Abstract
Hensin is a rabbit ortholog of DMBT1, a multifunctional, multidomain protein implicated in the regulation of epithelial differentiation, innate immunity, and tumorigenesis. Hensin in the extracellular matrix (ECM) induced morphological changes characteristic of terminal differentiation in a clonal cell line (clone C) of rabbit kidney intercalated cells. Although hensin is secreted in monomeric and various oligomeric forms, only the polymerized ECM form is able to induce these phenotypic changes. Here we report that hensin secretion and matrix assembly were inhibited by the peptidylprolyl cis-trans isomerase (PPIase) inhibitors cyclosporin A (CsA) and a derivative of cyclosporin A with modifications in the d-Ser side chain (Cs9) but not by the calcineurin pathway inhibitor FK506. PPIase inhibition led to failure of hensin polymerization in the medium and ECM, plus the loss of apical cytoskeleton, apical microvilli, and the columnar epithelial shape of clone C cells. Cyclophilin A was produced and secreted into the media to a much greater extent than cyclophilins B and C. Our results also identified the direct CsA-sensitive interaction of cyclophilin A with hensin, suggesting that cyclophilin A is the PPIase that mediates the polymerization and matrix assembly of hensin. These results are significant because this is the first time a direct role of peptidylprolyl cis-trans isomerase activity has been implicated in the process of epithelial differentiation.
Hensin is a rabbit ortholog of the human DMBT1 gene, which has been implicated in the etiology of many cancer forms and in innate immune defense (1-3). We previously established that the extracellular matrix (ECM)3 form of hensin induces, in a clonal cell line of rabbit kidney intercalated cells, terminal differentiation-like features including remodeling of the apical cytoskeleton, induction of apical endocytosis, and columnarization of the cell shape (1). In addition, hensin is present in a pattern compatible with ECM deposition in the mature columnar epithelia of the intestinal villus and prostate luminal cells but not in the less differentiated epithelia of the crypt of the small intestine and basal cells of the prostate, suggesting a general role for ECM hensin in terminal differentiation of epithelia. ECM hensin is critical for the adaptation of the cortical collecting duct to metabolic acidosis, during which an increase in the number of acid-secreting α-intercalated cells is observed (4). Hensin is a modular protein containing a signal peptide, SRCR (scavenger receptor, cysteine-rich) domains, SIDs (SRCR-interspersed domains), CUB (C1r/C1s Uegf Bmp1) domains, and a ZP (zona pellucida) domain (supplemental Fig. 1). SIDs are short (20-30 residues long) stretches that are rich in proline and threonine residues (5). Hensin is expressed in most epithelia in various alternately spliced forms. Its deletion in mice leads to embryonic lethality at the time of generation of the first columnar epithelium, the visceral endoderm (6).
We discovered that although hensin is secreted in monomeric form in the intercalated cell line when seeded at low density, it is found in many higher order oligomeric forms in the conditioned medium of the same cell line when seeded at superconfluent density (7). Moreover, the high density cells deposited a polymeric detergent-insoluble form of hensin in the ECM. Only this ECM form of hensin was found to be functionally active in the development of fully differentiated phenotype of this cell line. Galectin-3, a lectin that binds to β-galactoside, interacts with ECM hensin through protein-protein interactions and was implicated as playing a key role in the functional assembly of ECM hensin (8). However, the soluble monomeric and other oligomeric forms of hensin found in the conditioned medium were not associated with galectin-3, suggesting that other mechanisms are involved in the oligomerization and proper folding of hensin.
In many proteins containing proline, both de novo protein folding and the refolding processes following cellular membrane traffic necessitate isomerization of the preceding peptide bond to the cis form with side chains adjacent to each other instead of the sterically favored trans form with side chains opposite each other. Spontaneous isomerization of peptidylprolyl bonds is a slow process, which not only constitutes a rate-limiting step in protein folding but also occurs during the assembly of multidomain proteins (9). In addition to FKBP and parvulins, cyclophilins constitute a family of the enzyme class of peptidylprolyl cis-trans isomerases (PPIases) that accelerate peptidylprolyl cis-trans isomerization (10). A possible role of peptidylprolyl cis-trans isomerase activity in the extracellular matrix assembly of hensin came to light when we were investigating the role of cyclosporin A (CsA), a widely used immunosuppressant, in causing distal renal tubular acidosis (11, 12). Although the immunosuppressive property of CsA is attributed to its interaction with its cytosolic receptor cyclophilin A (CypA) to inhibit calcineurin, a serine-threonine phosphatase required for cytokine induction in response to stimulation of T cells, CsA also inhibits the PPIase activity of cyclophilins. Using microperfused rabbit kidney collecting tubules, we observed that treatment with CsA but not FK506 (a FKBP PPIase activity inhibitor that inhibits calcineurin in complex with FKBP) caused a form of distal renal tubular acidosis in these tubules concomitant with a decrease in the deposition of hensin in the ECM of these tubules, suggesting the role of PPIase activity in this process (11). In this study, using cell lines derived from rabbit kidney collecting ducts, we show that CsA regulates the extracellular matrix assembly of hensin and the differentiation features of kidney intercalated cells by inhibiting PPIase activity. In addition, our study demonstrates that CypA is the candidate peptidylprolyl cis-trans isomerase responsible for this activity in these cell lines. Our results, establish for the first time a direct role of cyclophilin-mediated PPIase activity in epithelial differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture—Stock cultures of clone C of β-intercalated cells established from rabbit kidney cortex were maintained at 32 °C (13, 14). The cells were trypsinized and seeded on polycarbonate filters (pore size, 0.4 μm; Costar Corp.) at a density of 2 × 104 cells/cm2 (low density) or 5 × 105 cells/cm2 (high density) and transferred to 40 °C to inactivate the T antigen.
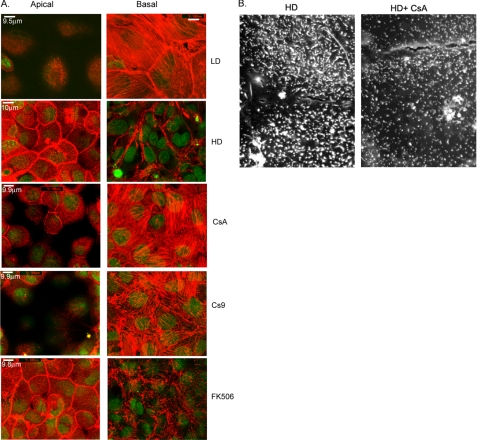
Immunocytochemistry—Clone C cells were plated at high density and cultured in the presence or absence of the reagent CsA, Cs9, FK506, or 6-(3,4-dichlorophenyl)-4-(N,N-dimethylaminoethylthio)-2-phenylpyriidine (LiE) for 2-5 days at 40 °C on Transwell filters. For F-actin staining, monolayers were fixed in 4% paraformaldehyde and stained with rhodaminephalloidin (R-415; Molecular Probes, Inc.). Stained monolayers were viewed using an LSM 510 Meta laser-scanning microscope equipped with Axiovert 200M (Carl Zeiss). Images were collected in 0.6-1-μm-thick optical sections and analyzed using a Zeiss LSM5 image browser and LSM-PC (LSM 410) software. The final images were processed with Adobe Photoshop® 6.0 software (Adobe Systems Inc.). In Fig. 1A, which shows apical versus basal actin staining, the top three apical sections and bottom three basal sections (0.6 μm each) were projected together.
FIGURE 1.
A, projection of three 0.5-μm confocal sections from the apical and basal surfaces of clone C monolayers cultured for 4-5 days under the conditions indicated in the labels (LD, low density; HD, high density; CsA, HD + CsA, Cs9, HD + Cs9; FK506, HD + FK506) and stained with rhodamine-phalloidin (F-actin, red) and SYTOX nuclear stain (green). Carl Zeiss LSM software (version 3.92) was used to obtain size markers, and all size markers are ∼10 μm. B, typical scanning electron microscopic images of clone C monolayers cultured at high density in the absence or presence of 10 μm CsA.
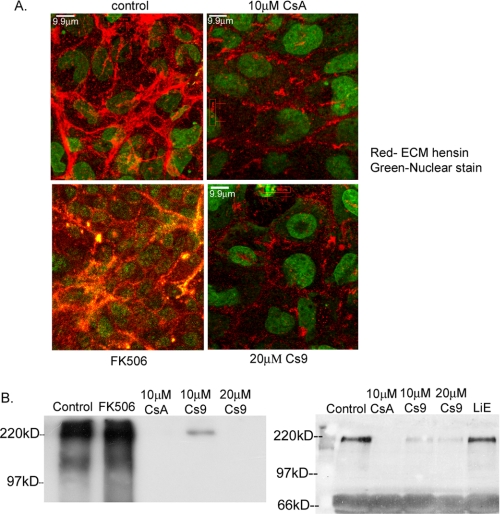
Guinea pig anti-hensin antibodies were obtained by immunizing fusion protein containing SRCR domains 5 and 6 of hensin as described previously (15). For visualization of extracellular matrix hensin staining, cells were incubated with guinea pig anti-hensin sera (1:50) followed by rhodamine-conjugated antiguinea pig antibody (Jackson ImmunoResearch, West Grove, PA), washed, and then fixed with cold methanol. Nuclear staining with SYTOX green dye (Molecular Probes) was performed after fixation, and the filters were then viewed using confocal microscopy as described above. A projection of three to four basal 0.6-μm sections is depicted in Fig. 2A.
FIGURE 2.
A, projection of basal section of monolayers cultured at high density for 4-5 days in the presence of various reagents and stained with anti-hensin antibodies (red) and nuclear stain (green). Monolayers were exposed to anti-hensin antibodies before fixation to access only the extracellular matrix hensin. B, anti-hensin immunoblots of the ECM fractions extracted from clone C cells cultured at high density in the presence of various reagents. Two blots show results from different experiments with different negative controls (FK506, left panel, and LiE, right panel).
Scanning Electron Microscopy—Filters were washed in phosphate-buffered saline and fixed with 2.5% glutaraldehyde in 100 mm phosphate buffer. The filters were postfixed for 1 h in 1% osmium tetroxide, 1.5% potassium ferricyanide and dehydrated through a graded ethanol series (50, 70, 85, 95, 100, 100, and 100%) for 10 min at each concentration. Afterward, cells underwent critical point drying using CO2 in an Omar SPC-1500 critical point dryer. The filters were cut from their holders and mounted on copper specimen supports, sputter-coated with ∼11 nm of gold-palladium in a sputtering unit (Pelco 91000; Ted Pella Inc.), and viewed at 20 kV in an electron microscope (JEOL 100 CX-II; JEOL USA, Inc.) equipped with an analytical scanning imaging device scanning unit. Images were recorded on Polaroid Type 55 positive/negative film.
Analysis of Shape and Height of Clone C Cells—All analyses of cell size and height were performed with a Microsoft Windows®-based version of Zeiss 410 laser-scanning microscope software (Carl Zeiss GmbH). For size measurements, individual cell boundaries of the merged phalloidin-SYTOX image from a field of view from each specific sample (i.e. HD + CsA, etc.) were delineated using a mouse-driven marker (via the Mark Area subfunction of the Area Measure function of this software), and the area obtained in μm2 was recorded. For measuring the height of the cells, the X/Z section of a sequence of images was used. 10-15 individual measurements on three different fields of view were recorded for each sample; the experiments were repeated five times.
Conditioned Media Preparation and Density Gradient Analysis—Clone C cells were seeded at high density and cultured in the presence or absence of 10 μm CsA, 20 μm Cs9, or 500 nm FK506 for 2 days at 40 °C and then washed with phosphate-buffered saline and cultured in serum-free culture medium for another 6 h. Conditioned medium from both the apical and basal compartments of the Transwell® filters from each sample was collected after 6 h and concentrated with Centricon® centrifugal filter units (Millipore, Billerica, MA), and protein concentration was determined using a BCA assay (Pierce). Concentrated conditioned media were loaded on 12 ml of 5-30% sucrose gradients (7) and ultracentrifuged at 100,000 × g for 16 h at 4 °C. Proteins in each fraction (1 ml) were precipitated by 10% trichloroacetic acid, dissolved in a sample buffer, and subjected to 10% SDS-PAGE followed by Western blotting with anti-hensin antibody.
Preparation of Extracellular Matrix—Clone C cells seeded at high density and cultured in the presence or absence of 10 μm CsA, 20 μm Cs9, or 500 nm FK506 for 2 days at 40 °C were extracted with lysis buffer (1% Triton X-100 and 1 mm calcium chloride) for 1 h at 4 °C in a rotary shaker. Filters were scraped in this solution with a cell scraper to remove cell extracts and loosely attached materials. The filters were then washed thoroughly with the same solution for another hour at 4 °C. Insoluble material remaining on these filters was extracted with ECM extraction buffer (4 m guanidine hydrochloride, 50 mm sodium acetate, pH 6.5, 5 mm EDTA, and 0.5% CHAPS) at 4 °C overnight (7, 16). ECM extracts were then dialyzed against 50 mm Tris-HCl, pH 8.0, at 4 °C overnight.
35S Labeling, Pulse-Chase, and Immunoprecipitation—Clone C cells were seeded at high density and cultured in the presence or absence of CsA, Cs9 and FK506 overnight at 40 °C. After cells were cultured in methionine- and cysteine-free minimum Eagle's medium (with or without inhibitors) for 3 h, they were pulse-labeled with 100 μCi/ml 35S (Expre35S35S protein labeling mix, PerkinElmer Life Sciences) in culture medium for 2 h. Pulse labeling was followed by chase with regular medium supplemented with 10 mm l-methionine. All Transwell® filters were incubated with exactly 3 ml of medium, and all 3 ml of medium were collected and mixed with 300 μl of cell lysis buffer (see below) at the indicated chase periods for immunoprecipitation. Cells were lysed in 1% SDS, 1 mm EDTA, 1% Triton X-100, and 10 mm Tris-HCl, pH 8.0, and boiled for 3 min. Insoluble materials were removed by a brief centrifugation (14,000 × g for 5 min at room temperature), and the total cell lysate from each sample was diluted 10-fold with 10 mm Tris-HCl, pH 8.0, and used for immunoprecipitation.
Conditioned medium and cell lysate samples were incubated with guinea pig anti-hensin serum (1:500 dilution) at 4 °C for 1 h followed by the addition of protein A-Sepharose CL-4B (Amersham Biosciences) at 4 °C for another 1 h. Beads were washed three times with a buffer containing 0.1% SDS, 0.1 mm EDTA, 0.1% Triton X-100, and 10 mm Tris-HCl, pH 8.0, boiled in sample buffer for 3 min, and then subjected to SDS-PAGE. Gels were fixed with 10% acetic acid, 10% methanol, soaked in Amplify™ solution (Amersham Biosciences), dried, and exposed to x-ray film at -80 °C for 2 days. The autoradiographs were scanned using an HP Scanjet 8300, and densitometry was performed using ImageJ (National Institutes of Health) software. The values were normalized to the highest densitometric value for high density control, set to 100. Statistical analysis of these normalized densitometric results was performed using GraphPad Instat3 (La Jolla, CA) software.
Cell Number Analysis—High density cells were cultured in the presence and absence of CsA, Cs9, and FK506 for 2 days and stained with SYTOX green nuclear stain. The stained cells were observed visually either in a fluorescent microscope or in a confocal microscope, and cells from each Transwell filter that occupied a 10,000 square μ area of eight different fields of view were counted. This procedure was repeated in six different filters obtained from at least three independent experiments.
His-tagged Recombinant CypA Cloning, Expression, and Purification in pET System—Total RNA was extracted from rabbit kidney cortex with an RNeasy Mini Kit (Qiagen, Inc., Valencia, CA). The cloning primers were designed based on the GenBank™ sequence data base (GenBank™ accession number AF139893). 5′-GGAATTCCATATGGTCAACCCCA-3′ and 5′-CCGCTCGAGTTGACACCTGTTGAG-3′ were used as the sense and antisense CypA primers, respectively. The full-length rabbit CypA cDNA was generated with the SuperScript™ III One-step RT-PCR system with Platinum Taq High Fidelity (Invitrogen) and cloned into Escherichia coli expression vector pET26b (Novagen, Madison, WI) by using NdeI and XhoI cloning sites. After the construct was confirmed by DNA sequencing, it was transformed into E. coli Rosetta 2(DE3) strain (Qiagen) and induced with 1 mm isopropyl-1-thio-d-galactopyranoside for 4 to 6 h. Protein was extracted from these cultures using standard methods and purified using Ni-NTA-agarose. Ni-NTA eluate was characterized with 15% SDS-polyacrylamide gels, and CypA identity was confirmed by immunoblotting with anti-CypA and anti-His tag antibody.
Hensin-recombinant CypA Binding Assay—Purified recombinant C-terminal His-tagged CypA (100 μg) was dialyzed against Tris-buffered saline to remove imidazole and immobilized on equilibrated Ni-NTA beads with a gentle rocking motion on a rotating platform for 1 h at 4 °C. After extensive washes with Tris-buffered saline, the Ni-NTA beads were incubated with or without 10 μm CsA for 1 h. Conditioned media from high density cultures (400 μg of total protein) were then added to these beads and incubated with gentle rocking on a platform at 4 °C for 1 h. After the beads were washed with Tris-buffered saline to remove nonspecific binding partners, CypA-binding proteins were eluted with elution buffer (50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole buffer, pH 8.0). The samples were examined by electrophoresis on 10% SDS-polyacrylamide gels followed by immunoblot with anti-hensin antibody.
Real-time PCR Assay—Total RNA was extracted from high density clone C cells cultured for 2 days in the presence or absence of CsA (10 μm), Cs9 (20 μm), or FK506 (500 nm) using the RNeasy® Mini Kit (Qiagen). After the RNA integrity was verified, first-strand cDNA was synthesized from 500 ng of total RNA utilizing a SuperScript® III first-strand cDNA synthesis kit (Invitrogen). Forward/reverse primers and fluorogenic probes (TaqMan®, fluorogenic 5′-nuclease chemistry) shown in Table 1 were designed using PrimerExpress™ software (Applied Biosystems, Foster City, CA) and were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). The relative abundance of β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Cyp isoforms, and hensin mRNA was determined by quantitative real-time PCR using the respective primer/probe sets and the ABI Prism 7000 Sequence Detection System and software. Plasmids containing a gene-specific sequence for rabbit β-actin, GAPDH, Cyp isoforms, or hensin were used to generate standard curves from which the relative mRNA copy number was calculated.
TABLE 1.
TaqMan real-time PCR primers and probes developed in this study
| Sequence 5′ to 3′ |
|---|
| β-Actin |
| Forward primer GACCGACTACCTCATGAAGATCCT |
| Reverse primer TGATGTCCCGCACGATCTC |
| Probe TACAGCTTCACCACCACGGCCG |
| GAPDH |
| Forward primer TGCACCACCACCAACTGCTTAG |
| Reverse primer GGTCTTCTGGGTGGCAGTGTGA |
| Probe TCATCCACGACCACTTCGGCATTGT |
| Hensin |
| Forward primer AACCACTGTAGGGCCCTCTTCAAA |
| Reverse primer AGGTAACCAGAAGGCACGGCTATT |
| Probe CCCAACAATGCCCTCTGTGTCTGGGA |
| CypA |
| Forward primer CACCTGACCATTCCTCTAGCTCA |
| Reverse primer ACAATTCAGGGCGTCACGA |
| Probe AGCGCCCTCCGCCCCCATCT |
| CypB |
| Forward primer CAGTTCTTCATCACCACAGTCAAGA |
| Reverse primer CCTCCAGAACTTTGCCAAACA |
| Probe CTGGCTGGACGGCAAGCACGT |
| CypC |
| Forward primer TTGATGTGAGGATCGGAGACAA |
| Reverse primer CTTGGGCACGACTTTTCCAA |
| Probe TGTCGGCAGAATTGTGATTGCCTT |
Electroblot Analysis of Cellulose-bound Hensin Peptides—The cellulose-bound peptide scan of the entire hensin sequence consisted of 12-mer peptides overlapping by 9 amino acids and anchored via a C-terminal (β-Ala)2 spacer on the membrane. The cellulose-bound peptide scan was prepared by standard automated spot synthesis (17, 18) using a pipetting robot (Abimed GmbH, Langenfeld, Germany). The standard SPOT synthesis protocol included coupling of Fmoc amino acids, preactivated as pentafluorophenyl esters. Fmoc protective groups were cleaved with 20% piperidine in dimethylformamide. The side chains of the amino acids were protected as follows: Pbf (arginine), Trt (asparagine, glutamine, histidine, cysteine), tBu (threonine, tyrosine serine), OtBu (aspartic and glutamic acids), Boc (lysine and tryptophan). After coupling, all unreacted amino groups were blocked with acetic anhydride. For analysis of CypA binding, 2 μm recombinant CypA in 20 mm Tris buffer, pH 8.2, was incubated with the membrane for 3 h. Bound protein was transferred to nitrocellulose membrane via electroblotting with a semidry blotter using a constant current of 0.8 mm/cm2. Protein transferred to the nitrocellulose was visualized by polyclonal rabbit antiserum raised against CypA using enhanced chemiluminescence detection (Amersham Biosciences). For studies of specific CsA effects, 30 μm CsA was added to 2 μm CypA in 20 mm Tris buffer, pH 8.2, and preincubated for 30 min prior to incubation of the blot.
Statistics—One-way ANOVA statistics (Instat3 GraphPad, La Jolla, CA) were applied to multiple group comparisons (e.g. comparing high density (HD) against HD + CsA, HD + Cs9, HD + FK506) with respect to height, area, and cell numbers. The Tukey-Kramer multiple comparisons test was used if the F value of the ANOVA was significant. Significance was asserted if the p value was less than 0.05. For nonparametric data, the Kruskal-Wallis test was used to compare multiple groups.
RESULTS
Inhibitors of PPIase Activity Prevent the Development of Differentiated Phenotype in Clone C Cells—We showed previously that an immortalized clonal cell line (clone C) derived from β-intercalated cells of the rabbit kidney exhibits distinctly different phenotypes when seeded at subconfluent density and superconfluent density (7). Low density cells (Fig. 1A, top row) exhibited basal actin stress fibers and no subapical actin cytoskeleton, whereas the high density (second row) cells had a subapical actin cytoskeleton and very few basal stress fibers. Scanning electron microscopy showed that the low density cells had sparse microvilli, whereas high density cells had exuberant apical surface infolding and microvilli. The cell shape of these two phenotypes was also different with high density cells being tall with a small cross-sectional area, whereas low density cells were low and flat. This columnarization and the remodeling of the apical cytoplasm was hensin-dependent; it could be induced by seeding low density cells on filters conditioned by high density cells and prevented by an antibody to hensin (7). However, only the polymerized ECM hensin induced the development of terminal differentiation-like features.
We tested whether CsA inhibited the development of ECM hensin-mediated actin cytoskeleton remodeling by culturing cells seeded at high density in the presence of 2, 5, and 10 μm CsA. We assayed their effect on actin cytoskeleton remodeling by performing rhodamine phalloidin (F-actin) staining on the monolayers after 4 days in culture and analyzing the results by scanning serial images of the monolayers from top to bottom using confocal microscopy. The cells cultured in the presence of 10 μm CsA almost resembled the low density phenotypes, with exuberant basal stress fibers but no apical actin cytoskeleton (Fig. 1A, third row), whereas the control high density cells had pronounced an apical actin cytoskeleton and very few basal stress fibers. High density cells treated with 2 and 5 μm CsA also had similar but weaker effects (data not shown).
As CsA inhibits both PPIase activity and the calcineurin pathway, we tested whether or not the observed inhibition in proper phenotype development of high density cells were due to the inhibition of PPIase activity by performing the following experiments. High density cells were cultured in the presence of 10 and 20 μm Cs9 (derivative 9 of CsA with modifications in the d-Ser side chain (O-[NH2(CH2)5NHC(O)CH2-]-d-Ser)2-CsA), a reagent that has been demonstrated to have a specific effect to inhibit only the PPIase activity of cyclophilins) (19). High density cells cultured in the presence of 20 μm Cs9 had a dramatic reduction in their apical actin cytoskeleton and a pronounced increase in basal stress fibers (Fig. 1A, fourth row). Concentrations of Cs9 higher than 20 μm were toxic to the cells, and 10 μm Cs9 produced distinct but weaker effects on actin cytoskeleton remodeling (data not shown). High density cells were also cultured in the presence of 500 nm, 1 μm, and 10 μm FK506, a reagent that inhibits the calcineurin activity of cyclophilins. We found that although concentrations higher than 1 μm FK506 were toxic to the cells and only about 10% of the cells were left on Transwell filters after day 4 (data not shown), 500 nm FK506 did not have a significant effect on cell growth (Table 2). At the same time, 500 nm FK506 also did not have any significant effect on the actin cytoskeleton of high density cells (Fig. 1A, bottom row). Scanning electron microscopy revealed that high density cells cultured in the presence of CsA for 4 days also had stunted and sparse microvilli compared with the numerous exuberant microvilli on the apical surface of control high density cells cultured in the absence of CsA (Fig. 1B).
TABLE 2.
PPlase inhibitors do not affect cell numbers
Clone C cells were cultured at high density for 2 days with the various treatments. Data are given as mean ± S.E. NS, not significant.
| Cell treatment | n | Cell number/104 μm2 | p value |
|---|---|---|---|
| High density | 6 | 45.99 ± 1.17 | |
| HD + 10 μm CsA | 6 | 45.54 ± 1.60 | NS |
| HD + 20 μm Cs9 | 6 | 45.53 ± 1.29 | NS |
| HD + 500 nm FK506 | 6 | 45.47 ± 0.82 | NS |
In addition to inhibiting the development of apical actin cytoskeleton in high density cells, PPIase inhibitors CsA and Cs9 also inhibited the columnarization of the high density cells. High density cells cultured in the presence of 10 μm CsA or 20 μm Cs9 were significantly shorter (5.7 ± 0.01 and 5.9 ± 0.11 μm, respectively, compared with the 9.2 ± 0.01 μm of high density cells), whereas cells cultured in the presence of the calcineurin inhibitor FK506 had no effect on cell height (Table 3). Additionally, the surface areas of high density cells cultured in the presence of PPIase inhibitors CsA and Cs9 were significantly larger (721 ± 4 and 657 ± 5 μm2) compared with the control high density cells (264 ± 3 μm2) or high density cells cultured with the calcineurin inhibitor FK506 (279 ± 3 μm2; see Table 3 for details).
TABLE 3.
PPlase inhibitors affect cell height and area
Clone C cells were cultured at high density for 2 days with the various treatments. Data are given as mean ± S.E. NS, not significant.
| Cell treatment | n | Height | p value | Area | p value |
|---|---|---|---|---|---|
| μm | μm2 | ||||
| High density | 5 | 9.18 ± 0.01 | 264.4 ± 2.9 | ||
| HD + 10 μm CsA | 5 | 5.74 ± 0.01 | p < 0.001 | 721.3 ± 4.2 | p < 0.001 |
| HD + 20 μm Cs9 | 5 | 5.92 ± 0.11 | p < 0.001 | 657.2 ± 4.8 | p < 0.001 |
| HD + 500 nm FK506 | 5 | 9.14 ± 0.01 | NS | 279.5 ± 2.7 | NS |
To test whether the observed inhibition in differentiation features in cells cultured in the presence of PPIase inhibitors was not because of changes in cell proliferation or cell death, we performed a statistical analysis of cell numbers of high density cells cultured for 2 days in the presence and absence of the PPIase inhibitors CsA (10 μm) and Cs9 (20 μm). The results shown in Table 2 indicate that there was no significant difference in cell numbers between the high density cells treated with or without the PPIase inhibitors, suggesting that the PPIase inhibitors inhibit only the appearance of differentiation features. We performed these studies on 2-day cell cultures instead of the 4- or 5-day cell culture used in differentiation experiments, because our previous results showed that ECM deposition of hensin occurs maximally around 2 days after seeding; hence, changes in cell size and shape that may affect cell numbers in cells cultured beyond the 2-day period may be attributed to the effect of ECM hensin. In summary, our results in this section establish that the PPIase inhibitors CsA and Cs9 inhibit the development of a fully differentiated phenotype in clone C cells.
Hensin Polymerization in the ECM Is Inhibited by PPIase Inhibitors—We then investigated whether the PPIase inhibitors inhibit differentiation primarily by inhibiting ECM polymerization of hensin by performing the following experiment. High density cells were cultured in the presence and absence of 10 μm CsA, 20 μm Cs9, and 500 nm FK506 for 2 days, and monolayers were immunostained for extracellular hensin followed by nuclear labeling. These experiments were performed three times; a typical projection of the basal sections corresponding to a 4-μm thickness is depicted in Fig. 2A. The results show a significant reduction in ECM hensin staining in cells cultured in the presence of CsA and Cs9 (Fig. 2A, right panels) but not in the presence of the calcineurin inhibitor FK506.
To confirm these results, we also compared the amount of hensin deposited in the ECM in high density cells cultured in the presence and absence of 10 μm CsA, 10 μm Cs9, 20 μm Cs9, and 500 nm FK506 by immunoblotting methods. ECM was extracted from 2-day cell cultures, and 80 μg of the ECM protein was immunoblotted with anti-hensin antibodies. The results depicted in Fig. 2B, left panel, show that whereas FK506 did not have any effect on the amount of ECM hensin, 20 μm Cs9 and 10 μm CsA completely inhibited ECM hensin deposition, and 10 μm Cs9 substantially decreased ECM hensin content. In another experiment, 40 μg of the ECM extract from high density cells cultured in the absence and presence of 10 μm CsA, 10 and 20 μm Cs9, and 10 μm LiE, a monocyclic heterocyclic compound that completely inhibits calcineurin activity at the concentration used,4 were immunoblotted with anti-hensin antibodies. The results shown in Fig. 2B, right panel, once again demonstrate that the specific calcineurin inhibitor LiE had no effect on ECM deposition of hensin, whereas the specific PPIase inhibitors Cs9 and CsA had inhibitory effects. Nonspecific protein bands at 66 kDa served as an internal protein loading normalization control. These results established that CsA and Cs9 indeed inhibit the polymerization of ECM hensin by inhibition of the cyclophilin PPIase activity.
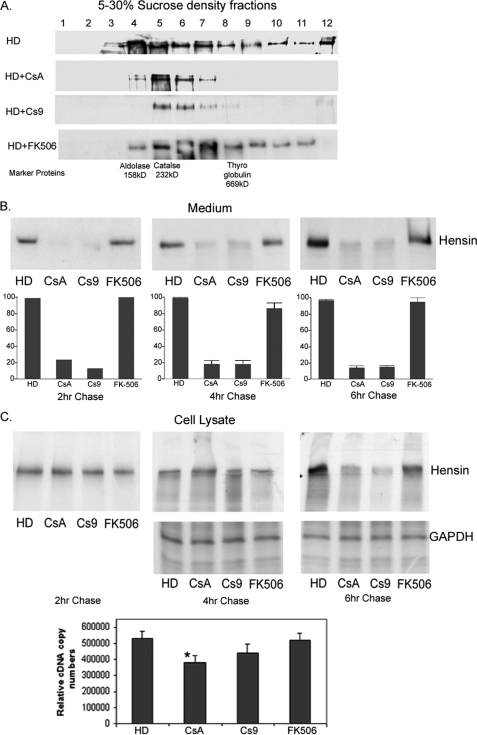
PPIase Inhibitors Block Hensin Secretion and Oligomerization in Clone C Cells but Do Not Significantly Alter Hensin Synthesis—We had previously demonstrated that the conditioned medium of high density cells exists in monomeric, dimeric, and other higher order oligomeric forms (7). To directly test whether PPIase inhibitors inhibit the oligomerization of hensin, serum-free conditioned media (normalized for 320 μg of total protein concentration) were fractionated using 5-30% sucrose gradients. Total protein from each fraction was precipitated with trichloroacetic acid and immunoblotted with anti-hensin antibodies. A mixture of marker proteins aldolase (158 kDa), catalase (232 kDa), and thyroglobulin (669 kDa) was also fractionated simultaneously to compare the molecular masses of various sucrose density fractions. Fig. 3A shows that conditioned media from HD cells and FK506-treated cells contained nearly one-third of the hensin bands in fractions 8 and above, indicating a significant amount of polymeric hensin. In contrast, in conditioned media of CsA- and Cs9-exposed cell cultures, the complete absence of hensin bands in fractions 8 and above demonstrated the absence of oligomers higher than trimers. Intriguingly, we also observed that the total amount of immunodetectable hensin was reduced in media from CsA- and Cs9-treated cell cultures.
FIGURE 3.
A, anti-hensin immunoblots of various sucrose density fractions of conditioned medium from clone C cells cultured at high density in the absence and presence of PPIase inhibitors CsA and Cs9. A control is cultured in the presence of FK506 (calcineurin inhibitor). The bottom row indicates where the high molecular mass marker proteins localize in the gradient fractions. B, autoradiographs of conditioned medium collected at various time points of chase immunoprecipitated with anti-hensin antibodies. High density cells were cultured in the presence or absence of CsA, Cs9, and FK506 overnight, pulse-labeled with 35S for 2 h, and chased at the indicated times. The bottom panel is a graphical representation of the statistical analysis of densitometric results obtained from three independent experiments (for 4- and 6-h chase). C, autoradiographs of total cell lysates obtained from 35S pulse-labeled cell cultures described above at various time points of chase immunoprecipitated with anti-hensin antibodies (top panel) and with anti-GAPDH antibody (synthesis control, middle panel). The bottom panel shows the relative hensin cDNA copy numbers obtained from quantitative RT-PCR analysis of RNA extracted from clone C cells cultured in the absence or presence of various PPIase inhibitor reagents as indicated in the labels (on left).
To test whether there was a decreased synthesis or secretion of hensin in the cells cultured in the presence of PPIase inhibitors, we carried out metabolic labeling studies. High density clone C cells cultured in the presence or absence of CsA, Cs9, and FK506 overnight were labeled metabolically with [35S]methionine and chased in the cell lysate and conditioned medium fractions for 4-6 h. Our results showed that the medium from CsA- and Cs9-treated cell cultures had significantly reduced intensity in the autoradiographs at both the 4- and 6-h time points compared with the high density control or FK506-treated cells (Fig. 3B, top panel). We repeated these experiments a total of three times, analyzed the autoradiographs by densitometry and found that hensin secretion was reduced by a significant 80-85% by treatment with CsA and Cs9, but not significantly altered by FK506 treatment (Fig. 3B, bottom panel). Additionally we also performed the chase at two hours and observed a similar result.
To test whether the reduction in secreted hensin observed in the CsA- and Cs9-treated cultures was caused by the effect of CsA and Cs9 on hensin synthesis, we analyzed equal amounts of the cell lysates of [35S]methionine-labeled cell cultures described above using immunoprecipitation with hensin followed by autoradiogaphy. Our results showed that the intensity of hensin bands in the CsA- and Cs9-treated cell lysates was similar to that in high density control cell lysates at 2 and 4 h of chase. However, there was a visible reduction in the intensity of hensin bands in CsA- and Cs9 treated cell lysates compared with the control cell lysates at the 6-h time point (Fig. 3C, top panel). A Kruskal-Wallis (nonparametric ANOVA) test of the densitometry results from three independent experiments confirmed this observation (data not shown). Cell lysates (obtained from 4- and 6-h chases) immunoprecipitated with GAPDH antibody were used as controls and showed that GAPDH synthesis was unaffected by CsA and Cs9 treatment in 6-h cell lysates (Fig. 3C, middle panel). This result indicates that although hensin is synthesized in equal amounts in PPIase inhibitor-treated cells compared with the control cells, it is degraded in PPIase inhibitor-treated cells by 6 h, suggesting that the PPIase inhibitors affect the stability of newly synthesized hensin. It is likely that inhibition of PPIase activity leads to improper folding of hensin, which is then targeted to the degradation process rather than to the normal trafficking pathway.
We also performed quantitative RT-PCR with hensin primers on high density cells cultured for 2 days in the absence or presence of CsA (10 μm), Cs9 (20 μm), and calcineurin pathway inhibitor FK506 (500 nm). The results depicted in Fig. 3C (bottom panel) indicate that the relative copy numbers of HD cells (530,000 ± 44,700) and HD cells cultured in the presence of FK506 (520,000 ± 44,700) were not significantly different from the number of cells cultured in the presence of the specific PPIase inhibitor Cs9 (440,000 ± 54,800) according to the nonparametric ANOVA test. Although the relative copy number of CsA-treated cells (380,000 ± 44,700) was smaller than the high density cell copy number by a statistically significant amount, our repeated metabolic labeling experiments described above did not show any significant decrease in hensin synthesis at the protein level following CsA treatment.
These results demonstrate that PPIase inhibitors specifically inhibit the secretion and higher order oligomerization of hensin. These studies also demonstrate that the stability of newly synthesized hensin is reduced by PPIase inhibitors. Taken together, these results suggest that PPIase inhibitors interfere with the proper folding of newly synthesized hensin.
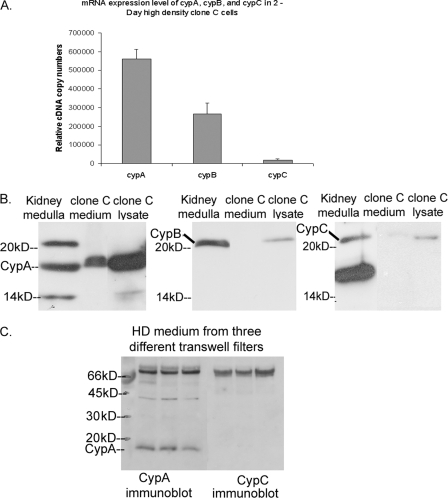
Characterization of Cyclophilin Isoforms in Clone C Cells—To determine the specific PPIase involved in regulating hensin oligomerization, we first characterized mRNA expression, protein expression, and secretion of cyclophilins A, B, and C in clone C cells. CypA is the main target of CsA, cyclophilin B (CypB) is a secreted cyclophilin with a signal sequence, and cyclophilin C (CypC) is abundantly expressed in the kidney (20, 21). Quantitative real-time PCR revealed that the relative copy number of CypA (560,000 ± 52,900) was 30-fold higher than that of CypC (18,300 ± 7,600) and more than twice the copy number of CypB (267,000 ± 57,700) (Fig. 4A). Western blotting of equal amounts of total protein from high density clone C cell lysates also revealed higher levels of CypA protein (18-kDa band) compared with CypB and CypC (Fig. 4B, third lane in all three panels). Western blotting of equal amounts (36 μg) of total protein isolated from rabbit kidney medulla, used as a positive control in this experiments, also established the presence each of these cyclophilins in rabbit kidney (Fig. 4B, first lane in all panels).
FIGURE 4.
A, relative cDNA copy numbers of CypA, CypB, and CypC obtained from the quantitative RT-PCR of RNA extracted from clone C cells as described under “Experimental Procedures.” B, immunoblot of protein lysates from rabbit kidney medulla and equal amounts of protein (36 μg) from clone C cell lysate and conditioned medium of high density cells with anti-CypA (left), anti-Cyp B (middle), and anti-Cyp C (right) antibodies. C, immunoblot of equal amounts of protein (41, 38, and 39 μg) from the conditioned medium of three independent Transwell filters probed with anti-CypA and anti-CypC antibodies.
To compare the steady-state secretion levels of cyclophilin isoforms by clone C cells, we purified equal volumes of conditioned medium from serum-free cell cultures using a size exclusion column and analyzed fractions 4 and 5 of the eluate (which contained the cyclophilin isoforms) by immunoblotting with CypA, CypB, and CypC antibodies. The results of these studies, depicted in the middle lanes of Fig. 4B, demonstrate the presence of significant amounts of CypA and the absence of CypB in the conditioned medium. There was a very weak band of CypC present in the conditioned medium. Western blot of conditioned medium corresponding to a total protein content of 600-800 μg confirmed that CypC was indeed present in the conditioned medium but in considerably lesser amounts than the CypA (data not shown). These findings suggest that CypA, being the abundant form of cyclophilin to be synthesized and secreted by the clone C cells, is the most likely candidate to mediate the hensin folding PPIase activity of clone C cells.
To confirm that CypA was the abundant secreted form of cyclophilin in clone C cells, equal volumes of conditioned medium from three different Transwell filters were trichloroacetic acid-precipitated, and exactly half of these samples were probed by both CypA and Cyp C antibodies. The results depicted in Fig. 4C unequivocally confirmed that CypA, but not CypC, was abundantly present in the conditioned medium.
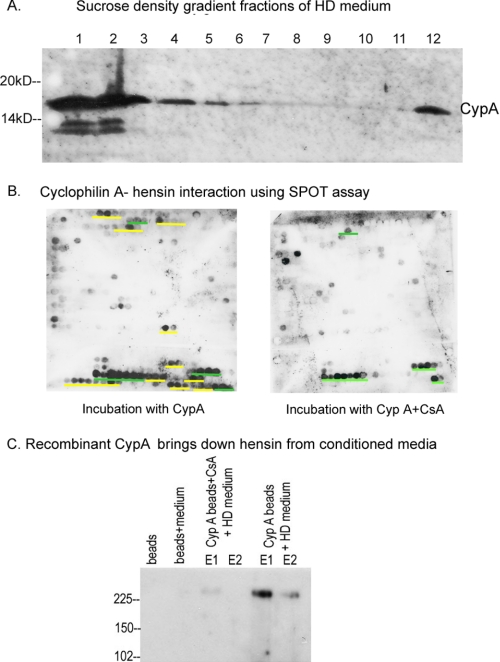
CypA Interacts with Specific Regions of Hensin and Is Present in High Molecular Mass Fractions of the Conditioned Medium—Interestingly, when we investigated the sucrose density fractions of the conditioned medium of clone C cells (obtained as described previously) by immunoblotting with anti-CypA antibodies, we found that CypA was present in higher molecular mass fractions, corresponding to molecular masses many times higher than CypA (Fig. 5A). This result prompted us to investigate further the direct binding of CypA with hensin.
FIGURE 5.
A, anti-CypA immunoblot of sucrose density fractions of conditioned medium from high density clone C cells. B, cellulose-bound hensin 12-mer peptides on a SPOT array were incubated with CypA either in the absence of CsA (left) or in the presence of CsA (right), transferred to nitrocellulose membranes, and immunoblotted with anti-CypA antibody. Yellow and green bars indicate the peptide regions of hensin that bound to CypA. Peptides in which interaction with CypA could not be competed off by preincubation with CsA are underlined in green, and those successfully competed off by preincubation with CsA (underlined in yellow) are thereby possible specific binding sites for the PPIase active site of CypA. See supplemental Fig. 2 for detailed sequence information of these peptides. C, conditioned medium was added to Ni-NTA beads coupled with recombinant CypA in the presence or absence of CsA and to the appropriate control beads. Bound protein was eluted off from the beads using imidazole elution buffer twice (elution 1 (lane E1) and elution 2 (lane E2)) and immunoblotted with anti-hensin antibody.
To characterize the specific interaction of CypA with hensin, we analyzed CypA binding to peptides organized in a peptide array synthesized on solid cellulose support (the SPOT method). A library of 12-mer hensin peptides covering the complete hensin sequence linked covalently to cellulose membrane with overlapping sequences of 9 amino acids from spot to spot was synthesized as described under “Experimental Procedures.” This peptide array cellulose membrane was incubated with 2 μm recombinant CypA, and the bound peptide regions were visualized using immunoblotting with anti-CypA antibody. The results of this study showed that CypA bound to 14 specific regions in the hensin primary structure, underlined in yellow and green (Fig. 5B, left blot). To analyze which peptides are targeted by the PPIase active site of CypA, we performed a competition binding experiment on the SPOT array after preincubating CypA with 30 μm CsA prior to the binding assay. This experiment showed that CsA abolished the binding of CypA to 10 of the CypA-interacting regions in hensin (underlined with yellow bars in Fig. 5B, left panel), indicating that these peptides are likely to interact with the PPIase active site of CypA. Seven of these ten specific CypA-binding hensin peptides included a proline residue (supplemental Fig. 2). The four CsA-insensitive binding peptides are underlined in green in Fig. 5B.
Additional verification of the direct interaction of CypA with hensin was accomplished by a pull down experiment. We expressed recombinant His-tagged CypA, immobilized it on Ni-NTA beads, and incubated these beads with conditioned medium from high density clone C cells. Following the incubation, the beads were washed and eluted twice, and the eluates were probed by immunoblotting with anti-hensin antibody. The results, depicted in Fig. 5C, show that the CypA-bound nickel beads pulled down hensin from the conditioned medium, demonstrating a direct interaction of CypA with hensin. This pulldown of hensin from conditioned medium by CypA beads was inhibited when the beads were preincubated with CsA, once again demonstrating the specific and direct interaction of hensin with CypA via its active site.
DISCUSSION
Our results have demonstrated that a specific PPIase inhibitor, Cs9 (a derivative of CsA with modifications in the d-Ser side chain), and CsA inhibited the development of a fully differentiated phenotype of clone C cells by inhibiting the polymerization of hensin in the ECM. Our results also show that inhibition of cyclophilin-mediated PPIase activity leads to the inhibition of hensin secretion and degradation of newly synthesized intracellular hensin, suggesting a critical rate-limiting role of cis-trans isomerization of a peptidylprolyl bond in the folding of hensin. Additionally, our studies suggest that the PPIase activity of CypA mediates the oligomerization of secreted hensin.
Our previous studies established that Cs9, is a specific inhibitor of PPIase activity (19). CsA exhibited an IC50 value of 3.7 nm for PPIase inhibition and an IC50 value of 100 nm for calcineurin inhibition. On the other hand, Cs9 did not show any inhibitory activity of calcineurin but exhibited an IC50 value of 3.2 nm to inhibit the PPIase activity of CypA. Our current studies demonstrate that the use of CsA and Cs9 did not alter cell proliferation properties or cell death but inhibited the development of fully differentiated features of clone C cells by preventing ECM hensin polymerization. Our new results also demonstrate that one mechanism by which PPIase inhibitors CsA and Cs9 inhibit ECM hensin polymerization is by inhibiting hensin secretion. Our metabolic labeling experiments showed that there was a significantly large decrease in the amount of secreted hensin in PPIase-inhibited samples at 2 and 4 h of chase, although there was no significant difference in the newly synthesized intracellular hensin, clearly establishing that PPIase inhibitors CsA and Cs9 inhibited hensin secretion. Further, the observation that at 6 h of chase the amount of intracellular hensin was substantially decreased in PPIase inhibitor-treated cells compared with the control cells suggests that newly synthesized hensin is degraded in PPIase inhibitor-treated cells. It is likely that in the absence of PPIase activity, newly synthesized hensin is misfolded and hence, instead of being targeted to the normal secretion pathway, is being targeted for degradation. The role of PPIase activity in protein folding and trafficking is well characterized. PPIase activity of Pin1 regulates the folding and processing of amyloid precursor protein (22), and cyclophilin-dependent PPIase activity regulates the intracellular transport of extracellular matrix metalloproteinase inducer protein CD147 (23).
Another mechanism by which PPIase inhibition may affect hensin polymerization is by inhibiting the oligomerization of secreted hensin in the conditioned medium. Our analysis of the hensin oligomers in the conditioned medium of PPIase inhibitor-treated and untreated cells showed that formation of higher order oligomers was completely inhibited in PPIase inhibitor-treated cells. Although there are 16 known isoforms of cyclophilins in humans (24), we explored the possible role of only CypA, CypB, and CypC in the oligomerization of secreted hensin, because these cyclophilins have already been described as acting extracellularly (25, 26), and thus exploring the role of other isoforms located either in the nucleus (CypE) or in the mitochondria (CypD) did not seem necessary in this context. Even though CypA is commonly viewed as a housekeeping gene in many studies, and is considered to be a cytosolic protein, recent studies indicate that CypA can be secreted by cells, and CypA protein exhibits several biological functions including immune modulation (27), response to oxidative stress (28), cell growth, tumorigenesis, and pathogenesis of vascular disease (29).
Our studies on CypA-hensin interaction revealed an interesting result. Hensin is a multidomain protein composed of eight SRCR domains, two CUB domains, and one ZP domain (supplemental Fig. 1). Most SRCR domains are separated by a short proline-rich amino acid stretch, previously termed the SID. Our results show that in the C-terminal domain of hensin, CypA binds to two proline-containing peptides in the ZP domain and one proline-containing peptide in the CUB2 domain. In the N-terminal region, one peptide that interacts with CypA is located in SID1 (a proline-rich intervening sequence before SRCR1), the peptide HFPS is located in SID2, and two peptides are located in the SRCR domains. The ZP domain has already been implicated in the polymerization of ECM proteins (30), and the SIDs, because of their proline-rich nature, would be potential substrates for CypA. Our results thus provide a starting point to investigate the mechanistic details of CypA-mediated peptidylprolyl cis-trans isomerization of hensin using mutagenesis studies.
It is important to note that two results, the specific interaction of CypA with hensin and the presence of CypA in the high molecular mass sucrose density gradient fractions, convincingly identify CypA as the primary PPIase mediating the oligomerization of secreted hensin. However, because CsA and Cs9 are not specific inhibitors of CypA alone (31), the identity of the cyclophilin involved in the folding and processing of newly synthesized hensin that aids in the normal transport and secretion of hensin is yet unknown. Cyclophilin B, the endoplasmic reticulum-resident isoform of cyclophilin, is present in our cell lines, and further studies to understand the specific cyclophilin isoforms involved in the folding and trafficking of hensin are under way in our laboratory.
There is growing evidence that CypA-mediated PPIase activity has a significant role in biological activity, particularly with respect to protein folding processes. CypA promotes the formation and infectivity of virions of the human immunodeficiency virus (HIV), in part by binding to and catalyzing the cis-trans isomerization of the N-terminal domain of the HIV-1 capsid (32). CypA is incorporated into HIV-1 virions and is thought to be essential for replication. Indeed, CsA binding to CypA prevents the interaction of CypA with HIV-1 and the replication of virions (33). CypA is secreted by vascular smooth muscle cells in response to an oxidative stress (28). It is likely that the PPIase activity of CypA is required for ERK1/2 activation in these cells, suggesting an important role for CypA in the pathogenesis of vascular diseases. The role of PPIase activity in the growth of colon cancer cells (34) and in breast epithelial proliferation has also been identified (35), but our finding that PPIase activity regulates epithelial differentiation is novel and significant.
Supplementary Material
Acknowledgments
We thank G. Fischer for discussions and critical reading of the manuscript and J. Stolar for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-20999 (to Q. Al-A.) and DK-50603 (to G. J. S.). This work was also supported by Grant-in-aid 0150138N from the American Heart Association (to G. J. S.) and by Deutsche Forschungsgemeinschaft Grant GK1026 (to C. S.-F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: ECM, extracellular matrix; SRCR, scavenger receptor cysteine-rich; SID, SRCR-interspersed domain; ZP, zona pellucida; PPIase, peptidylprolyl cis-trans isomerase; CsA, cyclosporin A; Cyp, cyclophilin; LiE, 6-(3,4-dichlorophenyl)-4-(N,N-dimethylaminoethylthio)-2-phenylpyriidine; Fmoc, N-(9-fluorenyl)methoxycarbonyl; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; RT, reverse transcription; Ni-NTA, nickel-nitrilotriacetic acid; ANOVA, analysis of variance; HD, high density; FKBP, FK506 (tacrolimus)-binding protein.
G. Fischer and J. Liebscher, unpublished results.
References
- 1.Takito, J., Yan, L., Ma, J., Hikita, C., Vijayakumar, S., Warburton, D., and Al-Awqati, Q. (1999) Am. J. Physiol. 277 F277-F289 [DOI] [PubMed] [Google Scholar]
- 2.Kang, W., and Reid, K. B. (2003) FEBS Lett. 540 21-25 [DOI] [PubMed] [Google Scholar]
- 3.Mollenhauer, J., Herbertz, S., Holmskov, U., Tolnay, M., Krebs, I., Merlo, A., Schroder, H. D., Maier, D., Breitling, F., Wiemann, S., Grone, H. J., and Poustka, A. (2000) Cancer Res. 60 1704-1710 [PubMed] [Google Scholar]
- 4.Schwartz, G. J., Tsuruoka, S., Vijayakumar, S., Petrovic, S., Mian, A., and Al-Awqati, Q. (2002) J. Clin. Investig. 109 89-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mollenhauer, J., Muller, H., Kollender, G., Lyer, S., Diedrichs, L., Helmke, B., Holmskov, U., Ligtenberg, T., Herbertz, S., Krebs, I., Madsen, J., Bikker, F., Schmitt, L., Wiemann, S., Scheurlen, W., Otto, H. F., von Deimling, A., and Poustka, A. (2002) Genes Chromosomes Cancer 35 242-255 [DOI] [PubMed] [Google Scholar]
- 6.Takito, J., and Al-Awqati, Q. (2004) J. Cell Biol. 166 1093-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hikita, C., Takito, J., Vijayakumar, S., and Al-Awqati, Q. (1999) J. Biol. Chem. 274 17671-17676 [DOI] [PubMed] [Google Scholar]
- 8.Hikita, C., Vijayakumar, S., Takito, J., Erdjument-Bromage, H., Tempst, P., and Al-Awqati, Q. (2000) J. Cell Biol. 151 1235-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, G., and Aumuller, T. (2003) Rev. Physiol. Biochem. Pharmacol. 148 105-150 [DOI] [PubMed] [Google Scholar]
- 10.Fanghanel, J., and Fischer, G. (2004) Front. Biosci. 9 3453-3478 [DOI] [PubMed] [Google Scholar]
- 11.Watanabe, S., Tsuruoka, S., Vijayakumar, S., Fischer, G., Zhang, Y., Fujimura, A., Al-Awqati, Q., and Schwartz, G. J. (2005) Am. J. Physiol. 288 F40-F47 [DOI] [PubMed] [Google Scholar]
- 12.Tsuruoka, S., Schwartz, G. J., Wakaumi, M., Nishiki, K., Yamamoto, H., Purkerson, J. M., and Fujimura, A. (2003) J. Pharmacol. Exp. Ther. 305 840-845 [DOI] [PubMed] [Google Scholar]
- 13.van Adelsberg, J., Edwards, J. C., Takito, J., Kiss, B., and Al-Awqati, Q. (1994) Cell 76 1053-1061 [DOI] [PubMed] [Google Scholar]
- 14.van Adelsberg, J., Edwards, J. C., Herzlinger, D., Cannon, C., Rater, M., and Al-Awqati, Q. (1989) Am. J. Physiol. 256 C1004-C1011 [DOI] [PubMed] [Google Scholar]
- 15.Takito, J., Hikita, C., and Al-Awqati, Q. (1996) J. Clin. Investig. 98 2324-2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijayakumar, S., Takito, J., Hikita, C., and Al-Awqati, Q. (1999) J. Cell Biol. 144 1057-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilpert, K., Winkler, D. F., and Hancock, R. E. (2007) Nat. Protoc. 2 1333-1349 [DOI] [PubMed] [Google Scholar]
- 18.Frank, R. (2002) J. Immunol. Methods 267 13-26 [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y., Erdmann, F., Baumgrass, R., Schutkowski, M., and Fischer, G. (2005) J. Biol. Chem. 280 4842-4850 [DOI] [PubMed] [Google Scholar]
- 20.Dornan, J., Taylor, P., and Walkinshaw, M. D. (2003) Curr. Top. Med. Chem. 3 1392-1409 [DOI] [PubMed] [Google Scholar]
- 21.Friedman, J., Weissman, I., Friedman, J., and Alpert, S. (1994) Am. J. Pathol. 144 1247-1256 [PMC free article] [PubMed] [Google Scholar]
- 22.Pastorino, L., Sun, A., Lu, P. J., Zhou, X. Z., Balastik, M., Finn, G., Wulf, G., Lim, J., Li, S. H., Li, X., Xia, W., Nicholson, L. K., and Lu, K. P. (2006) Nature 440 528-534 [DOI] [PubMed] [Google Scholar]
- 23.Yurchenko, V., Zybarth, G., O'Connor, M., Dai, W. W., Franchin, G., Hao, T., Guo, H., Hung, H.-C., Toole, B., Gallay, P., Sherry, B., and Bukrinsky, M. (2002) J. Biol. Chem. 277 22959-22965 [DOI] [PubMed] [Google Scholar]
- 24.Wang, P., and Heitman, J. (2005) Genome Biol. 6 226.1-226.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukrinsky, M. I. (2002) Trends Immunol. 23 323-325 [DOI] [PubMed] [Google Scholar]
- 26.Mi, Z., Oliver, T., Guo, H., Gao, C., and Kuo, P. C. (2007) Cancer Res. 67 4088-4097 [DOI] [PubMed] [Google Scholar]
- 27.Yang, F., Robotham, J. M., Nelson, H. B., Irsigler, A., Kenworthy, R., and Tang, H. (2008) J. Virol. 82 5269-5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, Z.-G., Melaragno, M. G., Liao, D.-F., Yan, C., Haendeler, J., Suh, Y.-A., Lambeth, J. D., and Berk, B. C. (2000) Circ. Res. 87 789-796 [DOI] [PubMed] [Google Scholar]
- 29.Yang, H., Li, M., Chai, H., Yan, S., Lin, P., Lumsden, A. B., Yao, Q., and Chen, C. (2005) J. Surg. Res. 123 312-319 [DOI] [PubMed] [Google Scholar]
- 30.Jovine, L., Qi, H., Williams, Z., Litscher, E., and Wassarman, P. M. (2002) Nat. Cell Biol. 4 457-461 [DOI] [PubMed] [Google Scholar]
- 31.Price, E. R., Zydowsky, L. D., Jin, M., Baker, C. H., McKeon, F. D., and Walsh, C. T. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 1903-1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosco, D. A., Eisenmesser, E. Z., Pochapsky, S., Sundquist, W. I., and Kern, D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5247-5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billich, A., Hammerschmid, F., Peichl, P., Wenger, R., Zenke, G., Quesniaux, V., and Rosenwirth, B. (1995) J. Virol. 69 2451-2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obama, K., Kato, T., Hasegawa, S., Satoh, S., Nakamura, Y., and Furukawa, Y. (2006) Clin. Cancer Res. 12 70-76 [DOI] [PubMed] [Google Scholar]
- 35.Wulf, G., Ryo, A., Liou, Y. C., and Lu, K. P. (2003) Breast Cancer Res. 5 76-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.