Abstract
Background
Since the full genome sequences of Saccharomyces cerevisiae were released in 1996, genome sequences of over 90 fungal species have become publicly available. The heterogeneous formats of genome sequences archived in different sequencing centers hampered the integration of the data for efficient and comprehensive comparative analyses. The Comparative Fungal Genomics Platform (CFGP) was developed to archive these data via a single standardized format that can support multifaceted and integrated analyses of the data. To facilitate efficient data visualization and utilization within and across species based on the architecture of CFGP and associated databases, a new genome browser was needed.
Results
The Seoul National University Genome Browser (SNUGB) integrates various types of genomic information derived from 98 fungal/oomycete (137 datasets) and 34 plant and animal (38 datasets) species, graphically presents germane features and properties of each genome, and supports comparison between genomes. The SNUGB provides three different forms of the data presentation interface, including diagram, table, and text, and six different display options to support visualization and utilization of the stored information. Information for individual species can be quickly accessed via a new tool named the taxonomy browser. In addition, SNUGB offers four useful data annotation/analysis functions, including 'BLAST annotation.' The modular design of SNUGB makes its adoption to support other comparative genomic platforms easy and facilitates continuous expansion.
Conclusion
The SNUGB serves as a powerful platform supporting comparative and functional genomics within the fungal kingdom and also across other kingdoms. All data and functions are available at the web site http://genomebrowser.snu.ac.kr/.
Background
As the number of sequenced genomes rapidly increases, search and comparison of sequence features within and between species has become an integral part of most biological inquires. To facilitate uses of the sequenced genomes, numerous bioinformatics tools have been developed; among these, genome browser plays an essential role by providing various means for viewing genome sequences and annotated features (e.g., chromosomal position and context of individual genes, protein/nucleotide sequences, structures of exon/intron, and promoters) via graphical and text interfaces. Widely utilized genome browsers include: (i) Ensembl http://www.ensembl.org/, which is specialized for mammalian genomics and comparative genomics [1], (ii) UCSC Genome Browser http://genome.ucsc.edu/, which archives genome sequences of 30 vertebrate and 24 non-vertebrate species [2], (iii) GBrowse http://gmod.org/wiki/Gbrowse, a widely-used component-based genome browser [3], and (iv) Map Viewer http://www.ncbi.nlm.nih.gov/projects/mapview at the National Center for Biotechnology Information (NCBI), which covers a large number of organisms [4]. A new genome browser based on the Google map engine, called the X::Map Genome Browser http://xmap.picr.man.ac.uk/[5], contains genomes of three mammalian species and is specialized for supporting microarray analyses based on the Affymetrix platform [6].
Since complete S. cerevisiae genome sequences were released in 1996, more than 90 fungal/oomycete species have been sequenced with many additional species being currently sequenced [7]. A few sequencing centers, such as the Broad Institute http://www.broad.mit.edu/ and the JGI http://www.jgi.doe.gov/, have sequenced most of the fungal genomes and provide their own genome browsers to support data visualization and utilization. Although they use standardized formats, such as fasta and gff3, for data presentation and distribution, each center uses its own data formats for sequences, annotation data, and other chromosomal information. In addition, some of the sequenced fungal genomes lack certain data, such as exon positions. These problems have hampered the integration and visualization of all available genome sequences via a single genome browser. As a solution for this problem, a group at Duke University http://fungal.genome.duke.edu/ installed an open-source browser called the GBrowse [3] after reannotating genome sequences of 42 fungal species from multiple sequencing centers through the use of their own annotation pipeline consisting of several gene prediction programs; large scale evolutionary analyses were conducted based on the archived genomes, demonstrating the usefulness of unified and standardized data formats [8].
A large number of sequenced fungal genomes have provided opportunities to compare genome sequences and features at multiple taxon levels, revealing potential mechanisms underpinning fungal evolution and biology [8-18]; however, due to the complexity and vast scale of the resulting data, presentation of these data in an easily accessible format is challenging. To overcome this limitation, both the database construction and the pipeline/tools for comparative analyses should be carefully designed. One good example is the e-Fungi project http://www.e-fungi.org.uk/[19], which archives genome sequences of 34 fungal and 2 oomycete species and supports various queries via the web interface. Comparative fungal genomics studies have been conducted using e-Fungi [9,11]. Yeast Gene Order Browser (YGOB; http://wolfe.gen.tcd.ie/ygob/) [20] archives genome sequences of the species belonging to the subphylum Saccharomycotina and provides a graphical gene order browser, which helps the dissection of evolutionary history of genome changes during yeast speciation [21]. Although these platforms provide useful tools and data, only certain fungal genomes are covered, and the function of user-friendly access to sequence information and graphical presentation of data are limited.
The Comparative Fungal Genomics Platform (CFGP; http://cfgp.snu.ac.kr/) was established to archive all publicly available fungal and oomycete genome sequences using a unified data format and to support multifaceted analyses of the stored data via a newly developed user interface named as Data-driven User Interface [7]. Currently, CFGP archives genome sequences of 92 fungal and 6 oomycete species (137 different datasets) and also carries genome sequences of 55 plant, animal and bacterial species (56 datasets). Taking advantage of the data warehouse and functionalities in CFGP, several databases specialized for certain gene families or functional groups have been constructed, one of which is the Fungal Transcription Factor Database (FTFD; http://ftfd.snu.ac.kr/) [22]. This database identified and classified all fungal transcription factors and provides a phylogenomic platform supporting analyses of individual transcription factor families [23]. In addition, Fungal Cytochrome P450 Database (FCPD; http://p450.riceblast.snu.ac.kr/) [24], Fungal Secretome Database (FSD; http://fsd.snu.ac.kr/; Choi et al., unpublished), Fungal Expression Database (FED; http://fed.snu.ac.kr/; Park et al., unpublished) have been constructed or are currently being constructed. The CFGP was also used to manage high-throughput experimental data and link them to corresponding genes [25,26] and to maintain the Phytophthora database http://www.phytophthoradb.org/[27].
To support comparative genomics analyses using CFGP and offer tools for versatile data visualization, we newly developed a genome browser named as the Seoul National University Genome Browser (SNUGB; http://genomebrowser.snu.ac.kr/). We chose to develop a new genome browser instead of adopting one of the existing browsers in part because the adoption required conversion of the data archived in CFGP into new formats, and the existing browsers do not support the integration of additional databases, such as the InterPro and customized homologous gene databases available through SNUGB. We also wanted to have a browser based on the architecture of CFGP and associated databases so that we would be able to quickly present updated contents in these resources and seamlessly integrate new tools for data processing, visualization, and/or utilization.
The SNUGB currently covers genome sequences and associated information for 92 fungal and 6 oomycete species (137 datasets), which is the largest among the available fungal genome browser services on the web. These 92 fungal species cover four phyla and one subphylum based on a recently revised fungal taxonomy framework [28] (Table 1, 2, and 3). It also houses genome sequences of 12 plant, 18 insect, and 3 nematode species and human genome sequences (38 datasets), to support comparison of fungal genomes with those in other kingdoms (Table 4). The taxonomy browser implemented in the SNUGB provides an easy means to access genome sequences of specific species via two ways. The SNUGB provides lists of putative orthologous genes of all fungal ORFs and a tool for comparison of genomic contexts of any orthologous genes among chosen species. In addition, SNUGB displays the InterPro terms assigned to each ORF as well as the genomic regions where expressed sequence tags (ESTs) are matched. With these functionalities, SNUGB will serve as a powerful platform supporting comprehensive fungal comparative genomics.
Table 1.
List and characteristics of the fungal genomes belonging to the subphylum Pezizomycota archived in SNUGB.
| Speciesa | Size (Mb) | # of ORFs | # of Exons | Cb | Ib | Eb | Source | Refs |
| Fungi (Kingdom)e | ||||||||
| Ascomycota (Phylum) | ||||||||
| Pezizomycotina (Subphylum) | ||||||||
| A: Botrytis cinerea T: Botryotinia fuckeliana | 42.7 | 16,448 | 43,358 | N | Y | N | BI | N |
| Sclerotinia sclerotiorum | 38.3 | 14,522 | 40,623 | N | Y | N | BI | N |
| Aspergillus clavatus | 27.9 | 9,121 | 27,959 | N | Y | N | BI | [17,44] |
| Aspergillus flavus | 36.8 | 12,604 | 40,971 | N | Y | N | BI | [16] |
| Aspergillus fumigatus AF293 | 29.4 | 9,887 | 28,164 | 8 | Y | N | TIGR | [45] |
| Aspergillus fumigatus A1163 | 29.2 | 9,929 | 29,094 | N | Y | N | TIGR | [44] |
| A:Aspergillus nidulans T:Emericella nidulans | 30.1 | 10,701 | 35,525 | 8 | Y | N | BI | [14] |
| Aspergillus niger ATCC1015 | 37.2 | 11,200 | 34,971 | N | Y | N | JGI | N |
| Aspergillus niger CBS513.88 | 34.0 | 14,086 | 50,371 | 8 | Y | N | NCBI | [38] |
| A: Aspergillus oryzae T: Eurotium oryzae | 37.1 | 12,063 | 35,319 | N | Y | N | DOGAN | [46] |
| Aspergillus terreus | 29.3 | 10,406 | 33,116 | N | Y | N | BI | [17] |
| A:Aspergillus fischerianus T: Neosartorya fischerid | 32.6 | 10,403 | N | N | N | N | BI | [44] |
| Penicillium chrysogenum | 32.2 | 12,791 | 40,441 | N | N | N | NCBI | [47] |
| Penicillium marneffei | 28.5 | 10,638 | 34,306 | N | N | N | TIGR | N |
| Coccidioides immitis RS | 28.9 | 10,457 | 36,137 | N | Y | N | BI | N |
| Coccidioides immitis H538.4 | 27.7 | 10,663 | 34,503 | N | Y | N | BI | N |
| Coccidioides immitis RMSCC 2394 | 28.8 | 10,408 | 34,807 | N | Y | N | BI | N |
| Coccidioides immitis RMSCC 3703 | 27.6 | 10,465 | 33,931 | N | Y | N | BI | N |
| Coccidioides posadasii Silveria | 27.5 | 10,125 | 33,520 | N | Y | N | BI | N |
| Coccidioides posadasii C735 | 26.7 | N | N | N | N | N | BI | N |
| Coccidioides posadasii CPA0001 | 28.7 | N | N | N | N | N | BI | N |
| Coccidioides posadasii CPA0020 | 27.3 | N | N | N | N | N | BI | N |
| Coccidioides posadasii CPA0066 | 27.7 | N | N | N | N | N | BI | N |
| Coccidioides posadasii RMSCC 1037 | 26.7 | N | N | N | N | N | BI | N |
| Coccidioides posadasii RMSCC 1038 | 26.2 | N | N | N | N | N | BI | N |
| Coccidioides posadasii RMSCC 1040 | 26.5 | N | N | N | N | N | BI | N |
| Coccidioides posadasii RMSCC 2133 | 27.9 | N | N | N | N | N | BI | N |
| Coccidioides posadasii RMSCC 3488 | 28.1 | 9,964 | 33,484 | N | Y | N | BI | N |
| Coccidioides posadasii RMSCC 3700 | 25.5 | N | N | N | N | N | BI | N |
| Paracoccidioides brasiliensis Pb01 | 33.0 | 9,136 | 37,310 | N | Y | N | BI | N |
| Paracoccidioides brasiliensis Pb03 | 29.1 | 9,264 | 31,468 | N | Y | N | BI | N |
| Paracoccidioides brasiliensis Pb18 | 30.0 | 8,741 | 33,239 | N | Y | N | BI | N |
| Blastomyces dermatitidis | 61.8 | N | N | N | N | N | WGSC | N |
| A: Histoplasma capsulatum G217B T: Ajellomyces capsulatus G217B | 41.3 | 8,038 | 26,711f | N | Y | N | WGSC | N |
| A: Histoplasma capsulatum G186AR T: Ajellomyces capsulatus G186AR | 29.9 | 7,454 | 24,562f | N | Y | N | WGSC | N |
| A: Histoplasma capsulatum NAm1 T: Ajellomyces capsulatus NAm1 | 33.0 | 9,349 | 32,844 | N | Y | N | BI | N |
| A: Histoplasma capsulatum H143 T: Ajellomyces capsulatus H143 | 39.0 | 7,365 | 25,164f | N | Y | N | BI | N |
| A: Histoplasma capsulatum H88 T: Ajellomyces capsulatus H88 | 37.9 | 7,428 | 25,356f | N | Y | N | BI | N |
| A: Arthroderma gypseum T: Microsporum gypseum | 23.3 | 8,876 | 28,624 | N | Y | N | BI | N |
| Microsporum canis | 23.3 | N | N | N | N | N | BI | N |
| Trichophyton equinum | 24.2 | N | N | N | N | N | BI | N |
| Ascosphaera apis | 21.6 | N | N | N | N | N | BGM | [48] |
| Uncinocarpus reesii | 22.3 | 7,798 | 24,094 | N | Y | N | BI | N |
| Chaetomium globosumd | 34.9 | 11,124 | N | N | N | N | BI | N |
| Epichloe festucae | 27.0 | N | N | N | N | N | OU | N |
| A: Fusarium graminearum PH-1 T: Gibberella zeae PH-1 | 36.6 | 13,321 | 37,549 | N | Y | N | BI | [37] |
| A: Fusarium graminearum GZ3639 T:Gibberella zeae GZ3639c | 15.1 | 6,694 | 11,692f | N | Y | N | BI | [37] |
| Fusarium oxysporum f. sp. lycopersici 4286 | 61.4 | 17,608 | 47,051 | 15 | Y | N | BI | N |
| A: Fusarium verticillioides 7600 T:Gibberella moniliformis 7600 | 41.9 | 14,199 | 39,058 | N | Y | N | BI | N |
| A: Fusarium solani MPVI T:Nectria haematococca MPVI | 51.3 | 15,707 | 48,387 | N | Y | N | JGI | N |
| A: Pyricularia oryzae 70–15 T:Magnaporthe oryzae 70–15 | 41.6 | 12,841 | 34,189 | 7 | Y | Y | BI | [49] |
| A: Pyricularia oryzae 70–15 chromosome 7 T:Magnaporthe oryzae 70–15 chromosome 7 | 4.0 | 1,122 | 3,289 | 1 | Y | N | [50] | |
| Cryphonectria parasitica | 43.9 | 11,184 | 33,090 | N | N | N | JGI | N |
| Neurospora crassa OR74A | 39.2 | 9,842 | 27,188 | 8 | Y | N | BI | [51] |
| Podospora anserina DSM980 | 35.7 | 10,596 | 24,437 | 9 | Y | N | IGM | [52] |
| Trichoderma atroviride IMI206040 | 36.1 | 11,100 | 32,563 | N | Y | N | JGI | N |
| A:Trichoderma reesei QM6a T: Hypocrea jecorina QM6a | 33.5 | 9,129 | 27,891 | N | Y | N | JGI | [53] |
| A:Trichoderma virens Gv29-8 T:Hypocrea virens Gv29-8 | 38.8 | 11,643 | 34,673 | N | Y | N | JGI | N |
| Talaromyces stipitatus ATCC 10500 | 35.6 | N | N | N | N | N | TIGR | N |
| Verticillium dahliae VaLs. 17 | 33.9 | 10,575 | 29,736 | N | N | N | BI | N |
| Verticillium albo-atrum VaMs. 102 | 32.9 | 10,239 | 28,842 | N | N | N | BI | N |
| Alternaria brassicicola | 32.0 | N | N | N | N | N | WGSC | N |
| A:Bipolaris maydis T:Cochliobolus heterostrophus C5 | 34.9 | 9,633 | 28,007 | N | N | N | JGI | N |
| Pyrenophora tritici-repentis | 38.0 | 12,169 | 32,717 | N | Y | N | BI | N |
| A: Septoria tritici T: Mycosphaerella graminicola | 41.9 | 11,395 | 30,629 | N | Y | N | JGI | N |
| A:Paracercospora fijiensis T:Mycosphaerella fijiensis | 73.4 | 10,327 | 25,289 | N | Y | N | JGI | N |
| A: Stagonospora nodorum T: Phaeosphaeria nodorum | 37.2 | 16,597 | 44,017 | N | Y | N | BI | [54] |
| Total | 2,844.0 | 637,006 | 1,755,655 | 8 | 43 | 1 | ||
aA indicates anamorph name and T presents teleomorph name of fungi.
bC means chromosomes, I indicates InterPro, and E presents EST.
cIncomplete coverage of genome information
dInsufficient exon/intron information
eTaxonomy based on [28]
fORFs and exons were predicted by AUGUSTUS 2.0.3 with species-specific training datasets [55].
'Y' indicates the existence of information in each field, and 'N' indicates the lack of information.
Table 2.
List and characteristics of the fungal genomes belonging to the subphyla Saccharomycotina and Taphrinomycotina archived in SNUGB.
| Speciesa | Size (Mb) | # of ORFs | # of Exons | Cb | Ib | Eb | Source | Refs |
| Fungi (Kingdom)e | ||||||||
| Ascomycota (Phylum) | ||||||||
| Saccharomycotina (Subphylum) | ||||||||
| Candida albicans SC5314 | 14.3 | 6,090 | 6,624 | N | Y | N | SGTC | [56,57] |
| Candida albicans WO-1 | 14.4 | 6,160 | 6,395 | N | Y | N | BI | N |
| Candida dubliniensisd | 14.5 | 6,027 | N | N | N | N | SI | N |
| Candida glabrata CBS138 | 12.3 | 5,165 | 5,249 | N | Y | N | CBS | [58] |
| A: Candida guilliermondii T: Pichia guilliermondii | 10.6 | 5,920 | 5,935 | N | Y | N | BI | N |
| Candida lusitaniae | 12.1 | 5,941 | 5,956 | N | Y | N | BI | N |
| Candida parapsilosis | 13.1 | 5,733 | 5,733 | N | Y | N | BI | N |
| Candida tropicalis | 14.7 | 6,258 | 6,292 | N | Y | N | BI | N |
| Candida tropicalisf | 2.1 | N | N | N | N | N | GS | [59] |
| Ashbya gossypii | 8.8 | 4,717 | 4,943 | 7 | Y | N | NCBI | [60] |
| Debaryomyces hansenii | 12.2 | 6,354 | 6,710 | 7 | Y | N | CBS | [58] |
| Debaryomyces hanseniif | 2.3 | N | N | N | N | N | GS | [61] |
| A: Candida sphaerica T: Kluyveromyces lactis | 10.7 | 5,327 | 5,457 | N | Y | N | GS | [58] |
| A: Candida sphaerica T: Kluyveromyces lactisf | 5.1 | N | N | N | N | N | GS | [62] |
| A: Candida kefyr T:Kluyveromyces marxianusf | 2.0 | N | N | N | N | N | GS | [63] |
| Kluyveromyces polysporus DSM70294 | 14.7 | 5,367 | 5,524 | N | Y | N | SIG | [64] |
| Kluyveromyces thermotoleransf | 2.2 | N | N | N | N | N | GS | [65] |
| Kluyveromyces waltii | 10.9 | 4,935 | 5,395 | N | Y | N | BI | [66] |
| Lodderomyces elongisporus | 15.5 | 5,802 | 5,856 | N | Y | N | BI | N |
| Saccharomyces bayanus MCYC 623 | 11.5 | 9,385 | 9,385 | N | Y | N | BI | [13] |
| Saccharomyces bayanus 623-6C YM4911 | 11.9 | 4,966 | 4,966 | N | Y | N | WGSC | [12] |
| Saccharomyces bayanus var. uvarumf | 4.5 | N | N | N | N | N | GS | [67] |
| Saccharomyces castellii | 11.4 | 4,677 | 4,677 | N | Y | N | WGSC | [12] |
| A: Candida robusta S288C T: Saccharomyces cerevisiae S288C | 12.2 | 6,692 | 7,042 | 16 | Y | N | SGD | [68] |
| A: Candida robusta RM11-1a T: Saccharomyces cerevisiae RM11-1a | 11.7 | 5,696 | 5,988 | N | Y | N | BI | N |
| A: Candida robusta YJM789 T: Saccharomyces cerevisiae YJM789 | 12.0 | 5,903 | 6,153 | N | Y | N | SI | [69] |
| Saccharomyces exiguusf | 2.0 | N | N | N | N | N | GS | [70] |
| Saccharomyces kluyveri | 11.0 | 2,968 | 2,968 | N | Y | N | WGSC | [12] |
| Saccharomyces kluyverif | 2.2 | N | N | N | N | N | GS | [71] |
| Saccharomyces kudriavzevii | 11.2 | 3,768 | 3,768 | N | Y | N | WGSC | [12] |
| Saccharomyces mikatae | 11.5 | 9,016 | 9,016 | N | Y | N | BI | [13] |
| Saccharomyces mikatae | 10.8 | 3,100 | 3,100 | N | Y | N | WGSC | [12] |
| Saccharomyces paradoxus | 11.9 | 8,939 | 8,939 | N | Y | N | BI | [13] |
| Saccharomyces servazziif | 2.0 | N | N | N | N | N | GS | [72] |
| Pichia angustaf | 4.5 | N | N | N | N | N | GS | [73] |
| Pichia stipitis | 15.4 | 5,839 | 8,428 | N | Y | N | JGI | [74] |
| Pichia sorbitophilaf | 3.8 | N | N | N | N | N | GS | [75] |
| A: Candida lipolytica T: Yarrowia lipolytica | 20.5 | 6,524 | 7,264 | 6 | Y | N | CBS | [58] |
| A: Candida lipolytica T: Yarrowia lipolyticaf | 4.6 | N | N | N | N | N | GS | [76] |
| Zygosaccharomyces rouxiif | 4.1 | N | N | N | N | N | GS | [77] |
| Taphrinomycotina (Subphylum) | ||||||||
| Pneumocystis cariniic, d | 6.3 | 4,020 | N | N | N | N | SI | N |
| Schizosaccharomyces japonicus | 11.3 | 5,172 | 10,321 | N | Y | N | BI | N |
| Schizosaccharomyces pombe | 12.6 | 5,058 | 9,869 | 3 | Y | N | GDB | [78] |
| Schizosaccharomyces octosporus | 11.2 | 4,925 | 10,168 | N | N | N | BI | N |
| Total | 424.6 | 176,444 | 188,121 | 5 | 28 | 0 | ||
aA indicates anamorph name and T presents teleomorph name of fungi.
bC means chromosomes, I indicates InterPro, and E presents EST.
cIncomplete coverage of genome information
dInsufficient exon/intron information
eTaxonomy based on [28]
fSequences from Random Sequence Tag (RST)
'Y' indicates the existence of information in each field, and 'N' indicates the lack of information.
Table 3.
List and characteristics of the genomes belonging to the phyla Basidiomycota, Chytridiomycota, and Microsporidia, the subphylum Mucoromycotina, and the phylum Peronosporomycota (oomycetes) archived in SNUGB.
| Speciesa | Size (Mb) | # of ORFs | # of Exons | Cb | Ib | Eb | Source | Refs |
| Fungi (Kingdom)e | ||||||||
| Basidiomycota (Phylum) | ||||||||
| Agricomycotina (Subphylum) | ||||||||
| Postia placenta | 90.9 | 17,173 | 116,596 | N | Y | N | JGI | N |
| T: Phanerochaete chrysosporium A: Sporotrichum pruinosum | 35.1 | 10,048 | 58,746 | N | Y | N | JGI | [79] |
| Coprinus cinereus | 36.3 | 13,544 | 72,887 | N | Y | N | BI | N |
| Laccaria bicolor | 64.9 | 20,614 | 111,290 | N | Y | N | JGI | [80] |
| A: Cryptococcus neoformans Serotype A T: Filobasidiella neoformans Serotype A | 19.5 | 7,302 | 43,325 | 20 | Y | N | BI | N |
| A: Cryptococcus neoformans Serotype B T: Filobasidiella neoformans Serotype B | 19.0 | 6,870 | 40,589 | N | Y | N | NCBI | N |
| A: Cryptococcus neoformans Serotype D B-3501A T: Filobasidiella neoformans Serotype D B-3501A | 18.5 | 6,431 | 40,942 | N | Y | N | SGTC | [41] |
| A: Cryptococcus neoformans Serotype D JEC21 T: Filobasidiella neoformans Serotype D JEC21 | 19.1 | 6,475 | 40,811 | N | Y | N | SGTC | [41] |
| Pucciniomycotina (Subphylum) | ||||||||
| Sporobolomyces roseus | 21.2 | 5,536 | 39,911 | N | Y | N | JGI | N |
| Puccinia graminis | 88.7 | 20,567 | 95,838 | N | Y | N | BI | N |
| Ustilaginomycotina (Subphylum) | ||||||||
| Malassezia globosa CBS7966 | 9.0 | 4,286 | 4,286 | N | N | N | PGC | [15] |
| Malassezia restricta CBS7877c | 4.6 | N | N | N | N | N | PGC | [15] |
| Ustilago maydis 521 | 19.7 | 6,689 | 11,589 | N | Y | N | BI | [81] |
| Ustilago maydis FB1 | 19.3 | 6,950 | 10,310f | N | Y | N | BI | [81] |
| Chytridiomycota (Phylum) | ||||||||
| Batrachochytrium dendrobatidis JEL423 | 23.9 | 8,818 | 38,551 | N | Y | N | BI | N |
| Batrachochytrium dendrobatidis JAM81 | 24.3 | 8,732 | 37,423 | N | Y | N | JGI | N |
| Mucoromycotina (Subphylum incertae sedis) | ||||||||
| Rhizopus oryzae | 46.1 | 17,467 | 57,981 | N | Y | N | BI | N |
| Phycomyces blakesleeanus | 55.9 | 14,792 | 71,502 | N | Y | N | JGI | N |
| Microsporidia (Phylum) | ||||||||
| Encephalitozoon cuniculi | 2.5 | 1,996 | 2,002 | N | Y | N | GS | [82] |
| Antonospora locustaed | 6.1 | 2,606 | N | N | N | N | JBPC | N |
| Stramenopila (Kingdom)e | ||||||||
| Peronosporomycota (Phylum) | ||||||||
| Phytophthora capsici | 107.8 | 17,414 | 45,661 | N | N | N | JGI | N |
| Phytophthora infestansd | 228.5 | 22,658 | N | N | N | N | BI | N |
| Phytophthora ramorum | 66.7 | 15,743 | 40,639 | N | Y | N | JGI | [83] |
| Phytophthora sojae | 86.0 | 19,027 | 53,552 | N | Y | N | JGI | [83] |
| Hyaloperonospora parasitica | 83.6 | 14,789 | 24,907 | N | Y | N | VBI | N |
| Pythium ultimum | 44.3 | N | N | N | N | N | N | |
| Total | 1,241.5 | 276,527 | 1,058,878 | 1 | 20 | 0 | ||
aA indicates anamorph name and T presents teleomorph name of fungi
bC means chromosomes, I indicates InterPro, and E presents EST.
cIncomplete coverage of genome information
dInsufficient exon/intron information
eTaxonomy based on [28]
fORFs and exons were predicted by AUGUSTUS 2.0.3 with species-specific training datasets [55].
'Y' indicates the existence of information in each field, and 'N' indicates the lack of information.
Table 4.
List and characteristics of the non-fungal genomes archived in SNUGB.
| Speciesa | Size (Mb) | # of ORFs | # of Exons | Cb | Ib | Eb | Source | Refs |
| Chloroplastida (Kingdom)e | ||||||||
| Streptophyta (Phylum) | ||||||||
| Arabidopsis thaliana | 119.2 | 28,581 | 150,369 | 5 | Y | N | TAIR | [33] |
| Carica papaya | 271.7 | N | N | N | N | N | PGSC | [84] |
| Glycine max | 996.9 | 62,199 | 281,102 | N | N | N | JGI | N |
| Lycopersicon esculentumc | 39.9 | 8,725 | 29,707 | N | Y | N | SOL | N |
| Medicago truncatula | 278.7 | 38,334 | 122,889 | 8 | Y | N | MTGSP | [85-87] |
| Oryza sativa var. Indicad | 426.3 | 49,710 | N | N | N | N | BGI | [88,89] |
| Oryza sativa var. Japonica | 372.1 | 66,710 | 319,140 | 12 | Y | N | IRGSP | [89,90] |
| Populus trichocarpa | 485.5 | 45,555 | 193,687 | N | Y | N | JGI | [91] |
| Ricinus communisd | 362.5 | 38,613 | N | N | N | N | TIGR | N |
| Selaginella moellendorffii | 212.8 | 22,285 | 124,645 | N | Y | N | JGI | N |
| Sorghum bicolor | 738.5 | 36,338 | 165,149 | 11 | Y | N | JGI | N |
| Vitis vinifera | 497.5 | 30,434 | 149,351 | 19 | Y | N | GS | [92] |
| Zea maysd | 2,314.7 | 420,732 | N | N | N | N | MGSP | N |
| Metazoa (Kingdom) | ||||||||
| Arthropoda (Phylum) | ||||||||
| Apis mellifera | 235.2 | 11,062 | 71,496 | N | N | N | HBGP | [93] |
| Acyrthosiphon pisum | 446.6 | N | N | N | N | N | BCM | N |
| Bombyx mori | 397.7 | 21,302 | 82,381 | N | N | N | BGI | [94] |
| Drosophila ananassae | 231.0 | 15,276 | 56,595 | N | N | N | FB | [95] |
| Drosophila erecta | 152.7 | 15,324 | 56,924 | N | N | N | FB | [95] |
| Drosophila grimshawi | 200.5 | 15,270 | 56,647 | N | N | N | FB | [95] |
| Drosophila melanogaster | 168.7 | 20,923 | 96,745 | N | N | N | FB | [96] |
| Drosophila mojavensis | 193.8 | 14,849 | 55,013 | N | N | N | FB | [95] |
| Drosophila persimilis | 188.4 | 17,235 | 59,116 | N | N | N | FB | [95] |
| Drosophila pseudoobscura | 152.7 | 16,363 | 57,864 | N | N | N | FB | [97] |
| Drosophila sechellia | 166.6 | 16,884 | 58,584 | N | N | N | FB | [95] |
| Drosophila simulans | 137.8 | 15,983 | 54,294 | N | N | N | FB | [95] |
| Drosophila virilise | 206.0 | 14,680 | 55,005 | N | N | N | FB | [95] |
| Drosophila willistoni | 235.5 | 15,816 | 56,641 | N | N | N | FB | [95] |
| Drosophila yakuba | 165.7 | 15,423 | 59,098 | N | N | N | FB | [95] |
| Glossina morsitans | 205.7 | N | N | N | N | N | TIGR | N |
| Nasonia vitripennis | 239.6 | 27,957 | 98,570f | N | N | N | BCM | N |
| Tribolium castaneum | 152.1 | 14,274 | 58,381f | N | N | N | BCM | N |
| Nematoda (Phylum) | ||||||||
| Caenorhabditis elegans | 100.3 | 26,902 | 175,232 | 7 | N | N | WB | [34] |
| Caenorhabditis briggsaed | 108.5 | 20,669 | N | N | N | N | WB | [98] |
| Caenorhabditis remanei | 145.4 | N | N | N | N | N | WB | N |
| Vertebrata (Phylum) | ||||||||
| Homo sapiens Celera assembly | 2,828.4 | 28,057 | 273,999 | N | N | N | NCBI | [99] |
| Homo sapiens HuRef assembly | 2,843.9 | 27,937 | 273,135 | N | N | N | NCBI | [100] |
| Homo sapiens NCBI Reference | 2,870.8 | 29,319 | 284,553 | N | N | N | NCBI | [100] |
| Homo sapiens | 3,665.5 | 43,570 | 452,099 | 29 | N | N | EM | [100] |
| Total | 21,241.0 | 1,294,281 | 4,142,169 | 7 | 8 | 0 | ||
aA indicates anamorph name and T presents teleomorph name of fungi
bC means chromosomes, I indicates InterPro, and E presents EST.
cIncomplete coverage of genome information
dInsufficient exon/intron information
eTaxonomy based on [101]
fORFs and exons were predicted by AUGUSTUS 2.0.3 with species-specific training datasets [55].
'Y' indicates the existence of information in each field, and 'N' indicates the lack of information.
Construction, content, and applications
Data processing via an automated pipeline and the function of Positional Database
Positional information of functional/structural units that are present on individual contigs/chromosomes, such as the start and stop sites of ORFs and exons/introns, was collected from the data warehouse of CFGP and stored in the Position Database of SNUGB. New types of data, such as Simple Sequence Repeats (SSRs) on the genome, can be easily added to the Positional Database for visualization via SNUGB. Along with the positional information, for each data, data type (e.g., ORFs), primary key, and any additional information were saved into the partitioned tables, which were designed for enhancing the speed of data retrieval. Through the primary key, SNUGB can display detailed information of each datum (e.g., sequences) stored at external sources. Considering the large number of available fungal genome sequences and those that are currently being sequenced, in addition to this data standardization scheme, a standardized pipeline for data extraction and management is needed to organize the data and to ensure orderly expansion of SNUGB.
The pipeline developed for SNUGB processes each genome data set via the following steps. Firstly, once whole genome sequences are deposited in the data warehouse of CFGP, the integrity of genome information, such as the position information of functional/structural units, is inspected. Several properties of the whole genome, such as the length and the GC content, are calculated. Secondly, the GC content, AT-skew, and CG-skew are calculated via 50-bp sliding windows with 20 bp steps. Thirdly, for each gene, three types of sequence information, including coding sequences (sequences from the start to stop codon without introns), gene sequences (sequences from the start to stop codon with introns), and transcript sequences (sequences from the transcription start site to end site without intron sequence), if transcript information is available, are generated based on the genome annotation information. Fourthly, all data generated in the previous steps are transferred into the Position Database to support graphical representation of these features. Fifthly, if the genome has chromosomal map information, including genetic map and optical map, this information is converted into a standardized format and stored in SNUGB for graphical representation via Chromosome Viewer. Lastly, after subjecting all ORFs in the genome through the InterPro Scan [29], the genomic position of each domain predicted by the InterPro Scan is calculated and stored into the Position Database.
Modular design of SNUGB facilitates its application
To facilitate the efficient implementation of SNUGB in diverse genomics platforms, a modular design was used for its application programming interface (API). Through API, a diagram showing genome features in a selected region can be created using only their chromosomal positions and display options. Four recent publications illustrate the utility of this design: T-DNA Analysis Platform (TAP; http://tdna.snu.ac.kr/) provides the GC content and AT skew around T-DNA insertion sites on the chromosomes of Magnaporthe oryzae via a mini genome browser supported by SNUGB [25]. The chromosomal distribution pattern of T-DNA insertion sites in M. oryzae http://atmt.snu.ac.kr/ was also displayed using SNUGB [26]. Fungal Cytochrome P450 Database (FCPD; http://p450.riceblast.snu.ac.kr/) [24] employs SNUGB to present the chromosomal distribution pattern and contexts of cytochrome P450 genes in fungal genomes. Two databases, FED http://fed.snu.ac.kr/ and FSD http://fsd.snu.ac.kr/, utilize SNUGB for presenting the genomic context of the region matched to EST and secreted proteins, respectively. Moreover, Systematical Platform for Identifying Mutated Proteins (SysPIMP; http://pimp.starflr.info/) [30] and Insect Mitochondrial Genome Database (IMGD; http://www.imgd.org; Lee et al., under revision) also adopted SNUGB for data presentation. These examples illustrate the utility of SNUGB.
Properties of the fungal/oomycete genomes archived in SNUGB
Among the 98 fungal/oomyvete species (137 genome datasets) covered by SNUGB, 77 species (111 genome datasets; 81%) belong to the phylum Ascomycota (Table 1 and 2), and 10 species (14 genome datasets; 10%) belong to the phylum Basidiomycota (Table 3). In contrast, the phyla Chytridiomycota and Micosporidia are represented only by one (2 datasets) and two species (both belong to the subphylum Mucoromycotina), respectively (Table 3). Six oomycete genomes, derived from Phytophthora, Hyaloperonospora, and Pythium species, are available for comparison with fungal genomes (Table 3). Although oomycetes belong to the kingdom Stramenophla and show closer phylogenetic relationships to algae and diatoms than fungi [31], due to their morphological similarities to fungi, they have been traditionally grouped with fungi.
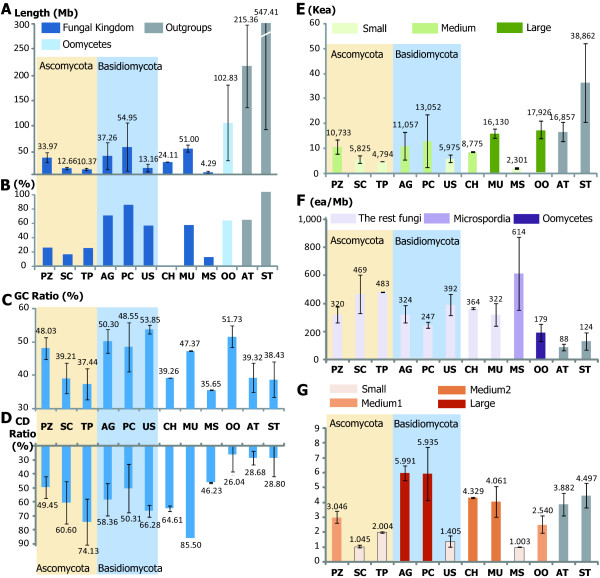
The datasets that cover the whole genome (121 out of the 137 datasets) were analyzed to investigate genome properties. The average size of the genomes, measured by adding lengths of all scaffolds together, is 31.42 Mb which is one-seventeenth of plant genomes (547.41 Mb in the phylum Streptophyta) and one-seventh of insect genomes (215.36 Mb in the phylum Arthropoda) (Figure 1A). The fungal/oomycete genome sizes ranged from 2.5 Mb (Encephalitozoon cuniculi) to 228.5 Mb (Phytophthora infestans); the genome of E. cuniculi is shorter than that of Escherichia coli (4.6 Mb) [32], while the genome of P. infestans is much larger than the genomes of Arabidopsis thaliana (119.2 Mb) [33] and Caenorhabditis elegans (100.5 Mb) [34], indicating no clear relationship between the genome size and the organismal complexity [35]. With regard to the average genome sizes in different taxon groups, the phylum Microsporidia, known as ancestral fungi, shows the smallest average size (4.28 Mb), while oomycetes show the largest at 102.83 Mb (Figure 1A). In the phylum Basidiomycota, which is large and very diverse, the degree of difference in average genome sizes within each of the represented subphyla is highest in the fungal kingdom: the ratios of standard deviation to the average length in three subphyla Agricomycotina, Pucciniomycotina, and Ustilaginomycotina are 71.95%, 86.93%, and 57.46%, respectively (Figure 1B). The subphylum Pucciniomycotina displays the largest size with large variation (Figure 1A and 1B), while two subphyla Saccharomycotina and Taphrinomycotina belonging to the phylum Ascomycota exhibit the relatively low degree of variations (Figure 1B), probably because only closely related species have been sequenced. Although the average genome sizes varied from group to group, ANOVA and TukeyHSD tests (P < 0.05) showed only the difference between fungi and oomycetes was significant (Figure 1A). The GC content of fungal genomes ranges from 32.523% (Pneumocystis carinii in subphylum Taphrinomycotina) to 56.968% (Phanerochaete chrysosporium in the subphylum Agricomycotina), while the GC content of plant and insect genomes ranges from 29.638% to 46.850% (Figure 1C). Although the coding regions exhibit higher GC contents than the rest of the genome, there is no relationship between the proportion of ORFs on the genome and the GC content of the whole genomes (linear regression; R2 = 0.04; Figure 1C and 1D).
Figure 1.
Characteristics of the 137 fungal and oomycetes genomes archived in SNUGB. In all graphs, the first six groups correspond to subphyla and the rests indicate phyla. Error bars indicate variation of data within each taxonomic group. The last two phyla were used as outgroup. In graphs A, E, F, and G, each color of bar indicates distinct group supported by Turkey HSD test. (A) Average genome size. (B) the ratio of variation of genome size to the average genome size. (C) Average GC ratio of each subphylum/phylum. (D) The percentage of coding regions to the genome length. (E) Average number of total ORFs. (F) The total number of ORFs per Mb (= ORF density). (G) The average exon number of each ORFs.
The number of total proteins encoded by each organism was once considered to reflect organism's characteristics [36]. Based on the size of total proteomes, all sequenced fungal and oomycete species were divided into three groups: The medium group contains the subphylum Pezizomycotina in Ascomycota and the subphyla Agricomycota and Puccinomycotina in Basidiomycota, the small group includes three subphyla Saccharomycotina, Taphrinomycotina, and Ustilagomycotina and the phylum Microsporidia, and the large group has the subphylum Mucoromycotina and the phylum Oomycota (ANOVA and TukeyHSD; P < 0.05; Figure 1E). This grouping shows that the number of total ORFs does not correlate with taxonomic positions at the phylum level, however, at the subphylum level, the correlation was high. For example, subphyla Saccharomycotina and Taphrinomycotina can be distinguishable from Pezizomycotina based on this character. The ORF density classified the sequenced species into three distinct groups, Oomycetes, Microsporidia and the rest, through ANOVA and TukeyHSD test (P < 0.05; Figure 1F). Taken together, these three indicators can be used to divide fungal subphyla/phyla. For example, the subphylum Pezizomycotina shows the medium-level of ORF number and ORF density, while the subphylum Saccharomycotina displays the low-level of ORF number but its ORF density is comparable to that of the subphylum Pezizomycotia. Both the number of ORFs and the ORF density are high for oomycetes, exhibiting a pattern different from fungi.
The number of exons per ORF was investigated, resulting in four groups (ANOVA and TukeyHSD test; P < 0.05; Figure 1G). With the exception of the subphylum Ustilagomycotina, the phylum Basidiomycota exhibits the highest number (~6). The subphyla Saccharomycotina and Mycoromycotina show the lowest value (nearly 1), indicating that almost all their genes do not have introns.
Comparison of genome sequences of multiple isolates within species
For 14 fungal species, two or more strains have been sequenced (Table 5). For some species, such as Fusarium graminearum, additional isolate(s) were sequenced only at a low coverage (e.g., 0.4× coverage for the second strain of F. graminearum); however, even such low-coverage provided some insights into the evolution of pathogenicity in this important cereal pathogen [37]. Except Aspergillus niger, Histoplasma capsulatum, and Paracoccidioides brasiliensis, all strains within same species showed less than 1 Mb variation in genome sizes (Table 5). It is possible that the 3.2 Mb difference between two A. niger strains is in part due to different sequencing coverage: the coverage of ATCC1015 was 8.9× while CBS513.88 was 7× http://genome.jgi-psf.org/Aspni1/Aspni1.info.html[38]. The differences among three P. brasiliensis genomes, ranging from 29.1 Mb to 33.0 Mb, may reflect their distinct phylogenetic positions [39]. The differences among five H. capsulatum genomes may be due to a combination of different levels of sequencing coverage http://www.broad.mit.edu/annotation/genome/histoplasma_capsulatum/Info.html and different geological origins [40]. Three isolates of H. capsulatum and P. brasiliensis showed approximately 1% difference in the GC content, whereas the degree of GC content variation among 11 strains of Coccidioides posadasii was only 0.5%. Four Cryptococcus neoformans strains, representing three different serotypes (A, B and D), showed around 0.3% variation in the GC content, and within a serotype (two serotype D strains) the difference was only 0.043% [41]. Isolates of Candida albicans, Saccharomyces bayanus, and Batrachochytrium dendrobatidis showed only 0.01% variation in the GC content. These intraspecific variations of genome properties can be compared in detail via SNUGB.
Table 5.
Basic properties of different strains of fungal genomes deposited in SNUGB.
| Species | # of Strains | Genome size (Mb) | GC content (%) |
| Fungi (Kingdom) | |||
| Ascomycota (Phylum) | |||
| Pezizomycotina (Subphylum) | |||
| Aspergillus fumigatus | 2 | 29.3 ± 0.1 | 49.672 ± 0.178 |
| Aspergillus niger | 2 | 35.6 ± 2.3 | 50.365 ± 0.012 |
| Coccidioides immitis | 4 | 28.3 ± 0.7 | 46.529 ± 0.514 |
| Coccidioides posadasii | 11 | 27.2 ± 0.9 | 46.839 ± 0.537 |
| Histoplasma capsulatum | 5 | 36.2 ± 4.7 | 43.400 ± 1.859 |
| Paracoccidioides brasiliensis | 3 | 30.7 ± 2.0 | 43.868 ± 0.930 |
| Fusarium graminearuma | 2 | 36.6 | 48.283 |
| Saccharomycotina (Subphylum) | |||
| Candida albicans | 2 | 14.4 ± 0.1 | 33.462 ± 0.010 |
| Saccharomyces cerevisiae | 3 | 11.9 ± 0.3 | 38.252 ± 0.090 |
| Saccharomyces bayanus | 2 | 11.7 ± 0.3 | 40.196 ± 0.011 |
| Saccharomyces mikataeb | 2 | 11.1 ± 0.5 | 37.920 ± 0.315 |
| Basidiomycota (Phylum) | |||
| Agricomycotina (Subphylum) | |||
| Cryptococcus neoformans | 4 | 19.2 ± 0.2 | 48.251 ± 0.316 |
| Ustilaginomycotina (Subphylum) | |||
| Ustilago maydis | 2 | 19.7 ± 0.0 | 53.995 ± 0.045 |
| Chytridiomycota (Phylum) | |||
| Batrachochytrium dendrobatidis | 2 | 24.1 ± 0.3 | 39.261 ± 0.011 |
| Chloroplastida (Kingdom) | |||
| Charophyta (Phylum) | |||
| Oryza sativa | 2 | 399.2 ± 38.4 | 43.530 ± 0.046 |
| Vertebrata (Phylum) | |||
| Vertebrata (Phylum) | |||
| Homo sapiens | 4 | 3,052.2 ± 409.3 | 40.878 ± 0.042 |
aOne of strains are incomplete whole genome sequences, so that standard deviation of genome length and GC content are not calculated.
bSame strain but different version of assembly
Update of SNUGB
The number of on-going fungal genome sequencing projects is approximately 40 http://fungalgenomes.org/wiki/Fungal_Genome_Links. 37 strains of S. cerevisiae and 25 strains of S. paradoxus were already sequenced and released by the Sanger institute http://www.sanger.ac.uk/Teams/Team118/sgrp/, indicating that more than 100 additional fungal genomes will be available soon. Next generation high throughput sequencing technologies, such as GS Flx, Solexa, and SOLiD [42,43], will further accelerate the rate of fungal genome sequencing, emphasizing the importance of frequently updating SNUGB. With the aid of the developed pipeline, SNUGB will be updated whenever new fungal genome sequences have been publicly released with annotation information. A notice for updated genomes will be posted on the SNUGB web site.
Functions and tools
Taxonomy browser
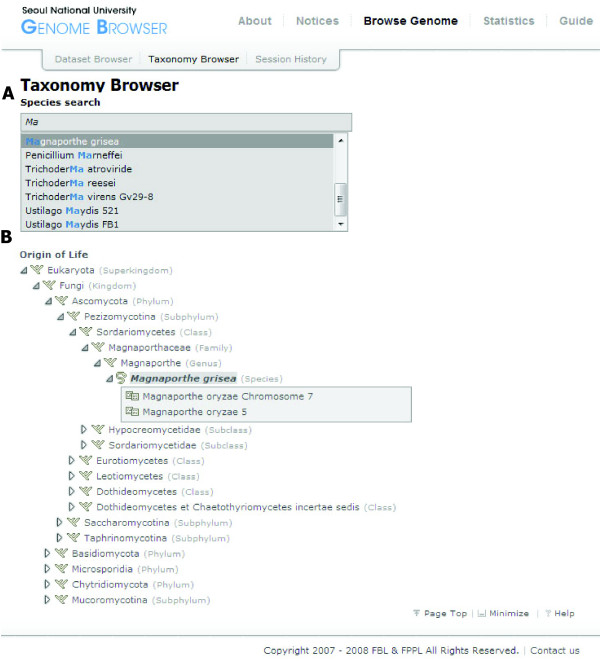
To support selection of species of interests based on their taxonomic positions, a web-based tool, named as the taxonomy browser, was developed. Considering an anticipated increase in comparing genome sequences and features across multiple species to investigate evolutionary questions at the genome scale, such a tool is necessary to provide an overview of the taxonomic positions of the sequenced species and their evolutionary relationships with other fungi to users of SNUGB and to assist them in selecting appropriate species for comparative analyses. The taxonomy browser provides two methods for accessing the data archived in SNUGB, one of which is text-search using species name (Figure 2A). When a user begins typing a species name in the text box, the full name will be completed automatically to assist a quick search of species. The other method is using the taxonomical hierarchy (i.e., tree of life). When a user clicks a specific taxon (e.g., phylum), taxonomy browser will present all subgroups within the chosen taxon for further selection (Figure 2B).
Figure 2.
Taxonomy browser. A screenshot of data generated using Taxonomy browser is shown. (A) Search interface by species name shows a list of species along with inserted string. (B) Taxonomical tree shows a lineage of the chosen species and its genome datasets deposited in SNUGB.
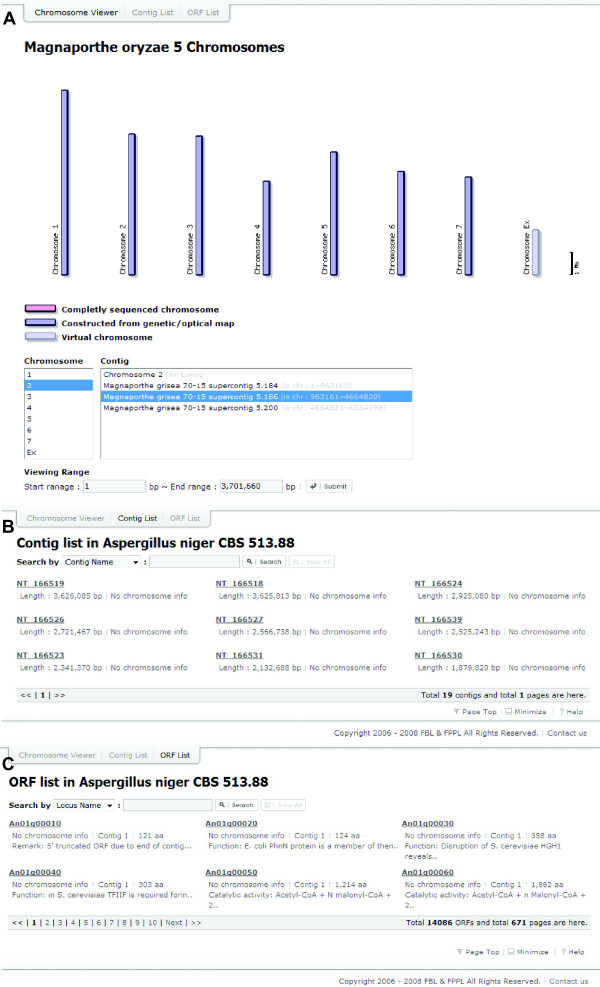
Chromosome viewer and Contig/ORF browser
Three different methods can be used to access genomic information. For those with chromosomal map data (21 species), their chromosomal maps can be displayed via Chromosome viewer (Figure 3A). The following color scheme was used to denote the level of completeness: i) chromosome constructed using genetic or optical map data (with gaps) as blue (Chromosomes 1 to 7 of M. oryzae; Figure 3A), ii) chromosome map based on a combination of sequences and genetic/optical map data as pink (e.g., chromosomes of A. niger), and iii) unassigned contigs (labeled as Chromosome Ex of M. oryzae; Figure 3A) as light blue. For the species without chromosomal map information, SNUGB provides the contig and ORF browsers, which display the name of contig and ORFs, respectively, and allow users to search them using their names (Figures 3B and 3C).
Figure 3.
Chromosome viewer, Contig Viewer, and ORF Viewer. (A) The chromosome viewer displays seven chromosomes of M. oryzae with a size indicator at the right side. At the bottom, the interface allows for jumping directly to a specific region by selecting chromosome/contigs and its position. (B) The contig viewer provides a list of contigs with its length. Through this interface, contigs can be searched by name. (C) The ORF viewer presents the names and lengths of ORFs with search function.
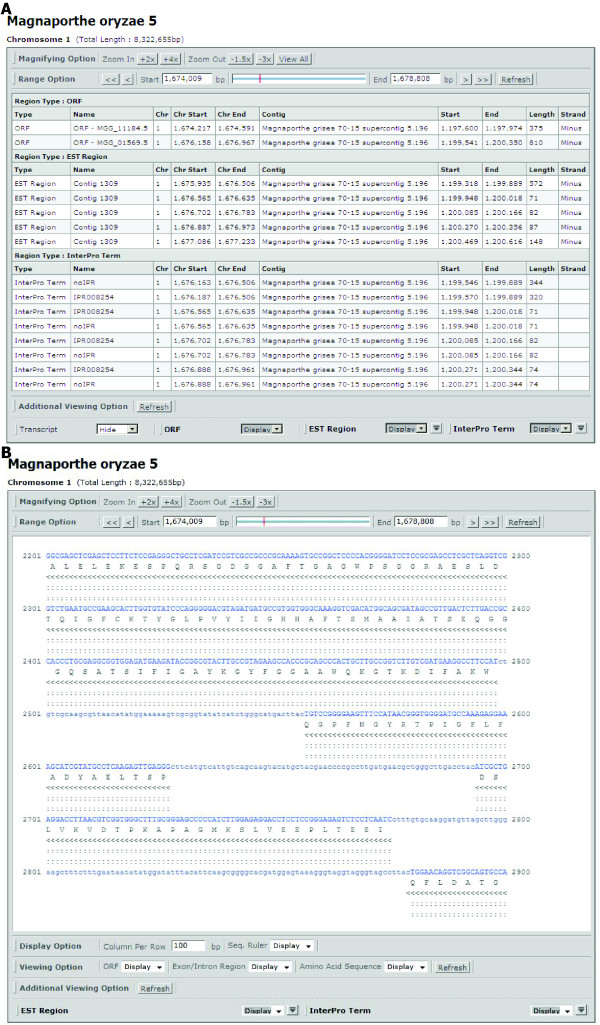
Graphical Browser with six different display formats
Gene annotation information in a selected area of chromosome or contig, such as transcripts, ORFs, and exon/intron structure, and InterPro domains [29], can be displayed through three formats: i) the 'single' format shows these features as bars; ii) the 'squish' format displays them via color-coded diagrams without description; and iii) the 'pack' format presents them as small color-coded icons with description (Figure 4A). These graphical formats were also used by UCSC Genome Browser [2]. In addition, the GC content and AT/CG skew information for individual chromosomes can be displayed via three formats: i) color-coded bar graph, ii) line, and iii) dotted lines along with a description of data (Figure 4B). For species with EST data (Table 1), the genomic region corresponding to each EST sequence can be displayed along with ORF and InterPro domains to help users identify predicted gene structure and expressed regions (see Figure 4A). Presentation of these data is supported by Fungal Expression Database http://fed.snu.ac.kr/.
Figure 4.
Six different display methods of the genome content and properties via Graphical browser. (A) The graphical browser in SNUGB shows the genome context via three different formats: bar, squash, and pack. At the bottom, ORFs, ESTs, and InterPro domains on chromosome 1 of M. oryzae are displayed. (B) Three graphic representations, including graph, line, and Single line (S. line), of the AT-skew, GC-skew, and GC content are shown.
Table browser and Text browser
Although graphical presentation of genomic features helps users view global patterns, the graphical browser does not provide sequences or a list of elements present in a chosen area. To provide such information, we developed two additional tools named as the table browser and the text browser. The table browser provides a list of the names and chromosomal/contig positions of all elements present in a selected region in the csv format, which can be opened using the Excel program (Figure 5A). The text browser provides sequences in a selected region. If ORFs exist in the region, exons and introns are presented using different colors and cases; this function is useful for designing primers and transferring selected sequences to a different data analysis environment (Figure 5B). Additionally, all InterPro domains present on each ORF are displayed as special characters under corresponding sequences so that putative functional domains can be easily recognized at the sequence level. The table and text browser can display sequences up to 50 kb.
Figure 5.
Table and Text browsers. (A) The table browser shows all ORFs, ESTs, and InterPro domains in a selected region as a list. (B) The text browser displays sequences showing exon/intron region as different colors and EST and InterPro domains.
Kingdom-wide identification of the putative orthologues of individual fungal proteins via BLAST and comparison of the genomic contexts and properties of homologous proteins among species via the Session History function
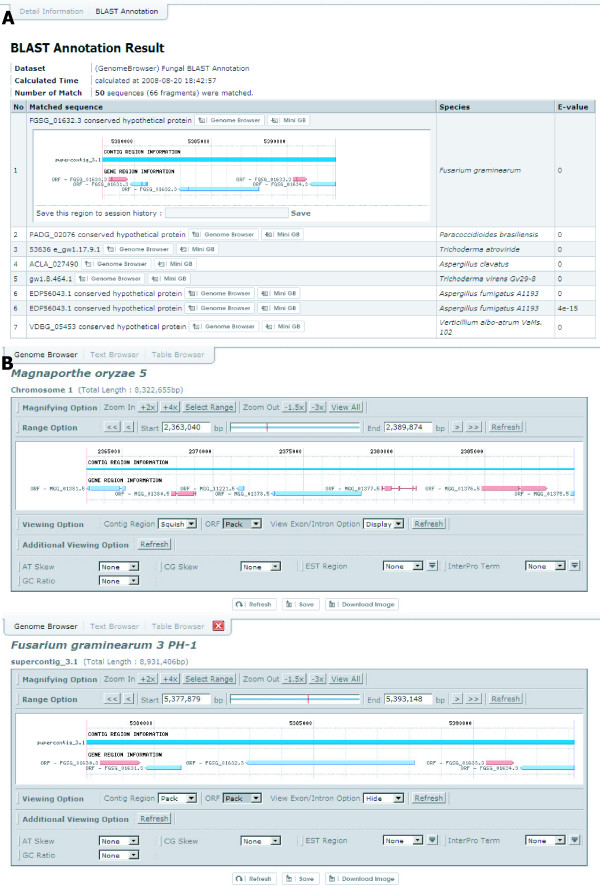
To identify putative orthologues of individual fungal proteins, BLAST searches with each of the 924,343 fungal proteins against all proteins were performed using the e-value of 1e-5 as the cut-off line. The 'BLAST annotation' tab shows a list of putative orthologues of a chosen gene product in other species with their BLAST e-values (see Figure 6A). To compare the genomic contexts around the orthologous genes between species or among multiple species, users can store the genomic contexts of the genes using the Session History function, in which the stored genomic contexts can be displayed in one screen (Figure 6B). In each session, other information, such as the GC content and InterPro terms, can also be presented to further support the comparison.
Figure 6.
BLAST annotation to catalog homologous proteins. (A) A result of 'BLAST annotation' is shown with the corresponding gene names, species names, and e-values of putative homologs. 'Genome Browser' button after gene name can display the genome context of the selected gene, and 'Mini GB' button will show genome contexts of the selected gene as a smaller size to provide a quick overview, supported by MiniGB. The session can be stored by clicking the save link inside the small SNUGB image. (B) Two independent sessions showing homologs of two genes, MGG_01378.5 and FGSG_01632.3, are shown. Clicking the red button X at the bottom will hide the session.
Additional functionalities of SNUGB
The 'flexible-range-select' function allows users to select a chromosomal segment by clicking a mouse at the start site and moving it over the desired segment; the selected area will be displayed as shaded box, and the subsequent click displays an enlarged view of the selected segment (Figure 3A). Through the 'high-resolution-diagram' function, users can obtain a high-resolution image (more than 3,000 pixels in width) showing various features on a whole chromosome, such as ORFs, InterPro terms, and GC content. This image can be downloaded as image file via both the graphical genome browser and the session-storage function.
Conclusion
The SNUGB supports efficient and versatile visualization and utilization of rapidly increasing fungal genome sequence data, as well as those from selected organisms in other kingdoms, to address various types of questions at the genome scale. Properties and features of the archived fungal genomes are available for viewing and comparison in SNUGB. The taxonomy browser helps users easily access the genomes of individual species and provides taxonomic positions of chosen species, and the chromosome map function shows the whole genome of selected species. The graphical browser, table browser, and text browser present a global view of genomic contexts in a selected chromosomal region and support analyses of sequences in the region. The 'BLAST annotation' provides lists of putatively orthologous proteins in the fungal kingdom and facilitates comparison of the genomic contexts of their genes across multiple species. The SNUGB also allows users to manage their own work histories via the SNUGB web site.
Availability and requirements
All data and functionalities in this paper can be freely accessed through the SNUGB web site at http://genomebrowser.snu.ac.kr/. The source code, a set of programs, and database structure of SNUGB will be publicly released in the future after finalizing packaging of SNUGB to be opened.
Abbreviations
PZ: the subphylum Pezizomycotina; SC: the subphylum Saccharomycotina; TP: the subphylum Taphrinophycotina; AG: the subphylum Agricomycotina; PC: the subphylum Pucciniomycotina; US: the subphylum Ustilagomycotina; CH: the phylum Chytridiomycota; MU: the subphylum Mucoromycotina; MS: the phylum Microsporidia; OO: oomycete (the phylum Peronosporomycota); AT: the phylum Arthropoda; ST: the phylum Streptophyta; BCM: Baylor College of Medicine; BGI: Beijing Genome Institute; BGM: Baylor College of Medicine; BI: Broad Institute; CBS: Center For Biological Sequences; DOGAN: Database Of the Genomes Analyzed at Nite; EM: Ensembl; FB: Flybase; GDB: GeneDB; GS: Genoscope; HBGP: Honey Bee Genome Project; IGM: Instituté de Génétique et Microbiologie; IRGSP: International Rice Genome Sequencing Project; JBPC: Josephine Bay Paul Center for Comparative Molecular Biology and Evolution; JGI: DOE Joint Genomic Institute; MGSP: Maize Genome Sequencing Project; MTGSP: Medicago Truncatula Genome Sequencing Project; OU: Oklahoma University; PGC: Procter & Gamble Co; PGSC: Papaya Genome Sequencing Consortium; SGTC: Stanford Genome Technology Center; SI: Sanger Institute; SIG: Trinity College Dublin: Smurfit Institute of Genetics; TAIR: The Arabidopsis Information Resource; VGI: Virginia Bioinformatics Institute; WB: Wormbase; WGSC: Washington University Genome Sequencing Center.
Authors' contributions
JP and YHL planed and managed this project, KJ designed the web site, KJ, JP, BP, KA, JYC, and JHC implemented various functions to SNUGB, JP, JYC, SIK, and DC processed genome sequences, and JP, SK and YHL wrote the manuscript.
Acknowledgments
Acknowledgements
We thank Donghan Kim and Wonho Song for collecting genome sequences from various web sites. This research was partially supported by grants from Crop Functional Genomics Center (CG1141) and a grant from Biogreen21 Project (20080401034044) funded by the Rural Development Administration to YHL. A USDA-NRI grant to SK (2008-55605-18773) also supported this work. KJ and JP thank graduate fellowships provided by the Ministry of Education through the Brain Korea 21 Project.
Contributor Information
Kyongyong Jung, Email: lulupon0@snu.ac.kr.
Jongsun Park, Email: starflr@snu.ac.kr.
Jaeyoung Choi, Email: amethyst1016@gmail.com.
Bongsoo Park, Email: pbsfree@gmail.com.
Seungill Kim, Email: ksi2204@gmail.com.
Kyohun Ahn, Email: dksrygns00@snu.ac.kr.
Jaehyuk Choi, Email: jaehyuk2@snu.ac.kr.
Doil Choi, Email: doil@snu.ac.kr.
Seogchan Kang, Email: sxk55@psu.edu.
Yong-Hwan Lee, Email: yonglee@snu.ac.kr.
References
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pedersen JS, Hsu F, Hinrichs AS, Harte RA, Diekhans M, Clawson H, Bejerano G, Barber GP, Baertsch R, Haussler D, Kent WJ. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–786. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoniewski MJ, Yates T, Dibben S, Miller CJ. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Park B, Jung K, Jang S, Yu K, Choi J, Kong S, Park J, Kim S, Kim H, Kim S, Kim JF, Blair JE, Lee K, Kang S, Lee YH. CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res. 2008;36:D562–571. doi: 10.1093/nar/gkm758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH. Comparative genomics and genome evolution in yeasts. Philos Trans R Soc Lond B Biol Sci. 2006;361:403–412. doi: 10.1098/rstb.2005.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS ONE. 2008;3:e2300. doi: 10.1371/journal.pone.0002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, Deangelis YM, Reeder NL, Johnstone KR, Leland M, Fieno AM, Begley WM, Sun Y, Lacey MP, Chaudhary T, Keough T, Chu L, Sears R, Yuan B, Dawson TL., Jr Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J. Whole genome comparision of A. flavus and A. oryzae. Med Mycol. 2006;44:9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- Wortman JR, Fedorova N, Crabtree J, Joardar V, Maiti R, Haas BJ, Amedeo P, Lee E, Jiang SVAB, Anderson MJ, Denning DW, White OR, Nierman WC. Whole genome comparison of the A. fumigatus family. Med Mycol. 2006;44:3–7. doi: 10.1080/13693780600835799. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS, Roy SW. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedeler C, Wong HM, Cornell MJ, Alam I, Soanes DM, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG, Paton NW. e-Fungi: a data resource for comparative analysis of fungal genomes. BMC Genomics. 2007;8:426. doi: 10.1186/1471-2164-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Park J, Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Kim S, Park B, Jung K, Kim S, Kang S, Lee YH. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24:1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- Park J, Kim H, Kim S, Kong S, Park J, Kim S, Han H, Park B, Jung K, Lee Y-H- A comparative genome-wide analysis of GATA transcription factors in fungi. Genomics & Informatics. 2006;4:147–160. doi: 10.1016/j.aei.2005.09.003. [DOI] [Google Scholar]
- Park J, Lee S, Choi J, Ahn K, Park B, Park J, Kang S, Lee YH. Fungal Cytochrome P450 Database. BMC Genomics. 2008;9:402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park J, Jeon J, Chi MH, Goh J, Yoo SY, Park J, Jung K, Kim H, Park SY, Rho HS, Kim S, Kim BR, Han SS, Kang S, Lee YH. Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae. Mol Microbiol. 2007;66:371–382. doi: 10.1111/j.1365-2958.2007.05918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S, Choi J, Park JY, Yi M, Yang S, Kwon MJ, Han SS, Kim BR, Khang CH, Park B, Lim SE, Jung K, Kong S, Karunakaran M, Oh HS, Kim H, Kim S, Park J, Kang S, Choi WB, Kang S, Lee YH. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- Park J, Park B, Veeraraghavan N, Jung K, Lee YH, Blair J, Geiser DM, Isard S, Mansfield MA, Nikolaeva E, Park SY, Russo J, Kim SH, Greene M, Ivors KL, Balci Y, Peiman M, Erwin DC, Coffey MD, Rossman A, Farr D, Cline E, Crünwald NJ, Luster DG, Schrandt J, Martin F, Ribeiro OK, Makalowska I, Kang S. Phytophthora Database: A Forensic Database Supporting the Identification and Monitoring of Phytophthora. Plant Dis. 2008;92:966–972. doi: 10.1094/PDIS-92-6-0966. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Koljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schussler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Nikolskaya AN, Orchard S, Orengo C, Petryszak R, Selengut JD, Sigrist CJ, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Park J, Ding G, Lee YH, Li Y. SysPIMP: the web-based systematical platform for identifying human disease-related mutated sequences from mass spectrometry. Nucleic Acids Res. 2009;37:D913–D920. doi: 10.1093/nar/gkn848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster H, Coffey M, Elwood H, Sogin ML. Sequence analysis of the small subunit ribosomal RNAs of three zoosporic fungi and implications for fungal evolution. Mycologia. 1990;82:306–312. doi: 10.2307/3759901. [DOI] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- AGI Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- CSC Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Thomas CA., Jr The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, Baker SE, Rep M, Adam G, Antoniw J, Baldwin T, Calvo S, Chang YL, Decaprio D, Gale LR, Gnerre S, Goswami RS, Hammond-Kosack K, Harris LJ, Hilburn K, Kennell JC, Kroken S, Magnuson JK, Mannhaupt G, Mauceli E, Mewes HW, Mitterbauer R, Muehlbauer G, Munsterkotter M, Nelson D, O'Donnell K, Ouellet T, Qi W, Quesneville H, Roncero MI, Seong KY, Tetko IV, Urban M, Waalwijk C, Ward TJ, Yao J, Birren BW, Kistler HC. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, Berg M van den, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d'Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, Hombergh JP van den, Hondel CA van den, Heijden RT van der, Kaaij RM van der, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, Maarel MJ van der, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, Vondervoort PJ van de, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Matute DR, Sepulveda VE, Quesada LM, Goldman GH, Taylor JW, Restrepo A, McEwen JG. Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J Clin Microbiol. 2006;44:2153–2157. doi: 10.1128/JCM.02540-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castaneda E, Da Silva Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancope-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol. 2003;12:3383–3401. doi: 10.1046/j.1365-294X.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D'Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJ, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Borevitz JO. The next generation of microarray research: applications in evolutionary and ecological genomics. Heredity. 2008;100:141–149. doi: 10.1038/sj.hdy.6800916. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Sequencing goes 454 and takes large-scale genomics into the wild. Mol Ecol. 2008;17:1629–1631. doi: 10.1111/j.1365-294X.2008.03699.x. [DOI] [PubMed] [Google Scholar]
- Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, Angiuoli S, Bussey H, Bowyer P, Cotty PJ, Dyer PS, Egan A, Galens K, Fraser-Liggett CM, Haas BJ, Inman JM, Kent R, Lemieux S, Malavazi I, Orvis J, Roemer T, Ronning CM, Sundaram JP, Sutton G, Turner G, Venter JC, White OR, Whitty BR, Youngman P, Wolfe KH, Goldman GH, Wortman JR, Jiang B, Denning DW, Nierman WC. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Berg MA van den, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martin JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, Klei IJ van der, van Peij NN, Veenhuis M, von Dohren H, Wagner C, Wortman J, Bovenberg RA. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Mol Biol. 2006;15:715–718. doi: 10.1111/j.1365-2583.2006.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- Thon MR, Pan H, Diener S, Papalas J, Taro A, Mitchell TK, Dean RA. The role of transposable element clusters in genome evolution and loss of synteny in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2006;7:R16. doi: 10.1186/gb-2006-7-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury JM, Segurens B, Poulain J, Anthouard V, Grossetete S, Khalili H, Coppin E, Dequard-Chablat M, Picard M, Contamine V, Arnaise S, Bourdais A, Berteaux-Lecellier V, Gautheret D, de Vries RP, Battaglia E, Coutinho PM, Danchin EG, Henrissat B, Khoury RE, Sainsard-Chanet A, Boivin A, Pinan-Lucarre B, Sellem CH, Debuchy R, Wincker P, Weissenbach J, Silar P. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barbote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Hane JK, Lowe RG, Solomon PS, Tan KC, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Galagan JE, Torriani SF, McDonald BA, Oliver RP. Dothideomycete Plant Interactions Illuminated by Genome Sequencing and EST Analysis of the Wheat Pathogen Stagonospora nodorum. Plant Cell. 2007;19:3347–3368. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, Cuomo C, Berriman M, Scherer S, Magee BB, Whiteway M, Chibana H, Nantel A, Magee PT. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Blandin G, Ozier-Kalogeropoulos O, Wincker P, Artiguenave F, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 16. Candida tropicalis. FEBS Lett. 2000;487:91–94. doi: 10.1016/S0014-5793(00)02287-0. [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, Wing RA, Flavier A, Gaffney TD, Philippsen P. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Lepingle A, Casaregola S, Neuveglise C, Bon E, Nguyen H, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 14. Debaryomyces hansenii var. hansenii. FEBS Lett. 2000;487:82–86. doi: 10.1016/S0014-5793(00)02285-7. [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M, Toffano-Nioche C, Artiguenave F, Duchateau-Nguyen G, Lemaire M, Marmeisse R, Montrocher R, Robert C, Termier M, Wincker P, Wesolowski-Louvel M. Genomic exploration of the hemiascomycetous yeasts: 11. Kluyveromyces lactis. FEBS Lett. 2000;487:66–70. doi: 10.1016/S0014-5793(00)02282-1. [DOI] [PubMed] [Google Scholar]
- Llorente B, Malpertuy A, Blandin G, Artiguenave F, Wincker P, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 12. Kluyveromyces marxianus var. marxianus. FEBS Lett. 2000;487:71–75. doi: 10.1016/S0014-5793(00)02283-3. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpertuy A, Llorente B, Blandin G, Artiguenave F, Wincker P, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 10. Kluyveromyces thermotolerans. FEBS Lett. 2000;487:61–65. doi: 10.1016/S0014-5793(00)02281-X. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Bon E, Neuveglise C, Casaregola S, Artiguenave F, Wincker P, Aigle M, Durrens P. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 2000;487:37–41. doi: 10.1016/S0014-5793(00)02276-6. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gu Z, David L, Petrov D, Jones T, Davis RW, Steinmetz LM. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon E, Neuveglise C, Lepingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. Genomic exploration of the hemiascomycetous yeasts: 6. Saccharomyces exiguus. FEBS Lett. 2000;487:42–46. doi: 10.1016/S0014-5793(00)02277-8. [DOI] [PubMed] [Google Scholar]
- Neuveglise C, Bon E, Lepingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett. 2000;487:56–60. doi: 10.1016/S0014-5793(00)02280-8. [DOI] [PubMed] [Google Scholar]
- Casaregola S, Lepingle A, Bon E, Neuveglise C, Nguyen H, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 7. Saccharomyces servazzii. FEBS Lett. 2000;487:47–51. doi: 10.1016/S0014-5793(00)02278-X. [DOI] [PubMed] [Google Scholar]
- Blandin G, Llorente B, Malpertuy A, Wincker P, Artiguenave F, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 13. Pichia angusta. FEBS Lett. 2000;487:76–81. doi: 10.1016/S0014-5793(00)02284-5. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H, Jin YS, Passoth V, Richardson PM. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- de Montigny J, Spehner C, Souciet J, Tekaia F, Dujon B, Wincker P, Artiguenave F, Potier S. Genomic exploration of the hemiascomycetous yeasts: 15. Pichia sorbitophila. FEBS Lett. 2000;487:87–90. doi: 10.1016/S0014-5793(00)02286-9. [DOI] [PubMed] [Google Scholar]
- Casaregola S, Neuveglise C, Lepingle A, Bon E, Feynerol C, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett. 2000;487:95–100. doi: 10.1016/S0014-5793(00)02288-2. [DOI] [PubMed] [Google Scholar]
- de Montigny J, Straub M, Potier S, Tekaia F, Dujon B, Wincker P, Artiguenave F, Souciet J. Genomic exploration of the hemiascomycetous yeasts: 8. Zygosaccharomyces rouxii. FEBS Lett. 2000;487:52–55. doi: 10.1016/S0014-5793(00)02279-1. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buee M, Brokstein P, Canback B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbe J, Lin YC, Legue V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kues U, Lucas S, Peer Y Van de, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouze P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HA, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho EC, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJ, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JK, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BW, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev IV, Rokhsar DS, Boore JL. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, Salzberg SL, Feng L, Jones MR, Skelton RL, Murray JE, Chen C, Qian W, Shen J, Du P, Eustice M, Tong E, Tang H, Lyons E, Paull RE, Michael TP, Wall K, Rice DW, Albert H, Wang ML, Zhu YJ, Schatz M, Nagarajan N, Acob RA, Guan P, Blas A, Wai CM, Ackerman CM, Ren Y, Liu C, Wang J, Wang J, Na JK, Shakirov EV, Haas B, Thimmapuram J, Nelson D, Wang X, Bowers JE, Gschwend AR, Delcher AL, Singh R, Suzuki JY, Tripathi S, Neupane K, Wei H, Irikura B, Paidi M, Jiang N, Zhang W, Presting G, Windsor A, Navajas-Perez R, Torres MJ, Feltus FA, Porter B, Li Y, Burroughs AM, Luo MC, Liu L, Christopher DA, Mount SM, Moore PH, Sugimura T, Jiang J, Schuler MA, Friedman V, Mitchell-Olds T, Shippen DE, dePamphilis CW, Palmer JD, Freeling M, Paterson AH, Gonsalves D, Wang L, Alam M. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S. Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol. 2005;137:1174–1181. doi: 10.1104/pp.104.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Crow JA, Heuer ML, Wang X, Cannon EK, Dwan C, Lamblin AF, Vasdewani J, Mudge J, Cook A, Gish J, Cheung F, Kenton S, Kunau TM, Brown D, May GD, Kim D, Cook DR, Roe BA, Town CD, Young ND, Retzel EF. Databases and information integration for the Medicago truncatula genome and transcriptome. Plant Physiol. 2005;138:38–46. doi: 10.1104/pp.104.059204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, Spannagl M, Monaghan E, Nicholson C, Humphray SJ, Schoof H, Mayer KF, Rogers J, Quetier F, Oldroyd GE, Debelle F, Cook DR, Retzel EF, Roe BA, Town CD, Tabata S, Peer Y Van de, Young ND. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA. 2006;103:14959–14964. doi: 10.1073/pnas.0603228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li S, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Wang J, Deng Y, Ran L, Shi X, Wang X, Wu Q, Li C, Ren X, Wang J, Wang X, Li D, Liu D, Zhang X, Ji Z, Zhao W, Sun Y, Zhang Z, Bao J, Han Y, Dong L, Ji J, Chen P, Wu S, Liu J, Xiao Y, Bu D, Tan J, Yang L, Ye C, Zhang J, Xu J, Zhou Y, Yu Y, Zhang B, Zhuang S, Wei H, Liu B, Lei M, Yu H, Li Y, Xu H, Wei S, He X, Fang L, Zhang Z, Zhang Y, Huang X, Su Z, Tong W, Li J, Tong Z, Li S, Ye J, Wang L, Fang L, Lei T, Chen C, Chen H, Xu Z, Li H, Huang H, Zhang F, Xu H, Li N, Zhao C, Li S, Dong L, Huang Y, Li L, Xi Y, Qi Q, Li W, Zhang B, Hu W, Zhang Y, Tian X, Jiao Y, Liang X, Jin J, Gao L, Zheng W, Hao B, Liu S, Wang W, Yuan L, Cao M, McDermott J, Samudrala R, Wang J, Wong GK, Yang H. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRGSP The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Peer Y Van de, Rokhsar D. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Consortium HGS. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Li G, Su J, Wang X, Li G, Zhang Z, Wu Q, Li J, Zhang Q, Wei N, Xu J, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan F, Huang X, Li Y, Fang L, Li C, Li D, Sun Y, Zhang Z, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Li J, Ye J, Chen H, Zhou Y, Liu B, Wang J, Ye J, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Li S, Wang J, Wong GK, Yang H. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Krasnow MA. The Drosophila genome sequence: implications for biology and medicine. Science. 2000;287:2218–2220. doi: 10.1126/science.287.5461.2218. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, Coulson A, D'Eustachio P, Fitch DH, Fulton LA, Fulton RE, Griffiths-Jones S, Harris TW, Hillier LW, Kamath R, Kuwabara PE, Mardis ER, Marra MA, Miner TL, Minx P, Mullikin JC, Plumb RW, Rogers J, Schein JE, Sohrmann M, Spieth J, Stajich JE, Wei C, Willey D, Wilson RK, Durbin R, Waterston RH. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]