Abstract
The voltage-gated sodium channel NaV1.5 is responsible for the initial upstroke of the action potential in cardiac tissue. Levels of intracellular calcium modulate inactivation gating of NaV1.5, in part through a C-terminal EF-hand calcium binding domain. The significance of this structure is underscored by the fact that mutations within this domain are associated with specific cardiac arrhythmia syndromes. In an effort to elucidate the molecular basis for calcium regulation of channel function, we have determined the solution structure of the C-terminal EF-hand domain using multidimensional heteronuclear NMR. The structure confirms the existence of the four-helix bundle common to EF-hand domain proteins. However, the location of this domain is shifted with respect to that predicted on the basis of a consensus 12-residue EF-hand calcium binding loop in the sequence. This finding is consistent with the weak calcium affinity reported for the isolated EF-hand domain; high affinity binding is observed only in a construct with an additional 60 residues C-terminal to the EF-hand domain, including the IQ motif that is central to the calcium regulatory apparatus. The binding of an IQ motif peptide to the EF-hand domain was characterized by isothermal titration calorimetry and nuclear magnetic resonance spectroscopy. The peptide binds between helices I and IV in the EF-hand domain, similar to the binding of target peptides to other EF-hand calcium-binding proteins. These results suggest a molecular basis for the coupling of the intrinsic (EF-hand domain) and extrinsic (calmodulin) components of the calcium-sensing apparatus of NaV1.5.
The cardiac voltage-gated sodium channel NaV1.5 mediates the voltage-dependent sodium ion permeability of excitable membranes. NaV1.5 is responsible for the initial upstroke of the action potential in the electrocardiogram. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein allows Na+ ions to pass in accordance with their electrochemical gradient. Upon repeated stimulation, channels convert to the third state known as the inactive state. Channels must pass from the inactive to the closed state before they can be opened again.
The structure of the channel is dominated by four membrane spanning domains (DI, DII, DIII, DIV), each containing six transmembrane helices linked by intracellular loops. The loop between DIII and DIV is of particular interest here because it is involved in the fast-inactivation of the channel (1). There are also substantial N- and the C-terminal domains, both located on the cytoplasmic side of the membrane. Several diseases are linked to the dysfunction of NaV1.5, such as long QT syndrome, Brugada syndrome, and idiopathic ventricular fibrillation (2-6), and some of these dysfunctions are due to mutations located in the C-terminal domain (7-9).
Substantial evidence has accumulated showing that inactivation gating of NaV1.5 is modulated in response to changes in the level of calcium (Ca2+) in the adjacent cytosol. Ca2+-dependent effects on NaV1.5 inactivation gating have been shown to play an important role in the progression of certain arrhythmia conditions. Therefore, elucidating the mechanism for the Ca2+-dependent modulation of channel function is an important objective for understanding the basis of the Brugada and Long QT syndromes and for the long range goal of finding suitable chemotherapeutic strategies to neutralize the damaging events caused by these cardiac defects.
Studies from our laboratory and others have shown that three regions of NaV1.5 as well as the ubiquitous intracellular Ca2+ sensor calmodulin are involved in generating the Ca2+-dependent effects on channel function (10, 11). In addition to the DIII-DIV loop noted above, the C-terminal domain contains two regions involved in Ca2+-dependent modulation of channel gating function. The first is an EF-hand domain, a structure closely associated with intracellular calcium sensing in a wide range of proteins including calmodulin. The second region of interest is an IQ motif. IQ motifs are calmodulin (CaM)2 binding domains most commonly associated with recruitment of CaM to specific sites so that it is able to respond rapidly to calcium signals. The fact that the NaV1.5 IQ motif has important functions for both the intrinsic sensing of Ca2+ by the EF-hand domain and the recruitment of CaM led us to propose a model in which the IQ motif serves as molecular switch that couples the two Ca2+-sensing modules (11).
In the present study we report the solution structure of the C-terminal domain EF-hand domain (CTD-EF) in NaV1.5 determined by NMR spectroscopy. Analysis of this structure reveals important features relating to the function of this domain and provides essential groundwork for determining why functional Ca2+ binding requires an additional 60 residues downstream from the EF-hand domain (11). The implications of these results are described with respect to the mechanism of action of the complex calcium-sensing apparatus in NaV1.5.
EXPERIMENTAL PROCEDURES
Expression and Purification—Recombinant human CTD-EF (Glu-1773—Ser-1865) was subcloned between BamHI and EcoRI restriction sites of an in-house pBG100 plasmid (Dr. L. Mizoue, Center for Structural Biology), which is derived from pET27. To facilitate purification, this vector codes for a His6 tag and a flexible linker containing a 3C protease cleavage site. The CTD-EF construct is expressed as a 97-residue protein that contains Gly-Pro-Gly-Ser fused to its N terminus after cleavage of the His6 tag.
Proteins were expressed in Escherichia coli BL21 (DE3) cells (Novagen) cells and grown at 37 °C up to 0.6 A600, then isopropyl 1-thio-β-d-galactopyranoside (1 mm) was added, and the culture was grown at 18 °C overnight. Production of unlabeled and 13C,15N-enriched proteins were carried out by growth on lysogeny broth or minimal medium, respectively. The minimal medium was supplemented with 15NH4Cl and glucose or [13C6]glucose as the sole nitrogen and carbon sources. Cell pellets were resuspended in 50 mm BisTris at pH 6.5, 200 mm NaCl, 10 mm MgCl2, 5 mm β-mercaptoethanol (BME), and 10 mm imidazole and lysed using sonication. The solution was centrifuged at 20,000 × g for 20 min. Supernatant was filtered and loaded onto a nickel affinity chromatography column (Amersham Biosciences). Nonspecifically bound proteins were removed by washing the column with 50 mm BisTris at pH 6.5, 200 mm NaCl, 10 mm MgCl2, 5 mm BME, and 10 mm imidazole, and the bound protein was eluted using a gradient to 50 mm BisTris at pH 6.5, 200 mm NaCl, 10 mm MgCl2, 5 mm BME, and 500 mm imidazole buffer. Fractions containing CTD-EF were pooled, and rhinovirus His6-tagged 3C protease was added. This solution was dialyzed at 4 °C overnight against 50 mm Bis-Tris at pH 6.5, 200 mm NaCl, 5 mm BME. The solution was then loaded onto a nickel affinity chromatography column to separate CTD-EF and the His6 tag. The flow-through was concentrated on a 3K-Centricon device to ∼0.5 ml and run over an S75 gel filtration column (Amersham Biosciences).
Peptide Synthesis—A 29-residue peptide (1897RRKHE EVSAM VIQRA FRRHL LQRSL KHAS1925) containing the IQ motif of NaV1.5 was synthesized (Sigma Genosys). Peptides were purified by reversed-phase high performance liquid chromatography and analyzed by mass spectrometry to confirm purity and molecular weight.
Isothermal Titration Calorimetry—Isothermal titration calorimetry (ITC) measurements were carried out with a VP-ITC MicroCalorimeter (MicroCal, Inc., Northampton, MA). Titration experiments were performed in 50 mm MES buffer at pH 6.5, 200 mm NaCl, and either 1 mm CaCl2 or 1 mm EDTA. Experiments were performed at 25 °C. Both protein and peptide were dialyzed against the same buffer. Peptide (25-28 μm) in the calorimetric cell was titrated by a series of 10-μl volume injections of CTD-EF (500-560 μm) with an interval of 240 s. The binding isotherms, ΔH versus molar ratio, were analyzed with a single-site binding model using MicroCal Origin software. The association constant, Ka (Kd = 1/Ka) was obtained directly from the fit.
NMR Sample Preparation—CTD-EF was dialyzed into 100 mm phosphate at pH 6.5, 200 mm NaCl, 5 mm BME, and 0.01% NaN3. The protein concentration was 1.2 mm for structure determination and 0.2 mm for titration studies. To minimize ionic strength effects on sensitivity, 300 μl of protein solution was loaded into a 4-mm NMR tube, and this tube was inserted into a 5-mm NMR tube containing 150 μl of D2O (12).
NMR Spectroscopy—All NMR data were recorded at 25 °C on Bruker DRX600 and DRX800 spectrometers. Data were processed with Topspin 2.0b (Bruker) and analyzed with Sparky (13). Backbone and sequential resonance assignments were obtained by the combined use of two-dimensional 15N,1H HSQC and three-dimensional HNCA, HNCACB, CBCA-(CO)NH, and HNCO experiments (for review, see Ref. 14). Aliphatic side chain resonance assignments were obtained from three-dimensional (H)CC(CO)NH, H(CCCO)NH, HCCH-TOCSY, HCCH-COSY, and HBHANH experiments. 1H chemical shift assignments of aromatic side chains were primarily based on two-dimensional homonuclear COSY, TOCSY, and NOESY experiments. To assign NOE-based distance restraints, a two-dimensional homonuclear NOESY experiment was recorded on an unlabeled sample, and three-dimensional 15N NOESY-HSQC and 13C NOESY-HSQC were recorded on a uniformly 13C,15N-enriched sample. The mixing time used in all NOESY experiments was 120 ms.
Heteronuclear 15N relaxation parameters (T1, T2, NOE) were measured at 500 MHz as described previously (15). The relaxation delays were 10, 25, 50, 100, 150, 250, 400, 600, 900, and 1200 ms for the T1 measurements and 11, 23, 34, 68, 79, 114, 136, and 170 ms for the T2 measurements. Steady-state {1H}-15N NOE data were obtained with and without 3 s of 1H saturation using a total recycle delay of 7 s. NOE values were extracted as the ratio of peak intensities with and without proton saturation for 88 well resolved resonances. The rate analysis functions in Sparky were used to analyze the T1 and T2 decay rates, which were computed using the two-parameter model in CurveFit (16) for 84 and 80 well resolved resonances, respectively. The isotropic rotational correlation time (τm) was approximated using the trimmed T1/T2 ratio computed for only well resolved resonances in the well structured regions of the protein (17).
The binding site of the IQ motif peptide to CTD-EF was determined by recording 15N,1H HSQC NMR spectra of CTD-EF as the peptide was titrated into the solution. Spectra were acquired for protein:peptide ratios of 1:0, 1:0.25, 1:0.5, 1:1, and 1:2. The overall chemical shift change of the 1H,15N peak was expressed as Δδ = [(ΔδHN)2 + (ΔδN/6.5)2)]1/2 (18).
Structure Calculations—The first stage of calculations used CYANA (19). The standard CYANA protocol of 7 iterative cycles of calculations was performed starting from a set of manually assigned NOEs. In each iteration 100 structures started from random torsion angle values were calculated with 10,000 steps of torsion angle dynamics-driven simulated annealing. 2131 NOE-based distance and 51 backbone ϕ angle restraints were used for the final calculations. The angle restraints were obtained from 1Hα, 13Cα, 13Cβ, 13C′, and 15N chemical shifts using PREDITOR (20) with a minimum range of ±40°. The 50 lowest CYANA target function conformers were selected for refinement by restrained molecular dynamics simulations in AMBER (21) using the generalized Born solvent model. The starting structures were energy minimized for 100 steps, then 20 ps of restrained molecular dynamics was performed with the following protocol: 1 ps heating the system from 0 to 600 K followed by 4 ps at 600 K with a coupling parameter TAUTP for heating and equilibration of 0.4; 15 ps of cooling to 0 K with 8 ps of slow cooling (loose coupling, TAUTP = 4.0-2.0) followed by 2 ps of faster cooling (TAUTP = 1.0) and a final 2 ps of very fast cooling (TAUTP = 0.5-0.05). The restraints were slowly ramped from 10 to 100% that of their final values over the first 3 ps. The representative ensemble corresponds to the 20 conformers with the lowest restraint and AMBER energy terms.
Docking of IQ Motif to CTD-EF—A model for the CTD-EF/IQ motif complex was generated using HADDOCK2 (22). The unstructured N-terminal 10 residues of the CTD-EF construct were removed from the coordinate files so as to not interfere with docking of the peptide. The central region (Glu-1901—Leu-1921) of the IQ motif was modeled as a helix with the N- and C termini in a random coil conformation in accord with secondary structure prediction (23) and available structures of IQ motif complexes (24,25). The average solvent accessibilities per residue in the ensemble of CTD-EF conformers were calculated using NACCESS (26). Residues with a solvent accessible surface higher than 40% and chemical shift perturbations more than 1 S.D. above the mean were designated as active for the HADDOCK calculations. The adjacent residues with solvent accessibility greater than 50% were designated as passive residues. In the first iteration of the calculation, an initial ensemble of 5000 rigid body docking models was generated. The 200 lowest energy models were selected for a second iteration with semi-flexible simulated annealing and were further refined in explicit water. The 10 lowest-energy models were selected from the most populated cluster with the lowest HAD-DOCK score and used for structural analysis.
Computational Analysis—Multiple sequence alignments were performed with T-Coffee (27) and MUSCLE (28) through the phylogeny web server (29). Graphical analyses of the structures and figure preparation were carried out with the programs MOLMOL (30) and PyMOL (31). The geometric quality and stereochemistry of the CTD-EF structural ensemble was assessed using PROCHECK-NMR (32). Electrostatic surface potentials were calculated with the Adaptive Poisson-Boltzmann Solver (APBS) software (33).
Electrophysiology Experiments—Site-directed mutagenesis was performed on the NaV1.5 α subunit cDNA and subcloned into the expression vector pCGI for bicistronic expression of the channel protein and green fluorescent protein as described (34). Cultured cells (tsA201) were transiently transfected with either wild-type or mutant (2X) cDNA (2.5 μg). Green cells were selected for electrophysiological analysis 24 h later. The cells were maintained on culture plates with Dulbecco's modified Eagle's medium with fetal bovine serum (10%) and 1% penicillin-streptomycin.
INa was recorded at 22 °C and analyzed as described (34). The voltage-clamp protocols used are shown in the legend to Fig. 5. A Boltzmann function (I = 1/(1 + exp(Vt - V½)/μ))) was fitted to the availability curves to determine the membrane potential eliciting half-maximal inactivation (V½), where μ is the slope factor. The pipette solution used to approximate zero [Ca2+]i contained 10 mm NaF, 100 mm CsF, 20 mm CsCl, 20 mm BAPTA, 10 mm HEPES, adjusted to pH 7.35 with CsOH. For high [Ca2+]i (1 μm free Ca2+), 1 mm BAPTA was used with 0.9 mm Ca2+. The extracellular (bath) recording solution contained 145 mm NaCl, 4 mm KCl, 1 mm MgCl2, 10 mm HEPES, and 1.8 mm CaCl2, adjusted to pH 7.35 with CsOH. To avoid the time-dependent shift of the INa availability curve commonly observed during patch clamp experiments, voltage-dependent inactivation was assessed within 2 min after rupture of the membrane. Patch clamp measurements are presented as the mean ± S.E. Comparisons were made using Student's t test, with p < 0.05 considered significant.
FIGURE 5.
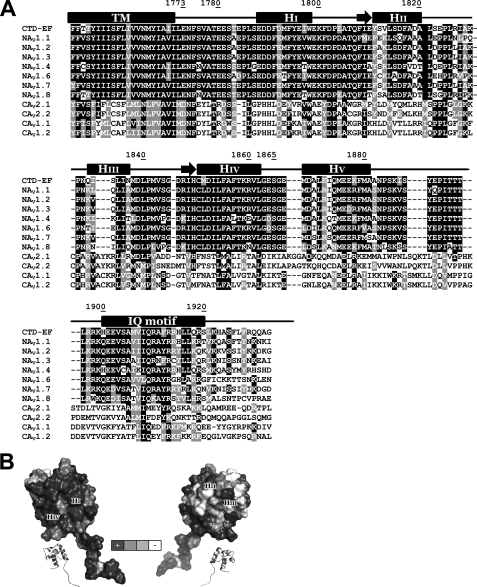
Conservation of residues in the C terminus of NaV and CaV channels. A, NaV and CaV sequence alignments. Sequences from top to bottom are shown for CTD-EF NaV1.5 (SWISSPROT Q14524), NaV1.1 (SWISSPROT P35498), NaV1.2 (SWISSPROT Q99250), NaV1.3 (SWISSPROT Q9NY46), NaV1.4 (SWISSPROT P35499), NaV1.6 (SWISSPROT Q9UQD0), NaV1.7 (SWISSPROT Q15858), NaV1.8 (SWISSPROT Q9Y5Y9), CaV2.1 (SWISSPROT O00555), CaV2.2 (SWISSPROT Q00975), CaV1.1 (SWISSPROT Q13698), and CaV1.2 (SWISSPROT Q13936). The residue numbers are indicated for CTD-EF along with secondary structure predicted based on sequence analysis and determined experimentally. B, surface representation of CTD-EF with residues shaded according to the extent of conservation in the NaV family with black indicating complete conservation and white indicating non-conservation.
Data Deposition—1H, 13C, and 15N chemical shift assignments are deposited in the BioMagResBank under accession number 16045. The coordinates of the final ensemble of 20 structures and the full list of NMR restraints are deposited in the Protein Data Bank under accession code 2KBI.
RESULTS
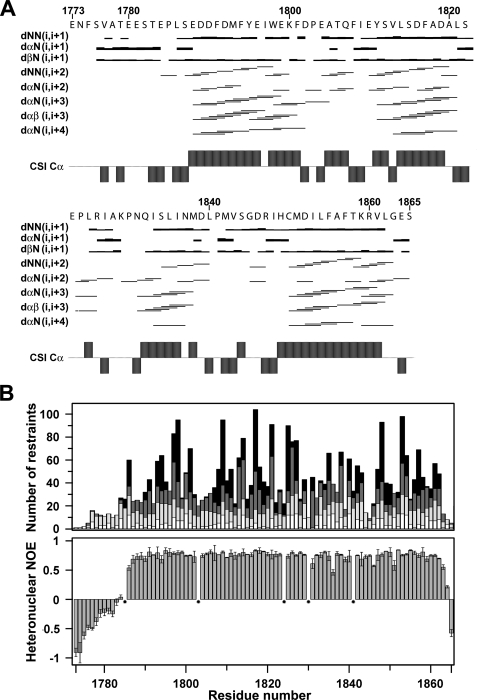
Structure of the EF-hand Domain in the NaV1.5 C Terminus—The three-dimensional solution structure of apoCTD-EF was determined by multidimensional heteronuclear NMR spectroscopy. To achieve this goal, the purification protocol and buffer conditions were extensively optimized relative to our previous report (11). 1H, 13C, and 15N resonance assignments (>98%) were obtained for all but the Cγ, Cδ, Cε, and, CδH resonances of Lys-1829 using the standard series of double- and triple-resonance backbone-based scalar correlated experiments described under “Experimental Procedures.” Of note, cis conformations for two of the five proline residues (Pro-1824, Pro-1830) were identified on the basis of characteristic Cγ and Cδ chemical shifts and Hα-Hα NOEs (35, 36). Qualitative analysis of the pattern of medium range NOEs revealed the presence of four α helices, which were fully consistent with analysis by the chemical shift index (37) (Fig. 1A).
FIGURE 1.
Distribution of NOEs and correlation with heteronuclear relaxation for the CTD-EF construct. A, distribution of sequential and medium-range NOEs. The NOEs were classified into strong, medium, and weak as indicated by thick, medium, and thin lines, respectively. Chemical shift index values for C-α atoms are indicated. B, histogram of all NOEs (top) and 15N NOEs (bottom). Intraresidue NOEs are in white, sequential NOEs are in light gray, medium-range NOEs are in dark gray, and long-range NOEs are in black. Prolines residues are indicated by stars.
Heteronuclear relaxation parameters were measured to characterize the backbone dynamics of CTD-EF, including steady-state {1H}-15N NOE, longitudinal 15N(T1), and transverse 15N(T2) relaxation times (supplemental Fig. S1). The average values of 0.74, 440 ms, and 77 ms, respectively, with small S.D. are consistent with the existence of a globular domain between residues 1785 and 1863. Clear trends to lower values for the NOE and T2 are clearly evident for the preceding 12 residues of CTD-EF, indicating that the N terminus of this construct is flexible. The trimmed T1/T2 ratio provides an estimate of 5.7 ns for the rotational correlation time, consistent with the CTD-EF tumbling as a monomeric species in solution.
The three-dimensional structure of CTD-EF was calculated using a two-step protocol that involves generation of initial structures using CYANA (19) and refinement by restrained molecular dynamics using AMBER (21) as described previously (38). In all, 2131 NOE-based distance restraints were generated (Table 1), and these were well distributed along the sequence except for the first 12 residues, which lack medium and long range restraints (Fig. 1B). In addition to NOEs, 51 backbone ϕ angle restraints were derived from chemical shifts, and the CYANA program provided stereo-specific assignments for 9 pairs of Leu and Val methyl groups and 23 pairs of Cβ protons. The final ensemble of 20 conformers was selected based on lowest restraint violation, and total molecular energy and is shown in Fig. 2A. Consistent with the large number of experimental restraints (>20/residue), this ensemble is well defined and exhibits low constraint violation energies, no distance violations >0.2 Å, no torsion angle violations >5°, and low molecular energies (Table 1). The structure has been refined to high precision with a root mean square deviation of 0.47 ± 0.07 Å for the backbone and 1.09 ± 0.10 Å for all heavy atoms (Table 1). The high quality of the structure is further reflected by PROCHECK analysis. For example, 99.4% of all backbone torsion angles occupy the favored regions of the Ramachandran plot.
TABLE 1.
Structural statistics for CTD-EF in Nav1.5
| Restraints for calculation | |
| Total NOE restraints | 2131 |
| Intraresidue | 466 |
| Sequential | 554 |
| Medium range | 526 |
| Long range | 585 |
| Dihedral angle restraint | 51 |
| Constraint violations, mean ± S.D. | |
| Distance violations | |
| 0.1 Å < d < 0.2 Å | 1.6 ± 1.0 |
| d > 0.2 Å | 0 |
| Average maximum distance violations (Å) | 0.13 ± 0.03 |
| Torsion angle violations >5.0° | 0 |
| Average maximum torsion angle violations | 0 |
| (degree) | |
| AMBER energiesa, mean ± S.D. (kcal mol−1) | |
| Restraint | 1.8 ± 0.4 |
| van der Waals | −741 ± 8 |
| Total molecular | −4021 ± 10 |
| Precision, root mean square deviation from the mean (Å), ordered regionsb | |
| Backbone | 0.47 ± 0.07 |
| All heavy atoms | 1.09 ± 0.10 |
| Ramachandran statisticsc(%) | |
| Most favored | 86.8 |
| Additionally allowed | 12.6 |
| Generously allowed | 0.4 |
| Disallowed (only for residues 1-16 and 55) | 0.2 |
FIGURE 2.
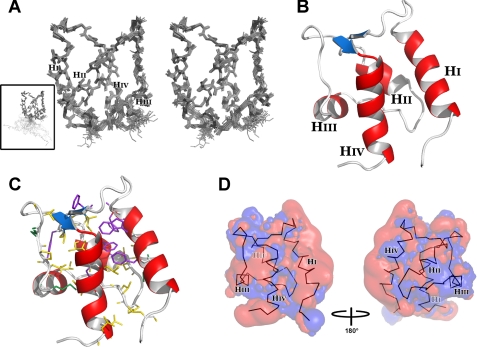
Three-dimensional solution structure of CTD-EF. A, stereo-view of the final ensemble of 20 conformers representing the solution structure, depicted with all backbone atoms. Inset, full-length CTD-EF. B, ribbon diagram with helices labeled and colored red and the cross-strand β-type interaction in blue. C, residues that contribute to the hydrophobic core with aromatic side chains in purple and aliphatic side chains in yellow and green. CTD-EF has the same orientation as in B. D, electrostatic field potentials of CTD-EF with negative charge in red and positive charge in blue. The backbone of CTD-EF is shown within the surface. The four helices are labeled. Contour levels for positive and negative isosurfaces were set to 1 kT and -2 kT, respectively.
The overall fold of CTD-EF is similar to other EF-hand domains, consisting of two helix-loop-helix EF-hand motifs and a short linker between them. The four helices include: I, Glu-1788—Trp-1798; II, Tyr-1811—Ala-1820; III, Gln-1832—Asn-1837; IV, Cys-1850—Leu-1862. The typical cross-strand β-like interaction between the two EF-hand loops is found involving residues Phe-1808—Glu-1810 and Arg-1947—His-1849 (Fig. 2B). The loop between helices III and IV is somewhat unusual; although it is the characteristic 12 residues in length, it possesses one proline (Pro-1841) and adopts a more extended conformation than is typical of other EF-hand domain loops. As noted above, the first 12 residues of the CTD-EF construct are unstructured. In the remainder of the discussion, CTD-EF will refer only to the globular EF-hand domain.
Although CTD-EF does not have substantial sequence similarity with well characterized EF-hand calcium-binding proteins such as calmodulin, troponin C, recoverin, or S100 proteins, it does have a typical EF-hand protein structure. The Vector Alignment Search Tool (VAST) (39) search finds three EF-hand proteins as the most similar: Ca2+-loaded troponin-C (PDB 1TOP), Ca2+-loaded domain VI of calpain (PDB 1ALV), and apoflagellar calcium-binding protein (FCaBP) (PDB 3CS1). Similarly, the Dali search algorithm (40) finds EF-hand proteins as the most similar, with scores of 7.8 for the zinc-loaded N terminus of Rat CaM (PDB 2PQ3) and for the N terminus of apoCaM (PDB 1F70), 7.7 for full-length apoCaM (PDB 1QX5), and 7.2 for human m-calpain (PDB 1KFU). Supplemental Fig. S2 shows comparisons of these structures, which have backbone root mean square deviations with respect to CTD-EF of 1.96, 2.42, 1.94, and 2.13 Å for EF-hand 3 of calpain domain VI, N-lobe of troponin-C, N-lobe of zinc-bound CaM, and N-lobe of apoCaM, respectively.
The surface properties of CTD-EF were evaluated to determine if any relevant functional insights could be obtained. CTD-EF contains a large number of hydrophobic residues, in excess of 50% of the sequence. As a result, the protein has a large internal hydrophobic core (Fig. 2C) and a significant amount of exposed hydrophobic surface. The latter is consistent with the domain functioning as part of the Ca2+-sensing regulatory apparatus of NaV1.5. The electrostatic field at the surface of the domain was calculated using APBS (33). CTD-EF has a net negative charge of -14 e (20 Asp/Glu, 6 Arg/Lys). Fig. 2D shows that the surface of CTD-EF is highly negatively charged. The substantial negative surface potential evident around Helix I is particularly significant and will be discussed in more detail below.
Ca2+ binding to the NaV1.5 C-terminal region has been demonstrated in a previous study, and the location of the EF-hand domain was predicted on the basis of a consensus 12-residue EF-hand calcium binding loop sequence spanning Glu-1788—Glu-1799 (41). A critical observation from determining the structure is that the location of the EF-hand domain is substantially shifted with respect to the prediction. Remarkably, none of the 12 consensus EF-hand residues is located in the first EF-hand loop, and rather, they form Helix I. Of the two putative Ca2+ binding loops found in the structure of CTD-EF, only Loop I possesses a sequence consistent with effective ion binding. Loop II does not have the pattern of characteristic acidic residues so it is highly unlikely to form a functional Ca2+ binding site. Although Loop I does not fit well to the consensus for a functional EF-hand, its sequence is similar to a putative Ca2+ binding loop in the EF-hand 5 of calpain domain IV, and in fact they form similar structures (supplemental Fig. S2). The loop in calpain was first described as aCa2+ binding loop (42, 43) and was later characterized as an important element in the association of the domains IV and VI of calpain (44).
Mutations in CTD-EF Alter Channel Function—Previous mutational analysis on the CTD of NaV1.5 was performed on residues in the sequence containing the consensus EF-hand Ca2+ binding loop (41). Our structure of the CTD-EF now reveals that these mutations were made in Helix I and not in the Ca2+ binding Loop I as was intended. To confirm the functional relevance of the globular EF-hand domain in the NaV1.5 C terminus, new mutations have been prepared in the putative Ca2+ binding Loop I and tested in patch clamp electrophysiology experiments on mutated channels.
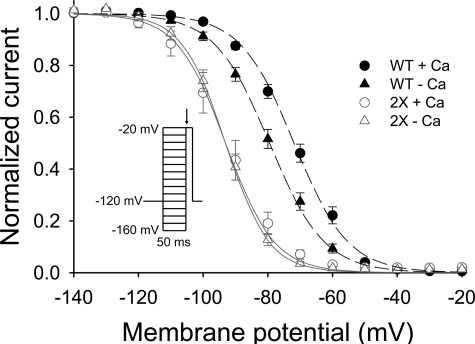
A series of Loop I mutations of acidic and polar residues in CTD-EF were designed based on the Loop I residues predicted to be involved in Ca2+ chelation. Results are included here for the double mutant NaV1.5-D1802A,E1804A (2X). As shown in Fig. 3, the steady-state availability curve at basal Ca2+ levels for the double mutant are shifted leftward relative to wild-type channels. Moreover, the mutant channels failed to exhibit the significant Ca2+-dependent shift observed in wild-type NaV1.5 that was first reported in our earlier study (41). These results support the proposed role of the NaV1.5 EF-hand domain in Ca2+-dependent regulation of channel function.
FIGURE 3.
Function of NaV1.5 is perturbed by mutation in Ca2+ binding Loop I of CTD-EF. Patch clamp experiments showing the availability curve of the wild-type and D1802A,E1804A mutant (2X) in the absence or presence of Ca2+. Voltage dependence of inactivation is shown for cells expressing wild-type (WT) or 2X Nav 1.5 in high [Ca2+]i (1 mm) or a nominally [Ca2+]i-free solution (20 mm BAPTA). The V½ values for wild-type NaV1.5 were -71.8 ± 1.3 mV (Δ; n = 17) in high [Ca2+]i and -79.4 ± 1.3 mV (Δ; n = 16) in low [Ca2+]i (p < 0.001). The V½ values for 2X NaV1.5 were -93.0 ± 2.7 mV (Δ; n = 10) in high [Ca2+]i and -93.1 ± 1.3 mV (Δ; n = 9) in low [Ca2+]i (p = note significant). Inset, voltage clamp protocol.
CTD-EF Binds the IQ Motif at a Surface between Helices I and IV—We have previously shown that the CTD EF-hand domain only binds Ca2+ with high affinity in a construct extended 60 residues downstream to include the IQ motif (41). Moreover, preliminary evidence had been obtained indicating the IQ motif binds to the EF-hand domain (11). To confirm and further characterize this interaction, isothermal titration calorimetry was used to quantify the binding affinity of an IQ motif peptide to CTD-EF. Fig. 4A shows binding isotherms generated upon the addition of apo and Ca2+-loaded CTD-EF to solutions of the IQ peptide. Fitting of this data to a standard single site binding equation provides dissociation constants (Kd) of 27.5 ± 2.3 and 37.9 ± 8.6 μm for apo and Ca2+-loaded CTD-EF, respectively.
FIGURE 4.
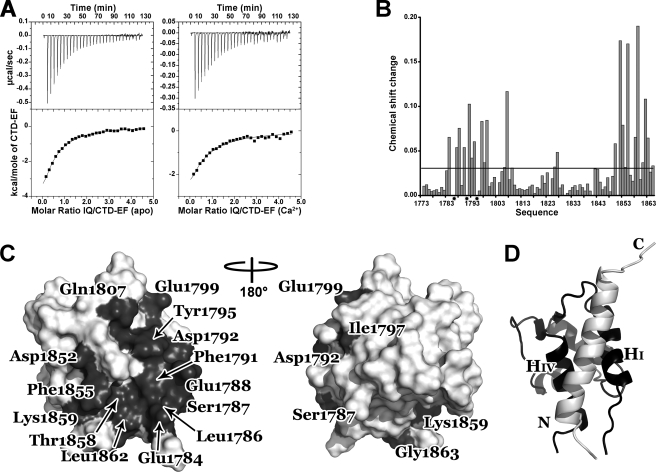
Characterization of the interaction between CTD-EF and the IQ motif. A, isothermal titration calorimetry of CTD-EF binding to the IQ motif peptide in the absence (left) and presence (right) of Ca2+. The upper panel shows heat release upon injection of CTD-EF into the IQ peptide solution. The lower panel shows integrated isotherms. B, changes in CTD-EF chemical shifts as the IQ peptide is titrated into the solution. The mean value of the chemical shift change is identified by the horizontal line. Stars indicate residues in “intermediate exchange.” C, interaction surface of CTD-EF. Residues with a chemical shift perturbation more than a half standard deviation above the average are colored dark gray and labeled. D, ribbon diagram of the model of CTD-EF (black) in complex with the IQ motif peptide (white). The N and C termini of the IQ motif and Helices I and IV of CTD-EF are labeled.
Having clearly demonstrated binding, NMR spectroscopy was then used to map the binding site of the IQ motif peptide on the structure of CTD-EF. Chemical shift perturbation analysis was performed by recording 15N,1H HSQC NMR spectra of CTD-EF as the peptide was titrated into the solution. Fig. 4B shows a plot of chemical shift perturbations induced by the binding of the peptide. The observed changes occurred mainly in the so-called “fast exchange” regime, which enables the resonance assignments to be transferred from the free protein to the peptide complex. Leu-1786, Phe-1791, and Tyr-1795 were the most significantly perturbed upon binding of the peptide; these residues exhibited “intermediate exchange,” and their signals were broadened beyond detection even at the first titration point. Mapping of the NMR chemical shift changes onto the structure of CTD-EF shows that the peptide binds to a surface between Helices I and IV (Fig. 4C). The putative binding interface includes a number of acidic residues that contribute to the strongly negative electrostatic surface potential in this region (Glu-1784, Glu-1788, Asp-1792, Glu-1799, Asp-1852) as well as hydrophobic residues present in or close to Helices I and IV (Leu-1786, Phe-1791, Tyr-1795, Ile-1797, Phe-1855, Leu-1862). The binding interface also includes two residues close to Helix I (Ser-1787, Gln-1807) and four other residues located in Helix IV (Cys-1850, Thr-1858, Lys-1859, Gly-1863).
To aid in understanding the nature of the binding interaction, the chemical shift perturbations were used as input to guide computational docking of the IQ motif peptide onto CTD-EF. A helical structure was generated for the IQ motif peptide based on both secondary structure prediction and the helical conformation of other IQ motifs in their complexes with EF-hand proteins (24, 25). Interestingly, the docking calculation consistently provided complexes with the peptide bound in one of two possible orientations with respect to the protein. A representation of the model of the complex is shown in Fig. 4D. Significant hydrophobic interactions were found to occur between residues Phe-1791, Tyr-1795, Leu-1854, Phe-1855 in CTD-EF and IQ residues Val-1907, Ile-1908, and Phe-1912. The IQ peptide also contains basic residues (Arg-1914, His-1915, Arg-1919, Lys-1922, and His-1923) that complement the strongly acidic character of the CTD-EF binding surface. Thus, the model reveals that the IQ motif readily fits into a binding pocket between Helices I and IV on the surface of CTD-EF with significant contributions from both hydrophobic and electrostatic interactions.
DISCUSSION
Babitch first predicted the presence of an EF-hand in the C-terminal region of calcium and sodium voltage-gated channels (45). We previously predicted the location of the EF-hand domain based on the presence of a consensus 12-residue EF-hand Ca2+ binding loop between Glu-1777 and Glu-1788 (41). Determination of the structure of the CTD-EF construct shows that, whereas an EF-hand domain does exist, it is offset from our previous prediction. Thus, despite close correspondence to the EF-hand Ca2+ binding loop consensus, the Glu-1777—Glu-1788 segment forms a helical element that is inconsistent with Ca2+ binding. Of the two putative Ca2+ binding loops in the structure, Loop II clearly lacks the residues needed to bind Ca2+, whereas Loop I has the acidic character required to effectively bind Ca2+ and in particular has the key glutamic acid residue at its C terminus that provides critical bidentate chelation of the Ca2+ ion.
The sequence of Ca2+ binding Loop I is similar to a putative Ca2+ binding loop in domain VI of calpain. Interestingly, calpain is also the closest structural homolog to CTD-EF and the structure of the respective loops are remarkably similar (supplemental Fig. S2). CTD-EF and EF-hand 3 of calpain domain VI have several common hydrophobic residues packing at the interface between Helices I and II, including Ile-1797, Phe-1801, Ile-1809, and Leu-1814 in CTD-EF, which correspond to calpain residues Ile-877, Phe-881, Ile-889, and Leu-894, respectively (supplemental Fig. S2). The conservation of these residues in the hydrophobic core presumably leads to their similarity in structure.
Much of the sequence of CTD-EF is conserved in NaV channels, and this conservation extends even to CaV channels, for which only a limited number of residues in Helix III are poorly conserved (Fig. 5A). Fig. 5B shows the conservation of the residues in CTD-EF of NaV channels plotted onto the structure. Residues from Helices I and IV (Fig. 5B, left) are the most highly conserved among the NaV channels, whereas residues 1810, 1812, 1813, 1819, 1822-1823, 1826, 1832-1834, and 1837 located on Helices II and III at the opposite side are the least conserved (Fig. 5B, right). The very high level of conservation of the EF-hand domain in the IQ motif binding pocket between Helices I and IV strongly implies an important role for CTD-EF in channel function.
The globular CTD EF-hand domains in NaV and CaV channels are positioned relatively close but not immediately adjacent to the cell membrane. The 97-residue construct used in this study includes the residues Glu-1773—Ser-1787 that link the globular CTD-EF domain to the last trans-membrane segment of NaV1.5 DIV. Our NMR relaxation studies (supplemental Fig. S1) show that these residues are disordered and highly dynamic, suggesting that they provide a flexible linker between the EF-hand domain and the last transmembrane segment of DIV and in turn to the pore and other structural elements within the channel. Notably, these residues are highly conserved in the NaV family, which suggests their functional relevance. We propose that the flexibility of this linker is important in NaV channels for enabling the EF-hand domain to alter its position with respect to the membrane as it functions.
Mutations within the CTD-EF domain have important effects on channel function. For example, D1790G and insertion of an aspartic acid between Tyr-1795 and Glu-1796 are associated with cardiac arrhythmia syndromes (46, 47) and presumably destabilize Helix I. Mutations of NaV1.1 residues Phe-1808, Trp-1812, and Phe-1831 (Phe-1794, Trp-1798, Phe-1812 in NaV1.5) in brain are associated with inherited epilepsies disease (48). These residues contribute to the very significant hydrophobic core of the EF-hand domain (Fig. 2C). Mutations of hydrophobic residues in NaV1.5 have also been shown to lead to perturbation of channel function (49). Such mutations would be expected to significantly destabilize the domain and may perturb its structure leading to alteration of channel function. We have shown here (Fig. 3) that mutations in putative Ca2+ binding Loop I affect the Ca2+ dependence of inactivation gating, although it is clear that a much more comprehensive analysis is needed to establish the precise role(s) of this domain in NaV1.5 function. The availability of the structure of CTD-EF represents a significant step forward toward performing these much-needed studies.
In previous studies we showed that high affinity binding of Ca2+ to the EF-hand domain is observed only in a construct that extends downstream to include the IQ motif and obtained evidence in support of an interaction between the EF-hand domain and the IQ motif (11). The isothermal titration calorimetry data showing binding of an IQ motif peptide to the CTD-EF with an affinity of ∼30 μm suggests physical interaction between these two structural elements is of functional importance. Note that the interaction of the IQ motif with CTD-EF should be even stronger in the intact channel because of the high local concentration due to covalent linkage.
How does the binding of the IQ motif enhance the ability of CTD-EF to bind Ca2+? One possibility is it provides one or more ligands that directly chelate the ion. Alternatively, it may induce or stabilize a conformation that is required for ion binding. The observation of significant chemical shift changes in residues located in Loop I of CTD-EF (e.g. Gln-1807) upon titration with the IQ motif peptide suggests changes in the structure in this region, which supports the second alternative. However, to fully elucidate the structural basis for the effect of IQ motif, structural analysis of a construct that includes both of these structural elements is required.
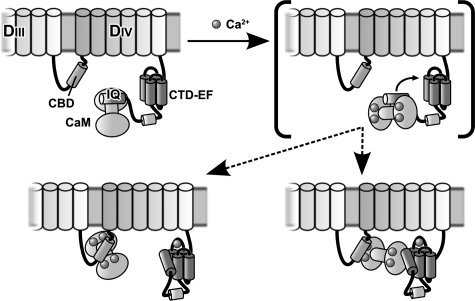
The structure of CTD-EF and direct demonstration of binding of the IQ motif to this domain is fully consistent with our previous proposal of a central role for the IQ motif as a molecular switch that integrates the action of intrinsic (CTD-EF) and extrinsic (calmodulin) Ca2+ sensors of NaV1.5 (11). Our results provide new evidence in support of the existence of a series of competing equilibria that couple Ca2+ binding by CTD-EF and calmodulin to interactions with the IQ motif (Fig. 6). In this model, under conditions of low concentration of Ca2+, calmodulin binds so strongly to the IQ motif that it effectively sequesters the IQ motif from interacting with the CTD-EF, which in turn inhibits Ca2+ binding to CTD-EF. As Ca2+ levels are raised, calmodulin actually binds the IQ motif less tightly (11), which increases the availability of the IQ motif to bind to CTD-EF. In addition, Ca2+ activates calmodulin to bind to other regions of NaV1.5, such as the calmodulin binding domain in the linker between DIII and DIV (11). These Ca2+-dependent binding sites compete with the IQ motif for calmodulin and, therefore, further increase the availability of the IQ motif to interact with CTD-EF. Our results contribute to the increasing realization that the response of NaV1.5 to intracellular Ca2+ signals levels is complex and involves calmodulin as well as multiple domains within the channel. Our model for the action of the Ca2+-sensing apparatus provides a valuable starting point for designing experiments to investigate function in vitro and in vivo and thereby obtain a better understanding of the molecular basis for the Ca2+ dependent modulation of inactivation gating of NaV1.5.
FIGURE 6.
Model for the action of the Ca2+-sensing apparatus in modulating inactivation gating of NaV1. 5. To simplify the diagram, the transmembrane helices are represented as cylinders on top of the cell membrane, and none of interconnecting loops is shown except for the critical linker between domains III and IV, which is known to mediate inactivation gating. The starting state (low Ca2+) is represented in the upper left panel. There are two alternatives for the final state (high Ca2+) shown in the lower panels. A bracket is drawn around the intermediate state in the upper right panel, which is shown to help explain how the apparatus works. When Ca2+ levels are raised, the interaction of CaM with the IQ motif (IQ) is altered (compare left and right upper panels). The binding of CaM to the IQ motif under high Ca2+ is weaker, which increases the availability of the IQ motif to interact with CTD-EF (lower panels). High Ca2+ also activates CaM interaction with other CaM binding domains (CBD) in NaV1.5 such as that in the linker between domains III and IV (lower panels). This may involve either a bridging interaction (lower right panel) or complete release of the IQ domain (lower left panel).
Supplementary Material
Acknowledgments
We thank Marie-Eve Chagot and Svetlana Stepanovic for expert technical assistance.
The atomic coordinates and restraints list (code 2KBI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM56307. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: CaM, calmodulin; CTD, C-terminal domain; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; BME, β-mercaptoethanol; MES, 4-morpholineethanesulfonic acid; HSQC, heteronuclear single quantum correlation; NOESY, nuclear Overhauser effect (NOE) spectroscopy.
References
- 1.Yu, F. H., and Catterall, W. A. (2003) Genome Biology http://genomebiology.com/2003/4/3/207 [DOI] [PMC free article] [PubMed]
- 2.Abriel, H., Cabo, C., Wehrens, X. H., Rivolta, I., Motoike, H. K., Memmi, M., Napolitano, C., Priori, S. G., and Kass, R. S. (2001) Circ. Res. 88 740-745 [DOI] [PubMed] [Google Scholar]
- 3.Wehrens, X. H., Abriel, H., Cabo, C., Benhorin, J., and Kass, R. S. (2000) Circulation 102 584-590 [DOI] [PubMed] [Google Scholar]
- 4.Priori, S. G., Napolitano, C., Gasparini, M., Pappone, C., Della Bella, P., Giordano, U., Bloise, R., Giustetto, C., De Nardis, R., Grillo, M., Ronchetti, E., Faggiano, G., and Nastoli, J. (2002) Circulation 105 1342-1347 [DOI] [PubMed] [Google Scholar]
- 5.Vatta, M., Dumaine, R., Antzelevitch, C., Brugada, R., Li, H., Bowles, N. E., Nademanee, K., Brugada, J., Brugada, P., and Towbin, J. A. (2002) Mol. Genet. Metab. 75 317-324 [DOI] [PubMed] [Google Scholar]
- 6.Chen, Q., Kirsch, G. E., Zhang, D., Brugada, R., Brugada, J., Brugada, P., Potenza, D., Moya, A., Borggrefe, M., Breithardt, G., Ortiz-Lopez, R., Wang, Z., Antzelevitch, C., O'Brien, R. E., Schulze-Bahr, E., Keating, M. T., Towbin, J. A., and Wang, Q. (1998) Nature 392 293-296 [DOI] [PubMed] [Google Scholar]
- 7.An, R. H., Wang, X. L., Kerem, B., Benhorin, J., Medina, A., Goldmit, M., and Kass, R. S. (1998) Circ. Res. 83 141-146 [DOI] [PubMed] [Google Scholar]
- 8.Smits, J. P., Eckardt, L., Probst, V., Bezzina, C. R., Schott, J. J., Remme, C. A., Haverkamp, W., Breithardt, G., Escande, D., Schulze-Bahr, E., LeMarec, H., and Wilde, A. A. (2002) J. Am. Coll. Cardiol. 40 350-356 [DOI] [PubMed] [Google Scholar]
- 9.Petitprez, S., Jespersen, T., Pruvot, E., Keller, D. I., Corbaz, C., Schlapfer, J., Abriel, H., and Kucera, J. P. (2008) Cardiovasc. Res. 78 494-504 [DOI] [PubMed] [Google Scholar]
- 10.Kim, J., Ghosh, S., Liu, H., Tateyama, M., Kass, R. S., and Pitt, G. S. (2004) J. Biol. Chem. 279 45004-45012 [DOI] [PubMed] [Google Scholar]
- 11.Shah, V. N., Wingo, T. L., Weiss, K. L., Williams, C. K., Balser, J. R., and Chazin, W. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3592-3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voehler, M. W., Collier, G., Young, J. K., Stone, M. P., and Germann, M. W. (2006) J. Magn. Reson. 183 102-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goddard, T. D., and Kneller, D. G. (2006) SPARKY 3, University of California, San Francisco
- 14.Cavanagh, J., Fairbrother, W. J., Palmer, A. G. I., and Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, Academic Press Inc., New York
- 15.Farrow, N. A., Zhang, O., Forman-Kay, J. D., and Kay, L. E. (1994) J. Biomol. NMR 4 727-734 [DOI] [PubMed] [Google Scholar]
- 16.Palmer, A. G. (1998) CurveFit, Columbia University, New York
- 17.Kay, L. E., Torchia, D. A., and Bax, A. (1989) Biochemistry 28 8972-8979 [DOI] [PubMed] [Google Scholar]
- 18.Mulder, F. A., Schipper, D., Bott, R., and Boelens, R. (1999) J. Mol. Biol. 292 111-123 [DOI] [PubMed] [Google Scholar]
- 19.Guntert, P. (2004) Methods Mol. Biol. 278 353-378 [DOI] [PubMed] [Google Scholar]
- 20.Berjanskii, M. V., Neal, S., and Wishart, D. S. (2006) Nucleic Acids Res. 34 63-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case, D. A., Cheatham, T. E., III, Darden, T., Gohlke, H., Luo, R., Merz, K. M., Jr., Onufriev, A., Simmerling, C., Wang, B., and Woods, R. J. (2005) J. Comput. Chem. 26 1668-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez, C., Boelens, R., and Bonvin, A. M. (2003) J. Am. Chem. Soc. 125 1731-1737 [DOI] [PubMed] [Google Scholar]
- 23.Rost, B., Yachdav, G., and Liu, J. (2004) Nucleic Acids Res. 32 321-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fallon, J. L., Halling, D. B., Hamilton, S. L., and Quiocho, F. A. (2005) Structure 13 1881-1886 [DOI] [PubMed] [Google Scholar]
- 25.Van Petegem, F., Chatelain, F. C., and Minor, D. L., Jr. (2005) Nat. Struct. Mol. Biol. 12 1108-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard, S., and Thornton, J. (1992-96) NACCESS, University of Manchester, U.K.
- 27.Notredame, C., Higgins, D. G., and Heringa, J. (2000) J. Mol. Biol. 302 205-217 [DOI] [PubMed] [Google Scholar]
- 28.Edgar, R. C. (2004) Nucleic Acids Res. 32 1792-1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., Dufayard, J. F., Guindon, S., Lefort, V., Lescot, M., Claverie, J. M., and Gascuel, O. (2008) Nucleic Acids Res. 36 465-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graph. 14 51-55 and 29-32 [DOI] [PubMed] [Google Scholar]
- 31.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA
- 32.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 33.Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10037-10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan, H. L., Bink-Boelkens, M. T., Bezzina, C. R., Viswanathan, P. C., Beaufort-Krol, G. C., van Tintelen, P. J., van den Berg, M. P., Wilde, A. A., and Balser, J. R. (2001) Nature 409 1043-1047 [DOI] [PubMed] [Google Scholar]
- 35.Schubert, M., Labudde, D., Oschkinat, H., and Schmieder, P. (2002) J. Biomol. NMR 24 149-154 [DOI] [PubMed] [Google Scholar]
- 36.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York
- 37.Wishart, D. S., and Sykes, B. D. (1994) J. Biomol. NMR 4 171-180 [DOI] [PubMed] [Google Scholar]
- 38.Maler, L., Sastry, M., and Chazin, W. J. (2002) J. Mol. Biol. 317 279-290 [DOI] [PubMed] [Google Scholar]
- 39.Gibrat, J. F., Madej, T., and Bryant, S. H. (1996) Curr. Opin. Struct. Biol. 6 377-385 [DOI] [PubMed] [Google Scholar]
- 40.Holm, L., and Sander, C. (1996) Science 273 595-603 [DOI] [PubMed] [Google Scholar]
- 41.Wingo, T. L., Shah, V. N., Anderson, M. E., Lybrand, T. P., Chazin, W. J., and Balser, J. R. (2004) Nat. Struct. Mol. Biol. 11 219-225 [DOI] [PubMed] [Google Scholar]
- 42.Minami, Y., Emori, Y., Imajoh-Ohmi, S., Kawasaki, H., and Suzuki, K. (1988) J. Biochem. (Tokyo) 104 927-933 [DOI] [PubMed] [Google Scholar]
- 43.Minami, Y., Emori, Y., Kawasaki, H., and Suzuki, K. (1987) J. Biochem. 101 889-895 [DOI] [PubMed] [Google Scholar]
- 44.Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003) Physiol. Rev. 83 731-801 [DOI] [PubMed] [Google Scholar]
- 45.Babitch, J. (1990) Nature 346 321-322 [DOI] [PubMed] [Google Scholar]
- 46.Abriel, H., Wehrens, X. H., Benhorin, J., Kerem, B., and Kass, R. S. (2000) Circulation 102 921-925 [DOI] [PubMed] [Google Scholar]
- 47.Clancy, C. E., and Rudy, Y. (2002) Circulation 105 1208-1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujiwara, T., Sugawara, T., Mazaki-Miyazaki, E., Takahashi, Y., Fukushima, K., Watanabe, M., Hara, K., Morikawa, T., Yagi, K., Yamakawa, K., and Inoue, Y. (2003) Brain 126 531-546 [DOI] [PubMed] [Google Scholar]
- 49.Glaaser, I. W., Bankston, J. R., Liu, H., Tateyama, M., and Kass, R. S. (2006) J. Biol. Chem. 281 24015-24023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.