Abstract
Background
The present study is aimed at identifying potential candidate genes as prognostic markers in human oral tongue squamous cell carcinoma (SCC) by large scale gene expression profiling.
Methods
The gene expression profile of patients (n=37) with oral tongue SCC were analyzed using Affymetrix HG_U95Av2 high-density oligonucleotide arrays. Patients (n=20) from which there were available tumor and matched normal mucosa were grouped into stage (early vs. late) and nodal disease (node positive vs. node negative) subgroups and genes differentially expressed in tumor vs. normal and between the subgroups were identified. Three genes, GLUT3, HSAL2, and PACE4, were selected for their potential biological significance in a larger cohort of 49 patients via quantitative real-time RT-PCR.
Results
Hierarchical clustering analyses failed to show significant segregation of patients. In patients (n=20) with available tumor and matched normal mucosa, 77 genes were found to be differentially expressed (P< 0.05) in the tongue tumor samples compared to their matched normal controls. Among the 45 over-expressed genes, MMP-1 encoding interstitial collagenase showed the highest level of increase (average: 34.18 folds). Using the criterion of two-fold or greater as overexpression, 30.6%, 24.5% and 26.5% of patients showed high levels of GLUT3, HSAL2 and PACE4, respectively. Univariate analyses demonstrated that GLUT3 over-expression correlated with depth of invasion (P<0.0001), tumor size (P=0.024), pathological stage (P=0.009) and recurrence (P=0.038). HSAL2 was positively associated with depth of invasion (P=0.015) and advanced T stage (P=0.047). In survival studies, only GLUT3 showed a prognostic value with disease-free (P=0.049), relapse-free (P=0.002) and overall survival (P=0.003). PACE4 mRNA expression failed to show correlation with any of the relevant parameters.
Conclusion
The characterization of genes identified to be significant predictors of prognosis by oligonucleotide microarray and further validation by real-time RT-PCR offers a powerful strategy for identification of novel targets for prognostication and treatment of oral tongue carcinoma.
Background
Cancer arising from the oral cavity accounts for approximately 1.6% of all cancers diagnosed in the United States with an incidence of 22,000 new cases per year [1]. Despite the advances in multimodality treatment, the overall prognosis for patients with oral squamous cell carcinoma (SCC) has remained unchanged in the past three decades. Furthermore, variability in the clinical course of patients with oral SCC remains unexplained and conventional clinicopathological parameters fail to answer all questions. Identification of novel prognostic factors may allow a rational selection of the most appropriate therapeutic options for individual patients. The cellular and molecular heterogeneity of oral SCC and the large number of genes potentially involved in oral carcinogenesis and progression emphasize the importance of studying multiple gene alterations on a global scale. Gene expression profiling by high-throughput technologies have proven to be valuable tools for prognostication of outcome and progression in human malignancies including head and neck cancer [2-10]. These technologies permit us to classify individual cancers and enhance our understanding of molecular cancer pathogenesis.
There are several distinct subsites within the oral cavity cancer including buccal mucosa, oral tongue, floor of mouth, gingiva, retromolar trigone and hard palate. Since they differ in their biological and clinical behaviors, the present study focused on one subsite – the oral tongue. This study utilized high-density oligonucleotide array to generate a molecular portrait of oral tongue SCC and to explore the correlations between gene expression patterns and clinically relevant parameters. We performed hierarchical clustering analysis, analyzed gene expression profiles by comparing primary tumor and their matched normal mucosa and compared different patient groups based on lymph node status and tumor stage to identify clinically significant genes. Data from the microarray analysis were then validated by real-time RT-PCR. The present study is the first to demonstrate the ability of gene expression profiling to predict clinical outcome in one cancer subsite within the oral cavity.
Methods
Tumor Selection
Following guidelines established by the Institutional Review Board at Memorial Sloan-Kettering Cancer Center (MSKCC), fresh tissue samples were sequentially collected after obtaining written informed consent from 49 patients undergoing therapeutic surgical resection for SCC of the oral tongue at the Head and Neck Service, MSKCC from January 28, 1998 to January 2, 2002. Post-operative adjuvant treatment was given to selected patients following the institutional protocol. In each case, the portion of tumor was resected near the advancing edge of the tumor to avoid its necrotic center. After excision, the tissues were immediately snap-frozen and stored in liquid nitrogen until use. Histologically normal mucosae of the upper aerodigestive tract, resected 5 cm away from the tumor area, were obtained in all cases and used as controls. Tumors were staged according to the AJCC/UICC TNM classification 5th edition [11]. "Node-positive cases" in this study refers to the presence of positive cervical nodes based on a histological diagnosis after a neck dissection, while the patients who experienced no metastasis for at least 12 months post-operatively were scored as "node-negative cases." The clinical and pathological characteristics of all patients analyzed in the study are summarized in Table 1.
Table 1.
Clinicopathological characteristics of the patients in the study and validation groups
| Parameter | Study (Array) | Validation | |
|---|---|---|---|
| All samples | TN paired | RT-PCR | |
| NUMBER % | |||
| Total | 37 | 20 | 49 |
| Age (yrs) | |||
| Median (Range) | 57 (36–97) | 62 (35–97) | 59 (35–97) |
| Gender | |||
| Male | 22 (59.5) | 12 (60) | 26 (53) |
| Female | 15 (40.5) | 8 (40) | 23 (47) |
| Therapy prior to surgery | |||
| None | 22 (59.5) | 12 (60.0) | 34 (69.4) |
| Yes | 15 (40.5) | 8 (40.0) | 15 (30.6) |
| Smoking history | |||
| Yes | 19 (51.4) | 11 (55.0) | 30 (61.2) |
| No | 18 (48.6) | 9 (45.0) | 19 (38.8) |
| Alcohol history | |||
| Yes | 14 (37.9) | 8 (40.0) | 20 (40.8) |
| No | 23 (62.1) | 12 (60.0) | 26 (59.2) |
| Unknown | 0 | 0 | 3 |
| Histological grading | |||
| Well-differentiated | 6 (16.2) | 3 (15) | 9 (18) |
| Mod-differentiated | 24 (64.9) | 13 (65) | 32 (65) |
| Poorly-differentiated | 7 (18.9) | 4 (20) | 8 (16) |
| Unknown | 0 | 0 | 0 |
| Lymph node involvement (Pathologic) | |||
| Negative | 26 (70.2) | 13 (65) | 25 (51) |
| Positive | 11 (29.8) | 7 (35) | 24 (49) |
| Unknown | 0 | 0 | 0 |
| TNM Stage (Pathologic) | |||
| I | 8 (21.6) | 4 (20) | 11 (22) |
| II | 12 (32.4) | 6 (30) | 12 (25) |
| III | 3 (8.1) | 3 (15) | 7 (14) |
| IV | 14 (37.9) | 7 (35) | 19 (39) |
| Unknown | 0 | 0 | 0 |
Oligonucleotide microarray analysis
Tumor and normal tissues from 37 of the 49 patients were used for the oligonucleotide microarray analysis. Twenty (TN paired) of the 37 patients had primary tumor samples and matched normal mucosa available for analysis. Total RNA from snap-frozen tissue samples from the 37 patients was extracted with TRIsol™ reagent (Gibco BRL) following the manufacturer's protocol and re-purified by the RNAeasy Mini-spin column (Qiagen). Five to 10 μg of total RNA was reverse transcribed in the presence of an oligo dT-T7 primer. The cDNA was used for in vitro transcription amplification reaction in the presence of biotinylated nucleotides. Fifteen μg of labeled cRNA was fragmented and then hybridized against the Affymetrix HG_U95Av2 oligonucleotide arrays (Affymetrix, Santa Clara, CA). The arrays were scanned using a Hewlett Packard confocal laser scanner and analyzed using MicroArray Suite 5.0 (Affymetrix).
The microarray data have been deposited in NCBIs Gene Expression Omnibus (GEO) http://www.ncbi.nlm.nih.gov/geo/ and can be accessed through GEO Series accession number GSE13601.
RNA preparation and real-time RT-PCR
RT-PCR of GLUT3, HSAL2, and PACE4 was performed on the larger cohort of 49 patients. Two μg of total RNA was reverse transcribed with MultiScribe™ Reverse Transcriptase (Applied Biosystems, Inc.). Gene specific primers were designed using the Primer3 Program. Sequences of PCR primer sets (in 5'-3' direction) were as follows: GLUT3 forward: TAGAAAGCCTGTTCCCCTCA, GLUT3 backward: GTGGCGGGATTACTTCAAAA; HSAL2 forward: CCCTCCTATTTCAGCCTCCT, HSAL2 backward: TCTTCAGTACCGGCACCTTC; PACE4 forward: CCTGTGTGACCCTCTGTCCT, PACE4 backward: GGTTCATCCACGCACTTTTT. The sequence of PCR primer sets for 18S rRNA were previously described [12]. Quantification of transcripts was performed by the ICycler Detection System (Bio-Rad Laboratories) using SYBR green detection. The relative quantification of a target gene in comparison to a reference (18S rRNA) was performed as described [13]. Unless otherwise stated, each assay included duplicate reactions for each sample and was repeated twice.
Statistical analysis
All correlation and outcome analysis was performed using the JMP statistical software package version 4.0.0 (SAS Institute, Inc.). Disease-free survival is defined as the time from surgery to the day of the first recurrence or death. Relapse-free survival was defined as time from surgery to the day of the first recurrence. Overall survival was defined as time from surgery to the day of death or last follow-up.
Results and discussion
Gene expression patterns in oral tongue SCC
We analyzed gene expression profiles in 20 patients with oral tongue SCC by comparison between primary tumor samples and their matched morphologically normal mucosa. Among 12,625 probe sets in the Affymetrix array, 77 probe sets had statistically significant difference (P < 0.05) between all tumors and their matched normal tissues. There were 60 probe sets representing 45 genes and 11 ESTs that were increased and 17 probe sets representing 9 genes and 8 ESTs that were decreased in tumors compared to normal controls. Table 2 lists the genes that were up-regulated or down-regulated along with the fold changes in gene expression in tumor compared to their normal counterparts. These include genes known to be relevant in oncogenesis such as cell proliferation, apoptosis, development, angiogenesis, invasion and metastasis as well as genes that have not been implicated in oral tongue carcinogenesis. Among the 45 over-expressed genes, MMP-1 encoding interstitial collagenase showed the highest level of increase (average: 34.18 fold). MMP-7 and MMP-12 were also found to be overexpressed. Matrix metalloproteinases (MMPs), a family of 23 human zinc-dependent extracellular endopeptidases involved in the degradation of extracellular matrix and basement membrane during tumor cell invasion, have been implicated in a number of different human tumors including head and neck SCC [14,15]. Not surprisingly, enhanced MMP-1 expression has been found to be associated with malignant progression as well as poor outcome in head and neck SCC [15-17]. Likewise, MMP-7 and MMP-12 have both been implicated in tumor aggressiveness in oral SCC [18]. Genes that have been shown to be involved in epithelial development and differentiation such as the cytokeratins KRT16 and KRT17 were found to be overexpessed. In their study investigating RNA from head and neck SCC and normal tissues, Villaret et al. (2000) found KRT6 and KRT16 to be the genes most commonly expressed [19]. Similarly, KRT16 has also been found to be highly expressed in squamous cell carcinoma of the skin [20]. In addition, those that play a role in angiogenesis, such as hypoxia-inducible factor (HIF-1α) and platelet-derived endothelial cell growth factor (ECGF1) were also overexpressed. Several transcripts were found to be significantly under-expressed or absent in tumor compared with matched normal tissues including those that encode for cell surface (CO-029), nuclear (ZAKI-4) and extracellular proteins (hSBP).
Table 2.
Genes differentially expressed between oral tongue tumor and case-matched histologically normal mucosae (TN paired, n = 20)
| Accession | Sequence identity (GenBank/EMBL) | Average log fold change |
|---|---|---|
| Nuclear proteins (transcription factors, DNA processing enzymes) | ||
| AJ001381 | Mutated allele of a myosin class I, myh-1c | 5.24 |
| D28364 | Annexin II | 4.61 |
| M92383 | Thymosin beta-10 | 3.75 |
| U22431 | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) | 3.47 |
| M86400 | Phospholipase A2 | 3.27 |
| L19779 | Histone H2A.2 | 2.99 |
| U10860 | Guanosine 5-monophosphate synthase | 2.71 |
| U12472, U21689 | Glutathione S-transferase (GST phi) | 2.70 |
| J04173 | Phosphoglycerate mutase (PGAM-B) | 2.69 |
| AL009179 | Histone H2B | 2.58 |
| D13748 | Eukaryotic initiation factor 4AI | 2.57 |
| S79639 | Exostosin 1 (EXT1) | 2.39 |
| AL049650 | snRNP (small nuclear ribonucleoprotein particle) protein B) | 2.35 |
| Ras-Like Protein Tc4 | 2.27 | |
| D26599 | Proteasome subunit HsC7-I | 2.04 |
| X82554 | SPHAR gene for cyclin-related protein | -1.89 |
| U40490 | Human nicotinamide nucleotide transhydrogenase | -3.53 |
| D83407 | ZAKI-4 in human skin fibroblast | -5.31 |
| Cytokines, growth factors and receptors | ||
| NM001953, M63193 | Platelet-derived endothelial cell growth factor (ECGF1) | 15.89 |
| M77349 | Transforming growth factor-beta induced gene product (BIGH3) | 6.17 |
| U73377 | p66shc (SHC) | 2.37 |
| M35252 | Tumor associated antigen CO-029 | -9.42 |
| Signaling molecules | ||
| AF054183 | GTP binding protein | 2.00 |
| U78525 | Eukaryotic translation initiation factor (eIF3) | 1.89 |
| Metabolic enzymes, transporters, ion channels | ||
| AF042498 | Rod photoreceptor CNG-channel beta subunit (RCNC2) | 6.73 |
| M94856 | Fatty acid binding protein homologue (PA-FABP) | 5.24 |
| M91670 | Ubiquitin carrier protein (E2-EPF) | 3.52 |
| J03626 | UMP synthase | 2.54 |
| D50840 | Ceramide glucosyltransferase | 2.54 |
| U89606 | Pyridoxal kinase | 2.41 |
| Accession | Sequence identity (GenBank/EMBL) | Average log fold change |
| X52851 | Cyclophilin | 2.22 |
| X97074 | Clathrin-associated protein | 2.07 |
| S81003 | L-UBC = ubiquitin conjugating enzyme | 2.04 |
| U09510 | Glycyl-tRNA synthetase | 1.96 |
| D25547 | PIMT isozyme I | 1.87 |
| AF052941 | DAP-kinase related protein 1 | -2.12 |
| D16294 | Mitochondrial 3-oxoacyl-CoA thiolase | -3.59 |
| Extracellular proteins | ||
| M13509 | Skin collagenase (MMP1) | 34.18 |
| X07820 | Stromelysin-2 (MMP7) | 15.89 |
| L10343 | Elafin | 9.95 |
| L23808 | Macrophage metalloelastase (MME, MMP12) | 7.67 |
| U29091 | Human selenium-binding protein (hSBP) | -5.41 |
| Others | ||
| AB018342 | KIAA0799 | 4.44 |
| D21261 | KIAA0120 | 2.59 |
| AB014515 | KIAA0615 | 2.42 |
| AB007889 | KIAA0429 | 2.09 |
| AA586894 | EST | 14.57 |
| AA010777 | EST | 14.12 |
| L05424 | EST | 5.76 |
| AI885852 | EST | 4.03 |
| Z98946 | EST | 3.42 |
| AC002073 | EST | 3.17 |
| AF053356 | EST | 1.73 |
| AB028994 | KIAA1071 | -6.08 |
| D42047 | KIAA0089 | -4.68 |
| AB007972 | KIAA0503 | -3.96 |
| AF007153 | EST | -5.35 |
| AI674208 | EST | -3.57 |
| AL080059 | EST | -3.12 |
| AL049381 | EST | -2.93 |
| AW021542 | EST | -2.60 |
Hierarchical clustering analysis
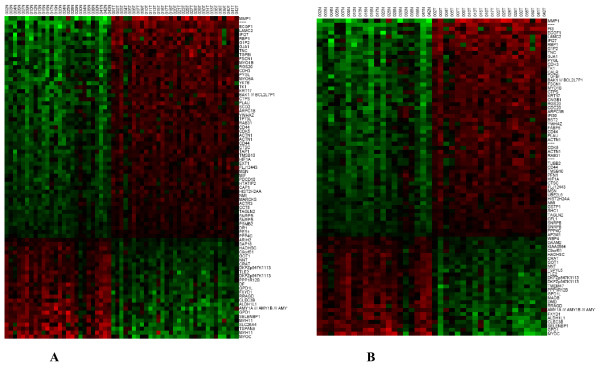
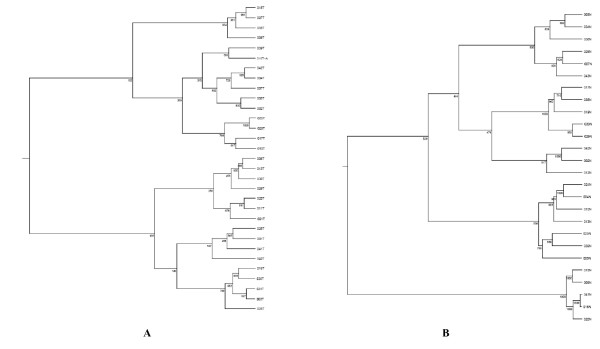
Four separate clustering analyses were performed: (1) All tumor and normal samples (TN, n = 37), (2) the tumor and their matched normal samples (TN paired, n = 20); (3) the tumor samples by themselves (T, n = 31); and (4) the normal samples by themselves (N, n = 26). The data were clustered using the standard hierarchical method with ward linkage and using the Pearson correlation to determine the distance function. The distance between samples was dist = (1-p)/2 where p is the correlation coefficient. Before clustering, the data was filtered to remove genes that were scored Absent (A) by the MAS5.0 software in 75% or more of the samples as they are likely to be measuring noise in the system. To assess the robustness of the clustering results, a resampling method was used to create 1000 replica datasets by adding Guassian noise to each point. These 1000 data sets were individually clustered and then a consensus tree was built from them. The number at each node in the tree indicates how often that subtree appears in the 1000 replica trees. The higher the number, the more robust is the subtree. The samples from the TN cluster set clearly segregated the tumor from the normal samples (Figure 1A). Similarly, TN paired set separated the tumor samples from their matched normal counterparts (Figure 1B). The two clusters exhibit nearly identical patterns of gene expression changes. As shown on Table 2, the genes that were overexpressed included those involved in tumor invasion, epithelial development and angiogenesis. In contrast, however, clustering analysis failed to show significant segregation of patients based on expression profiling in both the T and N cluster sets possibly due to the heterogeneous nature of the samples as well as the relatively small number of samples in this study (Figure 2).
Figure 1.
Hierarchical clustering of the gene expression data for the TN cluster set, N = 37. A; and the TN paired cluster set, N = 20. B. Approximately 12, 625 genes were clustered using the method described in the text. The genes shown represent the top 80 genes that were up-regulated (red) and down-regulated (green) in the sample sets. The normal tissue (N) and tumor tissue (T) are followed by their corresponding case numbers.
Figure 2.
Hierarchical clustering of tumor (T), N = 31. A; and normal (N), N = 26. B cluster sets based on the complete panel of 12,000 genes. Horizontal distance represents the correlation of the samples to one another. The numbers at each node represent frequency of the shown pattern using 1,000 iteration bootstrap resampling.
Assessment of correlation with lymph node status, stage and outcome
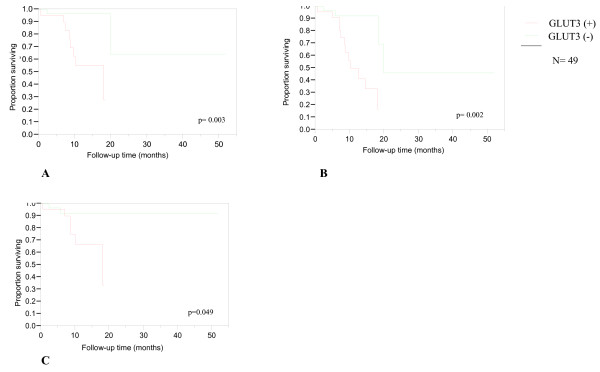
We grouped patients with pathologic stages I and II disease into an early-stage disease and grouped patients with pathologic stages III and IV disease into a late-stage disease category. Through statistical regression analysis, we identified genes whose expression differed in tumor versus normal mucosa and those whose expression was most different between the staging subgroups (Table 3A). The same analysis was performed to compare patients without cervical nodal metastasis (N0) to those with nodal disease (N1–N3) (Table 3B). We analyzed data from patients (n = 20) for which tumor and matched normal mucosae were available and not the larger subgroup of patients (n = 37) in order to obtain a more meaningful comparison of gene expression changes between tumor and normal mucosa. We selected three genes, GLUT3, HSAL2 and PACE4 for further analysis and validation in a larger cohort of 49 patients. We selected genes with known important roles in cellular functions and carcinogenesis and for which antibodies were available. We employed a two-step quantitative RT-PCR to validate expression changes identified by gene array analysis for the three selected genes in all 49 cases. We defined the cut-off value for over-expression as two-fold or greater relative to matched normal controls. Using these criteria, 30.6%, 24.5% and 26.5% of patients expressed high levels of GLUT3, HSAL2 and PACE4, respectively. We assessed the prognostic significance of expression of the selected genes and various clinicopathological parameters. Univariate analyses demonstrated that GLUT3 over-expression correlated with depth of invasion (P < 0.0001), tumor size (P = 0.024), pathological stage (P = 0.009) and recurrence (P = 0.038). HSAL2 was positively associated with depth of invasion (P = 0.015) and advanced T stage (P = 0.047). PACE4 expression failed to show correlation with clinicopathological parameters. Table 4 depicts the univariate analysis of GLUT3, HSAL2 and PACE4 expression and various clinicopathologic parameters. In survival studies, only GLUT3 showed a prognostic value with disease-free survival (P = 0.049), relapse-free survival (P = 0.002) and overall survival (P = 0.003) (Figure 3). Multivariate analysis with Cox's proportional hazards revealed that all parameters remained independent prognosticators in this group of patients.
Table 3.
Differentially expressed genes associated with cervical lymph node and pathologic stage status
| p-value | Number present | t-score | Accession: Definition/Description or EST name |
|---|---|---|---|
| 0.001% | 30 | 5.324 | M20681: Human glucose transporter-like protein-III (Glu3) |
| 0.006% | 10 | 4.525 | X71125: H. sapiens mRNA for glutamine cyclotransferase |
| 0.011% | 10 | 3.943 | X51757: HAP70B Human heat-shock protein HSP70B gene |
| 0.014% | 16 | 4.519 | AB028957: H. sapiens mRNA for KIAA1034 protein |
| 0.025% | 26 | 4.261 | Protein Phosphatase inhibitor homolog |
| 0.021% | 30 | 4.568 | Y12065: H. Sapiens mRNA for nucleolar protein hNop56 |
| 0.031% | 29 | 4.272 | AB015633: Homo sapiens mRNA for type II membrane protein |
| 0.031% | 8 | 4.517 | D78579: Homo sapiens mRNA for neuron derived orphan receptor |
| 0.031% | 17 | 3.764 | W25985: 17e6 |
| 0.044% | 31 | 3.792 | AF007140: Homo sapiens clone 23711 unknown mRNA |
| 0.061% | 31 | 3.659 | X86163: H. sapiens mRNA for B2-bradykinin receptor |
| 0.064% | 31 | 3.706 | A1935420: wo84c08 |
| 0.076% | 8 | 3.188 | AB020654: Homo sapiens mRNA for KIAA0847 protein |
| 0.076% | 7 | 3.246 | L23959: HUMDP1A Homo sapiens E2F-related transcription factor (DP-1) mRNA |
| 0.090% | 31 | 2.808 | AL09674: Homo sapiens mRNA: cDNA DKFZp58600223 |
| 0.090% | 21 | 3.544 | Z11697: Homo sapiens mRNA for HB 15 |
| 0.090% | 22 | 3.813 | X98834: H. sapiens mRNA for zinc finger protein, Hsal2 |
| 0.106% | 16 | 3.720 | M81750: H. sapiens myeloid cell nuclear differentiation antigen mRNA |
| 0.106% | 25 | 3.943 | S68134: CREM = cyclic AMP-responsive element modulator beta isoform |
| 0.125% | 31 | 3.302 | X98296: H. sapiens mRNA for ubiquitin hydrolase |
| 0.147% | 26 | 3.051 | U49392: Human allograft inflammatory factor-1 (AIF-1) mRNA |
| 0.193% | 7 | 3.269 | AL109669: Homo sapiens mRNA full length insert cDNA clone EUROIMAGE |
| 0.195% | 5 | 2.861 | M31166: HUMTSG14A Human tumor necrosis factor-inducible (TSG-14) mRNA |
| A | |||
| p-value | Number present | t-score | Accession: Definition/Description or EST name |
| 0.016% | 31 | 4.286 | AA402538: Soares ovary tumor NbHOT Homo sapiens cDNA |
| 0.035% | 22 | 3.690 | X98834: H. sapiens mRNA for zinc finger protein, Hsal2 |
| 0.035% | 26 | 3.501 | U49392: Human allograft inflammatory factory-1 (ALF-1) mRNA |
| 0.043% | 5 | 3.782 | X62055: H. sapiens PTP1C mRNA for protein-tyrosine phosphate |
| p-value | Number present | t-score | Accession: Definition/Description or EST name |
| 0.062% | 11 | 3.514 | AB023135: Homo sapiens mRNA for activation-inducible lymphocyte |
| immunomediatory molecule AILIM | |||
| 0.062% | 31 | 3.698 | A1828166: wk32h09 |
| 0.073% | 31 | 3.761 | U85773: Human phosphomannomutase (PMM2) mRNA |
| 0.085% | 6 | 3.501 | S67970: ZNF75 = KRAB zinc finger [human, lung fibroblast, mRNA |
| 0.104% | 16 | 3.709 | M81750: H. sapiens myeloid cell nuclear differentiation antigen mRNA |
| 0.104% | 31 | 3.824 | AA121509:zk88c10.s1 |
| p-value | Number present | t-score | Accession: Definition/Description or EST name |
| 0.104% | 21 | 3.023 | Z11697: Homo sapiens mRNA for HB15 |
| 0.104% | 31 | 3.952 | D67031: Homo sapiens ADDL mRNA for adducin-like protein |
| 0.123% | 29 | 3.565 | J02923: Human 65-kilodalton phosphoprotein (p65) mRNA |
| 0.123% | 21 | 3.271 | L10717: Homo sapiens T cell-specified tyrosine kinase mRNA |
| 0.145% | 26 | 2.955 | AL031228:dJ1033B10. 10/Membrane protein with histidine rich charge clusters |
| 0.145% | 24 | 3.512 | AB011102: Homo sapiens mRNA for KIAA0530 protein |
| 0.170% | 25 | 2.843 | M80482: Human subtilisin-like protein (PACE4) mRNA |
| 0.199% | 19 | 3.215 | AF072099: Homo sapiens immunoglobulin-like transcript 3protein variant 1 gene |
| 0.199% | 28 | 3.319 | J03037: Human carbonic anhydrase II mRNA |
| 0.199% | 31 | 2.979 | D21261: Human mRNA for KIAA0120 gene |
| 0.199% | 31 | 3.542 | L06797: HUMGPCR Human (clone L5) orphan G protein-coupled receptor mRNA |
| B | |||
The 20 patients were grouped into early-stage (Stage I and II disease) category and late-stage (Stage III and IV disease) category. A; Similarly, patients without cervical nodal metastasis (N0) were compared to those with nodal disease (N1–N3). B; Genes whose expression in tumor was different in normal mucosa and which were most different between the clinical stage and nodal disease subgroups were identified by statistical regression analysis (t test). The genes identified within the two subgroups are listed. Note that HSAL2 were identified in both subgroups. Only genes that were present in at least 5 samples were included.
Table 4.
Univariate analysis of clinicopathologic parameters and gene expression
| Parameter | GLUT3 | HSAL2 | PACE4 |
|---|---|---|---|
| p-value | |||
| DOI | p < 0.0001 | p = 0.015 | NS |
| Tumor size | p = 0.024 | NS | NS |
| pStage | p = 0.009 | NS | NS |
| Advanced T stage | NS | p = 0.047 | NS |
| Recurrence | p = 0.038 | NS | NS |
| N+ | NS | NS | NS |
DOI = depth of invasion; pStage = pathologic stage; NS = not significant; N+ = presence of lymph node metastasis
Figure 3.
Kaplan-Meier plot showing over-all survival. A; relapse-free survival. B; and disease-free survival. C in patients demonstrating GLUT3 over-expression.
Malignant cells show an increased glucose uptake in vitro and in vivo [21,22]. This process is thought to be mediated by glucose transporters (GLUTs), the expression and activity of which is regulated by oncogenes, growth factors and cytokines [23,24]. Studies of GLUT genes in human cancers have shown over-expression of GLUT1 and GLUT3 in cancers of various sites including the head and neck [25-28]. Recent studies in laryngeal carcinoma demonstrated an association between GLUT3 protein levels and poorer outcome [29]. HSAL2 is a member of a gene family that encodes a group of putative transcription factors. Evidence from various studies suggests that the HSAL gene family is necessary for normal embryonic development and genetic alterations can lead to human congenital defects and cancer [30,31]. HSAL2 is thought to have a role as a tumor suppressor gene in ovarian cancer [32]. The present study suggests the potential role of GLUT3 and HSAL2 in oral tongue SCC. Proprotein convertases (PC) are a family of serine endoproteases that play important roles in regulating cell function by converting proproteins to biologically active molecules such as neuropeptides and polypeptide hormones, protein tyrosine phosphatases, growth factors and their receptors, and enzymes including MMPs. Numerous members of the PC family have been associated with invasion and proliferation in various cancers including head and neck, breast and lung cancers [33-35]. PCs are thought to activate certain substrates that may play a significant role in carcinogenesis. Among these substrates are MMPs which are known to be involved in the degradation of extracellular matrix, a key process in the initiation of tumor microinvasion into the connective tissue. PACE4, a member of the PC family, activates membrane type MMPs (MT-MMPs). Bassi et al. demonstrated that PACE4 expression results in enhanced susceptibility to carcinogenesis in vivo [35]. In the present study, PACE4 failed to show clinical significance when validated by real-time RT-PCR likely due to the small sample size and heterogeneous nature of the specimens.
There is growing literature on the use of microarray technology to examine genomewide genetic expression changes associated with head and neck SCC development and to identify biomarkers as it relates to response to therapy and clinical outcome [5-10]. However, there is discordance among the studies. Furthermore, the biomarkers often lack predictive power. Two important limitations of the studies to date are the heterogeneity in subsites analyzed and the small number of study samples. Our study as well as that of others addressed the former by focusing only on oral tongue SCC [36-38]. Carinci et al. analyzed gene expression in 9 specimens with dysplasia, 8 with oral tongue SCC with no metastasis, and 11 with metastatic oral tongue SCC. Several genes were identified as potential markers of oral tongue progression and metastasis [36]. Shimada and colleagues identified 16 genes that were upregulated in 4 oral tongue SCC specimens. One gene, RabIA, a member of the Ras oncogene, was chosen and validated at the RNA and protein levels [37]. In a study by Zhou et al., 25 primary oral tongue SCC were classified based on lymph node status and the presence of extracapsular spread. Among the genes that were shown to be associated with metastasis included MMP-9 [38].
Our findings show overexpression of numerous genes that have been previously shown in other DNA microarray studies to have a potential role in the development of head and neck carcinogenesis; these include MMP-1 and KRT16. Furthermore, we have identified and validated GLUT3 and HSAL2 to be potential prognosticator of head and neck SCC. Although the present study, like the others, is based on a relatively small number of patients and the findings do not allow us to draw definitive conclusion regarding their biological importance, it is the first to use large scale transcriptional profiling for predicting survival outcome in oral tongue SCC.
Conclusion
The use of high-density oligonucleotide probe arrays to identify gene expression differences between oral tongue SCC and normal tissues provide powerful means to decode the molecular events involved in the genesis and progression of oral SCC. Although these initial findings will need to be validated in relationship to clinical parameters and outcome in larger patient cohorts, the characterization of genes identified to be significant predictors by oligonucleotide microarray analysis may provide novel targets for the prognostication and treatment of oral cavity cancer. Finally, a large multi-institutional study including specimens of uniform characteristics using independent techniques to verify gene expression at the RNA, DNA and protein levels will be vital in reaching our ultimate of goal of improving the care of head and neck cancer patients.
Abbreviations
SCC: squamous cell carcinoma; CGH: comparative genomic hybridization; RT: reverse transcription; PCR: polymerase chain reaction; RR: risk ratio; CI: confidence intervals; EST: expressed sequence tag.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CLE and PO performed all RNA extraction, RNA preparation and real-time RT-PCR experiments and correlation and outcome analysis. They also drafted the manuscript. ST, TW and YY collected clinical data on patients involved in the study. NS performed all statistical analysis of the microarray data. DLC and RG reviewed all the histologic specimens. JOB, DHK, SP, ARS, RJW, JMH and JPS assisted in the drafting of the manuscript. YR and BS supervised all work and aided in the drafting of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Cherry L Estilo, Email: estiloc@mskcc.org.
Pornchai O-charoenrat, Email: sipoc@mahidol.ac.th.
Simon Talbot, Email: sgtalbot@partners.org.
Nicholas D Socci, Email: soccin@mskcc.org.
Diane L Carlson, Email: carlsond@mskcc.org.
Ronald Ghossein, Email: ghosseir@mskcc.org.
Tijaana Williams, Email: williat3@mskcc.org.
Yoshihiro Yonekawa, Email: yoy2003@med.cornell.edu.
Yegnanarayana Ramanathan, Email: ramanaty@mskcc.org.
Jay O Boyle, Email: boylej@mskcc.org.
Dennis H Kraus, Email: krausd@mskcc.org.
Snehal Patel, Email: patels@mskcc.org.
Ashok R Shaha, Email: shahaa@mskcc.org.
Richard J Wong, Email: wongr@mskcc.org.
Joseph M Huryn, Email: hurynj@mskcc.org.
Jatin P Shah, Email: shahj@mskcc.org.
Bhuvanesh Singh, Email: singhb@mskcc.org.
Acknowledgements
This study was supported by grants from the American College of Surgeons. We thank Aarti Ravikumar for excellent literature review.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smnigal C, Thun MJ. Cancer Statistics 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:367–370. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Moch H, Schraml, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Path. 1999;154:981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallinen S-L, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi O-P, Kononen J. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Research. 2000;60:6617–6622. [PubMed] [Google Scholar]
- Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M, Gallagher GT, Varvares M, Cohen D, Kim D, Kent R, Donoff RB, Todd R, Yung CM, Warrington JA, Wong DT. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–6204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- Belbin TJ, Singh B, Barber I. Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res. 2002;62:1184–1190. [PubMed] [Google Scholar]
- Freier K, Joos S, Flechtenmacher C, Devens F, Benner A, Bosch FX, Lichter P, Hofele C. Tissue microarray analysis reveals site-specific prevalence of oncogene amplifications in head and neck squamous cell carcinoma. Cancer Research. 2003;63:1179–1182. [PubMed] [Google Scholar]
- Kuriakose MA, Chen WT, He ZM, Sikor AG, Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, Elango EM, Delacure MD. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61:1372–1383. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez E, Cheng C, Farwell DG, Ricks S, Agoff SN, Futran ND, Weymuller EA, Maronian NC, Zhao LP, Chen C. Transcriptional expression profiles of oral squamous cell carcinomas. Cancer. 2002;95:1482–1494. doi: 10.1002/cncr.10875. [DOI] [PubMed] [Google Scholar]
- O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, Muschel R. Gene expression signature predicts lymphatic metastatis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24:1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- American Joint Committee on Cancer. AJCC Cancer Staging Handbook. 5. Philadelphia: Lippincott-Raven; 1998. [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophy Methods. 2000;46:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueras AR, Pendas AM, Sanchez LM, Lopez-Otin C. Matrix metalloproteinases in cancer: from new functions to improved inhibition strategies. Int J Dev Biol. 2004;48:411–424. doi: 10.1387/ijdb.041811af. [DOI] [PubMed] [Google Scholar]
- O-charoenrat P, Rhys-Evan PH, Eccles SA. Expression of matrix metalloproteinases and their inhibitors correlate with invasion and metastasis in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2001;127:813–20. [PubMed] [Google Scholar]
- Jordan RC, Macabeo-Ong M, Shiboski CH, Dekker N, Ginzinger DG, Wong DT, Schmidt BL. Overexpression of matrix metalloproteinase-1 and -9 mRNA is associated with progression of oral dysplasia to cancer. Clin Cancer Res. 2004;10:6460–6465. doi: 10.1158/1078-0432.CCR-04-0656. [DOI] [PubMed] [Google Scholar]
- Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M, Saku T, Takagi R. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106:683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- Impola U, Uitto VJ, Hietanen J, Hakkinen L, Zhang L, Larjava H, Isaka K, Saarialho-Kere U. Differential expression of matrilysin-1 (MMP-7), 92 kD gelatinase (MMP-9), and metalloelastase (MMP-12) in oral verrucous and squamous cell cancer. J Pathol. 2004;202:14–22. doi: 10.1002/path.1479. [DOI] [PubMed] [Google Scholar]
- Villaret DB, Wang T, Dillon D, Xu J, Sivam D, Cheever MA, Reed SG. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope. 2000;110:374–381. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Medical Genomics. 2008;1(13) doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isselbacher KJ. Sugar and amino acid transport by cells in culture – differences between normal and malignant cells. N Engl J Med. 1972;286:929–933. doi: 10.1056/NEJM197204272861707. [DOI] [PubMed] [Google Scholar]
- Holm E, Hagmuller E, Staedt U, Schlickeiser G, Gunther HJ, Leweling H, Tokus M, Kollmar HB. Substrate balances across colonic carcinomas in humans. Cancer Res. 1995;55:1373–1378. [PubMed] [Google Scholar]
- Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras and src oncogenes. Science. 1987;235:1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- Hiraki Y, Rosen OM, Birnbaum MJ. Growth factors rapidly induce expression of the glucose transporter gene. J Biol Chem. 1988;263:13655–1366. [PubMed] [Google Scholar]
- Yamamoto T, Seino Y, Fukumoto H, Koh G, Yano H, Inagaki N, Yamada Y, Inoue K, Manabe T, Imura H. Biochem Biophys Res Commun. 1990. pp. 223–230. [DOI] [PubMed]
- Younes M, Brown RW, Stephenson M, Gondo M, Cagle PT. Overexpression of Glut1 and Glut3 in stage I nonsmall cell lung carcinoma is associated with poor survival. Cancer. 1997;80:1046–1051. doi: 10.1002/(SICI)1097-0142(19970915)80:6<1046::AID-CNCR6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younes M, Lechago LV, Somoano JR, Mosharaf M, Lechago J. Immunohistochemical detection of Glut3 in human tumors and normal tissues. Anticancer Res. 1997;17:2747–2750. [PubMed] [Google Scholar]
- Mellanen P, Minn H, Grenman R, Harkonen P. Expression of glucose transporters in head and neck tumors. Int J Cancer. 1994;56:622–629. doi: 10.1002/ijc.2910560503. [DOI] [PubMed] [Google Scholar]
- Baer S, Casaubon L, Schwartz MR, Marcogliese A, Younes M. Glut3 expression in biopsy specimens of laryngeal carcinoma is associated with poor survival. Laryngoscope. 2002;112:393–396. doi: 10.1097/00005537-200202000-00034. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C, Bink K, Schulz-Schaeffer W, Altmann M, Engel W. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics. 1999;62:212–222. doi: 10.1006/geno.1999.6005. [DOI] [PubMed] [Google Scholar]
- Li D, Tian Y, Ma Y, Benjamin T. p150 (Sal2) is a p53-independent regulator of p21(WAF1/CIP) Mol Cell Biol. 2004;24:3885–3893. doi: 10.1128/MCB.24.9.3885-3893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi DE, Mahloogi H, Al-Saleem L, Lopez De Cicco R, Ridge JA, Klein-Szanto AJ. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol Carcinog. 2001;31:224–232. doi: 10.1002/mc.1057. [DOI] [PubMed] [Google Scholar]
- Cheng M, Watson PH, Paterson JA, Seidah N, Chretien M, Shiu RP. Pro-protein convertase gene expression in human breast cancer. Int J Cancer. 1997;71:966–971. doi: 10.1002/(SICI)1097-0215(19970611)71:6<966::AID-IJC10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Mbikay M, Sirois F, Yao J, Seidah NG, Chretien M. Comparative analysis of expression of the proprotein convertase furin, PACE4, PC1 and PC2 in human lung tumours. Br J Cancer. 1997;75:1509–1514. doi: 10.1038/bjc.1997.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi DE, Lopez De Cicco R, Cenna J, Litwin S, Cukierman E, Klein-Szanto AJ. PACE4 expression in mouse basal keratinocytes results in basement membrane disruption and acceleration of tumor progression. Cancer Res. 2005;65:7310–7319. doi: 10.1158/0008-5472.CAN-05-1213. [DOI] [PubMed] [Google Scholar]
- Carinci F, Lo Muzio L, Piattelli A, Rubini C, Chiesa F, Ionna F, Palmieri A, Maiorano E, Pastore A, Laino G, Favia G, Dolci M, Pezzetti. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunol Pharm. 2005;18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N, Tanzawa H. Aberrant expression of RABIA in human tongue cancer. Bri J Cancer. 2005;92:1915–1921. doi: 10.1038/sj.bjc.6602594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Temam S, Oh M, Pungpravat N, Huang B-L, Mao L, Wong DT. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8:925–932. doi: 10.1593/neo.06430. [DOI] [PMC free article] [PubMed] [Google Scholar]