Abstract
The unusual adhesion G-protein-coupled receptors (aGPCRs) contain large extracellular N-terminal domains, which resemble cell-adhesion receptors, and C-terminal heptahelical domains, which may couple to G-proteins. These receptors are cleaved post-translationally between these domains into two fragments (NTF and CTF). Using the aGPCR latrophilin 1, we previously demonstrated that the fragments behave as independent cell-surface proteins. Upon binding the agonist, α-latrotoxin (LTX), latrophilin fragments reassemble and induce intracellular signaling. Our observations raised important questions: is the aGPCR signaling mediated by reassembled fragments or by any non-cleaved receptors? Also, can the fragments originating from distinct aGPCRs form hybrid complexes? To answer these questions, we created two types of chimerical constructs. One contained the CTF of latrophilin joined to the NTF of another aGPCR, EMR2; the resulting protein did not bind LTX but, similar to latrophilin, could couple to G-proteins. In another construct, the NTF of latrophilin was fused with the C terminus of neurexin; this chimera bound LTX but could not signal via G-proteins. Both constructs were efficiently cleaved in cells. When the two constructs were co-expressed, their fragments could cross-interact, as shown by immunoprecipitation. Furthermore, LTXN4C induced intracellular Ca2+ signaling only in cells expressing both constructs but not each individual construct. Finally, we demonstrated that fragments of unrelated aGPCRs can be cross-immunoprecipitated from live tissues. Thus, (i) aGPCR fragments behave as independent proteins, (ii) the complementary fragments from distinct aGPCRs can cross-interact, and (iii) these cross-complexes are functionally active. This unusual cross-assembly of aGPCR fragments could couple cell-surface interactions to multiple signaling pathways.
The unusual heptahelical receptors, called “adhesion G-protein-coupled receptors” (aGPCRs)3 (1) are thought to mediate cell-cell and cell-matrix interactions resulting in different cell guidance signals (for a review, see Ref. 2). One of the first members of this family to be isolated and studied was latrophilin (3, 4). Latrophilin 1 (4), or CIRL (5), is the major target of α-latrotoxin (LTX) and is implicated in regulating neurotransmitter release. Together with EMR1 (6) and CD97 (7), latrophilin has served as a model for uncovering many features common for all aGPCRs (4, 5, 8-14).
One outstanding characteristic of aGPCRs is the presence in their N termini of various cell-adhesion modules. The N-terminal domains are connected to heptahelical C termini that possess structures of typical GPCRs. In that sense, these proteins seem to be natural chimeras of cell-adhesion receptors and proper GPCRs (13, 15), giving rise to their modern name.
The second characteristic that makes aGPCRs unusual is their proteolytic cleavage, which occurs exactly between the cell-adhesion and GPCR domains. This proteolysis (which is probably autocatalytic (16, 17)) takes place inside the cell (13, 14) and, thus, is not associated with any signaling. In fact, full-size, non-cleaved aGPCRs are not normally found in tissues (3, 5, 8) and may never be delivered to the cell surface (14). (The full-size GPR56 reportedly present in mouse tissues (18) is apparently an unrelated protein (19).)
The proteolytic cleavage occurs upstream of the first transmembrane domain, at the conserved “GPCR proteolysis site” (GPS) (5), comprising 50-60 residues, which becomes unequally split between the resulting N-terminal fragment (NTF, or “α-subunit”) and the C-terminal fragment (CTF, or “β-subunit”) (13). Despite having no transmembrane regions, the NTF remains associated with the cell surface (8, 13). Initially, this was thought to be mediated by a non-covalent interaction of the NTF with the membrane-anchored CTF (13, 20). However, the two fragments have been found to behave as independent cell-surface proteins (14): they can be delivered to different domains of the plasma membrane, recycled separately, patched independently by specific antibodies, and individually solubilized. The nature of the NTF anchoring in the membrane is still under investigation.
Despite their independence in the membrane, the fragments are able to re-associate, and their reassembly, thought to be mediated by the GPS domain (13, 14, 21), plays an important role in the biology of these receptors. This re-association, at least in the case of latrophilin (14), is stimulated by the binding of the exogenous agonist (LTXN4C) to the NTF. Formation of tripartite complexes containing NTF, CTF, and LTXN4C is coupled to intracellular signaling and release of Ca2+ from intracellular stores (14).
Several important questions arise from these results. Is the signaling mediated by the non-covalent NTF·CTF complexes or by a low number of non-cleaved full-size receptors? Given the independence of the fragments and the conserved GPS structures, is it possible that fragments of distinct aGPCRs cross-associate? If so, are these hybrid complexes functionally active? Answers to these questions could have profound implications for the role of aGPCRs in the cell.
To test these hypotheses, we created two types of hybrid constructs, each containing only one fragment of latrophilin and designed to have two opposite phenotypes: one hybrid was able to bind the agonist LTXN4C but incapable of coupling to G-proteins, whereas the second hybrid could not bind the toxin but retained the latrophilin-like signaling capacity. The constructs, stably expressed in neuroblastoma cells, were efficiently cleaved. When the constructs were co-expressed, their fragments were able to cross-interact. Importantly, these non-covalent complexes were functional: they bound LTXN4C and mediated release of Ca2+ from intracellular stores. Finally, to prove the physiological relevance of this cross-interaction, we immunoprecipitated the NTF of latrophilin from mammalian brain and demonstrated that the CTF of another aGPCR, GPR56, co-precipitates with it. Thus, independent aGPCR fragments can cross-interact and signal, forming a dynamic, complex network of signal transduction pathways.
MATERIALS AND METHODS
Reagents and Biochemical Methods—General chemicals were of analytical grade and obtained from Sigma, unless otherwise stated. Rabbit antibodies against latrophilin NTF (termed RL1) and CTF (R4) were described in a previous study (22); anti-Myc and anti-V5 monoclonal antibodies (mAbs) were from AbD-Serotec, an antibody against the CTF of GPR56 was from Santa Cruz Biotechnology (Santa Cruz, CA). Production of LTXN4C, LTX-affinity chromatography, 125I-LTX binding, immunoprecipitation of Triton X-100-solubilized cells (or freshly homogenized rat brain), and Western blotting (WB) were performed as outlined elsewhere (14, 22-25). For SDS-gel electrophoresis of heptahelical CTFs, samples were prepared in a conventional SDS buffer but heated for 30 min at 50 °C and never boiled to avoid irreversible precipitation. Positive bands were visualized with a chemiluminescent substrate (Millipore) and captured using a gel image digitizer (LAS-3000, Fujifilm). Protein bands with optical density within the digitizer's linear response range were quantified relative to the initial content of the same protein, using Aida software (Raytek Scientific Ltd.). The mean proportion of each protein precipitated was calculated from multiple independent experiments.
Construction of Expression Vectors—All expression constructs were created using the pcDNA3.1 vector (Invitrogen) and verified by complete sequencing. A plasmid containing the full-length cDNA of EMR2, previously described (26), was a kind gift from H.-H. Lin (University of Oxford). Production of L-Lm and L-Nm was outlined in (14), where these constructs were referred to as LPH-A and LPH-D, respectively. The vL-Lm was made by replacing the HindIII/BamHI fragment of L-Lm with two fragments, which (i) were amplified on the L-Lm template using two pairs of primers (TAATACGACTCACTATAGG/CTCCGAATTCGACCTTGCATAGTCGGGTACATCGTAGGGGTAGCCTTGGGTAGCAGAGGTGACGAGG and CAGCGAATTCGGGTAAGCCTATTCCGAATCCCCTCCTCGGCTTAGATTCCACAGGCCTGAGCCGGGCTGGACTCCC/TGTGGCGTGCAGCCACATAGTCCTCCC); (ii) cleaved with HindIII/EcoRI and EcoRI/BamHI, respectively; (iii) joined at the introduced EcoRI site; and (iv) together encoded consecutive HA and V5 epitopes in-frame with the signal peptide. The vL-Nm was created from L-Nm in a similar manner. To obtain vE-Lm fusion construct, the EcoRI/KpnI fragment of vL-Lm was replaced with two fragments: one amplified on the EMR2 template between the primers CAGCGAATTCGGGTAAGCCTATTCCGAATCCCCTCCTCGGCTTAGATTCCACACAGGACTCCAGGGGCTGTGCCCGGT/CGTAACCGGTCATGAGGACGGCAAAGTTGGTCAGGTGGGTGCAA; and the other amplified on the vL-Lm template between the primers CATGACCGGTCGAGGGATCTACCAAGGCCGT/CACGGGGCTGTAGCACAGGGTTGGTCAGTAGG. The fragments were cleaved with EcoRI/AgeI and AgeI/KpnI, respectively, and joined at the introduced AgeI site. The vE-Lem construct was made similarly, except using the primer pair CAGCGAATTCGGGTAAGCCTATTCCGAATCCCCTCCTCGGCTTAGATTCCACACAGGACTCCAGGGGCTGTGCCCGGT/CGTAACCGGTCATGAGGACGGCAAAGCT for the amplification of the EMR2 NTF. Finally, the hybrid receptors vE-L and vE-Le were made exactly as vE-Lm and vE-Lem, but the PCR fragments were inserted into the respective sites of wild-type LPH not containing C-terminal epitopes (4, 22).
Cell Culture and Transfection—A neuroblastoma cell line (NB2a) was cultured as described before (14). Stable cell lines were generated using Escort III (Sigma)-aided transfection, Geneticin (Invitrogen) selection, and cell sorting (BD Biosciences).
Confocal Immunofluorescent Microscopy—All images were captured and processed using a laser-scanning confocal fluorescent microscope (LSM510/Axioplan 2, Zeiss). The following configurations were used in double-staining experiments: laser excitation, 488 and 543 nm; emission filters, 505-530 and >560 nm. For triple-staining experiments, the settings were as follows: laser excitation, 488, 543, and 633 nm; emission filters, 505-530, 560-615, and >650 nm. Antibodies used were: primary, rabbit RL-1 antibody, anti-Myc or anti-V5 mouse mAbs; secondary, goat-anti-rabbit or anti-mouse IgG labeled with Alexa Fluor-488 or -594 (Invitrogen), as indicated in the figure legends.
Antibody/LTX Patching of NTFs and CTFs on Live Cells—Transfected cells, grown for 48 h on poly-d-lysine-coated glass coverslips, were treated by one of the following methods. Method 1 (control): cells were incubated for 20 min at 0 °C, fixed with 4% paraformaldehyde for 10 min, stained with the RL-1 primary and anti-rabbit secondary antibodies, then permeabilized for 7 min with 0.1% Triton X-100 and stained with the anti-Myc primary and anti-mouse secondary antibodies. Method 2 (patching): live cells were processed at 0 °C by (a) incubating with the RL1 primary (30 min) and anti-rabbit secondary (20 min) antibodies, then fixing, permeabilizing, and staining for the CTF using anti-Myc mAbs, or (b) pre-treating cells with fluorescently labeled LTXN4C for 20 min, fixing, and immunostaining cells as in (a). Fluorescent LTXN4C was produced as in a previous study (14).
To quantify the NTF/CTF co-localization in the plasma membrane, 0.7-μm-thick confocal images were obtained near the cell's equator, where the membrane was perpendicular to the focal plane; then the distribution of each receptor fragment along the cell's perimeter was determined and Pearson's correlation coefficient r was calculated for the two resulting profiles, using,
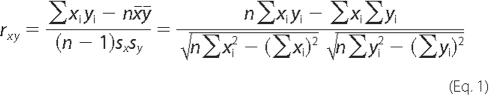
where 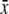 and
and
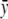 are the means of x and
y (second derivatives of the NTF and CTF fluorescence profiles,
respectively), sx and sy are the
standard deviations of x and y.
are the means of x and
y (second derivatives of the NTF and CTF fluorescence profiles,
respectively), sx and sy are the
standard deviations of x and y.
Measurements of Intracellular Ca2+—Stably transfected NB2a cells, starved of serum for 24 h, were loaded with Fluo-4 acetomethoxy ester (Invitrogen) using the manufacturer's protocol and equilibrated in buffer (145 mm NaCl, 5.6 mm KCl, 5.6 mm glucose, 1 mm MgCl2, 15 mm HEPES, 0.5 mg/ml bovine serum albumin, pH 7.4). To identify EMR2-expressing cells, samples were stained using the anti-V5 mAb labeled with Alexa Fluor-568 (Zenon labeling system, Invitrogen). 2 nm LTXN4C conjugated with Alexa Fluor-647 (Invitrogen) was added to the cells in the absence of Ca2+ and incubated for 15 min. Fields of view containing large numbers of transfected cells (red and/or blue) were identified using the LSM510 confocal microscope and a water-dipping objective (Achroplan, 40×, Zeiss), as specified for triple-staining above. Images were then acquired every 5 s, using 488 nm excitation and 505-550 nm emission filters. The initial baseline was recorded for 5 min, followed by the addition of 2 mm Ca2+ and further imaging for 60 min. Above-baseline Ca2+ fluorescence in individual cells was quantified using the LSM510 software.
RESULTS
Design and Nomenclature of Chimerical Receptors—To detect cross-interactions between the NTF and CTF synthesized as parts of different molecules and to avoid interference from any remaining non-cleaved receptors, new fusion constructs were designed based on the following main principles. First, homotypic fragments produced by the cleavage of different constructs must be immunologically distinct. Second, the hybrid proteins must possess only one of the two normal receptor functions, agonist binding or intracellular signaling, so that cross-reassembly of fragments can be detected as a result of functional complementation. Third, oligomerization of homotypic fragments originating from different constructs should be avoided.
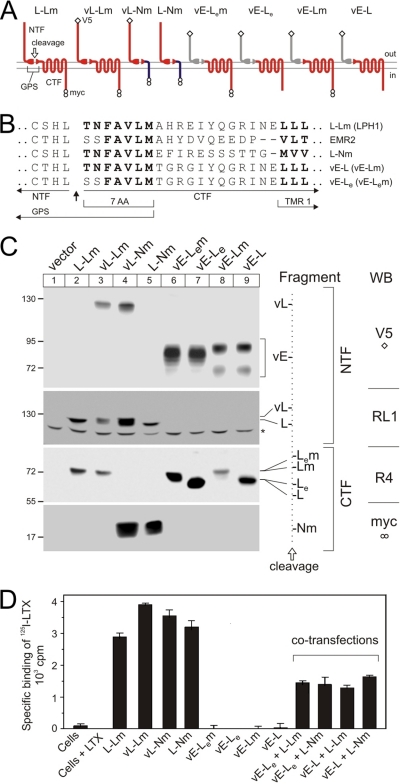
These goals have been achieved by creating several chimerical constructs (color-coded in Fig. 1A). All the fusion proteins contained an NTF and a CTF. The NTFs used were from either latrophilin (indicated in the construct name by L- and colored red in the diagrams) or EMR2 (E-, shaded gray). The choice of NTFs was dictated by the low sequence similarity between the NTFs of latrophilin and EMR2, and the fact that EMR2 is not found in the brain and, thus, is unlikely to interact normally with latrophilin. Similarly, two types of CTF were employed: one was from latrophilin (-L, red in diagrams) and the other was created by fusing the N-terminal seven amino acids (7AA) of the latrophilin CTF with the single TMR and cytosolic tail of neurexin I (-N, dark blue). We used the neurexin C terminus because of its inability to bind the CTF of latrophilin (14) or G-proteins (23). In agreement with this hybrid structure of the constructs, their nomenclature consisted of two letters corresponding to the type of NTF and CTF combined in a fusion protein, with the dash indicating the cleavage site.
FIGURE 1.
Design and expression of chimerical constructs. A, a diagram of the structural features and membrane topography of the chimerical constructs employed in this work. Letters and colors in the diagrams identify protein sequences from latrophilin 1 (L, red), α-neurexin (N, dark blue), or EMR2 (E, gray) (see also text). Split oval, cleaved GPS domain, whose C-terminal portion (seven amino acids, 7AA) can be from latrophilin (red) or EMR2 (gray, subscript e). Open diamond, V5 epitope; two circles, double-Myc epitopes. B, amino acid structures of the fusion proteins near the cleavage site. Dashes have been introduced for better alignment of the transmembrane domains (TMR1). C, Western blotting (WB) of NB2a cells stably expressing the fusion proteins as shown above. Antibodies used are specified on the right-hand side of the panel and described under “Materials and Methods.” Cells were produced and selected, grown in serum-free medium for 48 h, harvested, and extracted with Triton X-100 (see “Materials and Methods”). Extracts were separated in four equally loaded 6-12% SDS-polyacrylamide gels, transferred onto a polyvinylidene difluoride membrane, and the resulting blots were stained with antibodies indicated on the far right. The positions and molecular masses of protein markers are shown on the left. The identities of the fragments are explained on the right. Dotted line, cleavage position, separating the NTFs (left) and CTFs (right). Asterisk, a protein band stained nonspecifically with RL1. D, specific binding of LTX by NB2a cells stably expressing individual or co-transfected constructs (as specified below). Cells, prepared as in C, were incubated with 1 nm 125I-LTX and washed by centrifugation, and bound radioactivity was determined in a γ-counter. Nonspecific binding was measured in the presence of 100 nm LTX (Cells+LTX) and subtracted from the total binding values.
To enable post-translational cleavage, each fusion protein contained a GPS domain (Fig. 1A). After proteolysis, its N-terminal portion formed part of the respective NTF, whereas the C-terminal 7AA included in the CTF were either from latrophilin (TNFAVLM) or EMR2 (SSFAVLM) (Fig. 1B). The latter variant was indicated by a subscript letter “e” in the name of the CTF (color-coded as in Fig. 1A). For specific immunodetection, NTFs of some constructs were supplemented with an N-terminal V5 epitope, whereas some CTFs had two C-terminal Myc epitopes (indicated in construct names by v or m, respectively). Finally, for simplicity, the NTF and CTF originating from the same protein are called here homogeneric, whereas fragments produced by the cleavage of distinct protein constructs are termed heterogeneric. NTFs and CTFs are referred to as complementary.
The resulting hybrid proteins of the L-L type (Fig. 1A) essentially represented epitope-tagged latrophilin and were previously shown to bind LTX (22) and induce G-protein-mediated signaling (14). Constructs of the L-N type, also described previously (23), were designed to only bind LTX but not G-proteins. In contrast, the E-L or E-Le hybrids were meant to lack LTX binding but have the ability to signal via G-proteins.
Expression of Chimerical Receptors—When stably expressed in NB2a cells, all the fusion proteins were efficiently cleaved at their GPS domains to produce the respective NTFs and CTFs (Fig. 1C). No full-size proteins were normally detected. All the NTFs were glycosylated (data not shown).
It was interesting to compare E-L and E-Le (Fig. 1B). Surprisingly, their NTFs were differentially glycosylated: E-Le type gave rise to only one NTF band, while the E-L type produced two unequally modified variants of NTF (Fig. 1C, lanes 6 and 7, and 8 and 9, respectively). Given the identical amino acid sequences of the two NTFs, the only reason for their distinct glycosylation could be the two-residue difference in their CTFs (Fig. 1B). This result indicates that the CTF affects the glycosylation of the NTF before or after cleavage. In the latter case, the fragments must interact during NTF processing.
All the receptors were efficiently delivered to the cell surface (supplemental Fig. S1). Here, their ability to bind LTX fully corresponded to construct design. All receptors containing the NTF from latrophilin (L-L and L-N types) demonstrated high affinity for LTX (Fig. 1D). By contrast, the fusion proteins possessing the NTF from EMR2 (E-L and E-Le types) were unable to bind this agonist (Fig. 1D). Thus, the constructs described above conformed to the initial requirements and could be used to study any cross-interaction between the NTFs and CTFs of different fusion proteins.
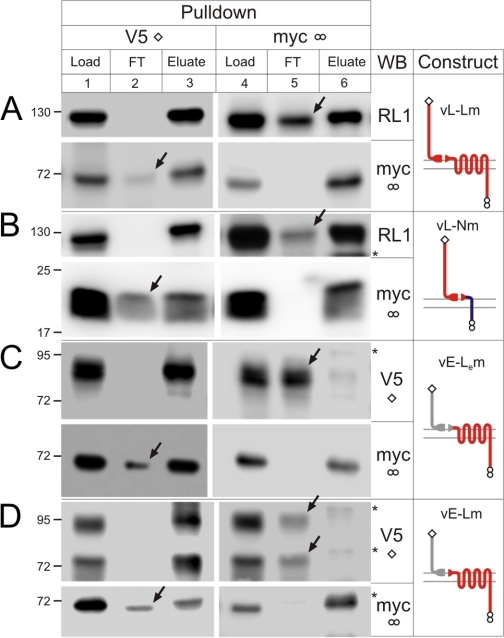
Interaction of Homogeneric Fragments—We first tested whether the complementary fragments from each individual protein were able to interact with each other. This was done by immunoprecipitation of detergent-solubilized cells expressing individual constructs, using antibodies against respective epitope tags.
As shown previously (5, 13, 14, 20), complementary homogeneric fragments could be co-immunoprecipitated when pulled down by either fragment (Fig. 2, Eluates). Interestingly, the NTFs of vE-Lem and vE-Lm brought down their respective CTFs equally well (Fig. 2, C and D, Eluate), despite the fact that their NTFs and CTFs corresponded to two structurally different parental receptors (EMR2 and latrophilin, respectively). This result indicated that the association of homogeneric NTFs and CTFs was not strictly sequence-specific, and, consequently, cross-interaction of heterogeneric fragments was possible.
FIGURE 2.
Co-immunoprecipitation of homogeneric fragments. Cells expressing individual constructs vL-Lm (A), vL-Nm (B), vE-Lem(C), or vE-Lm (D), were extracted with Triton X-100, and proteins were immunoprecipitated (as described under “Materials and Methods”), using the anti-V5 or anti-Myc mAbs (as indicated above). The initial extracts (Load), material not bound to affinity columns (flow through, FT), and the SDS-eluates (Eluate) from the columns were equally loaded onto SDS-gels and analyzed by WB as described in Fig. 1, using the antibodies indicated on the right. The positions and molecular masses of protein markers are shown on the left. Asterisks, protein bands stained nonspecifically. Arrows point to fragments incompletely precipitated with their complementary fragments and appearing in FT. The images shown are representative of n = 4-6 independent experiments.
These experiments also shed light on the possibility of cross-interaction of heterogeneric fragments. Any cross-interaction could theoretically occur only if some fragments were not engaged with their homogeneric partners. In fact, exhaustive pulldown of any fragment never resulted in a complete precipitation of its complementary counterpart (indicated by arrows in Fig. 2). The amount of co-precipitation varied among the constructs and was generally larger when the complexes were isolated via their NTFs (V5 pulldown) compared with CTFs (myc pulldown). The highest proportion of CTF precipitated (∼85%) was detected when vL-Lm was pulled down by its NTF (Fig. 2A, lane 3). For other constructs, 66-77% of CTFs came down with their NTFs (Fig. 2, B-D, lane 3). On the other hand, only a small fraction of the NTF from E-L type constructs (8-12%) precipitated with the respective CTF (Fig. 2, C and D, lane 6), whereas for L-L and L-N proteins, ∼56-73% of NTF was pulled down via the CTF (Fig. 2, A and B, lane 6).
These findings, which fully agreed with the previous observations for latrophilin (14) and GPR56 (supplemental Fig. 6C in Ref. 27), meant that each homogeneric fragment remained partially free from its counterpart and, therefore, was potentially available for cross-interaction with complementary heterogeneric fragments.
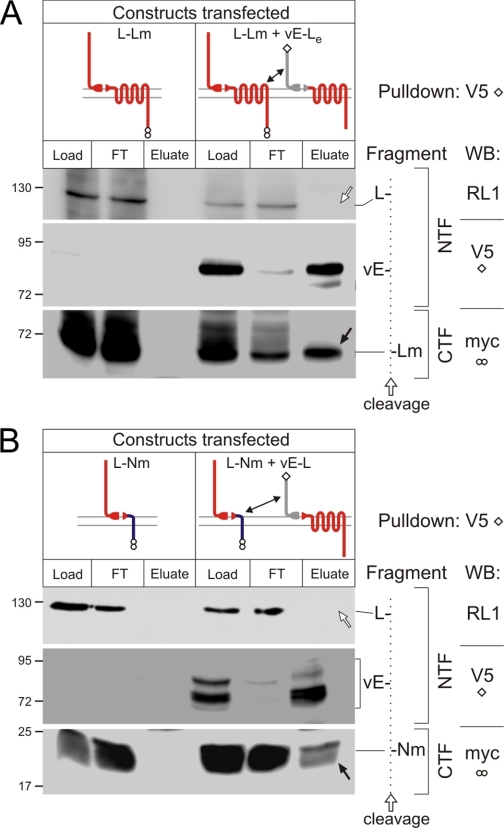
Cross-immunoprecipitation of Complementary Heterogeneric Fragments—We showed previously (Fig. 8B in Ref. 14) that the epitope-tagged fragments of vL-N and L-Nm constructs could be cross-precipitated, but only if they were co-expressed in the same cell and not if they were expressed separately and then solubilized and mixed. This was interpreted as evidence of the fragments' ability to interact on the cell surface and their adoption of a “closed” conformation on solubilization (14). Similarly, in our current work, when construct vE-Le was transfected into NB2a cells stably expressing the L-Lm receptor, the -Lm CTF could be specifically co-precipitated with the vE-NTF using anti-V5 antibodies (Fig. 3A, arrow).
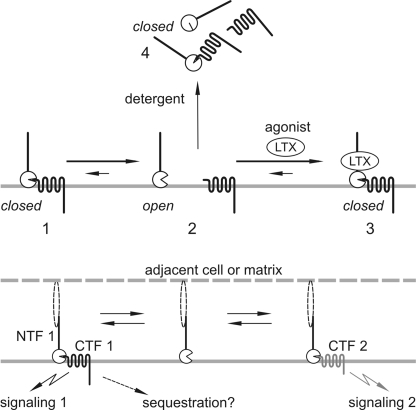
FIGURE 8.
NTF and CTF dynamics on the cell surface. Upper, relationships of homogeneric fragments. The newly made receptor is cleaved and delivered to the surface, where the fragments may form complexes (1) or stay largely independent (2). Upon agonist binding to the NTF, the equilibrium is shifted toward formation of ternary complexes (3). Solubilization of the membrane causes changes in the NTF structure that preclude further interactions; as the NTFs assume the “closed” conformation, they trap a certain amount of the CTFs; the binding is strong but sensitive to protein denaturation (4). Lower, hypothetical promiscuous interactions of the NTF from aGPCR 1 with the CTFs from aGPCR 1 and 2, leading to distinct signaling events 1 and 2.
FIGURE 3.
Cross-immunoprecipitation of co-expressed heterogeneric fragments. A and B, NB2a cells, expressing individual or co-transfected constructs (as indicated schematically at the top), were processed and immunoprecipitated using anti-V5 mAbs as described in Fig. 2, then analyzed by WB as specified on the far right. Fragment identities are explained on the right. Dotted line, cleavage position, separating the NTFs (left) and CTFs (right). The positions and molecular masses of protein markers are shown on the left. Filled arrows indicate fragments cross-precipitated with their complementary heterogeneric fragments; open arrows, lack of nonspecific precipitation of the NTF of latrophilin. A, cross-precipitation of heterogenerically expressed NTF of EMR2 and CTF of latrophilin. B, cross-precipitation of heterogenerically expressed NTF of EMR2 and the chimerical CTF of latrophilin/neurexin. Cross-interactions of the complementary heterogeneric fragments are shown by double arrows in the schemes above. Experiments in A and B were repeated four times.
However, the two constructs employed in this experiment had identical CTFs, differing only by the presence of epitopes. Hypothetically, oligomerization of -Le with -Lm in the cell membrane could lead to their co-precipitation without direct cross-interaction between vE- and -Lm. On the other hand, if this were the case, the untagged NTF (L-) should have also co-precipitated with the complex, which clearly did not occur (open arrow in Fig. 3A).
Nevertheless, to avoid any uncertainty, we then co-expressed two fusion proteins that had distinct N and C termini: cells stably expressing construct L-Nm were transfected with the vE-L receptor (Fig. 3B). Again, immunoprecipitation of vE-led to specific co-purification of the heterogeneric -Nm (arrow in Fig. 3B) but not the untagged L- (Fig. 3B, open arrow).
LTX Binds to Cross-complexes of Complementary Heterogeneric Fragments—The experiments described above dealt with the interaction of the NTF from EMR2 with the CTF from latrophilin (expressed either as homo- or heterogeneric fragments). The absence of a known ligand of EMR2 did not allow us to test whether such complexes were able to signal.
Fortunately, an exogenous agonist, LTXN4C, is available for latrophilin. This mutant toxin, which does not form membrane pores, has been shown to bind the NTF of latrophilin and induce its interaction with the homogeneric CTF, leading to intracellular Ca2+ signaling (14, 25).
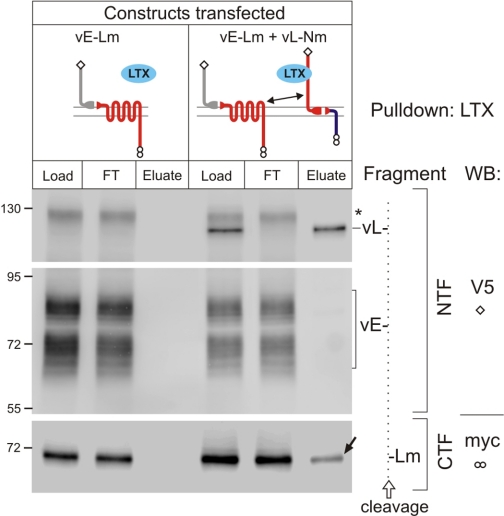
To verify that LTXN4C was also able to bind complexes of heterogeneric fragments, we co-expressed vE-Lm with vL-Nm in NB2a cells and carried out affinity chromatography of detergent-solubilized cells on a LTX column. As demonstrated in Fig. 4, toxin column exhausted the NTF of latrophilin (vL-) from the cell lysate, but also specifically isolated a substantial amount of the latrophilin CTF (-Lm) (arrow in Fig. 4). Importantly, the constructs used in this experiment contained four non-homologous fragments (vE-, vL-, -Lm, and -Nm). This excluded any possibility of the CTF of latrophilin being co-purified as part of an oligomeric complex (i.e. without cross-interaction).
FIGURE 4.
Cross-complexes of heterogeneric fragments isolated by LTX-affinity chromatography. Cells, expressing individual or co-transfected constructs (as indicated schematically at the top), were grown and processed as in Fig. 2, then passed through a LTX (blue oval) column and eluted with SDS. All fractions were equally loaded on SDS-gels and Western blotted as indicated on the far right. Fragment identities are explained on the right. Dotted line, schematic cleavage, separating the NTFs (left) and CTF (right). The positions and molecular masses of protein markers are shown on the left of the blots. Asterisk, uncleaved vE-Lm protein. The filled arrow marks the CTF of latrophilin co-purified with the heterogeneric NTF of latrophilin. The data are representative of n = 3 experiments.
Interestingly, in this experiment, some amount of the non-cleaved full-size vE-Lm was present (asterisk in Fig. 4). However, this did not bind to the toxin column, further proving that (i) neither the NTF of EMR2 (vE-) nor the CTF of latrophilin (-Lm) interact with LTX and (ii) molecular complexes of cleaved vL- and -Lm were formed. This result indicated that the fragments of latrophilin, synthesized as parts of different proteins, formed true cross-complexes, which were able to bind LTX.
LTX Induces Formation of Cross-complexes of Heterogeneric Fragments on the Cell Surface—Both in solution and on the cell surface, LTX is known to interact only with the NTF (12, 14), but not the CTF (14), of latrophilin (see also Figs. 1D and 4). Nevertheless, the toxin has been shown to bring together the homogeneric NTF and CTF, forming large patches on the cell surface that contain both fragments (14). However, in our previous work, such fragments were produced by the cleavage of the same parental protein and, thus, were not completely independent. In particular, some amount of the NTF and CTF might not have dissociated after cleavage. These complexes could theoretically serve as kernels promoting formation of larger aggregates.
Therefore, we designed an experiment to test the ability of LTX to cause the association of the truly independent fragments. To achieve this, two hybrid constructs, L-N and vE-Lm, were co-expressed in NB2a cells (Fig. 5A). In this pair, the NTF and CTF of latrophilin were synthesized as parts of different proteins and clearly had to be independent, at least immediately after cleavage. Their potential association on the cell surface was tested by the well known “antibody patching” technique (14). In this method, clumping of one protein on the cell surface is induced using antibodies or other ligands, and any changes in the distribution of a second protein indicate an interaction of the two proteins of interest.
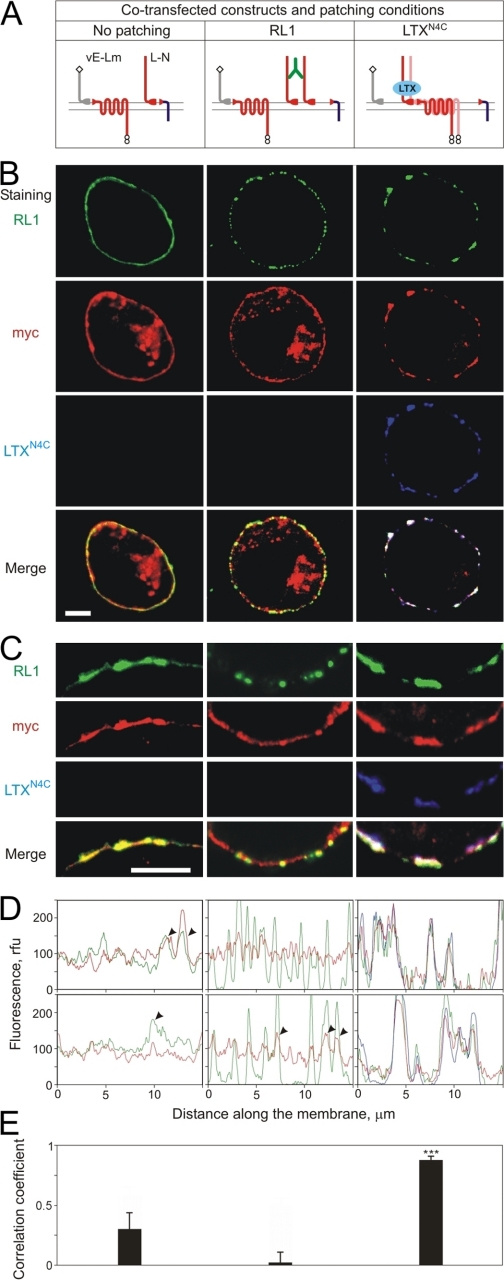
FIGURE 5.
Agonist, but not antibodies, induces parallel patching of heterogeneric fragments of latrophilin on the cell surface. A, a diagram of co-transfected constructs and patching conditions. B, NB2a cells, expressing co-transfected constructs (as indicated), were processed as described under “Materials and Methods.” The NTF of latrophilin was stained with the RL1 antibody (green) either after fixation (control, left column) or after respective patching (middle and right columns). LTXN4C used for agonist-induced patching was fluorescently labeled (blue). After patching, the cells were fixed, permeabilized, and counterstained for the CTF of latrophilin (red). Scale bar, 10 μm. C, higher magnification of cells, as in B. Scale bar, 5 μm. D, representative profiles of NTF and CTF fluorescence intensity distribution in respective cells (see “Materials and Methods”). Line color corresponds to the color of images in B and C. Rfu, relative fluorescence units. E, correlation coefficients for NTF and CTF distribution profiles (as in D) in respective cells; n = 3. Note the substantial increase in correlation of the NTF and CTF profiles after agonist patching, and low correlation of the NTF and CTF profiles after antibody patching. Each experiment was conducted n = 3 times, and 6-9 images were quantified for each condition.
In untreated control cells, both heterogeneric fragments of latrophilin were randomly distributed in the plasma membrane (Fig. 5, B-D, left panels). Because NTF and CTF of latrophilin had been produced by the cleavage of two different proteins, these fragments were co-localized only generally, and their intensity distribution profiles showed poor similarity (Fig. 5, D and E). On the other hand, some areas of the membrane (arrowheads in Fig. 5D) infrequently demonstrated an increased co-distribution of L- and -Lm. Was this a reflection of some interaction between these heterogeneric fragments?
To assess any possible molecular interactions between heterogeneric L- and -Lm, the NTF of latrophilin (L-) was subjected to cell-surface patching using the polyclonal anti-NTF antibody. This led to formation of distinct patches of green fluorescence on the membrane (Fig. 5, B-D, central panels). In most cell-surface areas (Fig. 5D, top), these green patches were not accompanied by any substantial changes in the distribution of the CTF of latrophilin (red). Accordingly, the correlation of the L- and -Lm intensity distribution profiles strongly deteriorated (Fig. 5, D and E). However, in some loci on the cell surface, 10-20% of the red fluorescence signal did appear to shift and form small peaks coinciding with the large peaks of green fluorescence (Fig. 5D, bottom). This confirmed the ability of heterogeneric complementary fragments of latrophilin to interact on the cell surface, at least to some extent.
We previously showed that the agonist of latrophilin, LTXN4C, can substantially increase the interaction of homogeneric fragments (14). In our current experiments, too, LTXN4C (blue) not only induced the patching of NTF of latrophilin (L-, green), but also caused a dramatic redistribution of the CTF of latrophilin (-Lm, red) (Fig. 5, B-D, right panels), forming large aggregates with the two heterogeneric fragments. Note that, with this treatment, almost all red fluorescence signal shifted to the patches, creating substantial areas of the membrane essentially devoid of both fragment (Fig. 5, B-D, right). These parallel changes in the localization of the heterogeneric L- and -Lm significantly increased the correlation of their distribution profiles (Fig. 5, D and E). These results unequivocally demonstrate not only that the NTF and CTF of latrophilin can interact even when they are produced by the cleavage of different proteins, but also that their association is stimulated by agonist binding to the NTF.
Cross-interacting Complementary Heterogeneric Fragments Mediate Agonist-induced Signaling—Were these complexes of LTXN4C and the heterogeneric NTF and CTF functional? We previously found that the interaction of LTXN4C with the NTF of latrophilin caused intracellular signaling that was mediated by the homogeneric CTF and, in the presence of extracellular Ca2+, led to an increase in the cytosolic Ca2+ concentration (14). We used the same approach here, except that latrophilin fragments in the new experiment were heterogeneric.
NB2a cells stably expressing the L-Nm and vE-Le constructs were selected for doubly positive cells by cell sorting. A clone stably transfected with latrophilin proper (L-Lm) served as a positive reference (14). Cells loaded with the fluorescent Ca2+ indicator Fluo-4 were pre-stained with fluorescent LTXN4C (blue), to detect the NTF of latrophilin (L-), and Zenon-labeled anti-V5 mAb (red), to identify those cells expressing the NTF of EMR2 (vE-) (see supplemental Figs. S2 and S3). This was necessary to find fields of view containing large numbers of receptor-expressing cells. Then all cells were synchronously stimulated by the addition of Ca2+ to the medium. Changes in Ca2+ fluorescence were recorded only from the cells stained positively for the respective receptor.
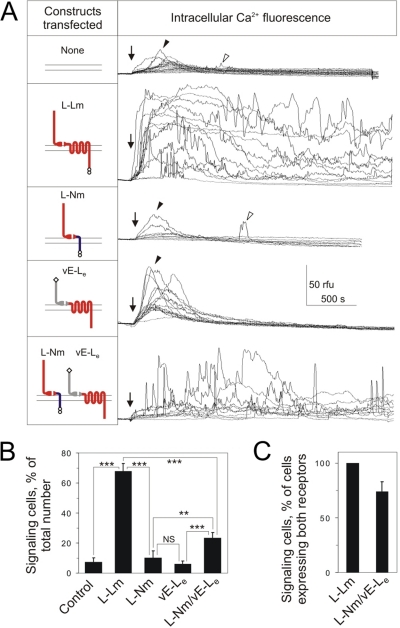
Fig. 6A demonstrates that, as expected, all cells remained quiescent on binding LTXN4C inaCa2+-free medium. Addition of extracellular Ca2+ (arrows in Fig. 6A) caused a transient increase in the cytosolic calcium concentration (filled arrowheads). In the vast majority of non-transfected control cells (Fig. 6A, top) and cells expressing L-Nm and vE-Le individually, this Ca2+ wave was followed by a quiet period, when the intracellular fluorescence did not deviate from the baseline. Only sporadically some of these cells displayed short, non-oscillating increases in cytosolic Ca2+ label (open arrowheads in Fig. 6A), apparently representing background signaling. In contrast, a large proportion of the positive control cells (L-Lm) showed massive intracellular Ca2+ signaling of an oscillatory type (Fig. 6A, second trace), which continued in most cells for the duration of the experiment. Importantly, the cells co-expressing the two hybrid receptors also demonstrated clear Ca2+ signaling (Fig. 6A, bottom trace), which resembled that of the positive control cells, being only smaller in amplitude.
FIGURE 6.
Functional activity of cross-complexes formed by heterogeneric latrophilin fragments. A, time course of intracellular Ca2+ fluorescence in individual NB2a cells, non-transfected or transfected with individual or double constructs (as shown on the left), preincubated with 2.5 nm LTXN4C and stimulated by 2 mm Ca2+ (arrows). Rfu, relative fluorescence units. Filled arrowheads, initial increase in intracellular [Ca2+]; open arrowheads, non-oscillatory Ca2+ spikes counted as signaling events. B, percentage of cells expressing one or both receptor constructs and demonstrating Ca2+ signaling in response to LTXN4C. Data are the means ± S.E.; **, p < 0.01; ***, p < 0.001; NS, non-significant (n = 4; the total number of cells recorded was: untransfected cells, 125; L-Nm cells, 337; vE-Le cells, 215; L-Lm cells, 195; co-transfected L-Nm+vE-Le cells, 461). C, percentage of cells co-expressing both receptors and demonstrating Ca2+ signaling in response to LTXN4C; data from B; number of signaling L-Lm cells taken as 100%.
The number of signaling cells was quantified in each stimulated culture. In concert with our previous data (14), prolonged signaling was detected in 67.7 ± 5.4% of the cells carrying latrophilin proper (L-Lm). Using the mildest quantification criteria (which counted even sporadic individual Ca2+ spikes, rarely occurring in control cells, as signaling events), the cells expressing the vE-Le fusion protein did not differ from non-transfected cells. Only 6.1 ± 2.1% of vE-Le and 7.2 ± 3% of control cells showed any signs of Ca2+ signaling (Fig. 6B). This was expected, because these cells did not bind LTX (see Fig. 1D). On the other hand, L-Nm cells, which bound large amounts of LTX (Fig. 1D), signaled only slightly and not significantly more than control cells (10.1 ± 4.6%) (Fig. 6), clearly indicating that the effect was only mediated by the CTF of latrophilin but not by the C terminus of neurexin. By contrast, 23.4 ± 3.5% of the co-transfected L-Nm/vE-Le cells demonstrated statistically significant, prolonged, and oscillatory Ca2+ signaling.
Did the stronger signaling in L-Lm cells (compared with cells co-expressing vE-Le and L-Nm) indicate preferential functional association of homogeneric, rather than heterogeneric, latrophilin fragments? Obviously, for quantitative comparison, cells expressing equal amounts of each fragment should be used. However, pre-staining of the doubly transfected cultures (as described above) demonstrated that most individual cells displayed different ratios of L-Nm and vE-Le on their surface. Furthermore, many cells contained only one type of receptor (supplemental Fig. S3), making signal transduction in such cells altogether impossible. To compensate for the presence of signaling-incapable cells, we used the LTXN4C and anti-V5 stained images (as in supplemental Figs. S2 and S3) to identify the number of cells expressing both receptors (not necessarily in equal proportions). The data in Fig. 6B were then normalized to the number of cells in which the staining for both NTFs was above the background. This figure (46.7% of co-transfected cells) was used to recalculate the percent of signaling cells among those co-transfected with the two constructs. As shown in Fig. 6C, the recalculated signaling ability of L-Nm/vE-Le cells constituted as much as 74% of signaling capacity of latrophilin-expressing cells.
Cross-immunoprecipitation of aGPCRs from Native Tissues—The data above prove convincingly that, in this model system, not only the fragments of unrelated aGPCRs interact, but also that their cross-complexes are functionally active. However, these effects could be due to overexpression or unnatural co-existence of chimerical constructs in the cultured cells. If distinct aGPCRs were much less abundant in vivo and not normally expressed in the same cell, then our findings would not be relevant for normal animal physiology.
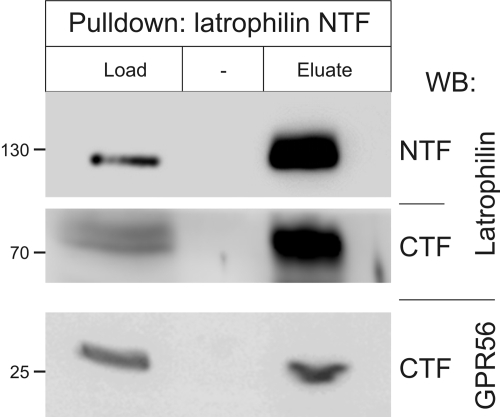
Overexpression was an unlikely reason for the functionality of the cross-complexes, because we only observed signaling (as in Fig. 6A) when receptor fragments were expressed at low levels and never when the constructs were over-abundant (individual traces in Fig. 6A). Nevertheless, we decided to test our theory in normal animal tissues. For this purpose, we studied the cross-interaction of latrophilin with GPR56, another aGPCR expressed in the brain (28). The NTF of latrophilin from solubilized rat brain was immunoprecipitated using the specific N-terminal antibody (Fig. 7, top). As expected, this led to co-precipitation of its homogeneric CTF (Fig. 7, middle). Importantly, the heterogeneric CTF of GPR56 was also clearly cross-precipitated (Fig. 7, bottom).
FIGURE 7.
Cross-immunoprecipitation of heterogeneric fragments from animal tissues. Homogenized rat forebrain was solubilized and immunoprecipitated using the anti-latrophilin NTF antibody (see “Materials and Methods”). Equal volumes of the starting material (Load) and immunoprecipitated sample (Eluate) were analyzed by WB (as in Fig. 1), using antibodies (as indicated on the right): against the NTF of latrophilin, the CTF of latrophilin and the CTF of GPR56 (n = 2). The positions and molecular masses of protein markers are shown on the left.
It is interesting that, unlike in experiments with cultured cells (Fig. 2), precipitation of NTF from the brain brought down only a small proportion of CTF of latrophilin (Fig. 7). If we take the efficiency of NTF precipitation as 100%, then only ∼14 ± 2.6% of CTF was pulled down, indicating that in the brain latrophilin fragments are mostly free from each other. Importantly, the efficiency of cross-precipitation of GPR56 CTF was relatively high (3.5 ± 0.4%), suggesting that in vivo ∼20% of all NTF·CTF complexes of latrophilin may contain the CTF of a distinct aGPCR.
DISCUSSION
“Split Personality” Receptors—We previously demonstrated an independent behavior of the fragments resulting from the cleavage of one aGPCR molecule (14). On the cell surface, such homogeneric fragments seem to be essentially free from each other, being independently distributed, patched, internalized, and solubilized (14). The proportion of truly associated, rather than simply co-localized, fragments is difficult to assess in overexpressing cells. On the other hand, more quantifiable methods, such as immunoprecipitation and affinity chromatography, demonstrate a higher degree of fragment association than cell-surface staining (14, 20). Nevertheless, substantial amounts of free fragments are still found in detergent solutions (Fig. 2). Considering that detergent appears to increase the degree of reassembly of the NTF and CTF (14),4 the actual proportion of free fragments in the cell membrane is probably much higher than that indicated by immunoprecipitation. This is further corroborated by our current experiments in normal tissues, where the co-immunoprecipitation of latrophilin fragments is much poorer (Fig. 7) than in overexpressing cell cultures (Fig. 2).
Interestingly, in immunoprecipitation assays, the proportion of NTF pulled down via CTF was substantially lower than the proportion of CTF precipitated with NTF (Fig. 2) (27). This can be explained by two possibilities: (i) the NTF-CTF interaction may depend on which fragment is bound to the substrate (immunoprecipitation matrix) or (ii) the NTF is much more abundant in the plasma membrane than the CTF. The former possibility seems to be less likely, because solubilized fragments usually lose their ability to interact even if they are produced homogenerically (Ref. 14 and data not shown). On the other hand, the CTF seems to be internalized much more readily than NTF in NB2a cells stably expressing latrophilin (14). Thus, the amount of NTF on the cell surface is likely to exceed that of the CTF. This peculiar observation strongly supports the independence of fragments and indirectly substantiates the idea of NTF being itself anchored in the membrane.
This independence of fragments must, obviously, have serious implications for ligand binding and signaling. Ligand-binding characteristics of the NTF change when it becomes separated from its CTF.5 This means that only a small proportion of the cell-surface NTF (that bound to the CTF) may be in the right conformation for any productive interaction with the endogenous ligand. The same may be true of the CTF, which may have its own ligands/agonists (as proposed in Ref. 14). Reciprocally, the signaling capacity of the CTF may be affected by complex formation with the NTF, whether with or without a bound ligand.
It might be surprising that, although endogenous ligands are known for a range of aGPCRs (27, 29-31, 31-33), no signaling has been linked to the binding of any endogenous ligand. Our findings, which postulate that the fragments of aGPCRs behave as independent proteins (Figs. 2 and 5), may explain some of the difficulties in identifying such intracellular signaling pathways.
Attraction of the Fragments—Nevertheless, the independence of the fragments is only partial or temporary, further increasing the complexity of this group of receptors. NTFs and CTFs are known to form strong complexes, and these can be readily isolated from detergent solutions (5, 8, 14). Likewise, fragments of soluble truncation mutants, which lack all transmembrane domains but possess the GPS region, can also be co-precipitated (13), albeit not quantitatively (14).
Interestingly, any free fragments remaining after membrane solubilization lose their ability to re-associate (14) (Fig. 2). Moreover, the NTF, solubilized separately from the CTF using a mild detergent perfluoro-octanoic acid, can no longer bind the CTF (14). Thus, it appears that the removal of the membrane by solubilization (or secretion of a soluble construct) somehow changes the NTF structure, closing its CTF-binding site (schematically presented in Fig. 8, upper). If the NTF happens to be close to a homo- or heterogeneric CTF, an almost irreversible NTF·CTF complex is formed. The level of such solubilization-induced re-assembly depends on the proximity of fragments in the plasma membrane, being substantially higher in overexpressing cells (Fig. 2) than in native tissues (Fig. 7). Given the substantial independence of the fragments on the cell surface, we hypothesize that the membrane (or its component/s) helps to maintain the open conformation of the NTF.
Some complex formation must also occur on the cell surface, because, when LTXN4C binds the NTF of latrophilin, its signaling effect can only be mediated by the heptahelical CTF (14) (Figs. 5 and 6). It is thought that LTX enhances the sporadic NTF-CTF interaction and, owing to its own dimeric nature, cross-links multiple reconstituted NTF·CTF complexes, forming large cell-surface patches containing both fragments. This re-association results in signaling to Ca2+ stores (14) (Fig. 6).
Mutagenesis studies (13, 14, 21) have shown that the interaction between the NTF and CTF most likely involves the 7AA at the N terminus of the CTF (see Fig. 1B). These residues are also very important for the cleavage of the full-size receptor, because bulky amino acid substitutions in this region block proteolysis (13). On the other hand, replacing the 7AA in latrophilin (TNFAVLM) with SSFAVLM from EMR2 not only permits cleavage but also supports the interaction of the fragments (Figs. 2C, 3A, and 6A). Similarly, the CTF of GPR56, with the 7AA sequence TYFAVLM, binds efficiently to the NTF of latrophilin (Fig. 7). These results suggest a relatively low sequence specificity both of the proteolytic system and of the CTF-binding site on the NTF. The sequence downstream of the 7AA (Fig. 1B) does not seem to contribute to either cleavage or re-association and can be replaced with an unrelated or artificial sequence, as in the L-Nm and vE-Le constructs (Fig. 1B), extending the previously published data (13, 14, 21).
Promiscuity of the Fragments—Intriguingly, these on-off, low specificity interactions between the complementary fragments suggest that heterogeneric NTF and CTF could, from time to time, form hybrid complexes.
This possibility has been clearly demonstrated in our work. Previously, we showed that fragments produced by the cleavage of differently tagged latrophilin constructs can cross-interact (14). Here, we extended these observations to heterogeneric fragments, i.e. fragments produced by proteolysis of different hybrid proteins (Figs. 3, 4 and 5). Fragment swapping between the hybrid fusion proteins alleviated any artifacts that could be caused by any incomplete receptor cleavage or oligomerization of homotypic fragments (as described in Ref. 34). Our results demonstrate that heterogeneric fragments not only can be cross-precipitated from detergent extracts (Figs. 3 and 4) but also can form cross-complexes on the cell surface (Figs. 5 and 6). Judging from the antibody patching experiments (Fig. 5D, middle) and receptor complexes isolated from live tissues (Fig. 7), from 10 to 20% of the CTF·NTF complexes in the plasma membrane can be heterogeneric.
It can be argued that, in immunoprecipitation (Fig. 3), LTX-affinity chromatography (Fig. 4), and cell-surface patching (Fig. 5), the amounts of cross-interacting heterogeneric fragments were relatively small (∼10%). However, the co-transfected cells expressed different (sometimes vastly) amounts of the hybrid constructs (e.g. supplemental Fig. S3), producing unequal quantities of free heterogeneric fragments and, hence, suboptimal conditions for their cross-interaction. From this point of view, it is significant that in a native tissue the ratio of homogeneric to heterogeneric complexes was ∼4:1 (Fig. 7). Obviously, the actual ratio in individual cells in vivo may be even higher, depending on the comparative expression of different aGPCRs in a given cell type, as well as the relative localization and turnover of their fragments. Finally, the fact that the fragments of different aGPCRs could form hybrid complexes provided the strongest evidence for the independent behavior of NTFs and CTFs.
Complex Signaling Network—The cross-complexes of heterogeneric fragments are not simple nonspecific aggregates formed because of detergent solubilization or membrane clumping. They are functional molecular assemblies in the plasma membrane, capable of transducing an intracellular signal (Fig. 6). This reaction starts with the binding of LTXN4C to the NTF of one hybrid receptor but is subsequently mediated by the CTF of latrophilin introduced as a fragment of another hybrid receptor. The CTF of latrophilin must be critically involved in this response, because the signaling does not occur in the absence of this fragment (Fig. 6) and because it closely resembles the normal reaction of latrophilin to toxin binding. The natural latrophilin signaling involves a G-protein (most likely Gαq), phospholipase C, and inositol-trisphosphate, which leads to release of Ca2+ from intracellular stores (9, 14, 25).
Our results unequivocally demonstrate that (i) aGPCR fragments can behave as independent proteins, (ii) complementary fragments from distinct aGPCRs can cross-interact, and (iii) the re-assembled cross-complexes of complementary heterogeneric fragments are functionally active. The consequences of this dynamic architecture are far-reaching (Fig. 8, lower). First, aGPCRs illustrate the novel idea that proteins may not always be constant and indivisible entities (although they often consist of multiple subunits), but that they may represent dynamic populations of independent subunits capable of various recombinations.
Second, intermixing populations of subunits might induce diverse intracellular reactions. Although the signaling pathways of most aGPCRs are poorly understood, it is known that latrophilin and GPR56 are coupled to Gαq (10, 35), whereas latrophilin can also bind Gαo (4). In addition, like many “regular” GPCRs, aGPCRs may be able to directly interact with plasma membrane proteins, for example, ion channels. In fact, GRP56 has been shown to form complexes with four-transmembrane protein CD81 (35). Different members of this family are likely to be linked to distinct intracellular or cell-surface interactions. Because aGPCRs can exchange their fragments (Fig. 7), the homo- and heterogeneric complexes formed by these fragments would couple cell-surface interactions, mediated by different NTFs, to multiple signaling pathways, mediated by different CTFs present in the same cell. Thus, aGPCR may be able to participate in a complex network of intra- and extracellular interactions. This diversification of cellular response to cell-cell and cell-matrix interactions may represent one of the mechanisms of cells' adaptability to complex environments.
Supplementary Material
Acknowledgments
We are grateful to H.-H. Lin for the gift of the EMR2 cDNA.
This work was supported by project grants from The Wellcome Trust (Grant GR074359) and the Biotechnology and Biological Sciences Research Council (Grant BB/D523078) (to Y. U. and C. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: aGPCR, adhesion G-protein-coupled receptor; CIRL, Ca2+-independent receptor for α-latrotoxin; CTF, C-terminal fragment; E-, NTF of EMR2; FT, flow-through; GPCR, G-protein-coupled receptor; GPS, G-protein-coupled receptor proteolysis site; L-, NTF of latrophilin in chimerical constructs; -L, CTF of latrophilin in constructs; LTX, α-latrotoxin; m, Myc epitope; mAb, monoclonal antibody; -N, C terminus of neurexin in chimerical proteins; NTF, N-terminal fragment; v, V5 epitope; WB, Western blotting.
J.-P. Silva and Y. Ushkaryov, unpublished data.
J.-P. Silva, V. Lelianova, and Y. Ushkaryov, manuscript in preparation.
References
- 1.Fredriksson, R., Lagerstrom, M. C., Lundin, L. G., and Schioth, H. B. (2003) Mol. Pharmacol. 63 1256-1272 [DOI] [PubMed] [Google Scholar]
- 2.Bjarnadottir, T. K., Fredriksson, R., and Schioth, H. B. (2007) Cell Mol. Life Sci. 64 2104-2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davletov, B. A., Shamotienko, O. G., Lelianova, V. G., Grishin, E. V., and Ushkaryov, Y. A. (1996) J. Biol. Chem. 271 23239-23245 [DOI] [PubMed] [Google Scholar]
- 4.Lelianova, V. G., Davletov, B. A., Sterling, A., Rahman, M. A., Grishin, E. V., Totty, N. F., and Ushkaryov, Y. A. (1997) J. Biol. Chem. 272 21504-21508 [DOI] [PubMed] [Google Scholar]
- 5.Krasnoperov, V. G., Bittner, M. A., Beavis, R., Kuang, Y., Salnikow, K. V., Chepurny, O. G., Little, A. R., Plotnikov, A. N., Wu, D., Holz, R. W., and Petrenko, A. G. (1997) Neuron 18 925-937 [DOI] [PubMed] [Google Scholar]
- 6.Baud, V., Chissoe, S. L., Viegas-Pequignot, E., Diriong, S., N′Guyen, V. C., Roe, B. A., and Lipinski, M. (1995) Genomics 26 334-344 [DOI] [PubMed] [Google Scholar]
- 7.Hamann, J., Eichler, W., Hamann, D., Kerstens, H. M., Poddighe, P. J., Hoovers, J. M., Hartmann, E., Strauss, M., and van Lier, R. A. (1995) J. Immunol. 155 1942-1950 [PubMed] [Google Scholar]
- 8.Gray, J. X., Haino, M., Roth, M. J., Maguire, J. E., Jensen, P. N., Yarme, A., Stetler-Stevenson, M. A., Siebenlist, U., and Kelly, K. (1996) J. Immunol. 157 5438-5447 [PubMed] [Google Scholar]
- 9.Davletov, B. A., Meunier, F. A., Ashton, A. C., Matsushita, H., Hirst, W. D., Lelianova, V. G., Wilkin, G. P., Dolly, J. O., and Ushkaryov, Y. A. (1998) EMBO J. 17 3909-3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman, M. A., Ashton, A. C., Meunier, F. A., Davletov, B. A., Dolly, J. O., and Ushkaryov, Y. A. (1999) Philos. Trans. R. Soc. Lond. B Biol. Sci. 354 379-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian, Y. M., Haino, M., Kelly, K., and Song, W. C. (1999) Immunology 98 303-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasnoperov, V., Bittner, M. A., Holz, R. W., Chepurny, O., and Petrenko, A. G. (1999) J. Biol. Chem. 274 3590-3596 [DOI] [PubMed] [Google Scholar]
- 13.Krasnoperov, V., Lu, Y., Buryanovsky, L., Neubert, T. A., Ichtchenko, K., and Petrenko, A. G. (2002) J. Biol. Chem. 277 46518-46526 [DOI] [PubMed] [Google Scholar]
- 14.Volynski, K. E., Silva, J. P., Lelianova, V. G., Atiqur, R. M., Hopkins, C., and Ushkaryov, Y. A. (2004) EMBO J. 23 4423-4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayflick, J. S. (2000) J. Recept. Signal. Transduct. Res. 20 119-131 [DOI] [PubMed] [Google Scholar]
- 16.Lin, H. H., Chang, G. W., Davies, J. Q., Stacey, M., Harris, J., and Gordon, S. (2004) J. Biol. Chem. 279 31823-31832 [DOI] [PubMed] [Google Scholar]
- 17.Wei, W., Hackmann, K., Xu, H., Germino, G., and Qian, F. (2007) J. Biol. Chem. 282 21729-21737 [DOI] [PubMed] [Google Scholar]
- 18.Iguchi, T., Sakata, K., Yoshizaki, K., Tago, K., Mizuno, N., and Itoh, H. (2008) J. Biol. Chem. 283 14469-14478 [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y., Fan, J., Yang, J., and Zhu, G. Z. (2008) Mol. Cell Biochem. 308 133-139 [DOI] [PubMed] [Google Scholar]
- 20.Qian, F., Boletta, A., Bhunia, A. K., Xu, H., Liu, L., Ahrabi, A. K., Watnick, T. J., Zhou, F., and Germino, G. G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16981-16986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, G. W., Stacey, M., Kwakkenbos, M. J., Hamann, J., Gordon, S., and Lin, H. H. (2003) FEBS Lett. 547 145-150 [DOI] [PubMed] [Google Scholar]
- 22.Volynski, K. E., Meunier, F. A., Lelianova, V. G., Dudina, E. E., Volkova, T. M., Rahman, M. A., Manser, C., Grishin, E. V., Dolly, J. O., Ashley, R. H., and Ushkaryov, Y. A. (2000) J. Biol. Chem. 275 41175-41183 [DOI] [PubMed] [Google Scholar]
- 23.Volynski, K. E., Capogna, M., Ashton, A. C., Thomson, D., Orlova, E. V., Manser, C. F., Ribchester, R. R., and Ushkaryov, Y. A. (2003) J. Biol. Chem. 278 31058-31066 [DOI] [PubMed] [Google Scholar]
- 24.Ashton, A. C., Volynski, K. E., Lelianova, V. G., Orlova, E. V., Van, R. C., Canepari, M., Seagar, M., and Ushkaryov, Y. A. (2001) J. Biol. Chem. 276 44695-44703 [DOI] [PubMed] [Google Scholar]
- 25.Capogna, M., Volynski, K. E., Emptage, N. J., and Ushkaryov, Y. A. (2003) J. Neurosci. 23 4044-4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, H. H., Stacey, M., Hamann, J., Gordon, S., and McKnight, A. J. (2000) Genomics 67 188-200 [DOI] [PubMed] [Google Scholar]
- 27.Xu, L., Begum, S., Hearn, J. D., and Hynes, R. O. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9023-9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haitina, T., Olsson, F., Stephansson, O., Alsio, J., Roman, E., Ebendal, T., Schioth, H. B., and Fredriksson, R. (2008) BMC Neurosci. 9 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamann, J., Vogel, B., van Schijndel, G. M., and van Lier, R. A. (1996) J. Exp. Med. 184 1185-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamann, J., Stortelers, C., Kiss-Toth, E., Vogel, B., Eichler, W., and van Lier, R. A. (1998) Eur. J. Immunol. 28 1701-1707 [DOI] [PubMed] [Google Scholar]
- 31.Stacey, M., Chang, G. W., Davies, J. Q., Kwakkenbos, M. J., Sanderson, R. D., Hamann, J., Gordon, S., and Lin, H. H. (2003) Blood 102 2916-2924 [DOI] [PubMed] [Google Scholar]
- 32.Kwakkenbos, M. J., Pouwels, W., Matmati, M., Stacey, M., Lin, H. H., Gordon, S., van Lier, R. A., and Hamann, J. (2005) J. Leukoc. Biol. 77 112-119 [DOI] [PubMed] [Google Scholar]
- 33.Park, D., Tosello-Trampont, A. C., Elliott, M. R., Lu, M., Haney, L. B., Ma, Z., Klibanov, A. L., Mandell, J. W., and Ravichandran, K. S. (2007) Nature 450 430-434 [DOI] [PubMed] [Google Scholar]
- 34.Davies, J. Q., Chang, G. W., Yona, S., Gordon, S., Stacey, M., and Lin, H. H. (2007) J. Biol. Chem. 282 27343-27353 [DOI] [PubMed] [Google Scholar]
- 35.Little, K. D., Hemler, M. E., and Stipp, C. S. (2004) Mol. Biol. Cell 15 2375-2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.