12-residue N-terminal His6 tag, 4 proline residues, and 2 isoleucine residues (Ile-176 and Ile-192, numbered based on the sequence of untagged full-length ArsR). The solution structure of ArsR-DBD was determined by NMR based on 1265 distance constraints, including 264 intraresidue, 329 sequential, 360 medium range, and 312 long range distance constraints, and 155 dihedral angle constraints calculated using TALOS (32) and J-coupling constants calculated from HNHA experiments. Table 1 summarizes the structural statistics for the calculations. The first 17 residues of the protein (including the 12 residues from the His6 tag) showed very few NOE interactions and chemical shifts, a finding that is characteristic of disordered peptides. Residues 128-225 form a well structured domain. The 20 structures presented in the final ensemble (Fig. 2A) conform to the average structure with atomic root mean square deviations (r.m.s.d.) about the mean coordinate positions of 0.55 (±0.07) Å for the backbone atoms and 1.22 (±0.12) Å for all the heavy atoms of residues 129-224. The structural quality of the ensemble of the calculated structures was analyzed by using PROCHECK-NMR. 98.4% of the residues fall into the allowed regions (84.3%, 11.4%, 2.7% in the most favorable, additionally allowed, and generously allowed regions, respectively) of the Ramachandran plot.
TABLE 1.
Structural statistics of the final ensemble of 20 energy-minimized structures of ArsR-DBD
| Number of NOE distance constraints | |
| All | 1265 |
| Intraresidue | 264 |
| Sequential (|i − j| = 1) | 329 |
| Medium range (2 ≤ |i − j| ≤ 5) | 360 |
| Long range (|i − j| > 5) | 312 |
| NOE violations present in all 20 structures greater than 0.2 Å | 0 |
| Hydrogen bonds distance restraintsa | 48 |
| Dihedral angle constraints | 155 |
| Mean r.m.s.d. from the average coordinate (Å)b | |
| Backbone atoms (N,Cα,C′) | 0.55 (±0.07) |
| Heavy atoms | 1.22 (±0.12) |
| Ramachandran plot (%) | |
| Most favored regions | 84.3 |
| Additional allowed regions | 11.4 |
| Generously allowed regions | 2.7 |
| Disallowed regions | 1.6 |
Two restraints per one hydrogen bond.
For residues 129-224.
FIGURE 2.
Structure of ArsR DNA-binding domain. A, a stereo view of the best-fit superposition of backbone Cα atoms of the final 20 energy-minimized structures of ArsR-DBD (residues 128-225). B, ribbon diagram of ArsR-DBD (residues 128-225). Secondary structural elements are labeled.
Overview of ArsR-DBD Structure—The ArsR-DBD is composed of an N-terminal four-stranded antiparallel β-sheet (β1-(Gly-129—Ala-131), β2-(Phe-134—Asp-137), β3-(Glu-142—Met-145), β4-(Lys-148—Asp-151)), three α-helices that form the core of the protein (α1-(Ala-155—Lys-166), α2-(Arg-173—Glu-179), α3-(Lys-190—Lys-205)), and a C-terminal β-hairpin (β6-(Ile-213—Val-216), β7-(Gly-220—Glu-224)) (Fig. 2B). In addition, the domain contains a short β-strand connecting helices α1 and α2 (β5-(Val-170—Ser-172)), which interacts with the C-terminal hairpin. The final topology of this domain, from the N to C terminus, is β1-β2-β3-β4-α1-β5-α2-α3-β6-β7. The structured regions of the 20 structures in the final ensemble (residues Gly-129—Ala-131, Phe-134—Asp-137, Glu-142—Met-145, Lys-148—Asp-151, Ala-155—Lys-166, Val-170—Ser-172, Arg-173—Glu-179, Lys-190—Lys-205, Ile-213—Val-216, and Gly-220—Glu-224) converge with a backbone r.m.s.d. value of 0.43 Å. As discussed further below, the ArsR-DBD contains a winged helix-turn-helix (wHTH) fold that is predicted to mediate binding of the protein to DNA. This wHTH fold is formed by the α2 and α3 helices, the loop connecting them, and the loop connecting β strands 6 and 7 as the “wing.”
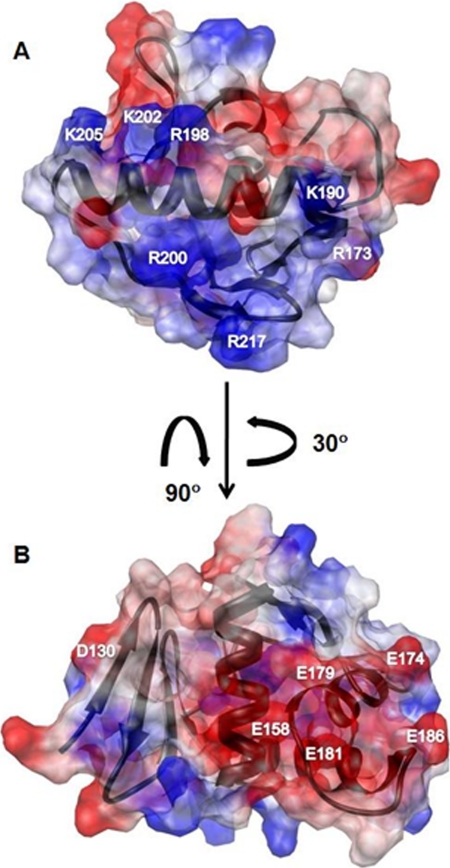
The electrostatic surface potential map of ArsR-DBD (Fig. 3) reveals two distinct regions with opposite charge distribution. A surface with a largely positive electrostatic potential (including residues Arg-173, Lys-190, Arg-198, Arg-200, Lys-202, Lys-205, and Arg-217) is shown in Fig. 3A. Based on comparisons with other RRs (41-43), this surface is predicted to bind to the negatively charged phosphate backbone of DNA. Fig. 3B displays a surface with a highly negative electrostatic potential (including residues Asp-130, Glu-158, Glu-174, Glu-179, Glu-181, and Glu-186), with the majority of the charge contribution from the residues at the C-terminal end of α2.
FIGURE 3.
Electrostatic surface potential maps of ArsR-DBD. Red indicates a negatively charged region, blue indicates a positively charged region, and white indicates a neutral or hydrophobic region. A, the molecule is oriented to display the predicted DNA binding surface. Residues contributing to the positive surface potential are Arg-173, Lys-190, Arg-198, Arg-200, Lys-202, Lys-205, and Arg-217. B, the molecule was rotated to reveal a negatively charged surface, attributable to residues Asp-130, Glu-158, Glu-174, Glu-179, Glu-181, and Glu-186.
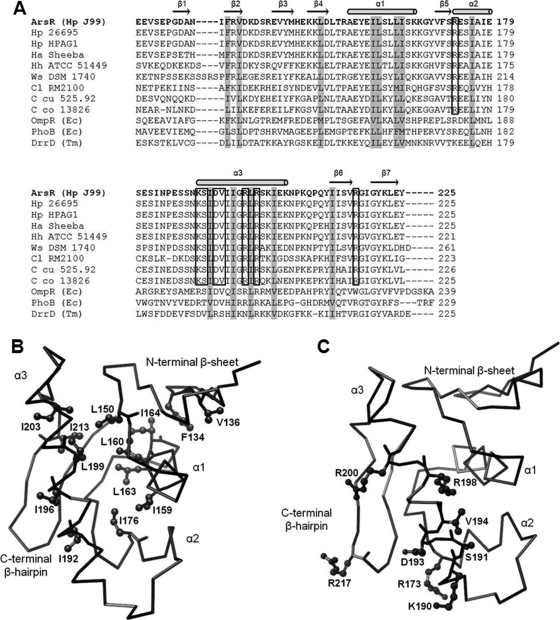
Structural Comparison of ArsR-DBD with Related Structures—A BLAST search of GenBank™ (44) using the ArsR amino acid sequence indicates that ArsR is most closely related to members of the OmpR/PhoB subfamily of response regulators, but the levels of amino acid sequence identity are fairly low (32% (OmpR), 28% (PhoB), and 32% (DrrD)) (Fig. 4A). Despite these low levels of sequence identity, an analysis of ArsR with DALI software and the Families of Structurally Similar Proteins data base (45) which catalogs known protein structures, showed that the ArsR-DBD structure was closely related to the structures of members of the OmpR/PhoB subfamily, including OmpR (PDB code 1opc) (43), PhoB (PDB code 1gxq) (41), and DrrD (PDB code 1kgs) (46), with z scores (depicting strength of structural similarity) of 3.8, 6.1, and 5.3, respectively. The secondary structural elements of ArsR-DBD are positioned by internally packed hydrophobic residues that stabilize the protein fold (Fig. 4B). These core hydrophobic residues of ArsR-DBD (corresponding to residues Phe-134, Val-l136, Leu-150, Ile-159, Leu-160, Leu-163, Ile-164, Ile-176, Ile-192, Ile-196, Leu-199, Ile-203, and Ile-213 in ArsR) are conserved across members of the OmpR/PhoB subfamily (Fig. 4, A and B) (47), which suggests that they are responsible for the shared structural features.
FIGURE 4.
Comparison of ArsR-DBD with related proteins. A, sequence alignment of ArsR-DBD with orthologs from closely related species and with closely related proteins from the OmpR/PhoB subfamily (see “Experimental Procedures” for a detailed description of the aligned sequences). Secondary structural elements of ArsR-DBD are displayed above the sequence with horizontal arrows indicating β strands and cylinders indicating α helices. Core hydrophobic residues that are conserved within the DBDs of the OmpR/PhoB subfamily are shaded gray. Surface-exposed residues in the wHTH motif that are identical in ArsR, and orthologs from closely related species are boxed. B, backbone Cα tracing of ArsR-DBD depicting side chains of conserved hydrophobic core residues. The hydrophobic residues conserved in ArsR-DBD and the DBDs of OmpR, PhoB, and DrrD are indicated in ball-and-stick style on the line tracing of the ArsR-DBD backbone. C, backbone Cα tracing of ArsR-DBD and side chains of conserved surface-exposed residues in the wHTH motif. Surface-exposed residues in the wHTH motif that are identical in ArsR and orthologs from closely related species are indicated in ball-and-stick style on the line tracing of the ArsR-DBD backbone.
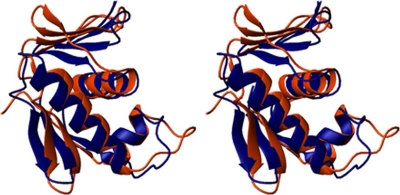
PhoB, an RR from E. coli, is a well studied prototype of the OmpR/PhoB subfamily. Despite the low 28% amino acid sequence identity of PhoB with ArsR, the DNA-binding domain of PhoB exhibits a high degree of structural similarity to the ArsR-DBD, as was shown by the DALI analysis (see above). Thus, we performed a detailed comparison of the ArsR and PhoB structures (Fig. 5). The arrangement of the three α-helical bundle is highly similar in the ArsR and PhoB DBDs and was used to superimpose the two structures. When tracing along the N, Cα, C′, and atoms, residues Ala-155—Lys-166 (α1), Arg-173—Glu-179 (α2), and Lys-190—Lys-205 (α3) of ArsR-DBD superimpose with residues of the PhoB-DBD helical core with an r.m.s.d value of 2.3 Å. The orientation of the loop between α2 and α3 (which is predicted to interact with subunits of RNA polymerase) and the length of the α3 helix (which is predicted to be involved in DNA recognition) are known to vary among members of the OmpR/PhoB subfamily, but these structural features are similar in ArsR and PhoB. However, a notable difference is that the N-terminal β-sheet in ArsR-DBD is rotated almost 45° with respect to the N-terminal β-sheet in PhoB-DBD. This antiparallel β-sheet functions as a platform for interactions with the N-terminal receiver domain in many OmpR/PhoB RRs (46, 48).
FIGURE 5.
Structural comparison of the DNA-binding domains of ArsR and PhoB. The images represent a stereo view of the superposition of the structures of ArsR-DBD (blue) and the DNA-binding domain of PhoB (orange) (PDB code 1gxq (41)). The superimposition was based on the N, Cα, and C′ atoms of the three core helices with an r.m.s.d. value of 2.3 Å.
Comparison of ArsR with Close Orthologs—A BLAST search using the ArsR amino acid sequence identified ArsR orthologs in all H. pylori strains for which genome sequences are available as well as in closely related species. A pair-wise sequence comparison of ArsR from H. pylori strains J99, 26695, and HPAG1 as well as strains from related species, H. acinonychis, H. hepaticus, Wolinella, and Campylobacter species, is shown in Fig. 4A. The ArsR proteins encoded by multiple H. pylori strains exhibited 99% pair-wise amino acid sequence identity. ArsR from another gastric Helicobacter species, H. acinonychis, also exhibited a high level of relatedness (94% amino acid identity) to H. pylori ArsR. ArsR from H. hepaticus, an intestinal Helicobacter species, was 71% identical to H. pylori ArsR. ArsR orthologs from Wolinella and Campylobacter were 54-67% identical to H. pylori ArsR (Fig. 4A). Comparisons of the ArsR-DBD orthologs from these species showed a high degree of conservation in core hydrophobic residues belonging to the major secondary structural elements of the protein. Furthermore, a high degree of sequence identity was maintained in the wHTH motif, most notably in the surface-exposed residues of the α3 recognition helix and the wing of the C-terminal β-hairpin (corresponding to residues Arg-173, Lys-190, Ser-191, Asp-193, Val-194, Arg-198, Arg-200, and Arg-217 in ArsR) (Fig. 4, A and C).
DISCUSSION
In this paper, we present a detailed three-dimensional structure of the ArsR DNA-binding domain, determined using NMR spectroscopy. The ArsR-DBD structure is most closely related to the structures of proteins classified in the OmpR/PhoB subfamily of response regulators. Like other members of this subfamily, the ArsR-DBD comprises two anti-parallel β-sheets flanking a core of three α-helices and contains a winged HTH motif that is predicted to bind DNA.
Despite similar tertiary structural organization, members of the OmpR/PhoB subfamily of RRs differ in their dependence on phosphorylation and oligomerization for transcriptional regulation. For example, upon phosphorylation, some RRs dimerize in solution, whereas others remain monomeric and subsequently dimerize after binding to specific DNA sequences (49-52). At present, relatively little is known about the effects of phosphorylation and oligomerization on ArsR activity.
The orientation of the N-terminal β-sheet in the ArsR-DBD differs markedly from the orientations of N-terminal β-sheets observed in previously solved effector domains of related RRs. Relative to the orientation of the N-terminal β-sheet in PhoB-DBD (41), the N-terminal β-sheet of ArsR-DBD is rotated about 45° to align in the direction of the α1 helix. As a consequence of this difference, we speculate that the interdomain interactions between the N-terminal receiver domain and the C-terminal DBD will differ in ArsR compared with what is observed in other members of the OmpR/PhoB subfamily.
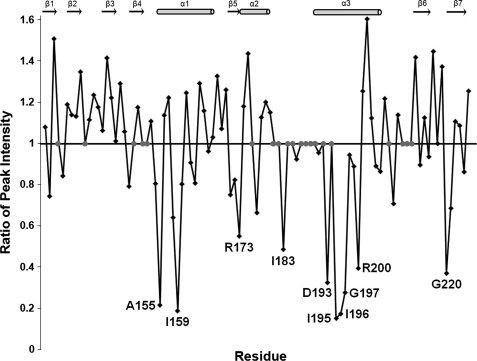
Based on the conservation of structural features in ArsR and members of the OmpR/PhoB subfamily of RRs, ArsR-DBD is predicted to interact with DNA in a manner similar to that of other members of the OmpR/PhoB subfamily. To test this hypothesis, the interaction of ArsR-DBD with DNA was analyzed by mapping spectral changes in 15N-labeled ArsR-DBD in the presence of an equimolar concentration of a 13-bp fragment of the promoter sequence of hp1408 (13), a member of the ArsRS regulon (see “Experimental Procedures” for details). Because OmpR/PhoB dimers bind ∼24 bp of DNA, it is predicted that this 13-bp fragment would bind only one molecule of ArsR-DBD. A comparison of the two-dimensional 1H,15N HSQC spectra of ArsR-DBD alone and combined with this DNA revealed changes in the intensities of peaks corresponding to specific amino acid residues (Fig. 6). This indicates moderately strong affinity between protein and DNA molecules, resulting in intermediate exchange regime in the NMR experiments. A detailed comparison of the peak intensities from the two spectra indicated that several residues exhibiting the most prominent reductions in peak intensities mapped to surface-exposed regions of the wHTH motif (residues Arg-173, Asp-193, and Arg-200). Additionally, several core residues of the wHTH motif and other elements of the protein that make close contact with the helices of the wHTH motif demonstrated diminished peak intensities (residues Ala-155, Ile-159, Ile-183, Ile-195, Ile-196, Gly-197, and Gly-220) (Fig. 6). These data combined with the analyses shown in Fig. 4 support our prediction that ArsR-DBD interactions with DNA involve molecular surfaces homologous to those identified for other members of the OmpR/PhoB subfamily.
FIGURE 6.
Analysis of HSQC spectra of ArsR-DBD alone and ArsR-DBD combined with a 13-bp dsDNA fragment derived from the promoter region of an ArsR target gene (hp1408). For each residue a ratio of peak intensity in the spectrum of ArsR-DBD with DNA to the peak intensity in the spectrum of ArsR-DBD without DNA is shown. Ratios substantially less than 1 indicate a decrease in peak intensity when the protein is combined with DNA. Residues for which intensity ratios were not available (due to absence of visible peaks or peak overlap) are depicted by gray symbols. Residues Ala-155, Ile-159, Arg-173, Ile-183, Asp-193, Ile-195, Ile-196, Gly-197, Arg-200, and Gly-220 showed the greatest decrease in intensity. ArsR-DBD secondary structure elements are shown above the plot.
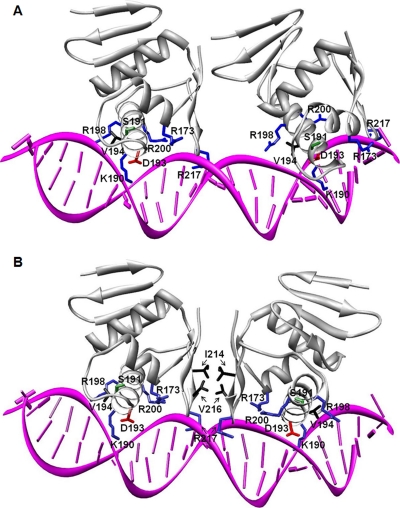
To date the only solved structure of an OmpR/PhoB subfamily member in complex with its target DNA binding site corresponds to the DBD of an E. coli RR, PhoB-DBD, on the pho box of the phoA operon promoter (41). Analysis of the electrostatic surface potential map of the PhoB-DBD revealed a positively charged surface in the region that binds DNA (41), and the electrostatic surface potential map of ArsR-DBD reveals a similar basic surface (Fig. 3A). We used the structure of the PhoB-DBD in complex with a target promoter sequence (41) as a template to generate a model of the ArsR-DBD binding to DNA (Fig. 7). Thus far, a consensus sequence for ArsR binding sites has not been identified, and therefore, to generate this model we retained the PhoB-specific binding site (pho box) as the target DNA; we did not attempt to model specific protein-DNA interactions when positioning ArsR-DBD on the DNA. Two ArsR-DBD domains were placed in tandem orientation (Fig. 7A) in the major groove of the target DNA fragment, ∼10 bp apart. The two ArsR-DBD molecules superimpose on the two PhoB-DBD molecules (tracing along the backbone N, Cα, C′ atoms of the corresponding α2 and α3 helices), both with an r.m.s.d. value of 2.1 Å.
FIGURE 7.
Modeling of the ArsR-DBD-DNA interaction. A, two ArsR-DBD molecules were positioned in tandem (head to tail) on a 23-bp direct-repeat dsDNA sequence derived from the crystal structure of the PhoB-DNA complex (PDB code 1gxp). Secondary structural elements of the proteins are shown as a gray ribbon. Residues in ArsR-DBD that potentially interact with DNA are presented as sticks with basic residues (Arg-173, Lys-190, Arg-198, Arg-200, Arg-217) in blue, acidic residue Asp-193 in red, polar uncharged residue Ser-191 in green, and hydrophobic residue Val-194 in black. B, two ArsR-DBD molecules positioned tail to tail on a 23-bp double-stranded DNA sequence containing an inverted repeat. Secondary structural and DNA binding elements are presented as in panel A. Residues within the C-terminal β-hairpin that could interact with each other in this orientation, Ile-214 and Val-216, are shown in black.
The PhoB-DBD binds to the direct repeat-containing pho box as a tandemly arranged (“head to tail”) dimer (41). However, several binding sites of ArsR that have been reported thus far (from promoters of genes arsR, ureA, ureI, amiE, amiF, rocF, hp1408, and hp1186 (carbonic anhydrase)) do not contain conserved symmetrical sequences (13, 17, 18, 20). The degeneracy of ArsR binding sites leaves open the possibility that ArsR may bind to DNA in a different manner than that observed with PhoB. To construct an alternative model of ArsR-DBD molecules arranged symmetrically on inverted repeat sequences, one-half of the dimeric PhoB-DBD-DNA complex was inverted and superimposed on the original copy of DNA using only the DNA backbone phosphates for alignment, with an r.m.s.d. value of 0.96 Å. Two ArsR-DBD molecules were superimposed on the PhoB-DBD molecules (tracing along the Cα atoms of the corresponding α2 and α3 helices) with r.m.s.d. values of 2.1 Å (Fig. 7B). In both orientations, residues Arg-173, Lys-190, Ser-191, Asp-193, Val-194, Arg-198, and Arg-200 from the ArsR HTH motif and residue Arg-217 from the “wing” between β-strand 6 and β-strand 7 of the C-terminal hairpin make contacts with the phosphate backbone and/or the bases of the target DNA. These interactions are supported by our NMR experimental data (Fig. 6). The functional groups of these ArsR residues are similar for the corresponding residues in PhoB, suggesting a conservation of their DNA recognition functions.
In the model shown in Fig. 7, residues Lys-190, Ser-191, Val-194, and Arg-198 protrude into the major groove of the DNA to make specific contacts with bases. The side chains of lysine and arginine and the hydroxyl group of Ser-191 provide potential hydrogen-bonding partners. In addition the methyl groups of valine can make specific van der Waals contacts with the methyl groups of thymines. The reported ArsR binding sites are A-T-rich (13, 17, 18, 20, 29), suggesting that the valine-thymine contacts may be important determinants of sequence-specific protein-DNA interactions. A distinctive feature of the “tail to tail” symmetric orientation of the ArsR-DBD-DNA complex model (Fig. 7B) is the interaction of hydrophobic patches on the two ArsR-DBD molecules formed by residues Ile-214 and Val-216 of each C-terminal β-hairpin. The surface-exposed hydrophobic residues on one ArsR-DBD molecule are stabilized by the hydrophobic residues exposed on the opposite ArsR-DBD molecule. This interaction could contribute to the overall stabilization of ArsR dimers on target DNA sequences.
In summary, the results of this study allow the classification of ArsR into a subfamily of DNA-binding proteins that contain a conserved wHTH motif and yet exhibit diversity in their interactions with DNA and diversity in interdomain and protein-protein interactions. The structure of the ArsR-DBD provides a basis for future experimental studies designed to understand these interdomain, protein-protein, and protein-DNA interactions.
Acknowledgments
We thank John Loh, Marcus Voehler, Donald Stec, and Young-Tae Lee for helpful discussions. UCSF Chimera package used to produce some of the molecular graphics images was obtained from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
The atomic coordinates and structure factors (code 2k4j) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The resonance assignments for ArsR-DBD were submitted to Biological Magnetic Resonance Bank ID, code 15801.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI39657. This work was also supported by the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TCS, two component system; ArsR, acid-responsive signaling regulator; ArsS, acid-responsive signaling sensor; DBD, DNA-binding domain; RR, response regulator; wHTH, winged helix-turn-helix; HSQC, heteronuclear single quantum correlation; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; NOESY, nuclear Overhauser effect (NOE) spectroscopy; r.m.s.d., root mean square deviation.
References
- 1.Parsonnet, J., Hansen, S., Rodriguez, L., Gelb, A. B., Warnke, R. A., Jellum, E., Orentreich, N., Vogelman, J. H., and Friedman, G. D. (1994) N. Engl. J. Med. 330 1267-1271 [DOI] [PubMed] [Google Scholar]
- 2.Marshall, B. J., and Warren, J. R. (1984) Lancet 1 1311-1315 [DOI] [PubMed] [Google Scholar]
- 3.Mueller, A., Falkow, S., and Amieva, M. R. (2005) Cancer Epidemiol. Biomark. Prev. 14 1859-1864 [DOI] [PubMed] [Google Scholar]
- 4.Nomura, A., Stemmermann, G. N., Chyou, P. H., Kato, I., Perez-Perez, G. I., and Blaser, M. J. (1991) N. Engl. J. Med. 325 1132-1136 [DOI] [PubMed] [Google Scholar]
- 5.Peterson, W. L. (1991) N. Engl. J. Med. 324 1043-1048 [DOI] [PubMed] [Google Scholar]
- 6.Ang, S., Lee, C. Z., Peck, K., Sindici, M., Matrubutham, U., Gleeson, M. A., and Wang, J. T. (2001) Infect. Immun. 69 1679-1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audia, J. P., Webb, C. C., and Foster, J. W. (2001) Int. J. Med. Microbiol. 291 97-106 [DOI] [PubMed] [Google Scholar]
- 8.Bury-Mone, S., Thiberge, J. M., Contreras, M., Maitournam, A., Labigne, A., and De Reuse, H. (2004) Mol. Microbiol. 53 623-638 [DOI] [PubMed] [Google Scholar]
- 9.Merrell, D. S., Goodrich, M. L., Otto, G., Tompkins, L. S., and Falkow, S. (2003) Infect. Immun. 71 3529-3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen, Y., Marcus, E. A., Matrubutham, U., Gleeson, M. A., Scott, D. R., and Sachs, G. (2003) Infect. Immun. 71 5921-5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schade, C., Flemstrom, G., and Holm, L. (1994) Gastroenterology 107 180-188 [DOI] [PubMed] [Google Scholar]
- 12.Beier, D., and Frank, R. (2000) J. Bacteriol. 182 2068-2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietz, P., Gerlach, G., and Beier, D. (2002) J. Bacteriol. 184 350-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh, J. T., and Cover, T. L. (2006) Infect. Immun. 74 3052-3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panthel, K., Dietz, P., Haas, R., and Beier, D. (2003) Infect. Immun. 71 5381-5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pflock, M., Dietz, P., Schar, J., and Beier, D. (2004) FEMS Microbiol. Lett. 234 51-61 [DOI] [PubMed] [Google Scholar]
- 17.Pflock, M., Finsterer, N., Joseph, B., Mollenkopf, H., Meyer, T. F., and Beier, D. (2006) J. Bacteriol. 188 3449-3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pflock, M., Kennard, S., Delany, I., Scarlato, V., and Beier, D. (2005) Infect. Immun. 73 6437-6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott, D. R., Marcus, E. A., Wen, Y., Oh, J., and Sachs, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7235-7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen, Y., Feng, J., Scott, D. R., Marcus, E. A., and Sachs, G. (2006) J. Bacteriol. 188 1750-1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, M. N., and Silhavy, T. J. (1981) J. Mol. Biol. 146 23-43 [DOI] [PubMed] [Google Scholar]
- 22.Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000) Annu. Rev. Biochem. 69 183-215 [DOI] [PubMed] [Google Scholar]
- 23.Mizuno, T. (1997) DNA Res. 4 161-168 [DOI] [PubMed] [Google Scholar]
- 24.Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., Nelson, K., Quackenbush, J., Zhou, L., Kirkness, E. F., Peterson, S., Loftus, B., Richardson, D., Dodson, R., Khalak, H. G., Glodek, A., McKenney, K., Fitzegerald, L. M., Lee, N., Adams, M. D., Hickey, E. K., Berg, D. E., Gocayne, J. D., Utterback, T. R., Peterson, J. D., Kelley, J. M., Cotton, M. D., Weidman, J. M., Fujii, C., Bowman, C., Watthey, L., Wallin, E., Hayes, W. S., Borodovsky, M., Karp, P. D., Smith, H. O., Fraser, C. M., and Venter, J. C. (1997) Nature 388 539-547 [DOI] [PubMed] [Google Scholar]
- 25.Eguchi, Y., and Utsumi, R. (2005) Trends Biochem. Sci. 30 70-72 [DOI] [PubMed] [Google Scholar]
- 26.Forsyth, M. H., Cao, P., Garcia, P. P., Hall, J. D., and Cover, T. L. (2002) J. Bacteriol. 184 4630-4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pflock, M., Kennard, S., Finsterer, N., and Beier, D. (2006) J. Biotechnol. 126 52-60 [DOI] [PubMed] [Google Scholar]
- 28.Schar, J., Sickmann, A., and Beier, D. (2005) J. Bacteriol. 187 3100-3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen, Y., Feng, J., Scott, D. R., Marcus, E. A., and Sachs, G. (2007) J. Bacteriol. 189 2426-2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sattler, M., Schleucher, J., and Griesinger, C. (1999) Progress in Nuclear Magnetic Resonance Spectroscopy 34 93-158 [Google Scholar]
- 31.Wishart, D. S., and Sykes, B. D. (1994) Methods Enzymol. 239 363-392 [DOI] [PubMed] [Google Scholar]
- 32.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 33.Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997) J. Mol. Biol. 273 283-298 [DOI] [PubMed] [Google Scholar]
- 34.Case, D. A., Darden, T. A., Cheatham, T. E., III, Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Merz, K. M., Pearlman, D. A., Crowley, M., Walker, R. C., Zhang, W., Wang, B., Hayik, S., Roitberg, A., Seabra, G., Wong, K. F., Paesani, F., Wu, X., Brozell, S., Tsui, V., Gohlke, H., Yang, L., Tan, C., Mongan, J., Hornak, V., Cui, G., Beroza, P., Mathews, D. H., Schafmeister, C., Ross, W. S., and Kollman, P. A. (2004) Amber 9, University of California, San Francisco
- 35.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 36.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graph. 14 51-55 and 29-32 [DOI] [PubMed] [Google Scholar]
- 37.Rocchia, W., Alexov, E., and Honig, B. (2001) J. Phys. Chem. B 105 6507-6514 [Google Scholar]
- 38.Rocchia, W., Sridharan, S., Nicholls, A., Alexov, E., Chiabrera, A., and Honig, B. (2002) J. Comput. Chem. 23 128-137 [DOI] [PubMed] [Google Scholar]
- 39.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco, A. G., Sola, M., Gomis-Ruth, F. X., and Coll, M. (2002) Structure 10 701-713 [DOI] [PubMed] [Google Scholar]
- 42.Hong, E., Lee, H. M., Ko, H., Kim, D. U., Jeon, B. Y., Jung, J., Shin, J., Lee, S. A., Kim, Y., Jeon, Y. H., Cheong, C., Cho, H. S., and Lee, W. (2007) J. Biol. Chem. 282 20667-20675 [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Hackert, E., and Stock, A. M. (1997) Structure 5 109-124 [DOI] [PubMed] [Google Scholar]
- 44.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2008) Nucleic Acids Res. 36 (database issue) 25-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm, L., Kaariainen, S., Rosenstrom, P., and Schenkel, A. (2008) Bioinformatics 24 2780-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckler, D. R., Zhou, Y., and Stock, A. M. (2002) Structure 10 153-164 [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Hackert, E., and Stock, A. M. (1997) J. Mol. Biol. 269 301-312 [DOI] [PubMed] [Google Scholar]
- 48.Robinson, V. L., Wu, T., and Stock, A. M. (2003) J. Bacteriol. 185 4186-4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiba, H., Nakasai, F., Mizushima, S., and Mizuno, T. (1989) J. Biochem. (Tokyo) 106 5-7 [DOI] [PubMed] [Google Scholar]
- 50.Fiedler, U., and Weiss, V. (1995) EMBO J. 14 3696-3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo, Y. L., Nara, F., Ichihara, S., Mizuno, T., and Mizushima, S. (1986) J. Biol. Chem. 261 15252-15256 [PubMed] [Google Scholar]
- 52.McCleary, W. R. (1996) Mol. Microbiol. 20 1155-1163 [DOI] [PubMed] [Google Scholar]