Abstract
The genetically encoded calcium indicator GCaMP2 shows promise for neural network activity imaging, but is currently limited by low signal-to-noise ratio. We describe x-ray crystal structures as well as solution biophysical and spectroscopic characterization of GCaMP2 in the calcium-free dark state, and in two calcium-bound bright states: a monomeric form that dominates at intracellular concentrations observed during imaging experiments and an unexpected domain-swapped dimer with decreased fluorescence. This series of structures provides insight into the mechanism of Ca2+-induced fluorescence change. Upon calcium binding, the calmodulin (CaM) domain wraps around the M13 peptide, creating a new domain interface between CaM and the circularly permuted enhanced green fluorescent protein domain. Residues from CaM alter the chemical environment of the circularly permuted enhanced green fluorescent protein chromophore and, together with flexible inter-domain linkers, block solvent access to the chromophore. Guided by the crystal structures, we engineered a series of GCaMP2 point mutants to probe the mechanism of GCaMP2 function and characterized one mutant with significantly improved signal-to-noise. The mutation is located at a domain interface and its effect on sensor function could not have been predicted in the absence of structural data.
The intracellular concentration of free calcium ions, [Ca2+], is a universal second messenger in excitable cells such as neurons and muscle cells (1). In neurons, action potential firing triggers Ca2+ influx through voltage-gated calcium channels (2), and synaptic input results in Ca2+ influx through neurotransmitter receptors (3). The timing of spikes and patterns of synaptic input can be inferred by observing intracellular [Ca2+] (4), allowing researchers to monitor the activity of individual neurons and neural populations by non-invasive imaging techniques, using calcium-sensitive dyes and proteins.
Genetically encoded calcium indicator proteins (also known as fluorescent calcium indicator proteins) offer the possibility of long-term in vivo imaging of neural network activity, with cell-type- and subcellular localization-specific targeting not easily possible with synthetic small molecule calcium indicators (5). In the decade since their first publication, several formats have been explored for genetically encoded calcium indicator construction, including both two-fluorescent protein (FP)2 sensors with a fluorescence resonance energy transfer signal change and single-FP intensity-modulated sensors (6). Genetically encoded calcium indicator-enabled calcium imaging has been used to detect single action potentials in awake mice (7, 8). GCaMP-derived single-FP sensors (9) have facilitated in vivo imaging of odor-evoked neural activity in Drosophila (10) and Caenorhabditis (11), visual-evoked tectal responses in zebrafish (12), and cardiac output in mouse (13).
The GCaMP sensor is composed of a circularly permuted enhanced GFP (cpEGFP) moiety attached to the calcium-binding protein calmodulin (CaM) and the CaM-binding peptide M13pep (from myosin light chain kinase) (Fig. 1A). The first GCaMP sensor was extremely dim and folded poorly at 37 °C, limiting its utility for imaging applications. Incremental rounds of improvement by grafting of GFP-stabilizing mutations and random mutagenesis resulted in GCaMP2 (13-15), with significantly improved folding and fluorescence characteristics. This calcium indicator was extensively characterized in acute mouse cortical brain slice, with the conclusion that although useful, GCaMP2 suffers from low baseline fluorescence and cannot reproducibly detect single action potentials due to poor signal-to-noise (6, 16).
FIGURE 1.
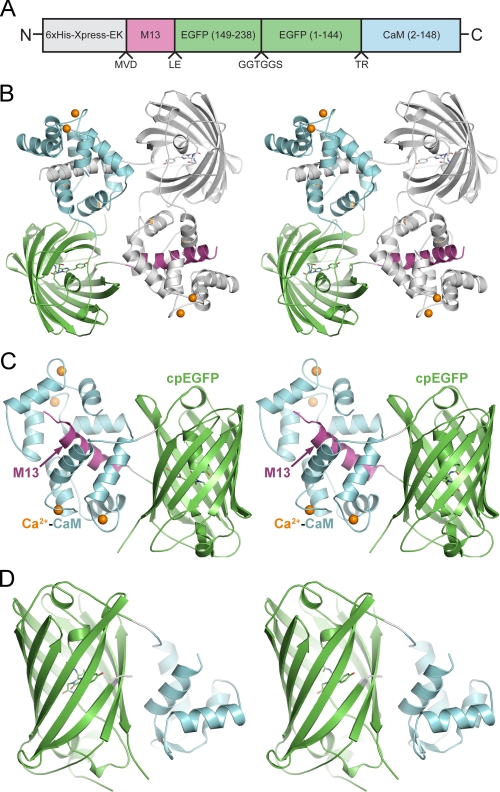
Crystal structures of GCaMP2. A, schematic of the primary amino acid sequence of GCaMP2 illustrating the domain organization. Domains are colored as depicted in B-D. Carets below the schematic show the positions of inter-domain linkers whose amino acid sequences are given. B, stereoview of the structure of the Ca2+-saturated domain-swapped GCaMP2 dimer, depicted as ribbons. One molecule of the dimer is colored by domain as in A, the other molecule is colored light gray. The EGFP chromophore is represented as sticks and calcium ions are shown as orange spheres. C, structure of Ca2+-saturated GCaMP2 monomer, represented as in B except the domains are labeled. D, structure of calcium-free GCaMP2, represented as in B and C. Note that the M13 peptide and the C-terminal half of CaM are not included in the model due to lack of electron density, suggesting their flexibility. This and other structure figures were prepared using PyMOL (Delano Scientific, San Carlos, CA).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The pRSETa vector containing GCaMP2 was a kind gift of Karel Svoboda (Janelia Farm Research Campus, Howard Hughes Medical Institute). GCaMP2 was expressed and purified as described previously (17). Briefly, BL21(DE3) cells containing pRSETa harboring gcamp2 or gcamp2 mutants were grown in ZYM-5052 medium (18) for 48 h at 25 °C with shaking at 200 rpm. After centrifugation, cell lysis, and clarification, proteins were purified from the cell-free extract by nickel-affinity chromatography. Protein purity over 95% was confirmed by SDS-PAGE analysis. Proteins were dialyzed into 20 mm Tris-HCl, 100 mm NaCl, 2 mm CaCl2, pH 8.0, and concentrated. Calcium-free samples were prepared identically, except the buffer contained 5 mm EGTA instead of CaCl2.
Crystallization and Data Collection—All GCaMP crystallization was carried out at 20 °C. All GCaMP protein samples for crystallization were in 20 mm Tris, 100 mm NaCl, 2 mm CaCl2, pH 8.0, except for the 8EF-apo mutant where the same buffer with 5 mm EGTA substituted for CaCl2 was used. All crystals used for data collection were grown using the hanging-drop vapor diffusion method in 24-well VDX plates. Crystallization of Ca2+-saturated dimeric GCaMP2 was described previously (17). The calcium-saturated K378W (at 5.6 mg/ml) and G87R (at 1.5 mg/ml) mutants crystallized ∼4 days after mixing with a precipitant solution consisting of 0.1 m magnesium formate dihydrate and 15% polyethylene glycol 3,350 using drop ratios of 2 μl of protein to 2 μl of precipitant for K378W and 1.5-2.5 μl for G87R. Ca2+-saturated monomeric GCaMP2 was crystallized identically to the K378W and G87R mutants using a drop ratio of 2 μl to 2 μl except that the drops were microseeded by streak seeding from K378W crystals immediately following setup. These crystals required more than 4 weeks to grow and had a distinct morphology. 8EF-apo GCaMP2 was crystallized after 1 week by mixing 2 μl of protein solution (9.5 mg/ml) with 2 μl of a precipitant solution consisting of 0.2 m lithium sulfate monohydrate, 0.1 m BisTris, pH 5.5, and 25% polyethylene glycol 3,350.
All crystals were cryoprotected for data collection by quickly (<10 s) soaking in the precipitant solution supplemented with 20% glycerol and then mounted in a nitrogen gas stream at 100 K or plunged into liquid nitrogen for storage and transport to synchrotron beamlines. All data were collected at 100 K in a N2 gas stream. X-ray diffraction data for the G87R mutant was collected in-house on a Rigaku RU-H3R rotating copper anode x-ray generator, equipped with a Saturn 92 CCD detector and X-stream 2000 low-temperature system. Data for Ca2+-saturated monomeric GCaMP2 was collected at the Advanced Light Source, beamline 8.2.2. Diffraction data from crystals of Ca2+-dimer, K378W, and 8EF-apo GCaMP2 were collected at the Advance Photon Source, beamline 31-ID.
X-ray diffraction data for G87R were integrated and scaled using d*TREK (19) from within the CrystalClear software package (Rigaku/Molecular Structure Corporation, Woodlands, TX). Data for Ca2+-saturated monomeric GCaMP2 were integrated and scaled in HKL2000 (20). Data from crystals of Ca2+-dimer, K378W, and 8EF-apo GCaMP2 were processed using Mosflm (21) and Scala (22).
Structure Solution, Model Building, and Refinement—All GCaMP2 structures were solved by molecular replacement using the program Phaser (23). The Ca2+-saturated dimer structure was solved as described previously (17) using the published coordinates of GFP (Protein Data Bank (PDB) entry 1EMA) and the coordinates of M13-bound calmodulin (PDB entry 1CDL) as search models. The G87R Ca2+-bound monomer mutant structure was solved by searching sequentially using the cpEGFP domain and CaM-M13 domains from the refined Ca2+-dimer structure and data between 29.3- and 2.8-Å resolution. Clear solutions were obtained in space group P41212 with translation function Z-scores of 43.1 and 21.3, respectively, for the two domains. Strong positive peaks in the difference map at the expected positions of the calcium ions in CaM (which were omitted from the MR model) indicated the correctness of the solutions. The K378W mutant crystals were isomorphous with those of the G87R mutant and the G87R model was used directly for rigid-body refinement against data from K378W crystals. The Ca2+-saturated monomeric GCaMP2 structure was solved using the refined K378W coordinates as a search model. A clear solution was obtained in space group P21212 with a translation function Z-score of 39.2 using data between 45.4- and 2.65-Å resolution. The 8EF-apo GCaMP2 calcium-free mutant structure was solved by searching for the cpEGFP domain from the Ca2+-dimer structure. A clear solution was obtained in space group C2 with a translation function Z-score of 18.9 using data between 31.9- and 2.8-Å resolution. Subsequently searching for the calcium-free N-terminal or C-terminal lobes of CaM (PDB code 1CFD (24)) did not reveal any clear solutions. Some positive difference density was present in the electron density maps calculated using the cpEGFP domain solution that suggested the position of the N-terminal lobe of CaM, which was placed manually into density and refined. The correctness of this CaM N-terminal lobe placement was indicated by additional positive difference density for the linker connecting cpEGFP and the CaM N-terminal lobe, which was subsequently built.
All models were improved by iterative cycles of model building in Coot (25) and positional refinement in REFMAC (26). Final GCaMP2 models have reasonable R-factors and model geometries, as illustrated in Table 1. A portion of the electron density map for each structure is provided in supplemental Fig. S1.
TABLE 1.
Data collection and refinement statistics
| GCaMP2 dimer (PDB 3EK7) | GCaMP2 monomer (PDB 3EK4) | GCaMP2-T116V-K378W (PDB 3EKH) | GCaMP2-T116V-G87R (PDB 3EK8) | 8EF-GCaMP2 (PDB 3EKJ) | |
|---|---|---|---|---|---|
| Data collection | |||||
| Radiation source | APS 31-IDa | ALS BL8.2.2b | APS 31-IDa | Copper anodec | APS 31-IDa |
| Wavelength (Å) | 0.9793 | 1.000 | 0.9793 | 1.5418 | 0.9793 |
| Space group | C2 | P21212 | P41212 | P41212 | C2 |
| Cell dimensions | |||||
| a, b, c (Å) | 126.13, 47.30, 68.94 | 60.49, 68.80, 117.26 | 121.64, 121.64, 97.32 | 120.82, 120.82, 97.35 | 211.87, 47.67, 42.99 |
| α, β, γ (°) | 90, 100.48, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 97.61, 90 |
| Resolution (Å) | 67.79-1.80 (1.90-1.80) | 50.00-2.65 (2.74-2.65) | 25.90-2.00 (2.11-2.00) | 29.30-2.80 (2.90-2.80) | 31.94-2.80 (2.95-2.80) |
| Rsym | 7.6 (42.2) | 7.5 (27.9) | 9.6 (60.1) | 18.4 (57.3) | 15.4 (61.7) |
| I/σI | 18.8 (5.1) | 20.44 (4.2) | 21.5 (4.7) | 8.6 (3.5) | 15.4 (3.2) |
| Completeness (%) | 98.5 (97.6) | 98.7 (92.1) | 100.0 (100.0) | 100.0 (100.0) | 98.9 (98.6) |
| Redundancy | 7.5 (7.6) | 3.1 (3.0) | 14.1 (13.5) | 11.94 (12.04) | 7.1 (7.1) |
| Refinement | |||||
| Resolution (Å) | 1.85 | 2.65 | 2.00 | 2.80 | 2.80 |
| Unique reflections | 33,937 | 14,650 | 49,718 | 18,310 | 10,580 |
| Rwork/Rfree | 0.189/0.241 | 0.222/0.280 | 0.194/0.224 | 0.224/0.266 | 0.210/0.283 |
| No. atoms (AU) | 3,333 | 2,799 | 3,472 | 3,182 | 2,430 |
| Protein | 3,097 | 2,767 | 3,185 | 3,154 | 2,425 |
| Ligand/ion | 4 | 3 | 10 | 4 | |
| Water | 198 | 29 | 277 | 24 | 5 |
| B-factors (Å2)d | |||||
| Protein | 25.4 | 44.0 | 33.4 | 38.8 | 31.1 |
| Ligand/ion | 19.4 | 75.3 | 38.7 | 41.4 | |
| Water | 32.2 | 29.4 | 37.1 | 23.3 | 12.9 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.018 | 0.010 | 0.011 | 0.011 | 0.016 |
| Bond angles (°) | 1.735 | 1.336 | 1.332 | 1.335 | 1.691 |
APS 31-ID, Advanced Photon Source, Beamline 31-ID.
ALS BL8.2.2, Advanced Light Source Beam Line 8.2.2.
Data collected on home source using Rigaku Rotating Copper Anode RUH3R.
B-factors were calculated on the STAN server using MOLEMAN2 (xray.bmc.uu.se/cgi-bin/gerard/rama_server.pl).
Size Exclusion Chromatography (SEC)—All SEC was carried out using a Superdex 200 10/300 GL column (GE Healthcare) at a flow rate of 0.5 ml min-1 in 20 mm Tris, 100 mm NaCl, 2 mm CaCl2, pH 8.0, for calcium-saturated samples or with 5 mm EGTA in place of CaCl2 for calcium-free samples. Molecular weights were estimated by comparison with elution volumes of standard proteins (Bio-Rad).
Sedimentation Velocity Analytical Ultracentrifugation—Analytical ultracentrifugation of GCaMP2 samples was carried out in a Beckman XL-I analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) within the Biophysics Instrumentation Facility at the Massachusetts Institute of Technology. Absorbance scans at 280 nm were collected on calcium-free (24 μm) and calcium-saturated (28 μm) GCaMP2 samples in two-sector cells within a four-hole AnTi-60 rotor at 42,000 rpm. Data were collected at 20 °C in the same buffers as the SEC experiments. Absorbance scans were modeled using a continuous c(s) distribution within Sedfit (27), correcting for buffer density and viscosity and using a partial specific volume of 0.7300 cm3 g-1. Molecular weight of observed species (Fig. 2B) was estimated based on the best-fit frictional ratio as determined by Sedfit for each sample.
FIGURE 2.
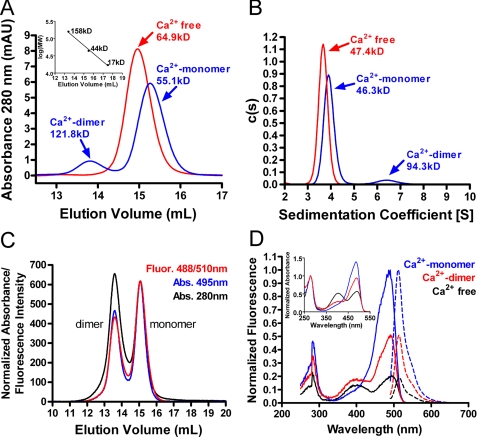
Solution studies of monomeric and dimeric GCaMP2. A, SEC of calcium-free (red) and calcium-bound (blue) GCaMP2 following affinity purification. Peaks are labeled with the assigned species and estimated molecular mass. The standard curve used to estimate Mr is in the inset. The predicted molecular mass for GCaMP2 monomer and dimer are 50.7 and 101.3 kDa, respectively. B, c(s) distribution analysis of sedimentation velocity analytical ultracentrifugation data for calcium-free (red) and calcium-bound (blue) GCaMP2 following affinity purification. Peaks are labeled as in B. C, SEC analysis of a nearly equimolar mixture of purified, Ca2+-saturated GCaMP2 monomer and dimer, monitoring protein absorbance at 280 nm (black), cpEGFP chromophore absorbance at 495 nm (blue), and cpEGFP fluorescence at 488/510 nm (red). Note the difference in relative chromophore absorbance and fluorescence of the dimer. D, fluorescence excitation (solid lines) and emission (dashed lines) spectra of isolated Ca2+-saturated GCaMP2 monomer (blue) and dimer (red), as well as calcium-free GCaMP2 (black). Absorbance spectra of these three species are shown in the inset.
Two-photon Laser Scanning Microscopy of Neurons Expressing GCaMP2—To measure intracellular [GCaMP2], acute brain slices containing neurons expressing GCaMP2 were prepared and imaged as previously described (6, 16). Purified GCaMP2 was diluted into pipette internal solution supplemented with 1 mm K2EGTA at 0.1, 1, and 10 μm concentrations. Each solution was drawn into a thin glass capillary (ID = 0.02 mm, Vitrocom number RT5002). Their fluorescence intensities were measured under two-photon excitation with identical parameters (910 nm excitation) to neuron imaging. 0.1 μm GCaMP2 was not bright enough to significantly exceed PMT dark current at laser powers used for neuronal imaging.
Intracellular GCaMP2 concentration in neurons with robust fluorescent responses to action potential firing was estimated by a linear extrapolation from the purified 10 μm GCaMP2 fluorescence intensity. Intracellular GCaMP2 was assumed to be in the apo state (6).
Generation and Screening of GCaMP2 Mutants—Mutants of GCaMP2 were prepared by site-directed mutagenesis (see supplemental Tables S1-S3) and confirmed by sequencing. Preliminary screening for variants with altered oligomerization equilibria (supplemental Table S1) was performed by passing 100-μl aliquots of cell-free extract from 200-ml cultures of overexpressed GCaMP2 over a Superdex 200 10/300 GL column while monitoring the absorption at 280 and 495 nm.
Spectrophotometric Analysis—Absorbance spectra were obtained in a Safire2 (Tecan) with UVStar 96-well plates (Greiner) for both the calcium-free (10 mm EGTA) and calcium-loaded state (10 mm CaCl2). For fluorescence spectra, Fluotrac 200 plates (Greiner) were used. Samples were diluted 10-fold in zero free calcium buffer (Invitrogen) (30 mm MOPS, 100 mm KCl, 10 mm EGTA, pH 7.2) for calcium-free spectra, and in 39 μm free calcium buffer (Invitrogen) (30 mm MOPS, 10 mm Ca-EGTA in 100 mm KCl, pH 7.2) for calcium-loaded spectra. For absorbance measurements, samples were dialyzed into 20 mm Tris, 100 mm NaCl, and EGTA or CaCl2 was added to a final concentration of 10 mm.
RESULTS
Structural Analysis of Dimeric Ca2+-GCaMP2—To understand the molecular mechanism of Ca2+-dependent fluorescence signal change and facilitate efforts to improve GCaMP properties, we set out to determine structures of the calcium-free and calcium-bound states of GCaMP2. Initial efforts to crystallize affinity purified, calcium-saturated GCaMP2 led to a single crystallization hit, as described previously (17). The structure was solved by molecular replacement, and the final model revealed a domain-swapped dimer of GCaMP2 in the crystals, with the M13 peptide of each monomer bound by the calcium-loaded CaM of the other (Fig. 1B and Table 1). The finding of a domain-swapped dimer structure was unexpected, given that GCaMP has previously been characterized as a monomer (9). We therefore characterized the oligomeric distribution of GCaMP2 in solution using several biophysical techniques.
Solution Characterization of GCaMP2—SEC analysis of affinity purified GCaMP2 in the absence of calcium showed a single peak close to the elution volume predicted for the monomeric form of GCaMP2, when compared with the elution volume of standard proteins (Fig. 2A). However, in the presence of saturating calcium, two peaks eluted. The two peaks were assigned as a monomeric form with a higher elution volume than the calcium-free monomer, and a smaller population of dimeric GCaMP2.
Analytical ultracentrifugation of GCaMP2 in the presence and absence of calcium gave results similar to SEC (Fig. 2B), as did dynamic light scattering (data not shown). These results indicate that calcium-bound GCaMP2 is more compact than calcium-free, consistent with data for calmodulin and M13pep (24, 28, 29). The observed dimeric structure, which preferentially crystallized in the initial screen, presumably represents the larger species in solution observed by SEC and analytical ultracentrifugation. After separation of the monomeric and dimeric calcium-saturated forms of GCaMP2 by SEC, neither showed significant presence of the other oligomeric state when re-analyzed days later, indicating that the kinetics of the monomer ↔ dimer equilibrium are very slow in the presence of saturating calcium.
The monomeric and dimeric forms of Ca2+-bound GCaMP2 have different absorption and fluorescence properties (Fig. 2C), suggesting an altered chromophore environment between the states. Monitoring absorbance at both 280 and 495 nm, as well as fluorescence at 488/510 nm during SEC separation of purified GCaMP2 monomer and dimer clearly indicated that the dimeric species had significantly lower chromophore absorption and fluorescence.
Fluorescence absorbance/excitation and emission scans of freshly separated monomeric and dimeric Ca2+-GCaMP2 indicated that apo, monomer, and dimer had nearly identical peak wavelengths for excitation and emission, but that the dimeric fraction was less fluorescent than the monomeric fraction, although both are substantially more fluorescent than the apo fraction (Fig. 2D). Comparison of the absorbance spectra of the three states shows that the chromophore environment changes dramatically upon Ca2+ binding: the protonated non-fluorescent state of the GCaMP2 chromophore is diminished upon Ca2+ binding, indicated by the smaller band at 400 nm in both Ca2+-saturated states, explaining the mechanism of the calcium-dependent fluorescence increase. The presence of a larger proportional amount of protonated non-fluorescent chromophore in the dimeric fraction compared with the monomeric fraction is in agreement with the SEC data (Fig. 2C), indicating that the monomeric GCaMP2 species is brighter than dimeric GCaMP2.
[GCaMP2] in Neurons—Calcium imaging experiments may be adversely affected by contribution from two competing species in situ. We thus attempted to quantitate the monomerdimer equilibrium present at physiologically relevant GCaMP concentrations. Two-photon laser scanning microscopy of in utero electroporated GCaMP2 under control of the CAG promoter was used to estimate sensor concentration in an acute slice of mouse somatosensory cortex, by comparison with a concentration series of purified Ca2+-free GCaMP2 protein in thin-window cuvettes (supplemental Fig. S2). Under these conditions, intracellular [GCaMP2] was found to be ∼10 μm. A previous report expressing GCaMP2 in transgenic mouse heart from a modified α myosin heavy chain promoter estimated the intracellular concentration to be 1.6 μm (13).
To understand what fraction of calcium-saturated GCaMP2 should be dimeric at these intracellular concentrations, we removed calcium from purified GCaMP2 protein with excess EGTA, prepared a dilution series of GCaMP2, added back excess calcium, and observed the proportion of monomeric and dimeric GCaMP2 using analytical SEC (supplemental Fig. S3). At the concentrations seen in the brain slice, only ∼5% of the GCaMP2 was present as a dimer, suggesting that the monomeric sensor predominates under the conditions of typical imaging experiments.
Engineering Monomeric GCaMP2—Given that the Ca2+-GCaMP2 dimer is presumably not the predominant form under imaging experimental conditions, we attempted to determine the structure of the monomeric form of Ca2+-GCaMP2. Preparative separation of the Ca2+-GCaMP2 monomer by SEC and immediate crystallization screening led only to crystals of the dimeric form that grew after several weeks.
To aid crystallization of the monomeric form, we made a series of single amino acid substitutions in GCaMP2 to selectively disrupt dimerization, based on visual inspection of the Ca2+-GCaMP2 dimer structure. Eleven single amino acid substitutions in GCaMP2 were chosen and produced by site-directed mutagenesis (supplemental Table S1). Six of these mutations appeared to decrease the amount of GCaMP2 dimer present in purified protein samples as analyzed by SEC (supplemental Fig. S4). Sparse-matrix crystallization screening was carried out with these six GCaMP2 mutants, and two of them (GCaMP2 G87R (gfp G174R) and GCaMP2 K378W (cam K75W), where gfp refers to green fluorescent protein and cam refers to calmodulin) crystallized with a unique morphology under conditions distinct from those used for dimer crystallization (a topology diagram and amino acid sequence displaying both GCaMP and EGFP/CaM numbering are provided in supplemental Figs. S5 and S6, respectively). Although the G87R and K378W mutations appear to decrease dimer formation, the ΔF/F0 for each was not significantly altered (supplemental Table S1) suggesting that the mutations did not disrupt sensor function.
X-ray diffraction data were collected from these new mutant crystals, and the structure was determined by molecular replacement using the cpEGFP and CaM-M13 domains from the dimer structure. The relative orientations of the cpEGFP and CaM-M13 domains revealed that the quaternary organization was different from the dimer structure, and electron density corresponding to the inter-domain linkers clearly showed that the structure represented the monomeric form of Ca2+-GCaMP2.
Structural Analysis of Monomeric Ca2+-GCaMP2—Subsequently, crystals of the native Ca2+-GCaMP2 monomer were grown by seeding drops of Ca2+-GCaMP2 protein with microcrystals of the GCaMP2 K378W monomer. Solution of the Ca2+-GCaMP2 monomer structure by molecular replacement revealed that although these crystals were grown by seeding with K378W microcrystals, they were distinct from the G87R, K378W, and GCaMP2 dimer crystals (see Table 1).
The Ca2+-GCaMP2 monomer structure is very similar to that of the K378W and G87R mutants (root mean square deviations values of 0.7 and 0.6 Å, respectively, for comparing all 353 common Cα atoms), but the C-terminal lobe of the CaM domain is largely disordered in the Ca2+-GCaMP2 monomer crystals and could not be modeled well. Therefore, further discussion of the Ca2+-GCaMP2 monomer structure is based on analysis of the native Ca2+-GCaMP2 monomer structure together with the K378W and G87R structures, in which the CaM C-terminal lobe is better defined.
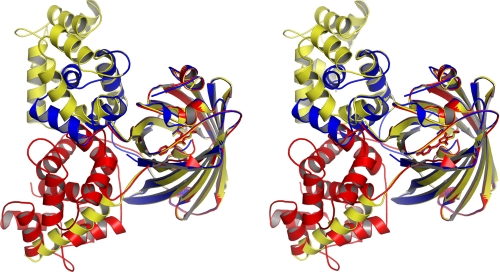
The domain organizations of the dimeric and monomeric forms of Ca2+-GCaMP2 are presented in Fig. 1, B and C. In the monomer structure, the Ca2+-CaM/M13pep complex makes more substantial contacts with the chromophore environment of cpEGFP, relative to the corresponding interaction in the dimer. Furthermore, the interaction surface of the Ca2+-CaM/M13pep with cpEGFP is quite different between the monomer and dimer (Figs. 3 and supplemental S7), with the former preferred in solution and the latter apparently favored by crystal packing.
FIGURE 3.
Stereoview of the structures of Ca2+-saturated GCaMP2-K387W monomer (red), Ca2+-saturated GCaMP2 dimer (yellow), and Ca2+-free 8EF-GCaMP2 (blue) superimposed using the cpEGFP domain. The proteins are represented as ribbons with the cpEGFP chromophore represented as sticks.
As described (9), the EGFP domain of GCaMP is circularly permuted by connecting the native N and C termini with a flexible linker (GGTGGS), and by opening one of the β-strands of the EGFP barrel by elimination of amino acids 145-148 (supplemental Fig. S5), resulting in new N and C termini at the side of the barrel, directly abutting the EGFP chromophore. The GCaMP2 structures reveal that circular permutation of EGFP at this location has very little effect on the overall tertiary structure of the barrel or loops. Structural superimposition of the cpEGFP domain of either monomeric or dimeric GCaMP2 with the native GFP structures (PDB codes 1GFL (30) or 1EMA (31)) shows root mean square deviations of only ∼0.4 Å when comparing all common 220 Cα atoms (supplemental Fig. S8A). The GGTGGS linker connecting the native EGFP N and C termini is disordered in all of the GCaMP2 crystals. The circular permutation of EGFP resulted, as expected, in an opening to the interior of the barrel due to the removal of four residues of the β-strand, although this opening is partially occluded in both Ca2+-bound structures by the Ca2+-CaM/M13pep complex. The opening measures ∼5 Å × 10 Å and allows appreciable solvent access to the p-hydroxybenzylideneimidazolinone chromophore. The phenolate moiety of the chromophore tyrosine points directly out of the opening, and is the most solvent-exposed portion of the chromophore. In the GCaMP2 dimer structure, large channels allow access from the surface of the protein, through this opening, to the chromophore. However, in the monomeric conformation the opening is mostly occluded by the “linker” connecting the N-terminal and C-terminal lobes of CaM, as well as the linker connecting cpEGFP with CaM. Solvent access is likely to lead to protonation of the phenolate oxygen (pKa ∼ 6.0 in EGFP and ∼7.1 in GCaMP), which in EGFP is stabilized in the fluorescent, deprotonated form by a large hydrogen bond network. This may partially explain the difference in calcium-saturated fluorescence between the monomer and dimer.
Each Ca2+-bound CaM-M13 complex present in both the monomer and the dimer Ca2+-GCaMP2 structures closely resembles the isolated CaM-M13 peptide complex structure determined previously (28), with the two lobes of CaM wrapping around and enveloping the helical M13 peptide. Structural superimposition of each CaM-M13 unit of GCaMP2 with this isolated human CaM-M13 complex structure (PDB code 1CDL) gives root mean square deviation values of 0.6 Å for the dimeric form, and 1.6 Å for the monomer when comparing all 161 common Cα atoms (supplemental Fig. S8B). The primary source of the larger structural deviation of monomeric GCaMP2 is the inter-lobe “linker helix” of CaM, which packs against the opening of the cpEGFP domain in the monomer structure but has fewer interactions in the dimer. This linker, which is known from previous structural work to adopt different conformations (24, 28, 29), is one α-helical turn longer in its N-terminal section and one turn shorter in the C-terminal section of the Ca2+-GCaMP2 monomer structure, relative to the dimer (supplemental Figs. S5 and S8B). This conformational change is a direct consequence of the interaction of the CaM domain with the cpEGFP domain, and has important implications for sensor functionality.
The cpEGFP domain is tethered in two ways to the calcium-responsive CaM in the calcium-saturated structures: the linker covalently connecting the C terminus of cpEGFP with the N terminus of CaM (cpEGFP-CaM linker), and the non-covalent interaction of CaM with M13pep, which is connected to the N terminus of cpEGFP by a short linker (M13-cpEGFP linker). Of these two linkers, the cpEGFP-CaM linker appears to be the more flexible, with ∼5 consecutive amino acids between secondary structure elements or anchoring interactions, and with only weak electron density. The M13-cpEGFP linker, conversely, is shorter, less flexible, and makes several interactions with both the cpEGFP and CaM domains. In addition to the linkers and the CaM-M13 interaction, inter-domain contacts are made directly between cpEGFP and CaM. In the GCaMP2 monomer structure, the N-terminal lobe and inter-lobe linker of CaM contact the side of the cpEGFP barrel on opposite sides of the opening, occluding ∼638 Å2 of the solvent-accessible surface of each domain at the interface (supplemental Fig. S7). Very few interactions are made between cpEGFP and the C-terminal lobe of CaM in the monomer. In the dimer, each CaM domain contacts both cpEGFP domains (Figs. 1B and supplemental S7); the most extensive contact interface is observed between the N-terminal lobe of CaM and the side of the cpEGFP barrel of the same GCaMP monomer. Approximately 625 Å2 of the solvent-accessible surface of each domain is buried at this interface. Two smaller patches of contact surface, totaling ∼249 Å2, are present between each lobe of the CaM domain and the side of the cpEGFP barrel from the other molecule of the GCaMP dimer.
Engineering and Structural Analysis of Calcium-free GCaMP2—The GCaMP sensor was designed on the assumption that in the absence of calcium, CaM would not associate with M13pep, allowing significant solvent access-mediated darkening of the EGFP chromophore (9). Understanding the mechanism of calcium-free fluorescence decrease could allow targeted mutagenesis to improve baseline fluorescence or total dynamic range. To observe the domain organization of GCaMP2 in the absence of calcium (apo-GCaMP2) we attempted crystallization in the presence of an excess calcium chelator (EGTA). All crystallization trials produced dimeric, Ca2+-bound GCaMP2, despite repeated attempts to remove calcium from buffers, protein, glass, and plasticware. We therefore engineered an 8-fold mutant of GCaMP2, with two mutations in each of the four Ca2+-binding EF hands in the CaM domain (T329G/E334Q, D359G/E370Q, D396G/E407Q, D432G/E443Q) that should disrupt calcium binding. This mutant, 8EF-GCaMP2, spectrally resembles apo-GCaMP2 and does not respond to Ca2+ (ΔF/F0 = 0.15 ± 0.04) (see supplemental Fig. S9 for spectra). Sparse-matrix crystallization screens with 8EF-GCaMP2 produced thin plate crystals that diffracted to 2.8 Å. The structure of 8EF-GCaMP2 was solved by molecular replacement using the cpEGFP domain from the previous structures, manual placement of the N-terminal lobe of apo-CaM into the remaining electron density, and rebuilding of the inter-domain linkers (Fig. 1D). No electron density was present for either the C-terminal lobe of CaM or the M13 peptide, indicating that in the absence of calcium the inter-domain flexibility is increased. The N-terminal lobe of CaM in 8EF-GCaMP2 closely resembles the “closed” calcium-free conformation observed in previous solution structures of apo-CaM (24, 29), with substantial changes in the inter-helical angles serving to sequester the hydrophobic target-binding cleft (Figs. 3 and supplemental S8). The first and last helices of the N-terminal lobe of CaM pack against the barrel of cpEGFP, creating nearly the same interface observed in the dimeric Ca2+-bound structure. In the absence of inter-domain interactions blocking the opening of cpEGFP, as observed in the calcium-saturated structures, solvent is allowed direct access to the EGFP chromophore, which appears to be stabilized in the protonated, non-fluorescent form (supplemental Fig. S9). The position of the M13 peptide is not constrained by tight binding to CaM; consequently no electron density is observed for the M13pep. However, the linker connecting M13pep with cpEGFP (Leu60-Glu61) is observed and adopts a significantly different conformation in apo-GCaMP2 compared with the monomer and dimer GCaMP2 structures (supplemental Fig. S10). In the calcium-free structure, this linker is incorporated into the β-strand leading into cpEGFP; the side chain of Glu61 hydrogen bonds to the backbone amide nitrogen of Arg81, stabilizing this conformation (supplemental Fig. S10).
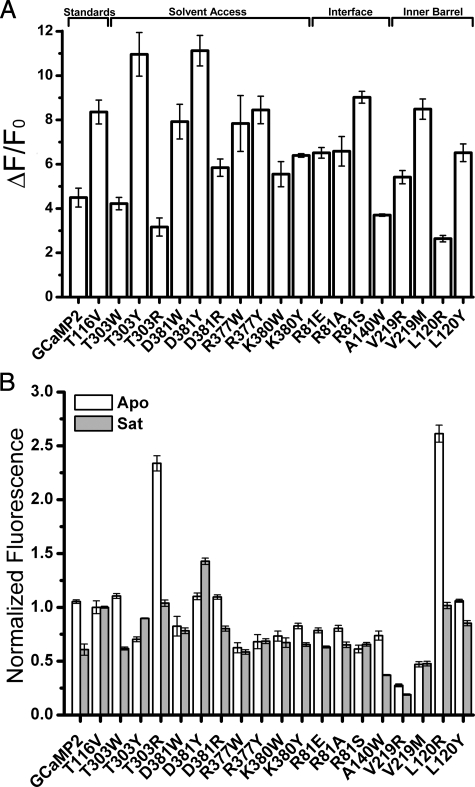
Structure-based Mutagenic Analysis of GCaMP2 Function—With the GCaMP2 crystal structures in hand, we set out to analyze GCaMP sensor function by site-directed mutagenesis to understand the mechanistic basis for the response to calcium. As GCaMP2 is largely monomeric under typical imaging conditions, the Ca2+-monomer structures served as the basis for mutagenesis studies to address function. All mutations were made in the context of GCaMP2-T116V (gfp T203V), a mutation previously described to increase the wild type EGFP excited-state proton transfer (32-34).3 We created groups of GCaMP2 mutants to test three hypotheses about GCaMP function: (a) that mutating CaM or the cpEGFP-CaM linker to block solvent access to the EGFP chromophore would improve brightness (“solvent access” mutants), (b) disrupting the cpEGFP-CaM interfaces seen in the crystal structure would decrease sensor performance (“interface” mutants), and (c) mutating EGFP β-barrel positions observed to be solvent-exposed in the GCaMP2 structure (but not in EGFP itself) due to the circular permutation would alter sensor function (“inner barrel” mutants). Eighteen mutations, through the three mutant classes, were created by site-directed mutagenesis and analyzed by measuring fluorescence of purified protein (Figs. 4 and supplemental S9).
FIGURE 4.
Fluorescent properties of GCaMP2 variants. A, ΔF/F0 for calcium binding of all GCaMP2 mutants listed in supplemental Table S3. All GCaMP2 mutants listed also contain the T116V mutation. B, normalized fluorescence of Ca2+-free (apo, white) and the Ca2+-saturated (Sat, gray) state of the GCaMP2 variants. Fluorescence of both states is normalized to the brightness of GCaMP2 T116V. Note that although GCaMP2 T116V-T303Y has a greater ΔF/F0 (panel A), the brightness of both Ca2+-free and Ca2+-saturated state is lower than for GCaMP2 T116V.
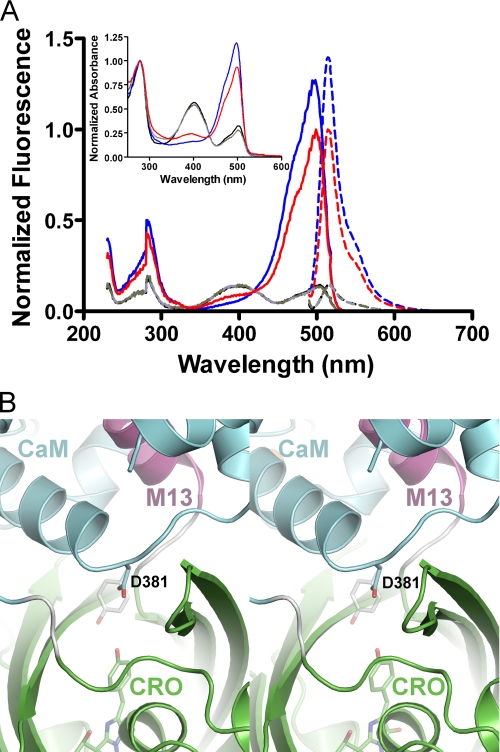
Two of the mutations predicted to block solvent access to the chromophore by introduction of a bulkier amino acid side chain near the cpEGFP opening were successful. GCaMP2-T303Y (in egfp-cam linker) and GCaMP2-D381Y (cam D78Y) showed significantly improved ΔF/F0 for Ca2+ binding (Fig. 4A). The D381Y (cam D78Y) mutation had a slightly brighter apo state (Fig. 4B), a substantially brighter Ca2+-bound state, and showed the highest dynamic range of all mutants tested (ΔF/F0, D381Y = 11.1; ΔF/F0, GCaMP2 = 4.5; ΔF/F0, T116V = 8.4). Absorbance and fluorescence scans showed that D381Y (cam D78Y) has a lower proportion of protonated chromophore in the Ca2+-bound state (Fig. 5A), thus improving signal-to-noise ratio. Position Asp381 of GCaMP2 is located on the flexible region of CaM between the two calcium-binding lobes and packs directly against the cpEGFP opening in the Ca2+-monomer structures (Fig. 5B). The side chain of a tyrosine at position 381 could extend into the opening of the cpEGFP barrel and come within a few angstroms of the chromophore (Fig. 5B).
FIGURE 5.
A rationally designed, improved GCaMP2 variant. A, fluorescence excitation (solid lines) and emission (dashed lines) spectra of Ca2+-saturated GCaMP2 T116V (red) and T116V/D381Y (blue), as well as their calcium-free forms (gray and black, respectively). Normalized absorbance spectra of each are shown in the inset. B, close-up stereo view of the Ca2+-saturated monomeric GCaMP2 structure, showing the location of aspartate 381 (D381) of CaM at the CaM/cpEGFP domain interface. GCaMP2 is displayed as ribbons colored by domain. The side chain of Asp381 and the cpEGFP chromophore are displayed as sticks. A model of a tyrosine side chain at position 381 is shown in semitransparent sticks to represent a possible conformation of the D381Y mutant and to illustrate the proximity of this side chain to the chromophore.
Disruption of the cpEGFP-CaM interfaces observed in the Ca2+-GCaMP2 crystal structures dramatically decreased saturated-state fluorescence (Fig. 4). Position Arg81 (gfp Arg168), from a β-strand lining the cpEGFP opening, was observed to form salt bridges with Glu61 of the M13-cpEGFP linker and Glu387 (cam Glu84) from the CaM inter-lobe linker, in addition to a hydrogen bond with the backbone carbonyl oxygen of Thr382 (cam T79). Mutation of Arg81 (gfp Arg168) to glutamate, alanine, or serine decreased Ca2+-saturated fluorescence by ∼25%. Mutation of Ala140 (gfp Ala227), near the center of the Ca2+-CaM/cpEGFP interface, to the bulky amino acid tryptophan resulted in a 60% decrease in Ca2+-saturated fluorescence. These disruptive mutations suggest that the domain interfaces observed in the Ca2+-monomer crystal structure are functionally relevant in solution as well.
Two positions in EGFP (gfp 62, gfp 207), normally tightly packed inside the barrel but partially exposed by circular permutation, were also selected for mutagenesis. Because both positions are near the chromophore, we hypothesized that mutations to larger side chains could significantly alter the chromophore environment. Mutation of Val219 (gfp Val62) to arginine or methionine dramatically decreased both apo and Ca2+-saturated fluorescence (Fig. 4B). Because Val219 (gfp Val62) is located only one turn of α-helix from the chromophore, it is possible that these mutations interfere with chromophore maturation or destabilize the cpEGFP structure. Mutation of Leu120 (gfp Leu207) to arginine dramatically increased apo fluorescence without significantly altering Ca2+-saturated fluorescence, whereas mutation to tyrosine increased apo fluorescence and slightly decreased Ca2+-saturated fluorescence. The mechanism by which L120R (gfp L207R) leads to a large increase in apo fluorescence is unknown, but the large functional variance of the two mutations tested suggest that this could be an interesting position for future mutagenic libraries.
DISCUSSION
The structures described here illustrate the molecular mechanisms by which the steric and electrostatic environment of the GCaMP2 chromophore is altered upon calcium binding. Large scale conformational changes that alter relative domain orientations serve to change the stereochemical functionality presented to the chromophore at the cpEGFP barrel opening, and to control the access of solvent molecules (consistent with Ca2+-dependent pKa decrease of the GCaMP chromophore (9)). The biophysical characterization in solution demonstrated that GCaMP2 is predominantly monomeric at physiological concentrations during imaging experiments in tissue cells, although the issue of dimerization remains relevant because future GCaMP mutations could inadvertently favor the dimeric form and complicate interpretation of imaging experiments. Earlier analysis of GCaMP (9) used protein concentrations too low and gel filtration of inadequate resolution to observe dimerization.
The crystal structures reveal some of the mechanisms by which mutations previously made to the GCaMP scaffold, both in the initial engineering and in later rounds of optimization, function to improve the sensor. The crystal structures of GCaMP2 demonstrate the close proximity of the M13pep-cpEGFP linker to the EGFP chromophore relative to the cpEGFP-CaM linker, which is consistent with the reported effect of mutation of these two regions on sensor response (9). During the original construction of GCaMP it was noted that positively charged side chains at position 61 within the M13-cpEGFP linker abolished responsiveness to calcium, whereas serine or threonine on that position led to photoisomerization (9). In the calcium-free 8EF-GCaMP2 structure, Glu61 occupies the same location as His148 in EGFP. The protonated Nδ atom of His148 is critical for the hydrogen bond network stabilizing the deprotonated state of the chromophore of EGFP (35, 36); Glu61 in the calcium-free 8EF-GCaMP2 could fulfill the same role and might preferentially stabilize the protonated state contributing to the decreased fluorescence observed in calcium-free GCaMP2. In the calcium-bound structures, Glu61 is pulled away from the cpEGFP barrel opening due to the interaction of M13pep with CaM and forms a salt bridge interaction with Arg81 from the outside of the cpEGFP barrel.
Small-probe contact dot analysis (37) of the EGFP crystal structure (PDB 1EMA) shows that Val163 is over-packed in the hydrophobic core, potentially clashing with Ile152. Mutation of EGFP-Val163 to Ala in GCaMP1.6 resulted in a more thermostable sensor (15), and the crystal structures of GCaMP2 show that Ala76 (gfp 163) eases this unfavorable interaction without creating a cavity. Mutation of EGFP-Ser175 in GCaMP1.6 to glycine similarly improved thermostability; in addition to improving packing within the EGFP domain, this change may allow backbone relaxation to facilitate interaction with the CaM inter-domain linker, critical for shielding the chromophore from solvent quenching. The GCaMP1.6 mutation GFP-A206K, intended to minimize EGFP dimerization, has an unexpected effect on the GCaMP sensor, as this position actually interacts with the CaM domain, rather than the EGFP domain of another sensor molecule. In the dimeric state, GCaMP2-Lys119 (gfp 206) forms an electrostatic interaction with GCaMP2-Glu309 (cam 6) within the GCaMP2 monomer. In the monomeric state, a similar electrostatic interaction occurs with GCaMP2-Asp305 (cam 2). Thus the actual effect of this mutation in GCaMP1.6 may be to stabilize EGFP-CaM contacts within the GCaMP monomer, rather than to disfavor EGFP dimerization. The GCaMP2 mutation GFP-D180Y appears to improve the β-strand propensity of the position regardless of Ca2+ binding and EGFP-CaM association. All five structures show that GCaMP2-Tyr93 (gfp 180) is solvent-exposed, with few interactions within the EGFP domain and none with other sensor components. The mutation V251I (gfp 93) in GCaMP2 increases packing interactions between two adjacent β-strands, particularly with GCaMP2-Ile101 (gfp 188). No interdomain contacts are observed at position 251 in the five crystal structures reported here, consistent with the calcium-independent stabilization observed with this mutation (13).
We have explored only a limited panel of mutations based on inspection of the crystal structures reported here. Future efforts to analyze additional point mutations and mutagenic libraries at positions shown to be of interest based on the crystal structures should lead to even better genetically encoded calcium indicators with further enhanced signal-to-noise capable of improved neural activity imaging. Additionally, we anticipate that the structural information gained from this work on GCaMP2 will aid rational design efforts to produce improved fluorescent protein-based sensors for other analytes.
Note Added in Proof—During the review process of this manuscript, Wang et al. (Wang, Q., Shui, B., Kotlikoff, M. I., and Sondermann, H. (2008) Structure (Lond.) 16, 1817-1827) reported on the structures of monomeric and dimeric GCaMP2. The results of that study are consistent with those described here.
Supplementary Material
Acknowledgments
We thank Karel Svoboda (HHMI Janelia Farm) and members of the Looger lab and Schreiter lab for helpful discussions. We also thank the beamline scientists of beamline 31-ID of the APS for x-ray diffraction data collection. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC02-05CH11231.
The atomic coordinates and structure factors (codes 3EK7, 3EK4, 3EKJ, 3EKH, and 3EK8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant 5P20RR016439-05 to the UPR Protein Research Center. This work was also supported by the University of Puerto Rico, Río Piedras Campus. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S10 and Tables S1-S3.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: FP, fluorescent protein; GFP, green fluorescent protein; EGFP, enhanced GFP; cpEGFP, circularly permuted EGFP; CaM, calmodulin; M13pep, myosin light-chain kinase target peptide; SEC, size exclusion chromatography; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MOPS, 4-morpholinepropanesulfonic acid; PDB, Protein Data Bank.
L. Tian, S. Hires, J. Akerboom, E. Schreiter, and L. Looger, manuscript in preparation.
References
- 1.Burgoyne, R. D. (2007) Nat. Rev. Neurosci. 8 182-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe, D. B., Johnston, D., Lasser-Ross, N., Lisman, J. E., Miyakawa, H., and Ross, W. N. (1992) Nature 357 244-246 [DOI] [PubMed] [Google Scholar]
- 3.Muller, W., and Connor, J. A. (1991) Nature 354 73-76 [DOI] [PubMed] [Google Scholar]
- 4.Yasuda, R., Nimchinsky, E. A., Scheuss, V., Pologruto, T. A., Oertner, T. G., Sabatini, B. L., and Svoboda, K. (2004) Sci. STKE 2004 pl5. [DOI] [PubMed] [Google Scholar]
- 5.Mank, M., and Griesbeck, O. (2008) Chem. Rev. 108 1550-1564 [DOI] [PubMed] [Google Scholar]
- 6.Hires, S. A., Tian, L., and Looger, L. L. (2008) Brain Cell Biol. 36 69-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace, D. J., Zum Alten Borgloh, S. M., Astori, S., Yang, Y., Bausen, M., Kugler, S., Palmer, A. E., Tsien, R. Y., Sprengel, R., Kerr, J. N., Denk, W., and Hasan, M. T. (2008) Nat. Methods 5 797-804 [DOI] [PubMed] [Google Scholar]
- 8.Mank, M., Santos, A. F., Direnberger, S., Mrsic-Flogel, T. D., Hofer, S. B., Stein, V., Hendel, T., Reiff, D. F., Levelt, C., Borst, A., Bonhoeffer, T., Hubener, M., and Griesbeck, O. (2008) Nat. Methods 5 805-811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakai, J., Ohkura, M., and Imoto, K. (2001) Nat. Biotechnol. 19 137-141 [DOI] [PubMed] [Google Scholar]
- 10.Wang, Y., Guo, H. F., Pologruto, T. A., Hannan, F., Hakker, I., Svoboda, K., and Zhong, Y. (2004) J. Neurosci. 24 6507-6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalasani, S. H., Chronis, N., Tsunozaki, M., Gray, J. M., Ramot, D., Goodman, M. B., and Bargmann, C. I. (2007) Nature 450 63-70 [DOI] [PubMed] [Google Scholar]
- 12.Naumann, E. A. (2005) Using G-CaMP 1.6 to Monitor Visually Evoked Synaptic Activity in Tectal Neurons in Vivo, PhD thesis, Universitat Konstanz, Konstanz, Germany
- 13.Tallini, Y. N., Ohkura, M., Choi, B. R., Ji, G., Imoto, K., Doran, R., Lee, J., Plan, P., Wilson, J., Xin, H. B., Sanbe, A., Gulick, J., Mathai, J., Robbins, J., Salama, G., Nakai, J., and Kotlikoff, M. I. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4753-4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez-Garcia, J., Matsushita, S., Mutoh, H., Nakai, J., Ohkura, M., Yokoyama, J., Dimitrov, D., and Knopfel, T. (2005) Eur. J. Neurosci. 22 627-635 [DOI] [PubMed] [Google Scholar]
- 15.Ohkura, M., Matsuzaki, M., Kasai, H., Imoto, K., and Nakai, J. (2005) Anal. Chem. 77 5861-5869 [DOI] [PubMed] [Google Scholar]
- 16.Mao, T., O'Connor, D. H., Scheuss, V., Nakai, J., and Svoboda, K. (2008) PLoS ONE 3 e1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez Guilbe, M. M., Alfaro Malave, E. C., Akerboom, J., Marvin, J. S., Looger, L. L., and Schreiter, E. R. (2008) Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 64 629-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier, F. W. (2005) Protein Expression Purif. 41 207-234 [DOI] [PubMed] [Google Scholar]
- 19.Pflugrath, J. W. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 1718-1725 [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., Minor, W., and Carter, C. W., Jr. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 21.Leslie, A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, Daresbury Laboratory, Daresbury, Warrington, UK
- 22.Collaborative Computational Project Number Four (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 23.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuboniwa, H., Tjandra, N., Grzesiek, S., Ren, H., Klee, C. B., and Bax, A. (1995) Nat. Struct. Biol. 2 768-776 [DOI] [PubMed] [Google Scholar]
- 25.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 27.Schuck, P. (2000) Biophys. J. 78 1606-1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meador, W. E., Means, A. R., and Quiocho, F. A. (1992) Science 257 1251-1255 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, M., Tanaka, T., and Ikura, M. (1995) Nat. Struct. Biol. 2 758-767 [DOI] [PubMed] [Google Scholar]
- 30.Yang, F., Moss, L. G., and Phillips, G. N., Jr. (1996) Nat. Biotechnol. 14 1246-1251 [DOI] [PubMed] [Google Scholar]
- 31.Ormo, M., Cubitt, A. B., Kallio, K., Gross, L. A., Tsien, R. Y., and Remington, S. J. (1996) Science 273 1392-1395 [DOI] [PubMed] [Google Scholar]
- 32.Jung, G., Wiehler, J., and Zumbusch, A. (2005) Biophys. J. 88 1932-1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kummer, A. D., Wiehler, J., Rehaber, H., Kompa, C., Steipe, B., and Michel-Beyerle, M. E. (2000) J. Phys. Chem. B 104 4791-4798 [Google Scholar]
- 34.Souslova, E. A., Belousov, V. V., Lock, J. G., Stromblad, S., Kasparov, S., Bolshakov, A. P., Pinelis, V. G., Labas, Y. A., Lukyanov, S., Mayr, L. M., and Chudakov, D. M. (2007) BMC Biotechnol. 7 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brejc, K., Sixma, T. K., Kitts, P. A., Kain, S. R., Tsien, R. Y., Ormo, M., and Remington, S. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2306-2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elsliger, M. A., Wachter, R. M., Hanson, G. T., Kallio, K., and Remington, S. J. (1999) Biochemistry 38 5296-5301 [DOI] [PubMed] [Google Scholar]
- 37.Word, J. M., Lovell, S. C., LaBean, T. H., Taylor, H. C., Zalis, M. E., Presley, B. K., Richardson, J. S., and Richardson, D. C. (1999) J. Mol. Biol. 285 1711-1733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.