Abstract
Noonan syndrome (NS), the most common single-gene cause of congenital heart disease, is an autosomal dominant disorder that also features proportionate short stature, facial abnormalities, and an increased risk of myeloproliferative disease. Germline-activating mutations in PTPN11, which encodes the protein tyrosine phosphatase SHP2, cause about half of NS cases; other causative alleles include KRAS, SOS1, and RAF1 mutants. We showed previously that knock-in mice bearing the NS mutant Ptpn11D61G on a mixed 129S4/SvJae X C57BL6/J background exhibit all major NS features, including a variety of cardiac defects, with variable penetrance. However, the cellular and molecular mechanisms underlying NS cardiac defects and whether genetic background and/or the specific NS mutation contribute to the NS phenotype remained unclear. Here, using an inducible knock-in approach, we show that all cardiac defects in NS result from mutant Shp2 expression in the endocardium, not in the myocardium or neural crest. Furthermore, the penetrance of NS defects is affected by genetic background and the specific Ptpn11 allele. Finally, ex vivo assays and pharmacological approaches show that NS mutants cause cardiac valve defects by increasing Erk MAPK activation, probably downstream of ErbB family receptor tyrosine kinases, extending the interval during which cardiac endocardial cells undergo endocardial-mesenchymal transformation. Our data provide a mechanistic underpinning for the cardiac defects in this disorder.
Keywords: cardiac development, human disease, signal transduction, protein tyrosine phosphatase, cardiac valves
The autosomal dominant genetic disorder Noonan syndrome (NS) is characterized by multiple, variably penetrant defects, most frequently proportional short stature, facial abnormalities, and congenital heart disease (1, 2). NS patients also exhibit a mild, typically self-limited, myeloproliferative disease, although, rarely, they develop the severe, invasive, and, if untreated, fatal malignancy juvenile myelomonocytic leukemia (JMML) (3). Gain-of-function mutations in PTPN11, which encodes the protein tyrosine phosphatase SHP2, cause ≈50% of NS cases (2). A component of most, if not all, receptor tyrosine kinase (RTK), cytokine receptor, and integrin signaling pathways, SHP2 is required for normal activation of the RAS/ERK MAPK cascade (4). These data, the autosomal dominant inheritance of NS, and the biochemical consequences of human disease-associated PTPN11 mutants on SHP2 activity, suggested that NS is caused by enhanced activation of the RAS/ERK pathway, a hypothesis solidified by the recent discovery that mutations in KRAS (5), SOS1 (6), or RAF1 (6) account for ≈1% to 2%, ≈20%, or ≈3% to 5%, respectively, of NS cases without PTPN11 mutations. Further support is provided by cardiofacialcutaneous syndrome (CFC), which shares several phenotypic features with NS and is caused by BRAF or MEK1/2 mutations (5).
Cardiac valves and parts of the atrial and ventricular septa arise from specialized structures in the atrioventricular (AV) and outflow track (OT) regions, termed “endocardial cushions,” wherein a complex interplay takes place between endocardium (EC), myocardium, and inward-migrating cardiac neural crest (NC) cells (7). Responding to signals from the underlying myocardium, cushion endocardial cells undergo endocardial-mesenchymal transformation (EMT). Transformed mesenchymal cells then proliferate and subsequently undergo a remodeling process that involves substantial apoptosis and cell-shape changes, ultimately yielding the mature valve leaflets. Cardiac NC cells also participate, although in mice these cells migrate only to the OT cushions and therefore contribute only to aortic and pulmonic valves (8).
Multiple signaling pathways orchestrate these complex events. TGFβ family members induce, whereas VEGF suppresses, EMT (7). The ErbB family receptor RTK ErbB3 is required for EMT and/or mesenchymal proliferation (7). Eph (9) and FGF (10) receptors also have been implicated in different stages of valve development in mice and chick, respectively. Signals from the EC also impinge on and help regulate myocardial development.
Previously, we reported that knock-in mice for the Ptpn11 mutation D61G (Ptpn11D61G/+ mice) display all major features of NS, including proportionate short stature, facial dysmorphia, and cardiac defects, such as valvular hyperplasia, atrial (ASD), ventricular (VSD), or AV septal defects, and double-outlet right ventricle (DORV) (11). The cardiac defects are incompletely penetrant (on a mixed 129S4/SvJae X C57BL6/J background), with half of Ptpn11D61G/+ embryos dying perinatally of the spectrum of cardiac defects and the rest exhibiting mild valvular hyperplasia during embryogenesis that eventually resolves. We found later that PTPN11D61G encodes the most activated SHP2 protein known to be associated with NS (12), leaving it unclear whether more weakly activated alleles would have similar phenotypic effects. Consistent with the notion that Ptpn11 mutants with different extents of Shp2 activation might cause different phenotypes, increasing Ptpn11D61G gene dosage (i.e., by generating Ptpn11D61G/D61G mice) enhances the penetrance of the valvuloseptal defects and results in severe myocardial thinning (11). The incomplete penetrance of the cardiac defects in Ptpn11D61G/+ mice could reflect strain-specific modifiers in the 129S4/SvEv and/or C57BL6/J background or result from stochastic events during valvuloseptal development.
Recently, Nakamura et al. reported that transgenic mice expressing the PTPN11 mutant Q79R under control of the β-MHC promoter have ventricular non-compaction and VSD (13), although these phenotypes were observed only in mice with high levels of transgene expression. In contrast, previous work showed that deletion of Nf1, which promotes enhanced Ras/Erk signaling, in EC, but not myocardial or NC cells, causes cardiac defects similar to those found in Ptpn11D61G/+ mice (14). Furthermore, deletion of Erk2, a downstream component of the RAS/ERK pathway, in NC cells causes VSD (15). Thus, multiple cell type(s) involved in valvulogenesis can be affected by altered Ras/Erk signaling, raising questions about the cell(s) in which Ptpn11 mutants act to cause NS cardiac defects. Also unclear is whether different Ptpn11 alleles and/or strain-specific modifiers contribute to the NS phenotype.
We have addressed these issues by generating and analyzing an allelic series of mutant Ptpn11 knock-in mice. Using these mouse models, we identify the cell types responsible for the facial and cardiac defects in NS and further characterize the mechanistic effects of NS mutants on cardiac valvulogenesis.
Results
Allelic Series of Mutant Ptpn11 Knock-In Mice.
To analyze the cell of origin for NS cardiac defects, we used standard gene-targeting techniques to generate mice (Ptpn11inD61Y/+) that inducibly express Ptpn11D61Y (Fig. 1A) (33), whose human counterpart is found as a somatic mutation associated with JMML and other neoplasms (16). In these mice, Ptpn11D61Y expression is prevented by a “STOP” cassette surrounded by lox P sites (flSTOP); therefore, this allele is normally silent but can be induced in a tissue-specific manner upon crossing to appropriate Cre recombinase (Cre)-expressing mice (Fig. 1A).
Fig. 1.
Mutant Shp2 expression in EC causes NS cardiac defects. (A) Schematic of mutant Ptpn11 loci. Locations of mutations and unique restriction sites are shown. (B) Phosphatase activity of recombinant WT, D61Y, D61G, and N308D Shp2 protein, from Keilhack et al. (12). (C) Sections of hearts from E13.5 WT, ecDY, ncDY, and mcDY embryos. Note the VSD (black arrow), thinned ventricular wall (red arrow), and hypertrophy of pulmonary (blue arrowhead) and AV valves (black arrowhead) observed only in ecDY hearts. (D) Stereologic analysis showing relative valve/cushion volumes of ecDY embryos compared with controls. Values are mean ± SD. n = 3. #, P < 0.05, by 2-tailed student's t test. (E) Efficiency of STOP cassette deletion was analyzed by PCR using outflow tract cushions from WT and ncDY embryos at E10.5 (Top), ventricles from WT and mcDY embryos at E9.5 (Middle) and EC from WT and ecDY at E9.5 (Bottom). The upper band shows the deleted locus; the lower band shows the WT locus.
To assess the effects of a NS allele that encodes a less-activated version of Shp2 than D61Y and D61G, we also generated knock-in mice bearing Ptpn11N308D (Figs. 1A and Fig. S1). Asn-308 is a hot spot for NS mutations, with PTPN11N308D, the most common NS allele, accounting for ≈25% of cases (2). The resultant allelic series (Ptpn11N308/+, Ptpn11D61G/+, and Ptpn11inD61Y/+; hereafter, ND, DG, and inDY, respectively) represents a wide array of Shp2 activation (Fig. 1B).
NS Cardiac Phenotypes Are Caused by Endocardial Expression of Mutant Ptpn11.
Global induction of DY expression (by crossing inDY to EIIa-Cre mice) resulted in embryonic lethality (data not shown). To rule out potential contributions of trophoblast-derived tissues to NS defects, we crossed inDY to Mox2-Cre, thereby inducing expression of D61Y in the epiblast. Similar to DG/+ and DG/DG embryos (11), all induced embryos showed cardiac defects, including valvular hyperplasia, ASD, VSD, DORV, and myocardial thinning (Fig. S2). These cardiac defects could arise from aberrant Shp2 function in EC, myocardium, and/or NC cells. To delineate the cell-type-specific effects of mutant Shp2, we crossed inDY mice to Tie2-Cre (a.k.a. Tek-Cre) mice, which express Cre in endothelial (and hematopoietic) cells, α-myosin heavy chain (αMHC)-Cre mice, which express Cre selectively in myocardial cells, and Wnt1-Cre mice, which express Cre in NC cells. Although PTPN11D61Y is found only in neoplastic disorders (i.e., not in germline NS), because expression of this allele in entire embryos caused qualitatively similar (although quantitatively more severe) versions of the DG/+ cardiac phenotype (Fig. S2), we expected tissue-specific expression of this allele would evoke the maximal cardiac defects possible for a gain-of-function Ptpn11 mutation.
DY expression in these cell types had dramatically different consequences. Endocardial-specific Cre-expression resulted in deletion of the flSTOP cassette in EC as early as embryonic day (E)9.5, and, like DY expression in the epiblast (Fig. S2), resulted in embryonic lethality (data not shown). At E13.5, ecDY embryos showed a spectrum of cardiac defects similar to those in DG/DG embryos: enlarged cushions (≈160% compared with control), VSD, DORV, and thinned myocardium (Fig. 1 C and D and data not shown). Enlarged cushions were evident as early as E11.5 (Fig. S3). In contrast, NC-specific DY (ncDY) expression caused no evident cardiac defects (Fig. 1C). We detected deletion of the flSTOP cassette in E10.5 OT cushions in ncDY mice, indicating that the DY allele was expressed in the inward-migrating cardiac NC cells during valvulogenesis (Fig. 1E). The apparently low deletion efficiency reflects the presence of the other cell types (e.g., EC, mesenchymal cells, myocardium) in OT cushions. Although ncDY mice had no obvious cardiac phenotype, they had facial defects quantitatively similar to (although more severe than) those in DG/+ mice (Fig. S4 and ref. 11), providing additional evidence that the DY allele was expressed effectively in NC cells.
The αMHC promoter often is viewed as a “late” cardiac promoter, expressed only in postnatal cardiomyocytes. Actually, however, it is transiently active at E9.5 and able to catalyze effective excision at this early developmental stage in cardiomyocytes derived from both the primary and secondary heart fields (refs. 17 and 18 and data not shown). Consistent with these published observations, ventricles from mcDY embryos showed ≈70% to 80% deletion of the flSTOP cassette, indicating that at least 70% to 80% of cardiomyocytes expressed the highly active DY allele by E9.5 (Fig. 1E). This value is almost certainly an underestimate of the deletion efficiency, because other cells (e.g., non-cushion EC, vascular endothelial cells, smooth muscle) are present in ventricle samples. Remarkably—and in contrast to a recent report (13)—myocardial-specific expression of mutant Shp2 had no obvious cardiac consequences (Fig. 1C). Thus, the valvuloseptal defects in NS result from mutant expression in EC, whereas the facial defects reflect aberrant Shp2 action in NC-derived cells. The myocardial thinning seen in ecDY, DG/DG, and severely affected ND/ND mice also is caused by EC expression of mutant Shp2 and presumably reflects Shp2-dependent signaling from EC to myocardium during this developmental stage.
Specific PTPN11 Mutations and Genetic Background Affect NS Cardiac Defects.
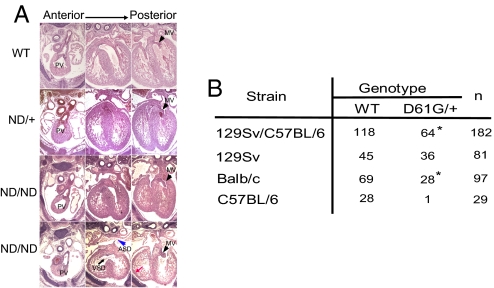
We also assessed cardiac development in ND/+ and ND/ND mice. At E13.5, all genotypes were obtained at expected Mendelian ratios (WT: 29, ND/+: 69, ND/ND: 20) from intercrosses between ND/+ and ND/+ mice. Histological analysis revealed that ND/ND embryos fell into 2 groups (Fig. 2A). Severely affected (5/8) embryos had multiple cardiac defects, including ASD and/or VSD, DORV, markedly enlarged AV valve primordia, and thinned myocardium. The 3 other ND/ND embryos and all ND/+ embryos had no apparent cardiac defects. Surviving ND/ND mice did display other NS features, such as growth defects, facial abnormalities, and hematologic abnormalities similar to those seen in surviving DG/+ mice (Fig. S5 and ref. 11). The abnormalities in ND/+ mice were less severe than those in ND/ND and DG/+ mice; hence, increasing Ptpn11N308D gene dosage enhanced the NS phenotype.
Fig. 2.
Ptpn11 allele and genetic background affect cardiac phenotype. (A) Representative transverse sections of WT, ND/+, and ND/ND embryos. Note the VSD (black arrow), ASD (blue arrowhead), thinned ventricular wall (red arrow), and enlarged AV valves (black arrowheads). The apparent ASD seen in the ND/ND embryo in the third set of panels is not seen in other sections from this embryo. (B) DG/+ mice were backcrossed onto 129Sv or BALB/c for 5 generations or onto C57BL/6 for 3 generations. Viable progeny at 3 weeks are indicated. Note the significantly deceased viability on C57BL/6 background. *, P < 0.05; χ2 test.
These data show conclusively that the specific Ptpn11 allele can impact the NS cardiac phenotype as well as other NS features significantly. Also, increasing degrees of Shp2 activation seem to have a hierarchical effect, with low-level activation being sufficient to cause postnatal growth defects, higher levels required for altered size at weaning and facial dysmorphia, and the highest levels needed to cause fatal cardiac and more severe hematologic defects.
DG/+ mice on a mixed 129S4/SvJae X C57BL/6 background are obtained at only ≈50% the expected Mendelian frequency, with the remaining DG/+ mice succumbing during late gestation or perinatally because of severe cardiac defects (11). The incomplete penetrance of these defects could reflect modifier alleles in one or the other strain or stochastic events. To distinguish between these possibilities, we backcrossed DG/+ onto 129S6/SvEv, Balb/c, and C57BL6/J backgrounds (Fig. 2B). After the fifth-generation backcross onto Balb/c, DG/+ mice were obtained at ≈50% the predicted Mendelian ratio. On 129S6/SvEv, DG/+ mice displayed nearly normal viability, whereas on the C57BL/6 background this mutant Ptpn11 allele almost always was lethal. We analyzed E13.5 D61G/+ embryos (6 of the 17 analyzed) from the fourth-generation backcross onto C57BL/6J. All had severe cardiac defects incompatible with viability, indicating that there are important strain-specific modifiers of the NS cardiac phenotype.
Abnormal Valvulogenesis in NS Results from Extended EMT Caused by Enhanced Erk Activation.
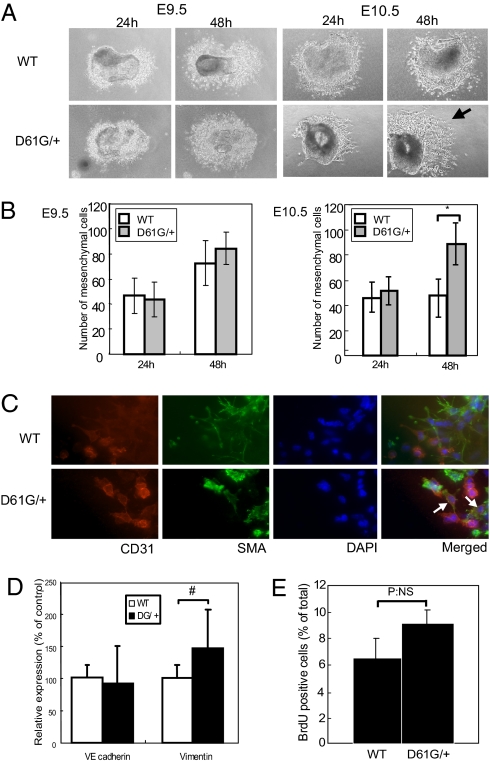
In mice, beginning at ∼E9.5, cushion EC undergoes EMT, and the resulting mesenchymal cells then proliferate substantially until ∼E13.5. Consequently, the enlarged cushions in E13.5 ecDY, DG/+, DG/DG, and ND/ND embryos could reflect increased EMT and/or increased mesenchymal cell proliferation. The early steps in valvulogenesis can be modeled ex vivo by monitoring the behavior of AV canal explants (19). Under normal conditions, cushion EC from E9.5 or E10.5 explants undergoes EMT, and the mesenchymal cells then migrate into the collagen gel. WT and DG/+ explants from E9.5 embryos gave rise to similar numbers of induced mesenchymal cells after 24 or 48 h of culture (Fig. 3 A and B). There also was no difference in the number of mesenchymal cells arising from DG/+ and WT explants from E10.5 embryos after 24 h. Although there was no further increase in mesenchymal cell generation from WT E10.5 embryo explants, the number of mesenchymal cells from DG/+ explants continued to increase over the succeeding 24 h.
Fig. 3.
Sustained EMT in D61G/+ EC cushions. (A, B) Explant assays using E9.5 (Left) or E10.5 (Right) EC from WT and DG/+ embryos. Mesenchymal cells were counted at 24 and 48 h. Note that EC from E10.5 DG/+ embryos produce more mesenchymal cells (arrow). Values represent mean ± SD. n = 8. *, P < 0.01, by 2-tailed student's t test. (C) Excess mesenchymal cells are caused by sustained EMT. Cushion explants at E10.5 were stained with the indicated antibodies and DAPI. CD31 and αSMA double-positive “transitional” cells (white arrows) are found only in DG/+ explants. (D) Quantitative RT-PCR of VE-cadherin and vimentin. Values represent mean ± SD. n = 8. #, P < 0.05, by 2-tailed student's t test. (E) Proliferation of cushion mesenchymal cells. Explants (7/genotype) exposed to BrdU for 6 h were immunostained with BrdU antibodies, and BrdU+ cells were counted. NS, not statistically significant.
The continuing increase in mesenchymal cells in DG/+ explants could reflect an extended period of EMT and/or increased mesenchymal cell proliferation. EC and mesenchymal cells can be distinguished by immunostaining for the markers CD31/PECAM (endothelial cells) and alpha-smooth muscle actin (αSMA), respectively. In explant cultures, transforming mesenchymal cells pass through a “transitional” stage characterized by retained CD31 expression, which results in CD31/αSMA double-positive cells (20). We detected such double-positive cells in explants from E10.5 DG/+ but not WT embryos after 24 h of culture (Fig. 3C, WT: 0, DG/+: 15 ± 5). Because simultaneous CD31/αSMA expression reflects newly born mesenchymal cells, these findings provide strong, direct evidence that the increase in mesenchymal cells generated from DG/+ explants reflects extension of the normal interval during which EMT occurs. Expression of vimentin, a marker of mesenchymal cells, also was increased at E11.5 cushions, whereas expression of VE-cadherin was comparable to WT (Fig. 3D). We assessed mesenchymal cell proliferation in explants by monitoring BrdU incorporation. Under these conditions, only ≈6% of WT cells incorporated label within 6 h. Although there was a nominal increase in BrdU incorporation in DG/+ mesenchymal cells, this did not reach statistical significance and, in any case, was too small to account for the increase in the number of mesenchymal cells in DG/+ explants (Fig. 3E). There was no apparent excess of apoptotic cells in WT explants (assessed by morphology) compared with DG/+ explants. We conclude that NS mutants cause abnormal valvulogenesis primarily by extending the interval over which cushion EC can undergo EMT, resulting in a substantially increased number of cushion mesenchymal cells. We cannot exclude an additional effect of mutant Shp2 on the migration of cushion mesenchymal cells out of the cushion explant.
Genetic studies strongly suggest that enhanced Ras/Erk signaling is the cause of NS and the related disorder CFC. Consistent with enhanced Erk activation during valvulogenesis (11), D61G/+ cushions (in vivo) show increased p-Erk immunostaining. Likewise, Erk activation (as measured by immunoblotting with p-Erk antibodies) was increased in cushions dissected from DG/+ embryos (Fig. 4 A and B). To test the functional significance of enhanced Erk activity, we monitored the effect of the Mek inhibitor U0126 on cushion explants. Even low doses (100 nM) of U0126 (IC50 ≈60–70 nM) inhibited excess EMT in DG/+ explants. Increasing the dose impaired normal EMT as well, consistent with an important role for Erk activity in normal valvulogenesis (Fig. 4C). Furthermore, analysis of a cross between Tie2-Cre and inDY:Erk1± mice indicates that decreasing Erk1 gene dosage by 50% (i.e., in ecDY:Erk1± mice) reduces the lethality of ecDY mice (data not shown).
Fig. 4.
D61G mutation enhances EMT by increasing Erk activation. (A, B) Increased Erk activation in DG/+ cushions. EC lysates at E10.5 were immunoblotted with anti-phospho-Erk (Upper) or anti-total Erk2 (Lower) antibodies. Data are quantified in (B). Values are mean ± SD of triplicates. n = 3. #, P < 0.05, by 2-tailed student's t test. (C) Effect of D61G mutant is blocked by Mek inhibitor. U0126 or DMSO as a control was added to explants after 24 h, and mesenchymal cells were counted 24 h later. Values are mean ± SD. n = 8. *. P < 0.01, assessed by 1-way ANOVA; Newman-Keuls multiple range test. (D) Effects of D61G mutation are blocked by ErbB family RTK inhibitors. The indicated inhibitors (PD173074: 60 nM, AG825: 10 μM, AG1478: 100 nM: Tarceva: 300 nM) or DMSO as a control were added to explants after 24 h. Mesenchymal cells were counted 24 h later. Values are mean ± SD. n = 8. Statistical significance (± inhibitor) was assessed by 1-way ANOVA; Newman-Keuls multiple range test (*, P < 0.01).
We used a pharmacological approach to identify potential signaling pathways that might mediate increased Erk activation (Fig. 4D). The FGF inhibitor PD173074 (IC50: ≈10–20 nM) (21) at doses that completely inhibit FGF receptor (FGFR) activation in fibroblasts (data not shown) had no effect on the number of mesenchymal cells in WT or D61G/+ explants. On the other hand, several different inhibitors of ErbB family RTKs (AG 825, AG1478, and Tarceva) blocked excessive mesenchymal cell production in D61G/+ explants to varying extents. These data suggest that excessive signaling from ErbB family RTKs (most likely ErbB2/3 and/or ErbB3/4) promotes increased Erk activation and excess EMT in D61G/+ and thereby contributes to NS cardiac defects.
Discussion
We previously generated a knock-in mouse model for the NS mutation Ptpn11D61G (11). D61G/+ mice demonstrate the major phenotypic features associated with NS, but the cellular and molecular basis for cardiac defects and whether genetic background and/or the specific NS mutation contribute to the NS phenotype remained unresolved. Using additional mouse models to address the cellular and molecular basis for NS phenotypes in more detail, we have obtained several insights into NS pathogenesis. By studying an inducible Ptpn11 allele, we find that endocardial expression of mutant Shp2 causes all cardiac defects in NS. Using an allelic series of knock-in mice, we find that both the specific Ptpn11 allele and genetic background affect syndromic phenotypes. There seems to be a hierarchy of phenotypic effects caused by specific Ptpn11 alleles, which is probably explained by increasing degrees of phosphatase activation. Finally, cushion explant assays suggest that NS alleles cause valvuloseptal defects by increasing Erk activation downstream of ErbB family RTKs, resulting, in turn, in an extended period of EMT by cushion EC.
Cardiac valve development is a complex process involving multiple cell types and signaling molecules (7). By crossing our inducible D61Y allele to tissue-specific Cre lines, we found that NS-associated cardiac defects are caused entirely by aberrant Shp2 action in EC (or at least endothelial-derived cells). Expression of this highly activated Shp2 protein evokes all the cardiac defects seen in severely affected DG/+ and ND/ND mice as well as the myocardial thinning seen in ND/ND and DG/DG embryos. Presumably, the myocardial thinning reflects aberrant signaling of EC to myocardium caused by mutant Shp2 expressed in EC. Neither myocardial (αMHC-Cre)- nor NC (Wnt-Cre)-evoked DY expression has apparent cardiac effects, even though the flSTOP cassette is excised in the proper time and place. In contrast, expression of mutant Shp2 in NC-derived tissues is responsible for the facial dysmorphia in NS.
Our data conflict with studies of transgenic mice that express high levels of the NS mutant Q79R under the control of the β-MHC promoter (13), which concluded that NS mutants act in myocardium. It is not clear how these disparate findings can be reconciled. Conceivably, even though the flSTOP cassette was deleted in 70% to 80%, if not all, developing mcDY cardiomyocytes, mutant Shp2 must be expressed in cardiomyocytes at earlier times to evoke these phenotypes. However, earlier expression of Cre in cardiomyocytes (using the Nkx 2.5-Cre line) also fails to evoke myocardial defects (data not shown). Furthermore, expression of DY only in EC causes all the cardiac defects (including thinned myocardium) seen in mice with global DG expression, so there is no need to invoke any other site of mutant Ptpn11 action. Another potential caveat is that we studied a Ptpn11 mutant (DY) associated only with neoplasia, not with NS per se. The mutant also is unlikely to have been misleading, given that increasing the dosage of either DG or ND (by generating DG/DG or ND/ND embryos) causes only quantitative, not qualitative, differences in NS phenotypes. Because DG and DY affect the same residue, it is difficult to imagine how endocardial DY expression could mistakenly phenocopy the cardiac defects of a bona fide NS allele (i.e., DG) when myocardium-specific expression did not cause cardiac defects.
Many disease-associated PTPN11 mutations have been identified, encoding SHP2 variants with a wide degree of catalytic activation (12). Differential SHP2 activation by different alleles might help explain the variable phenotypic spectrum in NS, yet patient surveys have failed to identify strong PTPN11 genotype/phenotype correlations (22, 23). Notably, family members with the same PTPN11 mutation can have significantly different presentations (24), indicating that modifier alleles may have a strong effect on phenotype. Conceivably the large number of NS alleles and the highly outbred human population obscure intrinsic differences in the effects of different PTPN11 alleles.
Indeed, different Ptpn11 alleles placed on the same genetic background definitely affect disease phenotype, probably because of differences in phosphatase activation. However, NS mutants are more activated because they are more “open” than WT Shp2 (12). We cannot be sure that greater protein tyrosine phosphatase activity, rather than another feature of the open state, accounts for phenotypes observed. This caveat is important given that PTPN11 mutants associated with LEOPARD syndrome, which shares features with NS, encode protein tyrosine phosphatase-inactive or -impaired SHP2 proteins (5).
D61G/+ mice on mixed background have incomplete penetrance of valvuloseptal defects and embryonic lethality (11). Our new data show that incomplete penetrance reflects strain-specific modifiers. Initial genomic scans using SNP panels have failed to identify strong modifier loci. More comprehensive analysis, using larger numbers of mice, will be required to determine whether cloneable modifiers exist or heterosis accounts for variable penetrance.
Using explant assays and marker analysis, we find that mutant Shp2 extends the normal interval during which EMT occurs. This extended period of EMT is accompanied and, based on the effects of Mek inhibition, probably is caused by enhanced Erk activation. Lakkis et al. reported that cushion explants from Nf1-deficient embryos also exhibit enhanced EMT (25), but the design of their experiments could not distinguish between extended EMT per se as opposed to excessive proliferation of cushion mesenchyme following transformation. Our results, and the similar phenotype of DG/+ (and DG/DG and ND/ND) and Nf1−/− embryos, strongly suggest that Nf1 deficiency also extends the interval of EMT. Krenz et al. reported that expression of PTPN11 Q79R in chick AV cushion explants causes increased Erk activation and excessive Erk-dependent mesenchymal cell proliferation without apparent effects on EMT (26). We did not detect excess proliferation in NS cushion explants. However, we previously observed an increased percentage of proliferating cells (by BrdU labeling) in EC cushions of DG/+ embryos later in development (E13.5) (11). Furthermore, addition of various growth factors (PDGF, FGF, neuregulin) to E10.5 explant cultures results in more extensive proliferation of cushion mesenchyme derived from WT embryos (T.A. and B.G.N., unpublished data). Thus, failure to observe excess proliferation in DG/+ cushion explant cultures could reflect a limitation of this assay rather than an intrinsic difference between different NS alleles and/or between mouse and chick cushions. We suspect that NS cardiac phenotypes are caused by both enhanced EMT and excess mesenchymal cell proliferation.
Our pharmacological studies provide new insight into the signaling pathways that mediate excess Erk activation in NS cushions. Several RTKs participate in early valvulogenesis. VEGF inhibits EMT (7), but SHP2 is a positive component of VEGF receptor signaling (27), making it hard to see how an SHP2 hypermorph could enhance EMT. Increased FGFR signaling also does not explain excessive cushion mesenchyme outgrowth in NS, given that the FGFR inhibitor PD173074 has no effect on D61G/+ cushion explants. The 4 members of the ErbB family are implicated in valvulogenesis at different developmental times. ErbB3 is a positive regulator of valvulogenesis (7), although another group failed to detect severe valvular defects in their ErbB3-deficient embryos (28). Unlike other family members, ErbB3 is kinase-dead and signals only upon heterodimerization with another ErbB RTK. The selective ErbB2 inhibitor AG825 (IC50 for ErbB2: 350 nM; IC50 for EGF receptor [EGFR]: ≈20 μM), at a concentration below the IC50 for EGFR (10 μM), blocks excessive mesenchyme production in D61G/+ explants. In addition, AG1478, which is a selective EGFR inhibitor (IC50 for EGFR: 3 nM; IC50 for ErbB2: > 100 μM) but also inhibits ErbB4 (29), inhibited excess mesenchyme production in D61G/+ explants, consistent with a possible enhancement of ErbB3/4 signaling as well. On the other hand, the EGFR inhibitor Tarceva (Erlotinib) also reduced excess mesenchymal outgrowth in NS explants. Although Tarceva in vitro is highly selective for the isolated EGFR kinase domain compared with that of ErbB2 (30), recent studies show that it has substantial (if not equal) potency against full-length ErbB2 in cells (31). Although we cannot exclude a role for increased EGFR signaling, our data, taken together, suggest that increased ErbB2/3 and/or ErbB3/4 action plays a key role in generating excess cushion mesenchyme in NS. Cushion explants contain myocardium, and myocardium is both an important site of ErbB RTK action and sends signals to EC required for EMT. Thus, the effects of ErbB inhibitors on mesenchymal outgrowth could be indirect. Aberrant Shp2 action in other RTK pathways involved in valvulogenesis, such as EphA signaling (9), also may contribute to the NS phenotype. Genetic approaches or a more purified cell system to study EMT are required to resolve these questions conclusively.
Finally, our results provide insight into the pathogenesis of NS caused by mutations in genes other than PTPN11, as well as NS-related disorders, particularly CFC. Presumably, NS phenotypes caused by SOS1, RAF1, or KRAS mutations, like those caused by PTPN11 mutations, involve similar cell types and, at least for the valvuloseptal defects, increased ERK activation. It remains to be determined why certain RAF1 mutations, unlike other NS alleles, cause hypertrophic cardiomyopathy. Like NS, CFC features facial, cardiac, and growth defects. Given that CFC is caused by mutations in BRAF, MEK1, or MEK2, it seems likely that these defects also result from increased ERK activation in the cell types affected by NS-associated PTPN11 alleles. Challenges for the future include elucidating the detailed mechanism by which enhanced ERK activity causes the shared phenotypes of NS, CFC, and other related disorders (5), the reasons why these similar disorders also manifest key differences, and determining whether manipulating ERK activity pharmacologically might have therapeutic benefit.
Materials and Methods
Ptpn11 Mutant Mice and Other Lines.
The generation of ND/+ and inDY mice is described in SI Text and ref. 33, respectively. Genotyping was carried out on tail DNA subjected to PCR and digestion with AgeI for the D61G and D61Y alleles or EcoRV for the N308D allele. Tie2-Cre, Wnt1-Cre, EIIA-Cre, and Mox2-Cre mice were obtained from the Jackson Laboratory. CMV-Cre and αMHC-Cre mice were provided by Klaus Rajewsky (Center for Blood Research) and Michael Schneider (Baylor College of Medicine), respectively. Unless indicated, all experiments used B6 × 129S6/SvEv hybrid mice. All studies were approved by the animal welfare committees at Harvard Medical School or the Ontario Cancer Institute.
Histology.
Embryos and tissues were fixed with Bouin's fixative or 4% paraformaldehyde and embedded in paraffin. Sections were stained with H&E. Stereologic assessment of cushion volume was performed by measuring all sections containing cardiac cushions and analysis using Image Pro Plus software (9).
Cushion Explant Assays and Quantitative RT-PCR.
AV explant cultures and immunostaining were performed as described (32). Where indicated, U0126, AG825, AG1478, PD173074 (Calbiochem) or Tarceva were added after 24 h of culture. For proliferation assays, explants were incubated in 20 μM 5′-bromo-2′-deoxyuridine (BrdU, Sigma) for 6 h, fixed with 70% ethanol, and stained with anti-BrdU antibodies (1:200). Fluorescent-conjugated secondary antibodies (Molecular Probes) were diluted 1:500. Nuclei were stained with DAPI and visualized on a Zeiss microscope. Quantitative RT-PCR was performed by TaqMan (Applied Biosystems), with expression normalized to GAPDH.
Protein Analysis.
Preparation of lysates and immunoblotting were performed as described (11). EC were dissected and frozen immediately. After genotyping, 10 EC were pooled and lysed. Polyclonal anti-Erk2 and anti-phospho-Erk1/2 antibodies were purchased from Santa Cruz Biotechnology and Cell Signaling Technology, respectively. All immunoreagents were used at the concentrations recommended by the manufacturers.
Supplementary Material
Acknowledgments.
We thank Drs. Klaus Rajewsky and Michael Schneider for mice, Dr. Ken Iwata (Oncogene Science, Inc.) for helpful discussions, and Dr. Kiyomi Araki (Ontario Cancer Institute) for technical support. This work was supported by National Institutes of Health Grants HL083273, R37CA49152, and R01CA114945 (to B.G.N.). T.A. was supported by the Leukemia and Lymphoma Society.
Note added in proof.
While our manuscript was in review, Krenz et al. (34) reported that EC expression of Ptpn11 Q79Q transgtene caused NS phenotypes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810053106/DCSupplemental.
References
- 1.Noonan JA. Noonan syndrome. An update and review for the primary pediatrician. Clinical Pediatrics. 1994;33:548–555. doi: 10.1177/000992289403300907. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: Genetics and pathogenesis. Annual Review of Genomics and Human Genetics. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 3.Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 2008;27:179–192. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- 4.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 5.Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 6.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: Novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong EJ, Bischoff J. Heart valve development: Endothelial cell signaling and differentiation. Circ Res. 2004;95:459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 9.Stephen LJ, Fawkes AL, Verhoeve A, Lemke G, Brown A. A critical role for the EphA3 receptor tyrosine kinase in heart development. Dev Biol. 2007;302:66–79. doi: 10.1016/j.ydbio.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 10.Sugi Y, et al. Fibroblast growth factor (FGF)-4 can induce proliferation of cardiac cushion mesenchymal cells during early valve leaflet formation. Dev Biol. 2003;258:252–263. doi: 10.1016/s0012-1606(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 11.Araki T, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 12.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T, et al. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117:2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitler AD, et al. Nf1 has an essential role in endothelial cells. Nat Genet. 2003;33:75–79. doi: 10.1038/ng1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newbern J, et al. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci USA. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tartaglia M, et al . Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 17.Gaussin V, et al. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange F, et al. Lineage and morphogenetic analysis of the cardiac valves. Circ Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- 19.Nakajima Y, et al. Extracellular fibrillar structure of latent TGF beta binding protein-1: Role in TGF beta-dependent endothelial-mesenchymal transformation during endocardial cushion tissue formation in mouse embryonic heart. J Cell Biol. 1997;136:193–204. doi: 10.1083/jcb.136.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850–855. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- 21.Mohammadi M, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tartaglia M, et al. PTPN11 mutations in Noonan syndrome: Molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenker M, et al. Genotype-phenotype correlations in Noonan syndrome. J Pediatr (Berlin) 2004;144:368–374. doi: 10.1016/j.jpeds.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Bertola DR, Pereira AC, de Oliveira PS, Kim CA, Krieger JE. Clinical variability in a Noonan syndrome family with a new PTPN11 gene mutation. Am J Med Genet A. 2004;130A:378–383. doi: 10.1002/ajmg.a.30270. [DOI] [PubMed] [Google Scholar]
- 25.Lakkis MM, Epstein JA. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development (Cambridge, UK) 1998;125:4359–4367. doi: 10.1242/dev.125.22.4359. [DOI] [PubMed] [Google Scholar]
- 26.Krenz M, Yutzey KE, Robbins J. Noonan syndrome mutation Q79R in Shp2 increases proliferation of valve primordia mesenchymal cells via extracellular signal-regulated kinase 1/2 signaling. Circ Res. 2005;97:813–820. doi: 10.1161/01.RES.0000186194.06514.b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha CH, Bennett AM, Jin ZG. A novel role of vascular endothelial cadherin in modulating c-Src activation and downstream signaling of vascular endothelial growth factor. J Biol Chem. 2008;283:7261–7270. doi: 10.1074/jbc.M702881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riethmacher D, et al. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature. 1997;389:725–730. doi: 10.1038/39593. [DOI] [PubMed] [Google Scholar]
- 29.Fukazawa R, et al. Neuregulin-1 protects ventricular myocytes from anthracycline-induced apoptosis via erbB4-dependent activation of PI3-kinase/Akt. J Mol Cell Cardiol. 2003;35:1473–1479. doi: 10.1016/j.yjmcc.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Kim TE, Murren JR. Erlotinib OSI/Roche/Genentech. Current Opinion in Investigational Drugs. 2002;3:1385–1395. [PubMed] [Google Scholar]
- 31.Schaefer G, Shao L, Totpal K, Akita RW. Erlotinib directly inhibits HER2 kinase activation and downstream signaling events in intact cells lacking epidermal growth factor receptor expression. Cancer Res. 2007;67:1228–1238. doi: 10.1158/0008-5472.CAN-06-3493. [DOI] [PubMed] [Google Scholar]
- 32.Rivera-Feliciano J, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan G, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009 doi: 10.1182/blood-2008-10-182626. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krenz M, et al. Role of ERK1/2 signaling in congenital valve malformations in Noonan syndrome. Proc Natl Acad Sci USA. 2008;105:18930–18935. doi: 10.1073/pnas.0806556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.