Abstract
Post-translational sumoylation, the covalent attachment of a small ubiquitin-like modifier (SUMO), regulates the functions of proteins engaged in diverse processes. Often associated with nuclear and perinuclear proteins, such as transcription factors, it is not known whether SUMO can conjugate to cell surface receptors for growth factors to regulate their functions. We show that the type I TGF-β receptor, TβRI, is sumoylated in response to TGF-β and that its sumoylation requires the kinase activities of both TβRI and the type II TGF-β receptor, TβRII. Sumoylation of TβRI enhances receptor function by facilitating the recruitment and phosphorylation of Smad3, consequently regulating TGF-β-induced transcription and growth inhibition. TβRI sumoylation modulates dissemination of transformed cells in a mouse model of TβRI-stimulated metastasis. Hence, TβRI sumoylation controls TGF-β responsiveness, with implications for tumor progression. Sumoylation of cell surface receptors may regulate other growth factor responses.
INTRODUCTION
TGF-β signaling plays key roles in cell growth, differentiation, apoptosis, development and tumorigenesis. The mechanisms that lead to receptor activation and gene expression responses to TGF-β are generally understood1. Binding of TGF-β to a complex of two type I and two type II kinase receptors, i.e. TβRI and TβRII, confers TβRI activation and consequent direct C-terminal phosphorylation of Smad2 and Smad3 by TβRI. The activated Smads then associate with Smad4 and translocate into the nucleus to regulate transcription of target genes. TGF-β signaling is modulated by other signaling pathways and post-translational modifications. Indeed, the function of the Smad proteins is controlled by phosphorylation, acetylation, ubiquitylation and sumoylation2,3.
Less is known about the regulation of TGF-β receptors by post-translational modification. Since the receptor complex is a central point for protein interactions, post-translational modifications could play key roles in the transduction of TGF-β signals. So far, phosphorylation and ubiquitylation of the type I receptor have been shown to post-translationally modify the receptors3–7. Thus, recruitment of E3 ubiquitin ligases, including Smurfs, by the inhibitory Smad6 or Smad7 to the TβRII/TβRI complex can lead to TβRI ubiquitylation and consequent degradation. We now demonstrate that SUMO proteins, which primarily modify nuclear proteins and regulate their function, are conjugated to TβRI receptors in a regulated manner. TβRI sumoylation modulates the function of the TGF-β receptors and helps define the cellular responses to TGF-β.
RESULTS
The type I TGF-β receptor TβRI is sumoylated
To examine the sumoylation of TβRI or TβRII, we expressed Flag-tagged rat TβRI or TβRII with myc-tagged SUMO-1. Cell lysate immunoprecipitations using anti-Flag antibodies, followed by western blotting detected myc-tagged, sumoylated TGF-β receptors. As shown in Figure 1a, SUMO was conjugated to TβRI, but not TβRII, resulting in a >20 kd shift, similarly to other sumoylated proteins, indicating that TβRI is post-translationally sumoylated in vivo. TβRI sumoylation was increased when the E2 conjugating enzyme Ubc9 was co-expressed with SUMO-1, suggesting that Ubc9 is involved in sumoylation of TβRI (Fig. 1b). Under conditions of Ubc9 overexpression and proportionally insufficient E3 SUMO ligase expression, up to three sumoylated TβRI forms were observed. Since only one SUMO can be linked to a Lys, we assume that, under these conditions, the initial, site-specific sumoylation can confer additional TβRI sumoylation at other sites.
Figure 1.
The type I TGF-β receptor TβRI is sumoylated. (a) TβRI, but not TβRII, is sumoylated. Lysates of COS cells, expressing Flag-tagged TβRI or TβRII and myc-tagged SUMO-1, were subjected to immunoprecipitations using anti-Flag, followed by western blotting with anti-myc to detect sumoylated TGF-β receptors. (b) Increasing expression of Ubc9 enhances TβRI sumoylation. COS cells, expressing Flag-tagged TβRI, myc-tagged SUMO-1 and increasing levels of Ubc9, were lysed and subjected to immunoprecipitation, followed by western blotting with the indicated antibodies. (c) In vitro sumoylation of TβRI. Immunopurified Flag-tagged TβRI was incubated with or without recombinant SUMO-1, the E1 enzyme Aos1/Uba2, and the E2 conjugating enzyme Ubc9. The reaction mixture was analyzed by western blotting with anti-Flag. (d) TGF-β induces sumoylation of endogenous TβRI. Lysates of Mv1Lu or MDA-231 cells, treated with or without TGF-β, were immunoprecipitated with anti-TβRI, and immunoblotted with antibody against SUMO-1. (e) TβRI, but not other type I receptors, is sumoylated. 293T cells ectopically expressing the indicated type I receptor, myc-tagged SUMO-1 and Ubc9, were lysed, and sumoylation was analyzed by western blotting.
We next evaluated whether TβRI can be sumoylated in vitro. Immunopurified TβRI was incubated with SUMO-1, the E1 activating SUMO enzyme, Aos1/Uba2, and Ubc9 in the presence of ATP. Western blotting detected a band with a size that was compatible with the attachment of a SUMO-1 protein to TβRI, and corresponded to the sumoylated TβRI in vivo. When the E1 or E2 enzyme, or SUMO-1, was absent, this band was not detected (Fig. 1c). This result suggests that TβRI was sumoylated in vitro.
Immunoprecipitation of TβRI from Mv1Lu or MDA-231 cells, treated with or without TGF-β, and immunoblotting with antibodies against SUMO-1, revealed that endogenous TβRI was sumoylated and that TGF-β induced TβRI sumoylation (Fig. 1d). These data indicate that receptor activation by TGF-β may induce sumoylation of TβRI.
We assessed if other type I TGF-β family receptors could be sumoylated. Each type I receptor was expressed in the presence or absence of SUMO-1 and Ubc9, and sumoylation was analyzed by immunoblotting. Whereas TβRI was sumoylated, other type I receptors were not (Fig. 1e).
TβRI kinase activity and phosphorylation are required for sumoylation of the TβRI receptor
To further characterize whether activation of TβRI affects its sumoylation, as apparent by the TGF-β-induced TβRI sumoylation (Fig. 1d), we compared the in vitro sumoylation efficacy of immunopurified wild-type TβRI and activated TβRI (caTβRI) with a Thr202 to Asp mutation (Thr202 in rat TβRI corresponds to Thr204 in human TβRI) resulting in elevated kinase activity8. As shown in Fig. 2a, caTβRI was sumoylated much more efficiently than wild-type TβRI, suggesting that TβRI activation, which normally occurs by TβRII-mediated phosphorylation in response to TGF-β, facilitates sumoylation of the receptor.
Figure 2.
The kinase activities of TβRI and TβRII are required for TβRI sumoylation. (a) Activated TβRI is more sumoylated than wild-type TβRI. In vitro sumoylation of immunopurified Flag-tagged wild-type and activated (ca) TβRI in the presence or absence of recombinant SUMO-1, Aos1/Uba2 (E1), and Ubc9. The reaction mixture was analyzed by western blotting for TβRI. (b) Effects of the TβRI kinase inhibitor and TβRI dephosphorylation on TβRI sumoylation. In vitro sumoylation was performed as in (a) with wild-type or activated (ca) TβRI, as indicated, in the presence or absence of the TβRI kinase inhibitor SB431542. The phosphates were removed from TβRI using lambda phosphatase, prior to in vitro sumoylation. (c) The kinase activities of TβRII and TβRI are required for efficient TβRI sumoylation. 293T cells co-expressed a cytoplasmic receptor chimera TβRII-RI, in which the TβRI cytoplasmic domain follows the TβRII cytoplasmic domain, or chimeras in which the TβRII and/or TβRI kinase activities were inactivated by point mutation (KR), with myc-tagged SUMO-1 and Ubc9. The chimera sumoylation was analyzed by western blotting. (d) In vitro sumoylation of immunopurified cytoplasmic receptor chimeras or each of the kinase-defective receptor chimeras, used in panel (c). (e) Diagram showing TGF-β-induced sumoylation of TβRI in the receptor complex.
Since the activated TβRI has elevated kinase activity and increased phosphorylation8, we examined whether increased TβRI sumoylation resulted from increased TβRI kinase activity or phosphorylation. In vitro sumoylation of wild-type TβRI or caTβRI was decreased in the presence of SB431542, a specific TβRI kinase inhibitor, although this was more easily detected using caTβRI (Fig. 2b). These data suggested that the TβRI kinase regulates TβRI sumoylation. Since TβRI did not phosphorylate SUMO-1, the E1 enzyme (Aos/Uba2) and Ubc9 (Supplementary Fig. S1a), these data suggested that TβRI autophosphorylation plays a role in its sumoylation. To determine whether TβRI phosphorylation regulates TβRI sumoylation, we removed the Ser/Thr phosphorylation from TβRI using lambda phosphatase prior to in vitro sumoylation. The absence or reduction of TβRI phosphorylation decreased the sumoylation of wild-type or activated TβRI (Fig. 2b). This again was more easily detected with activated than with wild-type TβRI, due to the difference in sumoylation level. These results suggest that increased kinase activity together with increased phosphorylation contribute remarkably to the efficiency of TβRI sumoylation.
TGF-β binding to TβRII results in stable complex formation of two TβRII and two TβRI receptors, in which TβRII phosphorylates the TβRI cytoplasmic domain and thereby activates the TβRI kinase1. The activated receptor complex allows for autophosphorylation of the TβRII and TβRI dimers. To determine the roles of the TβRII and TβRI kinases in TβRI sumoylation, we used a cytoplasmic chimera that fuses the TβRI cytoplasmic domain to the TβRII cytoplasmic domain9. In this complex, the TβRII kinase activates the TβRI kinase without the need to add TGF-β. The receptor chimera, expressed in the presence of SUMO and Ubc9, was sumoylated. Since TβRII is not sumoylated (Fig. 1a), the sumoylation site is within the TβRI cytoplasmic domain. Inactivation of the TβRI kinase by Lys230 to Arg mutation decreased the chimera sumoylation (Fig. 2c), consistent with the decreased TβRI sumoylation in the presence of SB431542 (Fig. 2b). Similar inactivation of the TβRII kinase by Lys277 to Arg mutation also decreased the chimera sumoylation, when compared with the wild-type, kinase-active version (Fig. 2c). Since the TβRII cytoplasmic domain is not targeted for sumoylation, this result indicates that TβRI cytoplasmic domain phosphorylation by the TβRII kinase plays an important role in the sumoylation of TβRI. Mutation of both kinase ATP binding sites in the chimera blocked sumoylation.
The requirement for both receptor kinase activities for sumoylation was also studied in vitro. The efficacies of in vitro sumoylation of wild-type, TβRI kinase-defective, TβRII kinase-defective and TβRI/II kinase-defective chimeras, immunopurified from transfected cells, were compared (Fig. 2d). Inactivation of the kinase functions of TβRII or TβRI strongly decreased the sumoylation in vitro, while inactivation of both kinases abolished the chimera sumoylation (Fig. 2d).
These observations indicate that the kinase activities of both TβRI and TβRII, and consequent phosphorylation of TβRI, are required for efficient TGF-β-induced sumoylation of TβRI in the receptor complex. This is consistent with the TGF-β-induced phosphorylation and consequent activation of TβRI by TβRII (Fig. 2e).
The TβRI receptor is sumoylated on lysine 389
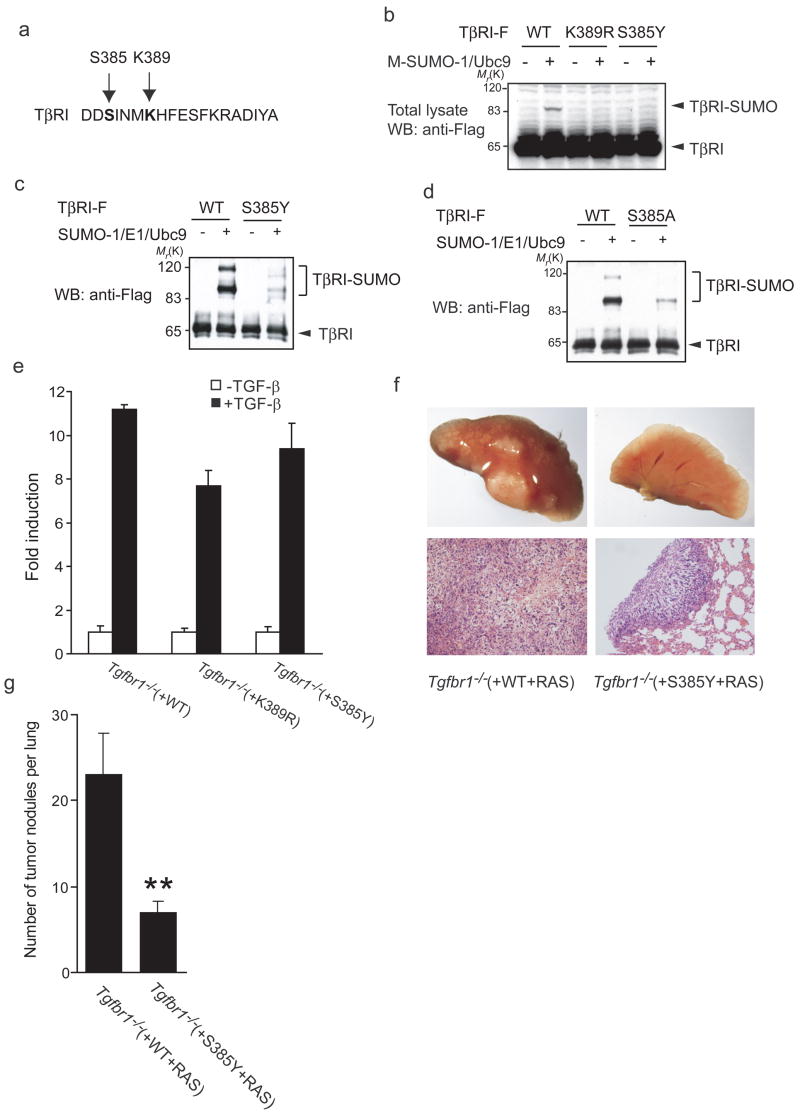
Sumoylation often occurs on lysine (K) within a consensus sequence ΨKx(D/E), in which Ψ represents a large hydrophobic residue10. Since this consensus sequence is absent in the TβRI amino acid sequence, each of the 18 lysines in the cytoplasmic domain was singly replaced by arginine, and the effect of each mutation on TβRI sumoylation was tested. Lysates of cells co-expressing each Flag-tagged mutant lysine TβRI with SUMO-1 and Ubc9 were subjected to immunoprecipitation using anti-Flag antibody, and western blotting. TβRI was not sumoylated when Lys389 residue was replaced by arginine, whereas arginine replacements of other lysines did not affect sumoylation of TβRI (Fig. 3a; data not shown), indicating that Lys389 is a major site for TβRI sumoylation. The Lys389 mutation also affected in vitro sumoylation, since no sumoylated TβRI was detected when Lys389 was replaced by arginine (Fig. 3b). These results indicate that Lys389 is the only residue targeted for sumoylation.
Figure 3.
The TβRI receptor is sumoylated on lysine 389. (a) Mutation of Lys389 abolishes TβRI sumoylation. 293T cells expressed Flag-tagged wild-type or mutant TβRI with the indicated lysine-to-arginine mutation, with myc-tagged SUMO-1 and Ubc9. Cell lysates were subjected to immunoprecipitations using anti-Flag, followed by western blotting for SUMO-1. (b) In vitro sumoylation of wild-type TβRI or K389R TβRI. Immunopurified TβRI was subjected to in vitro sumoylation followed by immunoblotting to detect sumoylation. (c) Proposed structure of the TβRI cytoplasmic domain. The Lys389, L45 loop, GS region and ATP binding site are indicated. N and C represent the N- and C-termini. (d) Sequence alignment of the TβRI sequence containing Lys389 and corresponding regions of other type I receptors.
The proposed structure of the TβRI cytoplasmic domain (Fig. 3c) predicts that Lys389 is located in the hinge between the αEF helix and the αF helix, and is exposed at the surface of the C lobe of the kinase domain. The sumoylation site faces the same orientation as the GS region, which is phosphorylated by TβRII upon TGF-β binding, and the L45 loop, which specifies the Smad interaction, albeit in a separate protein domain. The GS region and L45 loop, both located in the N lobe, interact with the Smad for phosphorylation by TβRI. The exposure of Lys389 at the protein surface predicts that SUMO conjugation strongly affects the cytosolic surface of TβRI, and may regulate the Smad binding to the L45 loop and GS domain of TβRI, and interactions of additional proteins with the receptor complex.
Sequence comparisons (Fig. 3d) show that Lys389 is not conserved in other TGF-β family type I receptors, with the exception of the activin receptor ActRIB/ALK-4. This is consistent with the absence of sumoylation of these type I receptors in vivo (Fig. 1e). The lack of ActRIB/ALK-4 sumoylation suggests that other determinants besides the target lysine are needed for sumoylation of TβRI. It is unlikely that this is due to the Ser versus Thr difference, four residues preceding the sumoylated Lys in TβRI compared to ActrIB/ALK-4 (Fig. 3d), since S385T replacement did not affect the in vitro sumoylation of TβRI (Supplementary Fig. S1b).
TβRI sumoylation regulates Smad interaction and activation
To evaluate whether sumoylation of the exposed Lys389 affects Smad activation, we examined the interaction of Smad3 with caTβRI. Since this interaction is hard to detect by immunoprecipitation, likely due to its low affinity and transient nature, we examined the interaction of the Smad3D407E mutant with caTβRI. The D407E mutation in the MH2 domain was identified in Smad2 in colorectal carcinoma, and affects the Smad interaction with TβRI and heteromerization with Smad411. Increased caTβRI sumoylation by co-expressing Ubc9 and SUMO-1 enhanced the TβRI interaction with Smad3D407E. In contrast, coexpression of SUMO and Ubc9 did not enhance the interaction of Smad3D407E with caTβRI carrying the sumoylation-resistant K389R mutation (Fig. 4a). We also incubated immobilized GST-Smad3D407E with a mixture of sumoylated and unsumoylated TβRI. Western blotting of purified GST-Smad3 - TβRI complexes showed preferential binding of Smad3 to sumoylated TβRI, compared with unsumoylated TβRI, even though the latter was in large excess (Fig. 4b). This result, and the data in Fig. 4a, indicates that sumoylation of TβRI enhances Smad3 recruitment and suggests that TβRI sumoylation enhances Smad activation.
Figure 4.
TβRI sumoylation regulates Smad activation and TGF-β responses. (a) Interaction of Smad3 with TβRI. 293T cells were transfected to co-express activated (ca) TβRI or its K389R mutant, with myc-SUMO-1 and Ubc9, and/or Smad3D407E. The lyates were subjected to immunoprecipitation, and analyzed by western blotting. (b) In vitro interaction of Smad3 with TβRI. Immobilized GST or GST-Smad3(D407E) were incubated with in vitro sumoylated and non-sumoylated Flag-tagged TβRI. Adsorbed proteins were subjected to western blotting for TβRI. The lower panel shows Coomassie blue staining of GST and GST-Smad3(D407E) used for the adsorption. (c) Tgfbr1−/− MEFs stably expressing wild-type or K389R TβRI, or transfected with an empty vector, were subjected to western blotting to assess the expression of TβRI. (d) Biotin-labeled cell surface proteins from the indicated MEFs were subjected to avidin precipitation. Precipitates were analyzed by western blotting to assess the cell surface expression levels of TβRI. (e) Wild-type and K389R TβRI have similar kinase activities. Wild-type or K389R TβRI were expressed in 293T cells, immunopurified and subjected to kinase reactions in the presence or absence of TβRI kinase inhibitor. (f, g) Lack of TβRI sumoylation confers a lower level of Smad3 (f) or Smad2 (g) activation. Tgfbr1−/− MEFs stably expressing wild-type or K389R TβRI were treated without or with TGF-β for the indicated time. The cell lysates were analyzed by western blotting. (h) Lack of TβRI sumoylation confers a lower level of Smad3-mediated transcription. Tgfbr1−/− fibroblasts stably expressing wild-type TβRI or K389R mutant TβRI were transfected with the Smad3-responsive (CAGA)12-luciferase reporter. Luciferase activities without or in response to added TGF-β were measured. The error bars represent mean ± s.d. (n = 2) (i) Lack of TβRI sumoylation decreases TGF-β-induced endogenous gene expression. Tgfbr1−/− fibroblasts stably expressing wild-type TβRI or K389R mutant TβRI were treated with or without added TGF-β. Smad7 mRNA was quantified using real-time PCR and normalized to RPL19 mRNA expression, which is not affected by TGF-β. The error bars represent mean ± s.d. (n = 3) (j) Lack of TβRI sumoylation confers a lower level of TGF-β-induced growth inhibition. Tgfbr1−/− fibroblasts stably expressing wild-type or K389R TβRI, or with an empty vector, were cultured without or with the indicated dose of added TGF-β for 3 days. The cell numbers were then counted. The error bars represent mean ± s.d. (n = 3). Full scans of a and f are shown in Supplementary Information, Fig. S4.
Since upon TGF-β binding, TβRI phosphorylates Smad2 and Smad3, we investigated whether sumoylation of activated TβRI affects Smad3 phosphorylation. TβRI-defective mouse embryonic fibroblasts (MEFs) derived from Tgfbr1−/− mice12 were retrovirally infected to express wild-type TβRI or sumoylation-resistant K389R TβRI. Stably selected cell populations, expressing either TβRI form at equal levels (Fig. 4c), showed equivalent cell surface levels of wild-type or mutant TβRI (Fig. 4d), suggesting that the K389R mutation did not affect cell surface transport or stability of TβRI. K389R TβRI also showed a similar phosphorylation level as wild-type TβRI, resulting primarily from the TβRI kinase activity, since SB431542 abolished this phosphorylation (Fig. 4e). Fractionation of cell lysates did not reveal differences in subcellular compartmentalization of wild-type versus mutant TβRI (Supplementary Fig. S1c). We then compared wild-type and K389R TβRI for their ability to phosphorylate Smad3 in response to TGF-β. In fibroblasts expressing wild-type TβRI, TGF-β induced Smad3 phosphorylation within 15 min, whereas, in cells expressing K389R TβRI, the Smad3 phosphorylation kinetics in response to TGF-β was slower, with first detection at 30 min. Furthermore, the overall level of Smad3 activation was lower in cells expressing K389R TβRI, compared to cells expressing wild-type TβRI (Fig. 4f). Similar results were seen with TGF-β-induced activation of Smad2 (Fig. 4g). Replacement of Lys393 in ActRIB/ALK-4, which is not sumoylated and corresponds to Lys389 in TβRI, with Arg did not affect Smad3 activation (Supplementary Fig. S1d).
The differences in level and kinetics of Smad2 and Smad3 phosphorylation by wild-type versus K389R TβRI, together with the results of the Smad3 interaction with TβRI, suggest that TβRI sumoylation enhances the Smad interaction with TβRI, allowing more efficient Smad2/3 phosphorylation and activation in response to TGF-β.
TβRI sumoylation regulates functional responses to TGF-β
Using Tgfbr1−/− fibroblasts ectopically expressing wild-type or K389R TβRI, we characterized the effect of TβRI sumoylation on Smad-mediated transcription, i.e. the functional consequence of Smad activation. We used a reporter in which tandem Smad binding sites control luciferase transcription. Cells expressing K389R TβRI showed reduced transcription from the Smad3-responsive promoter compared to cells expressing wild-type TβRI (Fig. 4h). We also compared the endogenous expression of the TGF-β-responsive Smad7 gene by RT-PCR. Cells expressing K389R TβRI showed reduced Smad7 mRNA expression in response to TGF-β compared to cells expressing wild-type TβRI (Fig. 4i). Similar results were obtained with two additional populations of Tgfbr1−/− fibroblasts ectopically expressing TβRI or K389R TβRI at similar levels (Supplementary Fig. S2). These results suggest that TβRI sumoylation defines the TGF-β-induced transcriptional regulation.
We also examined the contribution of TβRI sumoylation to the antiproliferative response to TGF-β. We seeded the fibroblasts expressing wild-type or K389R TβRI in parallel with the parental Tgfbr1−/− cells as control cells, and determined the proliferative response after adding TGF-β. Cells lacking TβRI were not affected in their proliferation by TGF-β, whereas those expressing wild-type TβRI responded with decreased proliferation, as assessed by cell number (Fig. 4j). In contrast to wild-type TβRI, cells expressing K389R TβRI showed a decreased growth inhibitory response to TGF-β. This result suggests that sumoylation regulates the TβRI-mediated antiproliferative response to TGF-β and renders the cells more responsive to TGF-β.
TβRI sumoylation enhances invasion and metastasis of Ras-transformed cells
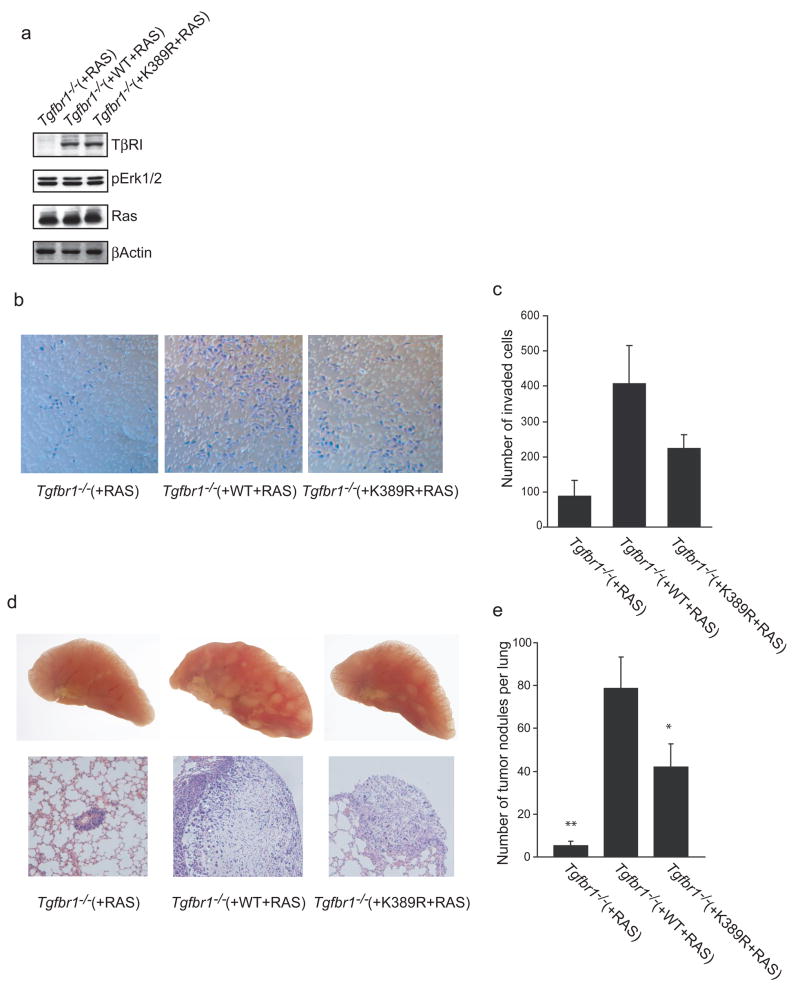
Since autocrine TGF-β signaling regulates cancer progression13,14 we postulated that resistance to sumoylation, while suppressing TGF-β growth inhibitory activities, affects tumor progression. To address this issue, the Tgfbr1−/− fibroblasts, carrying a control empty vector or ectopically expressing wild-type or K389R TβRI at similar levels, were transduced with a control vector or a vector expressing activated Ha-Ras (Leu-61) to generate tumorigenic cell populations. Mutant Ras was expressed and activated Erk MAP kinase to similar extents in all three Ras-transformed cell populations (Fig. 5a). Cells expressing activated Ras had a transformed phenotype, apparent from the altered cell morphology and loss of contact inhibition (data not shown).
Figure 5.
Lack of TβRI sumoylation decreases TGF-β-regulated invasion and metastasis. (a) Ras-transformed Tgfbr1−/− fibroblasts stably expressing wild-type or K389R TβRI, or with an empty vector, were subjected to western blotting for TβRI, Ras or phospho-ERK1/2 as marker of Ras activation. (b, c) TβRI-mediated TGF-β responsiveness of Ras-transformed cells promotes invasion, which is decreased by lack of TβRI sumoylation. Cells were seeded onto a Matrigel-coated Transwell filter and incubated for 24 h to allow invasion toward 10% serum. Cells that migrated through the filter were stained with crystal violet. The white stipples represent the pores in the filter. A representative picture and quantification of invaded cells are shown in panels b and c, respectively. Error bars represent mean ± s.d. (n = 4) (d, e) Ras-transformed Tgfbr1−/− fibroblasts expressing wild-type or K389R TβRI, or with an empty vector, were injected into the tail vein of nude mice. The lung tumor nodules were counted after three weeks. (d) Representative pictures of lungs from mice with Ras-transformed MEFs are shown in upper panels, and corresponding H&E-stained sections of tumor nodules at the same magnification (×10 objective) are shown in the lower panels. (e) Quantification of tumor nodules in the lungs. The error bars represent mean ± s.e.m. (n = 6). The single and double asterisks indicate P < 0.05 and P < 0.01, respectively, compared to wild-type TβRI.
We examined the invasion of the Ras-transformed cells using a modified Boyden chamber assay whereby cells migrate through Matrigel toward serum. Ras-transformed Tgfbr1−/− cells ectopically expressing wild-type TβRI showed a higher invasion activity compared to Ras-transformed Tgfbr1−/− cells lacking TβRI (Fig. 5b), indicating that the invasive capacity of Ras-transformed MEFs depends on TβRI signaling. Ras-transformed Tgfbr1−/− MEFs expressing K389R TβRI were less invasive than cells expressing wild-type TβRI, indicating that, in this system, lack of TβRI sumoylation impairs the TβRI-dependent invasion of transformed cells (Fig. 5b, c). These observations are consistent with the role of TβRI sumoylation in TGF-β-induced gene expression and growth inhibition (Fig. 4h–j).
Metastasis is a complex process, requiring cell growth, migration, invasion, intra- and extravasation, and cell survival in the circulatory system and at the metastatic site. Using a mouse tail vein injection model, autocrine TGF-β signaling was shown to enhance the ability of tumor cells to establish metastatic nodules within the lung15,16. To determine the roles of TβRI and TβRI sumoylation in the formation of metastatic nodules in this model, we compared the ability of the Tgfbr1−/− MEF derivatives to colonize the lung. Colonization of the lungs by MEFs was fully dependent on expression of activated Ha-Ras (Figs. 5d, e; data not shown). Ras-transformed Tgfbr1−/− cells gave rise to only few very small lung tumor nodules (Figs. 5d, e). Tumor cells were proliferative with a high incidence of apoptosis (Supplementary Fig. S3h). The tumors were morphologically heterogeneous, and large cells with massive nuclei were indicative of chromosomal instability (Supplementary Fig. S3e). Remarkably, Ras-transformed Tgfbr1−/−cells ectopically expressing wild-type or K389R TβRI developed numerous large metastatic nodules (Figs. 5d, e). MEFs expressing sumoylation-defective TβRI gave rise to fewer tumor nodules than cells expressing the wild-type receptor (Fig. 5e). These nodules were generally smaller than those from wild-type TβRI expressing MEFs (data not shown; Fig. 5d), although their histological appearance (Fig. 5d) and proliferative rates (Supplementary Fig. S3a) were similar. These results suggest that, in this model of TGF-β-mediated metastasis, TβRI sumoylation contributes to tumor progression by enhancing tumor cell extravasation, survival and/or growth at the metastatic site.
The Ser385Tyr TβRI mutation, implicated in metastatic cancer, confers sumoylation resistance
Mutations in TGF-β signaling mediators, including TGFBR1, have been associated with human cancers13. Among these, a missense mutation, S387Y, in TGFBR1 was enriched in breast and head-and-neck cancer metastases, compared to corresponding primary tumors17,18. This mutation confers diminished TGF-β signaling, and is the only mutation in TGFBR1 or TGFBR2, known to specifically associate with tumor metastases17. Since the corresponding residue in rat TβRI, i.e. Ser385, localizes close to the Lys389 sumoylation site of TβRI (Fig. 6a), we examined whether rat S385Y TβRI was sumoylated. In contrast to wild-type TβRI, this mutant was not sumoylated in cells overexpressing SUMO-1 and Ubc9 (Fig. 6b). In vitro, replacement of Ser385 with Tyr (Fig. 6c) or Ala (Fig. 6d) also strongly decreased sumoylation. In contrast, replacement of Ser385 with Thr, which may, similarly to Ser, be targeted for phosphorylation by Ser/Thr kinases, did not affect sumoylation (Supplementary Fig. S1b). These results indicate that this single amino acid substitution prevents TβRI sumoylation.
Figure 6.
The Ser385Tyr mutation impairs TβRI sumoylation and function. (a, b) Ser385Tyr TβRI is not sumoylated. Panel (a) shows the rat TβRI sequence with Lys389 as sumoylation site four amino acids away from Ser385, which is equivalent to Ser387 in human TβRI. (b) 293T cells co-expressing wild-type, K389R or S385Y TβRI, with Myc-tagged SUMO-1 and Ubc9, were lysed and analyzed by western blotting to detect sumoylation. (c, d) In vitro sumoylation of S385Y mutant TβRI (c) or S385A TβRI (d) in comparison with wild-type TβRI. Immunopurified TβRI was subjected to in vitro sumoylation followed by immunoblotting to detect sumoylation. (e) Tgfbr1−/− fibroblasts stably expressing wild-type, K389R or S385Y TβRI were transfected with the Smad3-responsive (CAGA)12-luciferase reporter. Luciferase activities without or in response to added TGF-β were measured. Error bars represent mean ± s.d. (n = 3). (f) Representative pictures of lungs from mice with Ras-transformed MEFs are shown in upper panels, and corresponding H&E-stained sections of tumor nodules at the same magnification (×10 objective) are shown in lower panels. (g) Quantification of tumor nodules in the lungs. Error bars represent mean ± s.e.m. (n = 6). Double asterisk indicates P < 0.01 compared to wild-type TβRI.
We examined whether the S385Y mutation decreased the TGF-β responsiveness, as observed with the sumoylation-defective K389R TβRI (Fig. 4). Using the Smad3-responsive reporter of Fig. 4h, cells expressing S385Y TβRI responded to TGF-β with a lower level transcription than cells expressing wild-type TβRI, but a higher level than cells expressing K389R TβRI (Fig. 6e), indicating that the decreased TGF-β responsiveness associated with S385Y TβRI correlates with impaired TβRI sumoylation. We also evaluated the S385Y TβRI mutation in the lung colonization tumor model. Ras-transformed MEFs expressing S385Y TβRI formed fewer and smaller metastatic nodules than cells expressing wild-type TβRI (Fig. 6f, g). While this difference in tumor nodule formation compares qualitatively with the K389R TβRI cells, the efficiency of metastatic nodule formation was more impaired in S385Y TβRI cells than in K389R TβRI cells. This quantitative difference between cells expressing K389R TβRI or S385Y TβRI, when compared with their differential activities in transcription assays, raises the possibility that the impaired TβRI sumoylation resulting from the S385Y mutation may be complemented with an additional defect of relevance to cancer progression.
DISCUSSION
We provide the first evidence that a cell surface polypeptide growth factor receptor is modified by sumoylation in a regulated, ligand-dependent manner. Conjugation of SUMO to TβRI depends upon TβRI activation by phosphorylation, which in turn is induced by binding TGF-β to the TβRII/TβRI complex. TGF-β receptor sumoylation enhances the recruitment and activation of Smad3, and consequently regulates TGF-β responses.
Little is known about sumoylation of transmembrane proteins and how this impacts their function. The ion channel proteins, K2P1 and Kv1.5, are sumoylated, and this modification regulates their inactivation19,20. Sumoylation of the kainate receptor subunit GluR6 is increased in response to kainate, and enhances endocytosis, resulting in a negative effect on synaptic transmission21. This raises the possibility that TβRI sumoylation affects receptor internalization, recycling and/or stability.
Interplay between different modifications provides a mechanism to regulate protein function. For example, phosphorylation, acetylation, methylation and ubiquitylation of histone H3 and H4 N-terminal sequences affect each other to control gene expression22. Stress-induced N-terminal phosphorylation of p53 regulates its C-terminal acetylation23. In addition, Smad7 acetylation inhibits its ubiquitylation that leads to degradation24. Here we show interplay between phosphorylation and sumoylation in regulating TβRI function. Thus, TGF-β-induced phosphorylation of TβRI by itself and TβRII is required for TβRI sumoylation to regulate its function. Phosphorylation-dependent sumoylation has been observed in transcription regulators, such as heat-shock factors, GATA-1 and MEF-2, and was shown to involve a ΨKxExxSP motif25. Within this motif, serine phosphorylation was suggested to contribute to sumoylation of lysine. Although TβRI lacks this motif, the phosphorylation-dependence of TβRI sumoylation is consistent with the requirements of the kinase activities of the receptor complex, and consequent TβRI phosphorylation, for TβRI sumoylation. How TβRI phosphorylation augments sumoylation needs to be determined.
TβRI sumoylation provides a new mechanism for functional regulation of TGF-β responses. By regulating Smad activation and downstream transcription responses, TβRI sumoylation may elaborate the TGF-β responses that drive cancer progression. Indeed, sumoylation is thought to play an important role in tumorigenesis since some oncogene proteins and tumor suppressors26–29 are sumoylated. Also, sumoylation of the reptin chromatin complex regulates the KAI1 metastasis suppressor gene and the invasive activity of cancer cells30. We here provide independent data to link sumoylation with tumor progression. Increased TGF-β1 expression by tumor cells and their microenvironment, and increased TGF-β signaling within tumor cells, are thought to be important factors in cancer progression16,18. In a model of TGF-β-dependent metastasis of cancer cells to the lung, TβRI sumoylation enhanced tumorigenesis. The proposed association of a sumoylation-resistant TβRI mutant with breast cancer metastasis14 seems at odds with our in vivo results using Ras-transformed Tgfbr1−/− fibroblasts. Differences in cellular origin of the tumors (highly malignant epithelial cells versus diploid embryonic fibroblasts31) and the use of an immune incompetent mouse model that addresses only a few components of the metastatic process may explain this discrepancy. Compared to MEFs, human metastatic tumors bear multiple mutations32, and have elevated levels of sumoylating and diminished levels of desumoylating enzymes27. Thus there may be a metastatic advantage to mutating the TβRI sumoylation site to prevent receptor over-activation. Retention of functional TGF-β receptors is advantageous to metastatic tumor spread15, thus human tumors with homozygous deletion of TGFBR1 or TGFBR2 are rare13,14 and present better prognosis for patient survival13,14. Mutation of the sumoylation site might more subtly alter TβRI activity, modulating a mechanism that contributes to metastasis, whilst retaining enough TβRI activity for tumor cell intra- and extravasation. Overall though, along with a previous report27, our data suggest a connection between sumoylation and cancer progression.
METHODS
Plasmids
The expression plasmids for N-terminally Flag- or Myc-tagged SUMO-1, Ubc9 or C-terminal Flag-tagged TβRI have been described33,34. The expression plasmid for the Flag-tagged TβRII-TβRI cytoplasmic chimera has also been described9. Site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA) generated plasmids encoding C-terminal Flag-tagged TβRI or TβRII-TβRI cytoplasmic chimera with amino acid substitutions, and mutations were confirmed by DNA sequencing. To generate retroviral vectors expressing Flag-tagged TβRI, the coding region was inserted into the HpaI site of LNCX35. The retroviral vector pBABE-H-Ras(Leu-61)-IRES-Puro36 was provided by R. Davis (University of Massachusetts Medical School, Worcester, MA). The expression plamid for Myc-tagged Smad3(D407E)11 was provided by K. Miyazono (University of Tokyo, Tokyo, Japan). Expression plasmids for HA-tagged ALK-1, ALK-3 or ALK-6 were generated by subcloning the coding regions from pcDNA3-HA-ALK1, -ALK3, and -ALK637,38, provided by P. ten Dijke (University of Leiden, The Netherlands), into the EcoRI/XhoI site of pRK5. A plasmid encoding Flag-tagged ALK-2 was made by inserting the coding region from pRK5-myc-ALK2 into the EcoRI/SalI site of pXFIF39. To generate a plasmid encoding Flag-tagged ALK-4, the coding region from pcDNA1-hALK437, provided by C.H. Heldin (Ludwig Institute for Cancer Research, Uppsala, Sweden) was PCR amplified and inserted into the EcoRI/SalI site of pRK540. The reporter plasmid (CAGA)12-luciferase41 was also provided by P. ten Dijke.
Cell Culture and transfections
Tgfbr1−/− MEFs12 were provided by S. Karlsson (Lund University Hospital, Lund, Sweden), immortalized and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). Cells were plated at 2 × 105 cells/well in 6-well plates and transfected with reporter and β-galactosidase plasmids using Lipofectamine Plus (Invitrogen, Carlsbad, CA). One day after transfection, cells were transferred to medium containing 0.2% FBS with or without TGF-β (1–5 ng/ml) for 16 h. Cell extracts were prepared and assayed for luciferase activity as described42. Luciferase activities were normalized to β-galactosidase activity from a cotransfected β-galactosidase plasmid.
Immunoprecipitations and immunoblotting
COS or 293T cells were harvested 48 h after transfection and lysed by brief sonication in lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% TritonX-100, 10% glycerol and protease inhibitor cocktail). Lysates were subjected to immunoprecipitation with anti-Flag M2 agarose (Sigma-Aldrich, St Louis, MO). Immune complexes were washed three times with lysis buffer and subjected to western blotting using anti-Flag or anti-Myc antibodies. To detect sumoylation of endogenous TβRI, Mv1Lu or MDA-231 cells were cultured for 3 h in DMEM with 0.2% FBS with or without TGF-β (10 ng/ml). Cells were washed twice with PBS, harvested and lysed by sonication in lysis buffer containing 10 mM N-ethylmaleimide. The lysates were precleared with mouse IgG and incubated with rabbit anti-TβRI antibody (Santa Cruz Biotech, Santa Cruz, CA) or IgG. Immune complexes were precipitated with protein G beads, and immunoblotted using mouse anti-SUMO-1 (Cell Signaling Technologies, Danvers, MA) or rabbit anti-TβRI antibodies (Santa Cruz Biotech).
In vitro sumoylation
293T cells were transfected with plasmids expressing Flag-tagged proteins. Lysates were subjected to immunoprecipitation with anti-Flag M2 agarose (Sigma-Aldrich), and immune complexes were eluted with Flag peptide (Sigma-Aldrich). The immunopurified proteins were incubated with 2 μg recombinant SUMO-1 (BIOMOL International, Plymouth Meeting, PA), 0.5 μg Aos1/Uba2 (BIOMOL International), and 0.1 μg Ubc9 (BIOMOL International) for 2 h in 20 μl 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 2 mM ATP. The reaction mixture was analyzed by western blotting with anti-Flag antibody (Sigma-Aldrich). The TβRI kinase was inhibited by adding 5 μM SB431542 (Sigma-Aldrich). TβRI was dephosphorylated by treatment with lambda protein phosphatase (New England Biolabs, Ipswich, MA) for 30 min.
GST adsorption assays
Immunopurified Flag-tagged TβRI was subjected to in vitro sumoylation. The mixture of sumoylated and unsumoylated TβRI was incubated with immobilized GST-Smad3(D407E) in 50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 0.1% NP40, 10% glycerol and protease inhibitor cocktail. After pull-down, precipitates were subjected to SDS-PAGE, followed by western blotting with anti-Flag antibody (Sigma-Aldrich) to detect TβRI.
Generation of stable cell lines
Tgfbr1−/− cells were infected with the LNCX-based retroviral vector expressing Flag-tagged wild-type or K389R TβRI, and stably infected cell populations were generated, as described43. The expression levels of TβRI were assessed by western blotting with anti-Flag antibody (Sigma-Aldrich).
To generate Ras-transformed cells, Tgfbr1−/− cell populations with an empty vector or expressing Flag-tagged wild-type or K389R TβRI were transduced with the retroviral vector pBABE-H-Ras(Leu-61)-IRES-Puro or control vector pBABE-Puro, and selected with 2 μg/ml puromycin. The expression levels of H-Ras(Leu-61) were examined by western blotting with anti-c-H-Ras antibody (Calbiochem, San Diego, CA), and phosphorylated Erk1/2 was detected by western blotting with anti-phospho-Erk1/2 antibody (Cell Signaling Technologies).
In vitro kinase assays
Immunopurified receptors were incubated at 30°C for 30 min in 10 mM HEPES-KOH, pH 7.5, 5 mM MgCl2 and 5 mM CaCl2 with or without 5 μM TβRI kinase inhibitor SB431542 (Sigma-Aldrich). The reaction mixture was subjected to SDS-PAGE, followed by autoradiography.
Cell surface TGF-β receptor biotinylation and precipitaiton
Tgfbr1−/− cells expressing wild-type or K389R TβRI, or transfected with empty vector were grown to confluence, and labeled with sulfo-NHS biotin (Pierce, Rockford, IL) at 4°C for 2 h. Cells were washed with 100 mM glyine and lysed by brief sonication in 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% TritonX-100, 10% glycerol and protease inhibitor cocktail. Biotinylated cell surface proteins were precipitated with avidin-immobilized beads (Pierce) and subjected to western blotting with anti-Flag antibody (Sigma-Aldrich) to detect cell surface TβRI.
TGF-β response analyses
To evaluate Smad3 phosphorylation, Tgfbr1−/− cells expressing wild-type or K389R TβRI were treated without or with 2.5 ng/ml TGF-β for 10, 30, 60 or 120 min, and cell lysates were analyzed by western blotting with anti-phospho-Smad3 (Cell signaling Technologies) or anti-Smad3 antibody (Invitrogen).
To perform cell growth inhibition assay, 5 × 104 Tgfbr1−/− cells expressing wild-type or K389R TβRI or having an empty vector were grown in DMEM with 10% FBS without or with TGF-β for 3 days, and the cell numbers were counted by hemocytometer.
To quantify Smad7 mRNA expression, Tgfbr1−/− cells expressing Flag-tagged wild-type or K389R TβRI were treated with or without TGF-β (2.5 ng/ml) for 4 h. RNA was isolated using RNeasy kit (Qiagen, Valencia, CA) and used as a template for reverse trancriptase. The Smad7 mRNA was quantified by real-time PCR using cyber-green (Invitrogen), and normalized against RPL19 mRNA. The primer sequences were: Smad7, 5′-TCTGGACAGTCTGCAGTTGG-3′ (forward) and 5′-TCCTGCTGTGCAAAGTGTTC-3′ (reverse); RPL19, 5′-GGAAGAGGAAGGGTACTGCC-3′ (forward) and 5′-GGATTCCCGGTATCTCCTGAG-3′ (reverse).
In vitro invasion assay
In vitro invasion assays were performed using Biocoat Matrigel Invasion chambers (BD Biosciences, San Jose, CA). 2.5 × 104 cells were seeded into the upper insert of chamber and incubated for 24 h, allowing invasion through Matrigel toward 10% serum. The invaded cells were fixed with 96 % ethanol and stained with 0.05% crystal violet.
Tumor formation
To perform lung tumor formation assays, 5 × 105 cells were injected into the tail vein of 8 weeks-old nude mice18. Three weeks post injection, the mice were labeled by intra-peritoneal injection of 100 mg/kg 5-bromo-2-deoxyuridine (BrdU). One hour post BrdU injection, the mice were sacrificed, lung tumor nodules were counted, and lungs were fixed in 4% PFA and processed for histological analysis of paraffin-embedded tissue sections.
Immunostaining for BrdU was performed using biotin-conjugated mouse anti-BrdU antibody (Alexis Biochemicals, San Diego, CA), detected using the Vectastain Elite ABC kit detection system (Vector Laboratories, Inc. Burlingame, CA).
Supplementary Material
Acknowledgments
This research was supported by grants RO1-CA63101 and R21-CA125190 to R.D. and PO1 AR050440 and RO1s CA116019 and HL078564 to R.J.A. from the National Institutes of Health, and a Scientist Development grant 0630322N to J.S.K from the American Heart Association.
References
- 1.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–93. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 2.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 3.Izzi L, Attisano L. Ubiquitin-dependent regulation of TGFβ signaling in cancer. Neoplasia. 2006;8:677–88. doi: 10.1593/neo.06472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol Cell. 2000;6:1365–75. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 5.Ebisawa T, et al. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–80. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 6.Kuratomi G, et al. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem J. 2005;386:461–70. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komuro A, et al. Negative regulation of transforming growth factor-β (TGF-β) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23:6914–23. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 8.Wieser R, Wrana JL, Massague J. GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng XH, Derynck R. Ligand-independent activation of transforming growth factor (TGF) β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J Biol Chem. 1996;271:13123–9. doi: 10.1074/jbc.271.22.13123. [DOI] [PubMed] [Google Scholar]
- 10.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 11.Goto D, et al. A single missense mutant of Smad3 inhibits activation of both Smad2 and Smad3, and has a dominant negative effect on TGF-β signals. FEBS Lett. 1998;430:201–4. doi: 10.1016/s0014-5793(98)00658-9. [DOI] [PubMed] [Google Scholar]
- 12.Larsson J, et al. Abnormal angiogenesis but intact hematopoietic potential in TGF-β type I receptor-deficient mice. EMBO J. 2001;20:1663–73. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy L, Hill CS. Alterations in components of the TGF-β superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 15.Oft M, Heider KH, Beug H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–52. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 16.Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4:487–94. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 17.Chen T, Carter D, Garrigue-Antar L, Reiss M. Transforming growth factor β type I receptor kinase mutant associated with metastatic breast cancer. Cancer Res. 1998;58:4805–10. [PubMed] [Google Scholar]
- 18.Chen T, et al. Novel inactivating mutations of transforming growth factor-β type I receptor gene in head-and-neck cancer metastases. Int J Cancer. 2001;93:653–61. doi: 10.1002/ijc.1381. [DOI] [PubMed] [Google Scholar]
- 19.Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 2005;121:37–47. doi: 10.1016/j.cell.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Benson MD, et al. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc Natl Acad Sci U S A. 2007;104:1805–10. doi: 10.1073/pnas.0606702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–5. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 24.Grönroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–93. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 25.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. Embo J. 1999;18:6455–61. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitylation and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–62. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A. 2002;99:2872–7. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bies J, Markus J, Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- 30.Kim JH, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8:631–9. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 31.Dore JJ, Jr, et al. Intracellular sorting differ between fibroblasts and epithelial cells. Mol Biol Cell. 2001;12:675–84. doi: 10.1091/mbc.12.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-β family signaling. J Biol Chem. 2003;278:27853–63. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 34.Feng XH, Filvaroff EH, Derynck R. Transforming growth factor-β (TGF-β)-induced down-regulation of cyclin A expression requires a functional TGF-β receptor complex. Characterization of chimeric and truncated type I and type II receptors. J Biol Chem. 1995;270:24237–45. doi: 10.1074/jbc.270.41.24237. [DOI] [PubMed] [Google Scholar]
- 35.Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–2. 984–6, 989–90. [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–11. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ten Dijke P, et al. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8:2879–87. [PubMed] [Google Scholar]
- 38.ten Dijke P, et al. Characterization of type I receptors for transforming growth factor-β and activin. Science. 1994;264:101–4. doi: 10.1126/science.8140412. [DOI] [PubMed] [Google Scholar]
- 39.Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–63. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graycar JL, et al. Human transforming growth factor-β3: recombinant expression, purification, and biological activities in comparison with transforming growth factors-β1 and -β2. Mol Endocrinol. 1989;3:1977–86. doi: 10.1210/mend-3-12-1977. [DOI] [PubMed] [Google Scholar]
- 41.Dennler S, et al. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–55. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choy L, Skillington J, Derynck R. Roles of autocrine TGF-β receptor and Smad signaling in adipocyte differentiation. J Cell Biol. 2000;149:667–82. doi: 10.1083/jcb.149.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.