Abstract
Alternative splice variants of fibroblast growth factor receptor 2 (FGFR2) IIIb, designated C1, C2, and C3, possess progressive reduction in their cytoplasmic carboxyl termini (822, 788, and 769 residues, respectively), with preferential expression of the C2 and C3 isoforms in human cancers. We determined that the progressive deletion of carboxyl-terminal sequences correlated with increasing transforming potency. The highly transforming C3 variant lacks five tyrosine residues present in C1, and we determined that the loss of Tyr-770 alone enhanced FGFR2 IIIb C1 transforming activity. Because Tyr-770 may compose a putative YXXL sorting motif, we hypothesized that loss of Tyr-770 in the 770YXXL motif may cause disruption of FGFR2 IIIb C1 internalization and enhance transforming activity. Surprisingly, we found that mutation of Leu-773 but not Tyr-770 impaired receptor internalization and increased receptor stability and activation. Interestingly, concurrent mutations of Tyr-770 and Leu-773 caused 2-fold higher transforming activity than caused by the Y770F or L773A single mutations, suggesting loss of Tyr and Leu residues of the 770YXXL773 motif enhances FGFR2 IIIb transforming activity by distinct mechanisms. We also determined that loss of Tyr-770 caused persistent activation of FRS2 by enhancing FRS2 binding to FGFR2 IIIb. Furthermore, we found that FRS2 binding to FGFR2 IIIb is required for increased FRS2 tyrosine phosphorylation and enhanced transforming activity by Y770F mutation. Our data support a dual mechanism where deletion of the 770YXXL773 motif promotes FGFR2 IIIb C3 transforming activity by causing aberrant receptor recycling and stability and persistent FRS2-dependent signaling.
Fibroblast growth factors (FGFs)2 compose a large family of structurally related growth factors (22 human members) that mediate cell proliferation, differentiation, migration, and angiogenesis (1–4). The activities of FGFs are mediated by their binding to a family of four receptor tyrosine kinases (RTKs), designated FGFR1–4 (5). FGFRs are composed of an extracellular domain that consists of two or three Ig-like domains, a single transmembrane domain, and an intracellular catalytic tyrosine kinase domain and flanking regulatory sequences (Fig. 1A).
FIGURE 1.
Loss of carboxyl-terminal sequences enhances FGFR2 IIIb transforming potency in RIE-1 epithelial cells and Rat-1 fibroblasts. A, carboxyl-terminal sequence comparison of three FGFR2 IIIb splice variants (C1, C2, and C3). Known or putative phosphorylated tyrosine residues are indicated. B, conservation of carboxyl-terminal tyrosines in FGFR family members. Of the five tyrosine residues present in the C1 but not C3 variant of FGFR2, only Tyr-770 (Tyr-766 in FGFR1) and Tyr-780 (Tyr-776 in FGFR1) are conserved in all four receptors. Receptor autophosphorylation of Tyr-770 of FGFR2 IIIb (analogous to Tyr-766 of FGFR1 and Tyr-760 of FGFR3) creates a recognition site for the Src homology 2 domain of PLCγ. C, loss of carboxyl-terminal sequences enhances FGFR2 IIIb-induced focus formation in RIE-1 epithelial cells. Mass populations of RIE-1 cells that stably express the indicated FGFR2 IIIb carboxyl-terminal splice variants were assayed for focus forming activity using a secondary focus formation assay. Cells were plated and allowed to grow for 21 days, and then the appearance of foci of transformed cells was monitored. The cultures were fixed and stained with crystal violet, and focus forming activity was quantitated. D, loss of carboxyl-terminal sequences enhances FGFR2 IIIb-induced soft agar growth of RIE-1 cells. Mass populations of RIE-1 cells that stably expressed the indicated FGFR2 IIIb variants were assayed for their ability to grow in soft agar. The number of colonies was quantitated after 21 days. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of three independent experiments. E, loss of carboxyl-terminal sequences enhances FGFR2 IIIb-induced focus formation in Rat-1 cells. Analyses were done similarly as described for RIE-1 cells. F, loss of carboxyl-terminal sequences enhances FGFR2 IIIb-induced soft agar growth of Rat-1 cells. Analyses were done similarly as described for RIE-1 cells.
An important feature and mode of regulation of FGFR2 function is that structural variants of FGFR2 are generated by numerous alternative gene splicing events that generate transcripts that encode proteins altered in both the extracellular and intracellular regions of the FGFR2 (5). To date, more than 20 alternative splicing variants of FGFR2 have been identified. The first major splicing event occurs in the second half of the third Ig-like domain (designated Ig-III domain). Tissue-specific inclusion of either exon IIIb or exon IIIc that encodes for the second half of the Ig-III domain generates either the epithelial cell-specific IIIb or mesenchymal cell-specific IIIc isoforms (6). This alternative splicing determines the ligand binding specificity of FGFR2. Although FGFR2 IIIb (also called KGFR) binds FGF7 (also called KGF) and FGF10, but not FGF2, FGFR2 IIIc (also called BEK) binds FGF2, but not FGF7 and FGF10 (7–10). Interestingly, although FGFR2 IIIb expression is restricted to epithelial cells (6), expression of the ligands for FGFR2 IIIb (FGF7 and FGF10) is restricted to mesenchymal cells (11–16), resulting in the creation of a paracrine signaling loop in epithelial-mesenchymal interactions that is likely to be critically for promoting FGFR2 IIIb activity in oncogenesis.
The second major splicing occurs in sequences that encode the carboxyl terminus of FGFR2. Three splice variants of FGFR2 IIIb that differ in their carboxyl-terminal sequences have been identified (designated C1, C2, and C3) (17). The C2-type carboxyl terminus is 34 amino acids shorter than the C1-type carboxyl terminus, and the C3-type carboxyl terminus is 19 amino acids shorter than C2-type carboxyl terminus (Fig. 1A). These sequence differences result in differential retention of tyrosine residues that may serve as sites of receptor autophosphorylation and docking sites for cytoplasmic signaling proteins.
A previous study found that expression of the C3 isoform was increased in gastric cancer cell lines (17). We also observed enhanced expression of C2 and C3 isoforms in a majority of human breast carcinoma cell lines when compared with nontransformed MCF-10A human mammary epithelial cells (18), suggesting that aberrant expression of the C2 or C3 splicing variants may contribute to cancer development. Furthermore, the C3 variant that lacks carboxyl-terminal sequences was shown to be more transforming than the IIIb C1 variant when expressed ectopically in NIH 3T3 fibroblasts and human mammary epithelial cells (17, 19). However, whether the C2 variant is more (or less) transforming than the C1 (or C3) variant has not been determined. Furthermore, the mechanistic basis for the enhanced transforming activity of the C3 variant remains to be elucidated.
Like other RTKs, FGFRs are activated by ligand-induced dimerization, causing stimulation of their intrinsic tyrosine kinase activity, tyrosine autophosphorylation, and recruitment of signaling proteins to specific phosphorylated tyrosine residues in their cytoplasmic carboxyl termini. The two best characterized downstream signaling components of FGFRs are phospholipase C-γ (PLCγ) and FGF receptor substrate 2 (FRS2) (1–4). In FGFR1, Tyr-766 in the carboxyl terminus is the major autophosphorylation site on FGFR1 and serves as a binding site for the Src homology 2 domain of PLCγ, resulting in tyrosine phosphorylation and activation of PLCγ (20, 21), leading to stimulation of phosphatidylinositol (PI) hydrolysis and the generation of diacylglycerol and inositol 1,4,5-trisphosphate (22). The Tyr-766 residue in FGFR1 is well conserved in all four FGFR family members and corresponds to Tyr-770 in FGFR2 IIIb (Fig. 1B). Recently, it was shown that a Y770F missense mutant of FGFR2 IIIb C1 cannot bind and phosphorylate PLCγ (23). However, whether this mutant fails to activate PI hydrolysis and whether altered PLCγ binding and activation contribute to FGFR2 transforming activity remain unresolved.
Unlike PLCγ, FRS2 association with FGFR is constitutive and independent of ligand stimulation and receptor phosphorylation (24). FRS2 is a docking protein that binds to a conserved sequence within the juxtamembrane domain of FGFRs. Ligand-stimulated activation of FGFRs leads to phosphorylation of multiple tyrosine residues in FRS2 (25). One well characterized FRS2 effector is the Grb2 adaptor protein that binds to tyrosine-phosphorylated FRS2 and forms a stable complex with the Sos Ras guanine nucleotide exchange factor, thus stimulating activation of the Ras small GTPase and the ERK MAPK cascade (25). In addition, Grb2 can recruit the docking protein Gab1, which is then tyrosine-phosphorylated by FGFR2 activation. The tyrosine-phosphorylated Gab1 recruits phosphatidylinositol 3-kinase (PI3K), resulting in activation of the AKT serine/threonine kinase (26).
One mechanism of negative regulation of RTK signaling involves ligand-mediated receptor internalization and lysosomal degradation (27, 28). Therefore, disruption of receptor internalization could lead to aberrant receptor activation and contribute to the development of human disease, including cancer. Interestingly, FGFR2 IIIb C1 contains a putative YXXΦ tyrosine-based sorting motif (where X is any amino acid and Φ is a bulky hydrophobic amino acid) (29) in carboxyl-terminal sequences deleted in the C3 variant, suggesting that the loss of the YXXΦ motif might impair receptor internalization and contribute to the enhanced transforming activity of the FGFR2 IIIb C3 variant.
To date, a detailed comparative analysis of the transforming potential of FGFR2 IIIb C1, C2, and C3 using the same cell system has not been done. In this study, we compared the transforming potency of FGFR2 IIIb C1, C2, and C3 splicing variants and found that the C3 variant is considerably more transforming than the C2 variant, and the C2 variant is modestly enhanced in transforming activity when compared with the weakly transforming C1 variant. Our results support a model where the potent transforming activity of the FGFR2 IIIb C3 splice variant is mediated, in part, by a mechanism involving loss of the 770YLDL motif, resulting in impaired receptor internalization and enhanced FRS2-dependent signaling.
EXPERIMENTAL PROCEDURES
Plasmid Expression Vectors—Human FGFR2 IIIb C1, C2, and C3 cDNA sequences were generated by PCR amplification from a T-47D breast cancer cell cDNA library and subcloned into the pBabe-puro retrovirus mammalian expression vector (5′-SalI and 3′-BamHI sites). The FGFR2 IIIb C2 (QST) cDNA sequence (deletion of FGFR2 IIIb C1 residues 789–822) was created by PCR amplification from the FGFR2 IIIb C1 cDNA sequence and subcloned into pBabe-puro (5′-SalI and 3′-BamHI sites). Additional cDNA sequences encoding missense mutations were created by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. Oligonucleotides designed to create the appropriate mutations to encode the indicated amino acid substitutions are as follows: 5′-CACAACCAATGAGGAATTCTTGGACCTCAGCCAACC-3′ (Y770F); 5′-GCCAACCTCTCGAACAGTTTTCACCTAGTTACCCTG-3′ (Y780F); 5′-CAGTATTCACCTAGTTTCCCTGACACAAGAAG-3′ (Y784F); 5′-CCAGACCCCATGCCTTTCGAACCATGCCTTCCT-3′ (Y806F); 5′-CATGCCTTCCTCAGTTTCCACACATAAACGGC-3′ (Y813F); 5′-GAGGAATTCTTGGACGCCAGCCAACCTCTCGAACAG-3′ (Y770F/L773A); 5′-CCAATGAGGAAGCCTTGGACGCCAGCCAACCTCTCG-3′ (Y770A/L773A); 5′-GAGGAATACTTGGACGCCAGCCAACCTCTCG-3′ (L773A); and 5′-GCACAAGCTGACCGCACGTATCGCCGCGCGGAGACAGG-3′ (K421A/P424A/L425A).
Cell Culture and Transformation Assay—Rat-1 and RIE-1 cells were maintained in Dulbecco's modified minimum essential medium supplemented with 10% fetal calf serum. For secondary focus formation assays, Rat-1 and RIE-1 cells were stably infected with pBabe-puro constructs encoding wild type FGFR2 IIIb isoforms and missense mutants of FGFR2 IIIb C1. Expression of ectopically introduced FGFR2 IIIb C1 proteins was determined by immunoblot analyses with anti-FGFR2 antibody (sc-122; Santa Cruz Biotechnology). After infection, cells were selected in growth medium supplemented with puromycin (2 μg/ml). Drug-resistant colonies were pooled and replated into 60-mm dishes and maintained in growth medium for 2–3 weeks. Cells were then fixed and stained with crystal violet. To determine anchorage-independent growth potential, Rat-1 cells stably expressing either the pBabe-puro empty vector or encoding various FGFR2 IIIb proteins were suspended in 0.4% bacto-agar in growth medium at 5 × 104 cells per 60-mm dish. After 2–3 weeks, colonies were stained with 2 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, and the average number of colonies on duplicate dishes was calculated. For conditioned medium assays, Rat-1 fibroblast conditioned medium was collected and passaged through a 0.45-μm filter. RIE-1 cells stably infected with the pBabe-puro empty vector, or encoding WT or Y770F mutant FGFR2 IIIb C1, were grown either in conditioned medium or regular growth medium, replenished every 2 days, for 3 weeks before cells were fixed and stained with crystal violet.
Immunofluorescence Microscopy—Rat-1 cells that stably express either wild type or mutant FGFR2 IIIb proteins were serum-starved for 20 h and then incubated with either vehicle (bovine serum albumin (BSA)) or 50 ng/ml FGF7 (R & D Systems) for 40 min at 37 °C to allow internalization. Cells were then fixed using 4% paraformaldehyde in phosphate-buffered saline (PBS), washed, and permeabilized (0.1% Triton X-100). Next, cells were immunostained with rabbit polyclonal FGFR2 antibody (sc-122; Santa Cruz Biotechnology) to determine FGFR2 IIIb subcellular location by Zeiss 510 LSM confocal microscope.
Cleavable Biotin Internalization Assay—Internalization of FGFR2 IIIb was quantitatively measured as described previously (30) with the following modifications. Rat-1 cells that stably express either wild type or mutant FGFR2 IIIb proteins were serum-starved for 20 h. The cells were then washed three times with ice-cold PBS containing Ca2+ and Mg2+ (Ca2+/Mg2+ PBS). For surface biotinylation, cells were incubated with thiolcleavable sulfo-NHS-S-S-biotin (0.5 mg/ml; Pierce) for 1 h on ice. Cells were then washed three times with ice-cold Ca2+/Mg2+ PBS and stimulated with FGF7 at 37 °C for the indicated times to allow internalization. Cells were then put back on ice and washed three times with ice-cold Ca2+/Mg2+ PBS. To remove the residual surface biotin, cells were washed three times, for 15 min each, with ice-cold glutathione (GSH; membrane-nonpermeable reducing agent) cleavage solution (50 mm glutathione, 75 mm NaCl, 1 mm EDTA, 1% BSA, and 0.75% 10 n NaOH). Cells were further washed twice on ice with Ca2+/Mg2+ PBS and incubated for 15 min with 5 mg/ml iodoacetamide (in Ca2+/Mg2+ PBS with 1% BSA; quenching buffer) to quench free sulfo-reactive groups. Cells were then washed five times, lysed, and immunoprecipitated with Neutravidin beads (Pierce) followed by immunoblotting with FGFR2 antibody. Immunoblot band intensity was quantified by ImageJ analysis (National Institutes of Health).
Receptor Down-regulation Assays—Rat-1 cells that stably express either wild type or mutant FGFR2 IIIb proteins were incubated with 50 ng/ml FGF7 for the indicated times at 37 °C to induce ligand-induced receptor degradation. Immunoblotting analyses with FGFR2 antibody were performed to determine the level of remaining FGFR2 protein. Band intensity was quantified by ImageJ analysis.
Signaling Analyses—To determine FGFR2 tyrosine autophosphorylation, cells were lysed and analyzed for tyrosine-phosphorylated FGFR2 by immunoprecipitation with an anti-FGFR2 antibody followed by immunoblotting with an anti-phosphotyrosine antibody (05-321, clone 4G10; Upstate Biotechnology, Inc.). To determine PLCγ activation, cells were lysed and then analyzed for tyrosine-phosphorylated PLCγ by immunoprecipitation with anti-PLCγ antibody (sc-7290, clone E-12; Santa Cruz Biotechnology) followed by immunoblotting with anti-phosphotyrosine antibody. PI hydrolysis assays were performed to measure PLCγ activity as we have described previously (31). Briefly, Rat-1 cells stably expressing either wild type FGFR2 IIIb C1 or 770F mutant were labeled with myo-[3H]inositol in inositol-free medium. Then cells were stimulated with FGF7, vehicle (BSA, negative control), thrombin (positive control), or lysophosphatidic acid (positive control) for 20 min in medium supplemented with 10 mm LiCl. Accumulation of [3H]inositol phosphates was quantitated as described previously (31).
To determine FRS2 activation, cells were lysed and then analyzed for tyrosine-phosphorylated FRS2 by immunoprecipitation with anti-FRS2 antibody (sc-8318; Santa Cruz Biotechnology) followed by immunoblotting with anti-phosphotyrosine antibody. To determine Gab1 activation, cells were lysed and analyzed for tyrosine-phosphorylated Gab1 by immunoprecipitation with anti-Gab1 antibody (sc-9049; Santa Cruz Biotechnology) followed by immunoblotting with anti-phosphotyrosine antibody. To determine MEK, ERK, or AKT activation, cells were lysed and then analyzed for active MEK, ERK, or AKT by immunoblot analyses with antibody specifically to detect phosphorylated MEK1/2 (9121; Cell Signaling Technology), ERK (9106; Cell Signaling Technology), or AKT (9271; Cell Signaling Technology). Total ERK and AKT levels were determined by anti-ERK (9102; Cell Signaling Technology) and anti-AKT antibodies (9272; Cell Signaling Technology), respectively.
To determine FRS2 binding to FGFR2, cells were lysed and immunoprecipitated with anti-FRS2 antibody followed by immunoblotting with anti-FGFR2 antibody. The same membrane was stripped and reprobed with the anti-FRS2 antibody to determine the protein levels of endogenous FRS2.
RESULTS
Loss of Carboxyl-terminal Sequences Enhances FGFR2 IIIb Transforming Potency in Rat-1 Fibroblasts and RIE-1 Epithelial Cells—To determine the role of carboxyl-terminal sequences in FGFR2 IIIb transformation, we compared the transforming potency of FGFR2 IIIb C1, C2, and C3 variants by examining two aspects of growth transformation, loss of density-dependent growth inhibition, and acquisition of anchorage-independent growth potential. For these analyses, we established Rat-1 rat fibroblasts and RIE-1 rat intestinal epithelial cells stably expressing the FGFR2 IIIb C1, C2, or C3 variants. First, we compared the ability of the three FGFR2 IIIb variants to induce loss of density-dependent growth inhibition by quantitating the appearance of foci of multilayered cells in confluent cultures. Cells stably transfected with the empty vector or expressing the C1 variant exhibited no focus forming activity. In contrast, we found that cells expressing the C2 variant exhibited weak focus forming activity, whereas the C3 variant caused significantly greater focus forming activity than the C2 variant in RIE-1 (Fig. 1C) as well as Rat-1 cells (Fig. 1E).
Second, we compared the ability of three FGFR2 IIIb variants to promote anchorage-independent growth by soft agar assay. Consistent with their relative focus-forming potencies, the C3 variant induced more soft agar colonies than the C2 variant, and the C2 variant induced more soft agar colonies than the C1 variant in RIE-1 (Fig. 1D) as well as Rat-1 cells (Fig. 1F). A similar hierarchy of transforming potency, as measured by soft agar colony formation, was seen when the FGFR2 IIIb isoforms were stably expressed in MCF-10 human breast epithelial cells (data not shown). These data indicate that multiple carboxyl-terminal sequences function as negative regulators of FGFR2 IIIb transforming activity, with the loss of residues within the region spanning 769–788 causing the most significant activation of transforming activity.
Mutation of Y770F Alone Activates FGFR2 IIIb C1 Transforming Activity in Rat-1 Fibroblasts—Next, we determined the critical residue losses that contribute to the enhanced transforming activity of FGFR2 IIIb variants lacking carboxyl-terminal sequences. The carboxyl-terminal domain tyrosine residues of RTKs are sites of autophosphorylation and critical for their cytoplasmic signaling and growth regulatory activities. The C3 variant lacks five tyrosine residues (Tyr-770, Tyr-780, Tyr-784, Tyr-806, and Tyr-813), whereas the C2 variant lacks two tyrosine residues (Tyr-806 and Tyr-813) present in the C1 isoform (Fig. 1A). We hypothesized that the loss of specific tyrosine residue(s) might account for the enhanced transforming activity of the C2 and (or) C3 variants. To address this possibility, we introduced phenylalanine substitutions at each of the five tyrosine residues (Y770F, Y780F, Y784F, Y806F, and Y813F) of the carboxyl terminus of the weakly transforming C1 variant. Because loss of multiple tyrosine residues might be required for enhanced transforming activity, we also generated double (F2, Y806F/Y813F), triple (F3, Y784F/Y806F/Y813F), quadruple (F4, Y770F/Y784F/Y806F/Y813F), and quintuple (F5, Y770F/Y780F/Y784F/Y806F/Y813F) mutants. We then established mass populations of Rat-1 cells that stably expressed wild type and mutant FGFR2 IIIb C1 proteins and compared their transforming potency in soft agar assays.
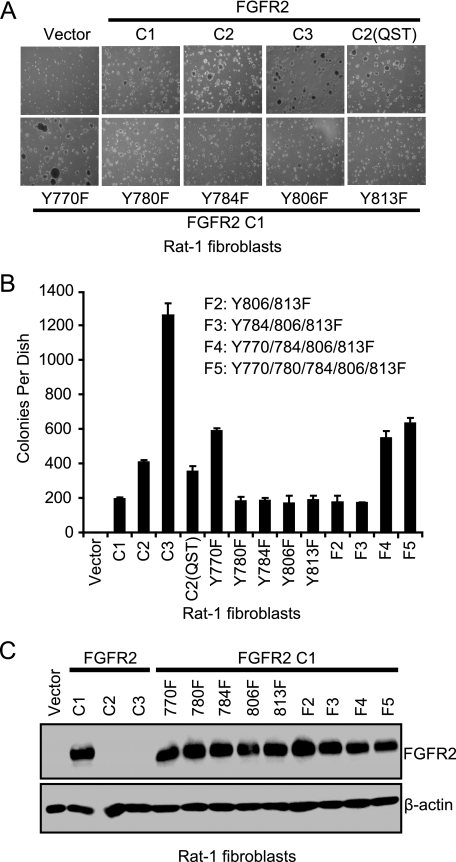
We found that the Y770F mutation alone promoted colony formation in soft agar (∼600 colonies per dish), whereas receptors with the Y780F, Y784F, Y806F, or Y813F mutations showed the same weak colony forming activity (∼200 colonies per dish) as cells expressing the WT FGFR2 IIIb C1 receptor (Fig. 2, A and B). In addition, we found that the F2 double or F3 triple mutant receptors showed the same transforming potency as WT FGFR2 IIIb C1 (Fig. 2B), whereas the F4 quadruple and F5 quintuple mutants that included the Y770F mutation promoted colony formation in soft agar to the same extent as the Y770F single mutant (Fig. 2B). We found that all FGFR2 IIIb C1 mutants were expressed at a similar level as WT FGFR2 IIIb C1 (Fig. 2C), indicating that the differences in transforming activity were not because of differences in steady-state levels of protein expression.
FIGURE 2.
Mutation of Y770F alone activates FGFR2 IIIb C1 transforming activity in Rat-1 fibroblasts. A, Rat-1 cells that stably expressed the indicated FGFR2 IIIb proteins were assayed for their ability to grow in soft agar. Cells were suspended in 0.4% soft agar and photographed 21 days after plating. The C2(QST) indicates a truncation mutant of C1 that has same length as C2 but contains the 779Q/783S/787T sequence. B, number of colonies was quantitated after 21 days. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of two independent experiments. C, Rat-1 cells that stably express the indicated FGFR2 IIIb proteins were assayed for their FGFR2 IIIb protein expression levels by immunoblot analyses with FGFR2 antibody against a peptide sequence in the FGFR2 IIIb C1 carboxyl-terminal sequence. This sequence has been deleted in the FGFR2 IIIb C2 and FGFR2 IIIb C3 variants; therefore, we cannot determine the level of expression of these two isoforms with this antibody. Blot analysis with anti-β-actin was done to verify equivalent total protein loading.
Because the anti-FGFR2 antibody recognizes a carboxyl-terminal epitope that is deleted in C2 and C3, we could not exclude the possibility that different transforming potency of the C2 and C3 mutants may be due, in part, to different levels of expression. As an indirect evaluation of receptor expression, we did find that FGF7 stimulation of the cells expressing the C1, C2, and C3 variants showed comparable levels of phosphorylated FRS2 (Fig. 7A), suggesting that the three isoforms were expressed at comparable levels. Together, these results indicate that loss of the Tyr-770 residue, but not other tyrosine residues (Tyr-780, Tyr-784, Tyr-806, and Tyr-813), contributes to the increased transforming potency of the C3 variant. However, because the C2 variant retains Tyr-770 (Fig. 1A), the loss of this residue is not the basis for the increased transforming activity of the C2 variant.
FIGURE 7.
Loss of the Tyr-770 causes sustained steady-state activation of FRS2 but not MEK-ERK or Gab1-AKT pathways. A, loss of Tyr-770 causes sustained activation of FRS2 in the absence of FGF7 stimulation. Rat-1 cells stably expressing the indicated FGFR2 IIIb proteins were grown to confluence and stimulated with either vehicle or 50 ng/ml FGF7 for 20 min. Then cells were lysed, and FRS2 activity was determined by immunoprecipitation (IP) with anti-FRS2 antibody followed by immunoblotting (IB) with anti-phosphotyrosine (P-Tyr) antibody. Total cell lysates were analyzed to determine stable FGFR2 IIIb protein expression levels by immunoblot analyses with anti-FGFR2 antibody. B, Rat-1 cells stably expressing the indicated FGFR2 IIIb proteins were stimulated with either vehicle or 50 ng/ml FGF7 for 20 min. MEK, ERK, or AKT activation was determined by immunoblot analyses using antibody that recognizes activated, phosphorylated forms of MEK, ERK, or AKT, respectively. To determine Gab1 activity, cells were lysed and immunoprecipitated with anti-Gab1 antibody followed by immunoblot analysis with phosphotyrosine antibody. C, MEK and PI3K activities are required for enhanced transforming potency by Y770F mutation. Rat-1 cells stably expressing the indicated FGFR2 IIIb were suspended in 0.4% soft agar. The cells were incubated for 3 weeks in the absence or presence of 10 μm LY294002 (PI3K inhibitor) or 30 μm U0126 (MEK inhibitor). Cells were photographed about 3 weeks after plating. Colonies per dish were counted 3 weeks after plating. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of two independent experiments.
It is notable that the C2 variant lacks 34 carboxyl-terminal residues found in the C1 variant, but additionally, it is also divergent from the C1 variant for three amino acids at positions 779, 783, and 787 (Fig. 1A). Although C1 contains 779Q/783S/787T, C2 contains 779P/783C/787P. Therefore, we speculated that differences in these three amino acids might contribute to the increased transforming potency of the C2 variant. To address this possibility, we generated a truncation mutant of C1 that has same length as C2 but contains the 779Q/783S/787T sequence (designated C2 (QST)) rather than the 779P/783C/787P sequence. We found that this C2 (QST) mutant exhibited a similar transforming potency as the C2 variant (Fig. 2B), indicating that the loss of the carboxyl-terminal 34 amino acids (from 789 residue to 822 residue), and not differences in 779, 783, and 787 residues, is responsible for increased transforming potency of the C2 variant.
Mutation of Y770F Does Not Activate FGFR2 IIIb C1 Transforming Activity in RIE-1 Epithelial Cells—In contrast to our observations in Rat-1 cells, Moffa and Ethier (32) showed that mutation of Tyr-770 alone was not sufficient to enhance FGFR2 IIIb C1 growth transforming activity when assayed in HME mammary epithelial cells. One possible explanation for their different conclusion is that fibroblasts, but not epithelial cells, express the ligand (KGF/FGF7) for FGFR2 IIIb (11, 13, 16). Therefore, FGFR2 IIIb can be activated in fibroblasts but not epithelial cells by formation of an autocrine signaling loop (11, 13, 33, 34). Thus, we examined whether the Y770F mutation enhanced FGFR2 IIIb C1 transforming activity when expressed in RIE-1 epithelial cells. Consistent with the previous report (32), we found that the Y770F mutation did not increase FGFR2 IIIb focus forming activity in RIE-1 epithelial cells (Fig. 3A). To exclude the possibility that different expression levels of proteins contributed to different transforming activities, we determined all FGFR2 IIIb C1 mutants were expressed at a similar level as WT FGFR2 IIIb C1 (Fig. 3B).
FIGURE 3.
Mutation of Y770F does not activate FGFR2 IIIb C1 transforming activity in RIE-1 epithelial cells. A, RIE-1 cells that stably expressed the indicated FGFR2 IIIb proteins were assayed for their ability to grow in soft agar. The number of colonies was quantitated after 21 days. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of two independent experiments. B, RIE-1 cells that stably express the indicated FGFR2 IIIb proteins were assayed for their FGFR2 IIIb protein expression levels by immunoblot analyses with anti-FGFR2 antibody that recognizes a peptide sequence in the FGFR2 IIIb C1, but not C2 or C3, carboxyl-terminal sequence. C, mutation of Tyr-770 causes ligand-dependent transformation of RIE-1 cells. RIE-1 cells stably infected with the empty vector or encoding WT or Y770F mutant FGFR2 IIIb C1 were grown in the regular growth medium or Rat-1 fibroblasts conditioned medium for 3 weeks (replenished every 2 days) before cells were fixed and stained with crystal violet.
Breast carcinoma cells expressing FGFR2 IIIb in situ will be exposed to FGFR2 IIIb ligands secreted by adjacent mesenchymal stromal tissue (16). Therefore, we determined whether the ligands secreted by fibroblasts can enhance the transforming potency of the Y770F mutant when expressed in RIE-1 epithelial cells. To address this question, conditioned medium from Rat-1 fibroblast was collected and added to RIE-1 cells stably expressing the WT or Y770F mutant receptor. Although Rat-1 fibroblast conditioned medium did not enhance the focus forming activity of empty vector control and WT receptor-expressing cells, it promoted the focus forming activity of the Y770F mutant (Fig. 3C). These observations suggest that both loss of Tyr-770 and paracrine stimulation by fibroblast-expressed ligands are required for the enhanced transforming activity of the Y770F mutant receptor in RIE-1 epithelial cells. These results also provide a mechanistic explanation for the negative observations of Moffa and Ethier (32) in mammary epithelial cells.
To emphasize the importance of epithelial (FGFR2 IIIb)-stromal (FGF7) interactions, for the remaining studies we used Rat-1 fibroblasts as our model cell system. This allowed us the ability to study how FGFR2 IIIb may function in epithelial cells when exposed to FGFR2 IIIb ligands expressed by surrounding stromal fibroblasts. Additionally, because Rat-1 cells do not express endogenous FGFR2 IIIb receptors, our analyses of mutant receptor function were not complicated by any activity contributed by endogenous receptor activity.
Leu-773 but Not Tyr-770 of the 770YXXL Motif Is Required for Ligand-induced FGFR2 IIIb C1 Internalization and Degradation—Next, we sought to determine how mutation of the Tyr-770 residue enhanced FGFR2 IIIb C3 transforming activity. Interestingly, the Tyr-770 residue and flanking sequences (770YLDL) correspond to a putative YXXΦ motif (Φ= bulky hydrophobic residue) that is known to be the major determinant for endocytosis of many transmembrane proteins (29). However, a previous study found that a Y770F mutation did not impair FGFR2 IIIb internalization, arguing against this possibility (23). Because there is evidence that the tyrosine residue of the YXXΦ sorting motif can be substituted with aromatic amino acids such as phenylalanine or tryptophan (35–37), we postulated that if Tyr-770 is substituted with an alanine instead of phenylalanine, the YXXΦ sorting signal might be disrupted. Alternatively, it is also possible that both the Tyr-770 and Leu-773 residues of 770YXXL motif might be required for FGFR2 internalization and that concurrent mutation of both residues will be required to disrupt its function.
To determine whether the Tyr-770 and/or Leu-773 residues are required for FGFR2 IIIb C1 internalization, we generated mutant cDNA sequences encoding four FGFR2 IIIb C1 missense mutants (Y770F, Y770F/L773A, Y770A/L773A, and L773A). Because steady-state protein levels may not reflect the kinetics of receptor internalization and turnover, we compared the ability of WT and mutant receptors to undergo ligand-stimulated internalization. For these analyses, Rat-1 cells that stably expressed WT or mutant FGFR2 IIIb C1 proteins were stimulated with either vehicle (BSA) or 50 ng/ml FGF7 and immunostained with anti-FGFR2 antibody to determine the subcellular localization of receptors. In unstimulated cells, the WT and all four mutant receptors were found associated with the plasma membrane as well as in the intracellular compartment (Fig. 4A). After FGF7 stimulation, no plasma membrane-associated WT receptor was seen, with the majority internalized to punctate vesicular structures (Fig. 4A). Similar to the WT receptor, the majority of the Y770F mutant receptor was also translocated to a punctate vesicular structure after FGF7 stimulation. However, all three mutant receptors containing the L773A substitution (Y770F/L773A, Y770A/L773A, and L773A) showed impaired ability to internalize and retain significant plasma membrane association after FGF7 stimulation (Fig. 4A).
FIGURE 4.
Mutation of the Leu-773 but not Tyr-770 residue of the 770YXXL motif causes impaired ligand-induced FGFR2 IIIb C1 internalization and degradation and sustained activation of FGFR2 IIIb. A, mutation of the Leu-773 residue of the 770YXXL motif reduces ligand-stimulated FGFR2 IIIb C1 dissociation from the plasma membrane. Rat-1 cells that stably express the indicated FGFR2 IIIb proteins were serum-starved for 20 h and then incubated with serum-free medium supplemented with either vehicle (BSA) or 50 ng/ml FGF7 for 40 min at 37 °C. The cells were fixed, permeabilized, and immunostained with anti-FGFR2 antibody to determine FGFR2 IIIb subcellular location by confocal microscopy. B, mutation of the Leu-773 residue of the 770YXXL motif reduces FGFR2 IIIb C1 internalization. Following surface biotinylation with thiol-cleavable sulfo-NHS-S-S-biotin, cells were treated with FGF7 (50 ng/ml) for the indicated time intervals to allow internalization of biotinylated FGFR2 IIIb and processed as described under “Experimental Procedures.” The amount of internalized receptor was determined by immunoblotting with FGFR2 antibody (left panel) and quantified as a percentage of the amount of total surface labeling (Total) that was measured in the absence of a biotin cleavage step (right panel). Data shown are the average of two independent experiments with the bars indicating standard deviation. C, mutation of Leu-773 of the 770YXXL motif reduces the rate of ligand-induced FGFR2 IIIb C1 degradation. Rat-1 cells that stably express either wild type or mutant FGFR2 IIIb proteins were incubated with 50 ng/ml FGF7 for the indicated times at 37 °C to induce ligand-induced receptor degradation. Immunoblotting analyses with FGFR2 antibody were performed to determine the level of remained FGFR2 protein (left panel). The receptor remaining indicates the percentage of FGFR2 IIIb remaining in FGF7-stimulated cells when compared with untreated control cells (right panel). The data shown are the average of two independent experiments with the bars indicating standard deviation. D, loss of the Leu but not Tyr residue of the 770YXXL motif causes sustained activation of FGFR2 IIIb. Rat-1 cells stably expressing the indicated FGFR2 IIIb proteins were stimulated with either vehicle or 50 ng/ml FGF7 for 20 min. To determine FGFR2 autophosphorylation, cells were lysed and immunoprecipitated (IP) with anti-FGFR2 antibody followed by immunoblotting (IB) with anti-phosphotyrosine (P-Tyr) antibody.
To quantitatively determine FGFR2 IIIb internalization, we labeled cell surface FGFR2 IIIb with a cleavable biotin, and then measured the extent of translocation of the biotinylated FGFR2 IIIb to intracellular compartments. Consistent with immunostaining analyses, we found that the Y770F mutation did not impair receptor internalization, whereas all three mutant receptors containing the L773A substitution (Y770F/L773A, Y770A/L773A, and L773A) caused ∼50% inhibition of internalization when compared with the WT and Y770F mutant receptors (Fig. 4B). Together, these data indicate that the Leu-773 but not Tyr-770 residue is critical for FGFR2 IIIb C1 internalization. In general, after ligand-stimulated internalization, receptors translocate to the lysosomal compartment and are degraded (29). Thus, we next determined the role of Tyr-770 and Leu-773 residues in ligand-induced receptor degradation. For these analyses, Rat-1 cells that stably express WT or YXXL mutant receptors were stimulated with FGF7, and the amount of total FGFR2 IIIb protein remaining was monitored for 0–6 h (Fig. 4C). FGF7 stimulation caused a significant decrease in both WT and Y770F mutant receptors (∼70% decrease in receptor protein compared with the untreated control cells after 4 h of FGF7 stimulation). In contrast, the three mutant receptors containing the L773A substitution (Y770F/L773A, Y770A/L773A, and L773A) exhibited a decreased rate of FGF7-induced degradation (∼30% decrease in receptor protein compared with the untreated control cells after 4 h of FGF7 stimulation). These data indicate that the Leu-773 but not Tyr-770 residue is critical for ligand-induced FGFR2 IIIb C1 degradation.
Loss of the Leu but Not Tyr Residue of the 770YXXL Motif Causes Sustained Activation of FGFR2 IIIb—To determine whether the impairments in receptor internalization and degradation were correlated with aberrant activation of FGFR2 IIIb, Rat-1 cells that stably express WT or mutant receptors (Y770F, Y770F/L773A, Y770A/L773A and L773A) were stimulated with either vehicle or FGF7 for 20 min. Then cells were lysed and immunoprecipitated with FGFR2 antibody followed by immunoblot analyses with phosphotyrosine antibody to detect FGFR2 IIIb autophosphorylation.
FGF7 stimulation caused comparable increased tyrosine phosphorylation of the WT and all four mutant receptors (Fig. 4D). In the absence of FGF7, both the WT and Y770F mutant receptors showed low levels of tyrosine phosphorylation. In contrast, all three internalization defective mutants lacking the Leu-773 residue (Y770F/L773A, Y770A/L773A, and L773A) showed significantly higher levels of tyrosine-phosphorylated FGFR2 IIIb when compared with the WT and Y770F mutant receptors (Fig. 4D). Taken together, these data suggest that loss of the Leu-773 residue results in sustained activation of FGFR2 IIIb that may be caused by decreased receptor internalization and degradation.
Loss of the Tyr and Leu Residues of the 770YXXL Motif Enhances FGFR2 IIIb Transforming Activity by Distinct Mechanisms—We next determined whether loss of the Leu-773 residue, to cause impaired receptor down-regulation and sustained FGFR2 IIIb activation (Fig. 4), was also associated with an enhanced FGFR2 IIIb C1 transforming activity. We found that the L773A mutation alone was sufficient to enhance the anchorage-independent growth potential of FGFR2 IIIb (Fig. 5A). Because the Y770F mutation did not impair receptor down-regulation and did not induce sustained FGFR2 IIIb activation (Fig. 4), yet was associated with enhanced FGFR2 IIIb transforming activity (Fig. 2), the Y770F and L773A mutations appear to cause distinct consequences on receptor function that promote transformation. If so, we would expect that the concurrent mutation of the Tyr-770 and Leu-773 residues should cause synergistic enhancement of FGFR2 IIIb transforming activity of FGFR2 IIIb C1. As expected, we found that both the Y770F/L773A and Y770A/L773A double mutants (∼10-fold more soft agar colonies than WT) exhibited ∼2-fold higher transforming activity when compared with either the Y770F (∼6-fold more soft agar colonies than WT) or L773A (∼4-fold more soft agar colonies than WT) single mutant (Fig. 5A). To exclude the possibility that different expression level of proteins could have an effect on transforming potency, we determined all FGFR2 IIIb proteins tested above were expressed at comparable levels (Fig. 5B). Taken together, these data indicate that concurrent loss of the Tyr and Leu residues of the 770YXXL motif cooperates to enhance the transforming activity of the FGFR2 IIIb C3 isoform.
FIGURE 5.
Mutation of the Tyr and Leu residues of the 770YXXL motif cooperates to enhance FGFR2 IIIb C1 induction of anchorage-independent growth transformation. A, FGFR2 IIIb C1 770YXXL motif mutants exhibit enhanced colony formation in soft agar. Rat-1 cells that stably express the indicated FGFR2 IIIb proteins were suspended in 0.4% soft agar and allowed to grow for 14 days before the number of colonies was quantitated. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of three independent experiments. B, wild type and 770YXXL motif mutants of FGFR2 IIIb C1 are stably expressed at comparable levels. Rat-1 cells that stably expressed the indicated FGFR2 IIIb proteins were assayed for their FGFR2 IIIb protein expression levels by immunoblot analyses with anti-FGFR2 antibody. Total cell lysates were also blotted with anti-β-actin to verify equivalent total protein.
Loss of Tyr-770 Impairs FGFR2 IIIb C1 Activation of PLCγ— Although the Y770F mutation did not impair receptor down-regulation (Fig. 4), the Y770F mutation did enhance the transforming activity of FGFR2 IIIb (Fig. 5A). These observations suggest that some other mechanism(s) contributes to the enhanced transforming activity of the Y770F mutant. A recent study showed that a Y770F missense mutant of FGFR2 IIIb C1 cannot bind and phosphorylate PLCγ (23). However, in contrast to our observations, this study found that the Y770F mutation impaired, rather than enhanced, ligand-stimulated mitogenic activity (23).
To resolve these apparently conflicting observations, we decided to more thoroughly evaluate the role of Tyr-770 in FGFR2 IIIb regulation of PLCγ signaling and transforming activity. First, we determined whether mutation of Tyr-770 and other 770YXXL motif residues caused decreased tyrosine phosphorylation of PLCγ. For these analyses, Rat-1 cells stably expressing WT and mutant FGFR2 IIIb C1 (Y770F, Y770F/L773A, Y770A/L773A, and L773A), as well as the C2 and C3 variants, were stimulated with FGF7, and the level of PLCγ tyrosine phosphorylation was determined. As expected the WT FGFR2 IIIb C1 and C2 variants that contain Tyr-770, and not the C3 that lacks Tyr-770, showed increased FGF7-stimulated levels of tyrosine-phosphorylated PLCγ (Fig. 6A). Moreover, we found that all three FGFR2 IIIb C1 mutants that lack the Tyr-770 residue (Y770F, Y770F/L773A, and Y770A/L773A) failed to exhibit elevated tyrosine-phosphorylated PLCγ, whereas the L773A mutant retained the ability to stimulate tyrosine phosphorylation of PLCγ (Fig. 6A). Finally, whether the Tyr-770 residue is required for PI hydrolysis was not determined by the previous study (23). We found that FGF7 stimulation induced marked [3H]inositol phosphate response in cells stably expressing the wild type and not Y770F mutant FGFR2 IIIb C1 receptor (Fig. 6B). Taken together, these data suggest that Tyr-770 is required for activation of PLCγ by FGFR2 IIIb, and the loss of capacity to promote inositol lipid hydrolysis might contribute to the enhanced transforming activity of the C3 variant that lacks Tyr-770.
FIGURE 6.
Loss of Tyr-770 but not Leu-773 impairs FGFR2 IIIb C1 activation of PLCγ. A, mutation of Tyr-770 but not Leu-773 impairs phosphorylation of endogenous PLCγ. Rat-1 cells stably expressing indicated FGFR2 IIIb proteins were serum-starved and stimulated with either vehicle or 50 ng/ml FGF7 for 30 min. Tyrosine-phosphorylated and total PLCγ protein was determined by immunoprecipitation (IP) with anti-PLCγ antibody followed by immunoblotting (IB) with anti-phosphotyrosine (P-Tyr) antibody or with anti-PLCγ antibody, respectively. Data shown are representative of three independent experiments. B, mutation of Tyr-770 impairs FGF7-stimulated formation of inositol phosphates. Rat-1 cells stably expressing the indicated wild type (WT) or mutant FGFR2 IIIb C1 proteins were serum-starved and stimulated with vehicle (basal), or with 100 ng/ml FGF7, 0.1 μm thrombin, or 10 μm lysophosphatidic acid (LPA) (positive controls), and [3H]inositol phosphate accumulation was measured to assess PLCγ activity. Data shown are representative of three independent assays.
Loss of Tyr-770 Causes Sustained Activation of FRS2 but Not Ras-MAPK or PI3K-AKT Pathways—PLCγ-mediated signaling is generally considered to promote mitogenesis and growth (38–40). Thus, the fact that the Y770F mutation enhances FGFR2 IIIb C1 transforming activity, yet results in the loss of PLCγ activation, argues that mutation of Tyr-770 may affect other signaling activities that promote growth transformation. One possible explanation for this paradoxical observation is that loss of PLCγ binding caused by the Y770F mutation may indirectly promote the activation of other FGFR2 IIIb effector pathways. Aside from PLCγ, the FRS2 proteins represent the next best characterized effectors of FGFR signaling (1–4).
To determine whether loss of Tyr-770 altered FRS2 activity, Rat-1 cells stably expressing WT or mutants of FGFR2 IIIb C1 as well as the C2 and C3 variants were lysed and immunoprecipitated with anti-FRS2 antibody followed by immunoblot analyses using anti-phosphotyrosine antibody. We found that mutant receptors with the Tyr-770 mutation (Y770F, Y770F/L773A, and Y770A/L773A) showed elevated basal FRS2 tyrosine phosphorylation in the absence of exogenous FGF7 stimulation (Fig. 7A). Consistent with these observations, elevated basal FRS2 tyrosine phosphorylation was also seen for the C3 isoform that lacks Tyr-770 but not the C2 isoform that retains Tyr-770 (Fig. 7A). The sustained activation of FRS2 observed in the absence of FGF7 seemed likely to be caused by receptor stimulation by endogenous ligands secreted by Rat-1 fibroblasts. After stimulation with FGF7 for 20 min, all three FGFR2 IIIb splice variants as well as the mutants of C1 showed increased levels of robustly tyrosine-phosphorylated FRS2 (Fig. 7A), indicating that all the receptor variants retained efficient coupling to FRS2.
Because we observed sustained FRS2 activation in Tyr-770 mutants in the absence FGF7 stimulation (Fig. 7A), we next determined whether this was associated with sustained activation of FRS2-mediated downstream signaling pathways activated transiently by ligand stimulation. We first examined whether loss of Tyr-770 led to activation of Ras and the Raf-MEK-ERK MAPK pathway. For these analyses, we utilized immunoblot analyses with a phospho-specific MEK1/2 or phospho-specific ERK1/2 antibodies. After stimulation with FGF7 for 20 min, MEK was phosphorylated in WT and all mutants to a similar extent. Similarly, FGF7 stimulation caused robust phosphorylation of ERK in WT and all mutants to a similar extent. In the absence of FGF7, surprisingly, none of the FGFR2 IIIb proteins tested above caused an increase in the steady-state levels of activated MEK or ERK (Fig. 7B).
Next, we determined whether mutation of Tyr-770 led to sustained activation Gab1 and the PI3K-AKT pathway. For these analyses, cells were lysed and immunoprecipitated with Gab1 antibody followed by immunoblot analyses with phosphotyrosine antibody to determine Gab1 activity. In addition, we utilized immunoblot analyses with a phospho-specific antibody that recognizes the phosphorylated and activated forms of AKT to determine AKT activity. FGF7 stimulation for 20 min caused Gab1 phosphorylation in WT and all mutants to a similar extent. FGF7 stimulation also induced a similar level of AKT phosphorylation in WT and mutant receptors. However, in the absence of FGF7 stimulation, no increased steady-state activation of Gab1 or AKT was detected for cells expressing WT or mutant receptors (Fig. 7B). These data suggest that whereas loss of Tyr-770 caused sustained FRS2 activation, it did not induce sustained activation of the ERK or Gab1-PI3K-AKT pathways.
Although we found that loss of Tyr-770 did not cause an increase in steady-state activation of the Ras-ERK or PI3K-AKT signaling pathways, it is possible that both pathways are still persistently activated but down-regulated by feedback mechanisms present during persistent signaling. Thus, we determined whether the activities of these two pathways were still important for Y770F transforming activity. We found that treatment with either the U0126 MEK inhibitor or the LY294002 PI3K inhibitor inhibited the anchorage-independent growth of cells expressing Y770F (Fig. 7C), indicating that both MEK-ERK and PI3K activities contribute to the enhanced transforming activity caused by the Y770F mutation.
Loss of Tyr-770 Enhances FRS2 Binding to FGFR2—Next, we decided to determine how loss of the PLCγ-binding site (Tyr-770) leads to sustained tyrosine phosphorylation of FRS2. One possible explanation for these observations is that PLCγ binding to FGFR2 may interfere with complex formation between FRS2 and FGFR2. By abolishing PLCγ binding, the Y770F mutation might relieve steric hindrance for FRS2 and facilitate its binding to the FGFR2. Consequently, loss of PLCγ binding by the Y770F mutation may indirectly enhance FRS2 activity. To examine this possibility, we determined whether the loss of the PLCγ-binding site (Tyr-770) enhances FRS2 association with the FGFR2. For these analyses, Rat-1 cells stably expressing WT or mutants of FGFR2 IIIb C1 (Y770F, Y770F/L773A, Y770A/L773A, and L773A) were lysed and immunoprecipitated with anti-FRS2 antibody followed by immunoblot analyses using anti-FGFR2 antibody. Previously, Ong et al. (24) reported that FGFR1 interacts with FRS2 constitutively, independent of receptor activation. Consistent with this study, we also found that FRS2 constitutively associated with the wild type and all mutants of FGFR2 examined. However, we observed ∼1.7-fold increase of FRS2 binding to FGFR2 in mutants lacking Tyr-770 (Y770F, Y770F/L773A, Y770A/L773A) when compared with the WT or L773A mutant (Fig. 8). These data indicate that loss of Tyr-770 but not Leu-773 enhances FRS2 association with FGFR2, and this enhanced FRS2 association with FGFR2 may lead to sustained receptor activation of FRS2.
FIGURE 8.
Loss of Tyr-770 enhances FRS2 binding to FGFR2. A, Rat-1 cells stably expressing the indicated FGFR2 IIIb proteins were lysed and FRS2-FGFR2 binding was determined by immunoprecipitation (IP) with anti-FRS2 antibody followed by immunoblotting (IB) with anti-FGFR2 antibody. To determine total endogenous FRS2 protein, the same membrane was reprobed with anti-FRS2 antibody. Total cell lysates were analyzed to determine stable FGFR2 IIIb protein expression levels by immunoblot analyses with anti-FGFR2 antibody. Data shown are representative of three independent experiments. B, quantitation of data shown in A. FGFR2-FRS2 binding indicates the relative band intensity of mutant FGFR2 bound to FRS2 when compared with the wild type FGFR2 bound to FRS2. Data shown represent as the average of three independent experiments with the bars indicating standard deviation.
FRS2 Binding Is Required for Increased FRS2 Tyrosine Phosphorylation and Enhanced Transforming Activity by the Y770F Mutation—Next, we determined whether FRS2 binding is required for the increased tyrosine phosphorylation and transforming activity caused by the Y770F mutation. For these analyses, we generated mutants of FGFR2 IIIb C1 that cannot bind FRS2. Previously, it was demonstrated that FRS2 binds constitutively to the juxtamembrane region of FGFR1 (24). They also determined that mutation of the Lys-419, Pro-422, and Leu-423 residues in the juxtamembrane region strongly diminished interaction between FGFR1 and the FRS2 (24). Because the Lys-419, Pro-422, and Leu-423 residues in the juxtamembrane region of FGFR1 are well conserved in FGFR2 IIIb, to generate a FRS2-binding deficient mutant, we constructed a mutant of FGFR2 IIIb (K421A/P424A/L425A, designated ΔFRS2) that is analogous to the FGFR1 mutant (K419A/P422A/L423A).
To determine whether FRS2 binding is required for sustained activation of FRS2 by Y770F mutation, we introduced mutations in the FRS2 binding region (ΔFRS2) in the Tyr-770 mutant (designated as Y770F/ΔFRS2). Rat-1 cells stably expressing WT, Y770F, or Y770F/ΔFRS2 were lysed and immunoprecipitated with anti-FRS2 antibody and followed by immunoblot analyses using anti-phosphotyrosine antibody. Consistent with our observation in Fig. 7A, we found that the Y770F mutation enhanced FRS2 tyrosine phosphorylation in the absence of FGF7 stimulation, and this activation was abolished by the loss of FRS2 binding (Fig. 9A). These observations indicate that FRS2 binding to FGFR2 is essential for sustained activation of FRS2 by the Y770F mutant receptor.
FIGURE 9.
FRS2 binding to FGFR2 is required for increased FRS2 tyrosine phosphorylation and enhanced transforming activity by the Y770F mutation. A, FRS2 binding is required for enhanced FRS2 tyrosine phosphorylation by Y770F mutation. Rat-1 cells stably expressing the indicated proteins were analyzed for FRS2 activity by immunoprecipitation (IP) with either anti-FRS2 antibody or anti-phosphotyrosine antibody followed by immunoblotting (IB) with either anti-phosphotyrosine or anti-FRS2 antibody, respectively. Total cell lysates were analyzed by immunoblot analyses with anti-FGFR2 antibody to verify equivalent expression of the C1 variants. ΔFRS2 corresponds to the K421A/P424A/L425A mutations in the FRS2-binding site. B, FRS2 binding is required for increased FGFR2 IIIb C1 transforming activity by the Y770F mutation. Rat-1 cells stably expressing the indicated FGFR2 IIIb proteins were suspended in 0.4% soft agar and allowed to grow for about 21 days before the number of colonies was quantitated. Data shown are the average of duplicate dishes, with the bars indicating standard deviation, and are representative of three independent experiments.
Finally, we determined whether loss of FRS2 binding and activation was associated with a reduction in the transforming activity of the Y770F mutant. When we introduced the ΔFRS2 mutation into the Y770F mutant, the resulting Y770F/ΔFRS2 mutant showed a similar weak transforming activity as seen with WT C1 (Fig. 9B). This observation indicates that the enhanced transforming potency caused by the Y770F mutation is dependent on FRS2 binding and activity.
DISCUSSION
Although missense mutations of FGFR2 are found in human cancers (41), another mechanism of FGFR2 activation in cancer may involve alternative gene splicing and enhanced expression of the structurally and functionally distinct C2 and C3 variant receptors. In this study, we compared the transforming potency of the C1, C2, and C3 splicing variants of FGFR2 IIIb and found that progressive loss of carboxyl-terminal sequences is correlated with increased transforming potency. Our mutation analyses of the 770YXXL motif suggest that the absence of this motif contributes to the highly transforming nature of the C3 splice variant by decreased receptor internalization and increased receptor expression activation and by enhanced activation of FRS2 (Table 1 and Fig. 10). Thus, the preferential expression of the C3 isoform in cancer cells represents another mechanism for altered receptor signaling that contributes to oncogenesis.
TABLE 1.
Consequences of mutation of the YXXL motif on FGFR2 IIIb C1 function
|
Activity
|
FGFR2 III
C1a
|
|||
|---|---|---|---|---|
| WT | Y770F | L773A | Y770F/L773A | |
| FGFR2 transforming activityb | ± | ++ | ++ | ++++ |
| FGFR2 internalization ratec | ++ | ++ | + | + |
| FGFR2 expression levelsc | + | + | ++ | ++ |
| FGFR2 autophosphorylationb | + | + | ++ | ++ |
| PLCγ phosphorylationb | + | – | + | – |
| FRS2 phosphorylationb | ± | + | + | ++ |
| FRS2-FGFR2 bindingb | + | ++ | + | ++ |
| Phospho-MEK1/2b | – | – | – | – |
| Phospho-ERK1/2b | – | – | – | – |
| Gab1 phosphorylationb | – | – | – | – |
| Phospho-AKTb | – | – | – | – |
Data were compiled from this study
Data were determined at steady state
Data were transient and FGF7-stimulated
FIGURE 10.
Loss of the 770YXXL motif in FGFR2 IIIb C3 enhances transforming activity by altered receptor recycling and signaling. Based on our observations with missense mutations in the 770YXXL motif, we propose a model where the absence of the 770YXXL motif contributes to the enhanced transforming potency of FGFR2 IIIb C3. First, loss of the Leu-773 residue causes impaired ligand-stimulated receptor internalization and degradation. Impairment in receptor down-regulation may further induce sustained activation of FGFR2 IIIb. Second, loss of the Tyr-770 results in a loss of PLCγ binding, which may relieves steric hindrance for FRS2 association, facilitating enhanced FGFR2 binding. The increased FRS2 binding to FGFR2 may lead to increased FRS2 activity and enhance transformation. Together, our results suggest a mechanism where loss of Tyr and Leu residues of 770YXXL motif cooperates to increase FGFR2 C3-mediated transformation.
In light of observations by us and others (18), it would seem most logical to study the signaling and transforming function of the epithelial cell-restricted FGFR2 IIIb splice variants in epithelial cells. However, in situ tumor epithelial cells are associated with mesenchymal stromal tissue, which secrete ligands that cause persistent paracrine stimulation of FGFR2 IIIb in epithelial cells. Hence, we speculated that a more accurate approach for evaluating FGFR2 IIIb function would be in fibroblast cells, where secreted fibroblast cell-derived ligands will foster a persistent autocrine stimulation of FGFR2 IIIb. As discussed below, our observations found differences in FGFR2 IIIb signaling and transforming activity when evaluated under conditions of transient or sustained ligand stimulation in epithelial and fibroblast cells, thus supporting the physiologic signification of our studies of FGFR2 IIIb function in fibroblasts with regards to FGFR2 IIIb function in epithelial cell-derived tumors in vivo.
We determined that loss of the Tyr-770 alone significantly enhanced the transforming activity of FGFR2 IIIb C1. This observation was unexpected because the Y770F residue is a PLCγ-binding site, and PLCγ is generally considered as a growth-promoting protein. Because this residue also corresponds to a putative YXXL tyrosine-based sorting motif, we evaluated the consequences of mutation of the 770YXXL motif on receptor internalization. Surprisingly, we found that loss of Leu-773 but not Tyr-770 residue impaired ligand-induced receptor internalization and degradation. Interestingly, impairment of receptor internalization was correlated with sustained activation of FGFR2 IIIb. We also found that concurrent mutation of the Tyr-770 and Leu-773 residues cooperatively enhanced transforming activity. Furthermore, we found that loss of the PLCγ-binding site (Tyr-770) caused sustained FRS2 activation by enhancing FRS2 association with FGFR2. Mutation of the FRS2-binding site impaired FGFR2 IIIb transforming activity, supporting a necessary role for FRS2 in FGFR2 IIIb receptor activation by loss of the 770YXXL motif. These results support a model where the enhanced transforming activity of FGFR2 IIIb C3 is because of two distinct mechanisms caused by the deletion of sequences that include the 770YXXL motif, altered receptor trafficking and signaling (Fig. 10).
In contrast to our observations, a previous study found that a Y770F mutation impaired, rather than promoted, ligand-stimulated FGFR2 IIIIb C1-mediated cell proliferation (23). Their results are more consistent with other observations, where PLCγ activation has been generally considered to promote, rather than antagonize, growth. For example, overexpression of PLCγ was shown to promote growth transformation of NIH 3T3 mouse and rat 3Y1 fibroblasts (40, 42). In addition, the analogous Y760F mutation in tumor-derived constitutively activated mutants of FGFR3 (the K650E missense and the TEL-FGFR3 fusion protein) was shown to abolish PLCγ activation and inhibited their transforming activities in Ba/F3 mouse pro-B cells (43). A similar conclusion was made for platelet-derived growth factor receptor activation of PLCγ, where selective loss of PLCγ activation resulted in loss of transforming activity (39). However, in contrast to these observations, it was shown that the Y766F mutation in FGFR1 abolished ligand-stimulated PLCγ activation but not mitogenesis, as measured by DNA synthesis in L6 myoblasts (20, 44), suggesting that PLCγ activity is not required for FGFR1-mediated growth stimulation. These different conclusions for PLCγ regulation of cell proliferation may reflect distinct functions of FGFR isoforms. Additionally, they may reflect cell context variations in PLCγ biological function as well as the different biological assays utilized. Another possible explanation for these conflicting observations is that this tyrosine residue may regulate PLCγ-independent functions that regulate cell growth. For example, it was shown that the PLCγ-binding site on PDGFR can serve as a binding site for ubiquitin ligase Cbl that negatively regulates PDGFR signaling (45).
In contrast to our study, where we found that a Y770F mutation caused sustained FRS2 tyrosine phosphorylation, Ceridono et al. (23) found that this same mutation diminished FRS2 tyrosine phosphorylation. We speculate that our different observations may reflect the different consequences of transient versus sustained ligand stimulation. In the previous study, transient (10 min) exogenous FGF7 stimulation was evaluated, whereas our analyses evaluated the consequences of sustained stimulation of FGFR2 by endogenous ligands. In support of this possibility, our evaluation of FGFR2 IIIb activation of FRS2 signaling found differences in transient versus persistent ligand stimulation. There is considerable evidence that the duration of signaling can elicit different biological and signaling outcomes (46–48). For example, although we found that transient exogenous FGF7 stimulation increased FRS2 tyrosine phosphorylation in WT and Y770F mutant receptors to a similar extent, sustained endogenous ligand stimulation increased FRS2 tyrosine phosphorylation only in the Y770F mutant but not WT receptor (Fig. 7A).
Although the Y770F mutant receptor induced sustained activation of FRS2, and we showed that loss of FRS2 binding impaired transformation, we were surprised to find that increased FRS2 activation was not associated with increased steady-state activation of the ERK or AKT signaling pathways (Fig. 7B). However, this result is not entirely surprising because FRS2 is also known to facilitate negative regulatory components to dampen FGFR signaling (2, 49). For example, ligand stimulation of FGFR1 results in ERK-mediated phosphorylation of FRS2 at threonine residues, leading to attenuation of ERK activation (50). Ligand stimulation of FGFR normally causes a rapid and transient activation of ERK, and prevention of ERK phosphorylation of FRS2 results in prolonged ERK activation (50). Finally, our finding that pharmacologic inhibition of MEK-ERK signaling or PI3K activation also reduced Y770F receptor transforming activity suggests that these two FRS2 effector pathways do contribute to transformation. Nevertheless, it remains possible that FRS2-mediated growth transformation may involve other signaling activities that remain to be identified.
In summary, our studies identified a mechanistic basis for the potent transforming activity of the C3 isoform of FGFR2 IIIb that involves two distinct mechanisms regulated by the 770YXXL motif. Although the Y770F/L773A mutant showed significantly higher transforming activity when compared with the WT receptor, the C3 variant was still more transforming than the Y770F/L773A mutant. Thus, our future studies will focus on defining additional mechanisms for the enhanced transforming potency of the C3 variant. Although we found that the C2 variant was more transforming than the C1 variant, the loss of the 770YXXL motif does not appear to account for C2 transforming activity, and therefore, we have not found a clear mechanistic basis for the enhanced transforming potency of the C2 variant. Finally, FRS2 possesses multiple tyrosine phosphorylation sites and can interact with other signaling proteins that may contribute to ERK and AKT independent mechanisms that promote transformation. Thus, our future studies will include the identification of the critical signaling proteins important for FRS2-mediated growth transformation.
Acknowledgments
We thank Wendy Salmon and Michael Chua (University of North Carolina Michael Hooker Microscopy Facility) and Misha Rand for assistance in manuscript and figure preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA69577 (to C. J. D.) and GM65533 (to T. K. H.) (USPHS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; RTK, receptor tyrosine kinase; KGF, keratinocyte growth factor; PLCγ, phospholipase C gamma; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase; FRS2, FGF receptor substrate 2; PBS, phosphate-buffered saline; BSA, bovine serum albumin; PI, phosphatidylinositol; WT, wild type; ERK, extracellular signal-regulated kinase; MEK, MAPK/ERK kinase.
References
- 1.Dailey, L., Ambrosetti, D., Mansukhani, A., and Basilico, C. (2005) Cytokine Growth Factor Rev. 16 233-247 [DOI] [PubMed] [Google Scholar]
- 2.Eswarakumar, V. P., Lax, I., and Schlessinger, J. (2005) Cytokine Growth Factor Rev. 16 139-149 [DOI] [PubMed] [Google Scholar]
- 3.Grose, R., and Dickson, C. (2005) Cytokine Growth Factor Rev. 16 179-186 [DOI] [PubMed] [Google Scholar]
- 4.Mohammadi, M., Olsen, S. K., and Ibrahimi, O. A. (2005) Cytokine Growth Factor Rev. 16 107-137 [DOI] [PubMed] [Google Scholar]
- 5.Itoh, N., and Ornitz, D. M. (2004) Trends Genet. 20 563-569 [DOI] [PubMed] [Google Scholar]
- 6.Orr-Urtreger, A., Bedford, M. T., Burakova, T., Arman, E., Zimmer, Y., Yayon, A., Givol, D., and Lonai, P. (1993) Dev. Biol. 158 475-486 [DOI] [PubMed] [Google Scholar]
- 7.Dell, K. R., and Williams, L. T. (1992) J. Biol. Chem. 267 21225-21229 [PubMed] [Google Scholar]
- 8.Igarashi, M., Finch, P. W., and Aaronson, S. A. (1998) J. Biol. Chem. 273 13230-13235 [DOI] [PubMed] [Google Scholar]
- 9.Miki, T., Bottaro, D. P., Fleming, T. P., Smith, C. L., Burgess, W. H., Chan, A. M., and Aaronson, S. A. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 246-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yayon, A., Zimmer, Y., Shen, G. H., Avivi, A., Yarden, Y., and Givol, D. (1992) EMBO J. 11 1885-1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finch, P. W., Rubin, J. S., Miki, T., Ron, D., and Aaronson, S. A. (1989) Science 245 752-755 [DOI] [PubMed] [Google Scholar]
- 12.Ohuchi, H., Nakagawa, T., Yamamoto, A., Araga, A., Ohata, T., Ishimaru, Y., Yoshioka, H., Kuwana, T., Nohno, T., Yamasaki, M., Itoh, N., and Noji, S. (1997) Development (Camb.) 124 2235-2244 [DOI] [PubMed] [Google Scholar]
- 13.Rubin, J. S., Osada, H., Finch, P. W., Taylor, W. G., Rudikoff, S., and Aaronson, S. A. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 802-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamasaki, M., Miyake, A., Tagashira, S., and Itoh, N. (1996) J. Biol. Chem. 271 15918-15921 [DOI] [PubMed] [Google Scholar]
- 15.Yan, G., Fukabori, Y., Nikolaropoulos, S., Wang, F., and McKeehan, W. L. (1992) Mol. Endocrinol. 6 2123-2128 [DOI] [PubMed] [Google Scholar]
- 16.Mason, I. J., Fuller-Pace, F., Smith, R., and Dickson, C. (1994) Mech. Dev. 45 15-30 [DOI] [PubMed] [Google Scholar]
- 17.Itoh, H., Hattori, Y., Sakamoto, H., Ishii, H., Kishi, T., Sasaki, H., Yoshida, T., Koono, M., Sugimura, T., and Terada, M. (1994) Cancer Res. 54 3237-3241 [PubMed] [Google Scholar]
- 18.Cha, J. Y., Lambert, Q. T., Reuther, G. W., and Der, C. J. (2008) Mol. Cancer Res. 6 435-445 [DOI] [PubMed] [Google Scholar]
- 19.Moffa, A. B., Tannheimer, S. L., and Ethier, S. P. (2004) Mol. Cancer Res. 2 643-652 [PubMed] [Google Scholar]
- 20.Mohammadi, M., Dionne, C. A., Li, W., Li, N., Spivak, T., Honegger, A. M., Jaye, M., and Schlessinger, J. (1992) Nature 358 681-684 [DOI] [PubMed] [Google Scholar]
- 21.Mohammadi, M., Honegger, A. M., Rotin, D., Fischer, R., Bellot, F., Li, W., Dionne, C. A., Jaye, M., Rubinstein, M., and Schlessinger, J. (1991) Mol. Cell. Biol. 11 5068-5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee, S. G. (2001) Annu. Rev. Biochem. 70 281-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceridono, M., Belleudi, F., Ceccarelli, S., and Torrisi, M. R. (2005) Biochem. Biophys. Res. Commun. 327 523-532 [DOI] [PubMed] [Google Scholar]
- 24.Ong, S. H., Guy, G. R., Hadari, Y. R., Laks, S., Gotoh, N., Schlessinger, J., and Lax, I. (2000) Mol. Cell. Biol. 20 979-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouhara, H., Hadari, Y. R., Spivak-Kroizman, T., Schilling, J., Bar-Sagi, D., Lax, I., and Schlessinger, J. (1997) Cell 89 693-702 [DOI] [PubMed] [Google Scholar]
- 26.Ong, S. H., Hadari, Y. R., Gotoh, N., Guy, G. R., Schlessinger, J., and Lax, I. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6074-6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bache, K. G., Slagsvold, T., and Stenmark, H. (2004) EMBO J. 23 2707-2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dikic, I., and Giordano, S. (2003) Curr. Opin. Cell Biol. 15 128-135 [DOI] [PubMed] [Google Scholar]
- 29.Bonifacino, J. S., and Traub, L. M. (2003) Annu. Rev. Biochem. 72 395-447 [DOI] [PubMed] [Google Scholar]
- 30.Fabbri, M., Fumagalli, L., Bossi, G., Bianchi, E., Bender, J. R., and Pardi, R. (1999) EMBO J. 18 4915-4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown, H. A., Lazarowski, E. R., Boucher, R. C., and Harden, T. K. (1991) Mol. Pharmacol. 40 648-655 [PubMed] [Google Scholar]
- 32.Moffa, A. B., and Ethier, S. P. (2007) J. Cell. Physiol. 210 720-731 [DOI] [PubMed] [Google Scholar]
- 33.Miki, T., Fleming, T. P., Bottaro, D. P., Rubin, J. S., Ron, D., and Aaronson, S. A. (1991) Science 251 72-75 [DOI] [PubMed] [Google Scholar]
- 34.Zheng, J., Saksela, O., Matikainen, S., and Vaheri, A. (1995) J. Cell Biol. 129 843-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canfield, W. M., Johnson, K. F., Ye, R. D., Gregory, W., and Kornfeld, S. (1991) J. Biol. Chem. 266 5682-5688 [PubMed] [Google Scholar]
- 36.Wu, Z., and Simister, N. E. (2001) J. Biol. Chem. 276 5240-5247 [DOI] [PubMed] [Google Scholar]
- 37.Wernick, N. L., Haucke, V., and Simister, N. E. (2005) J. Biol. Chem. 280 7309-7316 [DOI] [PubMed] [Google Scholar]
- 38.Chang, J. S., Noh, D. Y., Park, I. A., Kim, M. J., Song, H., Ryu, S. H., and Suh, P. G. (1997) Cancer Res. 57 5465-5468 [PubMed] [Google Scholar]
- 39.DeMali, K. A., Whiteford, C. C., Ulug, E. T., and Kazlauskas, A. (1997) J. Biol. Chem. 272 9011-9018 [DOI] [PubMed] [Google Scholar]
- 40.Smith, M. R., Court, D. W., Kim, H. K., Park, J. B., Rhee, S. G., Rhim, J. S., and Kung, H. F. (1998) Carcinogenesis 19 177-185 [DOI] [PubMed] [Google Scholar]
- 41.Pollock, P. M., Gartside, M. G., Dejeza, L. C., Powell, M. A., Mallon, M. A., Davies, H., Mohammadi, M., Futreal, P. A., Stratton, M. R., Trent, J. M., and Goodfellow, P. J. (2007) Oncogene 26 7158-7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang, C. P., Lazar, C. S., Walsh, B. J., Komuro, M., Collawn, J. F., Kuhn, L. A., Tainer, J. A., Trowbridge, I. S., Farquhar, M. G., and Rosenfeld, M. G. (1993) J. Biol. Chem. 268 19312-19320 [PubMed] [Google Scholar]
- 43.Chen, J., Williams, I. R., Lee, B. H., Duclos, N., Huntly, B. J., Donoghue, D. J., and Gilliland, D. G. (2005) Blood 106 328-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters, K. G., Marie, J., Wilson, E., Ives, H. E., Escobedo, J., Del Rosario, M., Mirda, D., and Williams, L. T. (1992) Nature 358 678-681 [DOI] [PubMed] [Google Scholar]
- 45.Reddi, A. L., Ying, G., Duan, L., Chen, G., Dimri, M., Douillard, P., Druker, B. J., Naramura, M., Band, V., and Band, H. (2007) J. Biol. Chem. 282 29336-29347 [DOI] [PubMed] [Google Scholar]
- 46.Marshall, C. J. (1995) Cell 80 179-185 [DOI] [PubMed] [Google Scholar]
- 47.Murphy, L. O., and Blenis, J. (2006) Trends Biochem. Sci. 31 268-275 [DOI] [PubMed] [Google Scholar]
- 48.Sharrocks, A. D. (2006) Curr. Biol. 16 R540-R542 [DOI] [PubMed] [Google Scholar]
- 49.Schlessinger, J. (2004) Science 306 1506-1507 [DOI] [PubMed] [Google Scholar]
- 50.Lax, I., Wong, A., Lamothe, B., Lee, A., Frost, A., Hawes, J., and Schlessinger, J. (2002) Mol. Cell 10 709-719 [DOI] [PubMed] [Google Scholar]