Abstract
Background
Altered patterns of gene expression mediate the effects of particulate matter (PM) on human health, but mechanisms through which PM modifies gene expression are largely undetermined.
Objectives
We aimed at identifying short- and long-term effects of PM exposure on DNA methylation, a major genomic mechanism of gene expression control, in workers in an electric furnace steel plant with well-characterized exposure to PM with aerodynamic diameters < 10 μm (PM10).
Methods
We measured global genomic DNA methylation content estimated in Alu and long interspersed nuclear element-1 (LINE-1) repeated elements, and promoter DNA methylation of iNOS (inducible nitric oxide synthase), a gene suppressed by DNA methylation and induced by PM exposure in blood leukocytes. Quantitative DNA methylation analysis was performed through bisulfite PCR pyrosequencing on blood DNA obtained from 63 workers on the first day of a work week (baseline, after 2 days off work) and after 3 days of work (postexposure). Individual PM10 exposure was between 73.4 and 1,220 μg/m3.
Results
Global methylation content estimated in Alu and LINE-1 repeated elements did not show changes in postexposure measures compared with baseline. PM10 exposure levels were negatively associated with methylation in both Alu [β = −0.19 %5-methylcytosine (%5mC); p = 0.04] and LINE-1 [β = −0.34 %5mC; p = 0.04], likely reflecting long-term PM10 effects. iNOS promoter DNA methylation was significantly lower in postexposure blood samples compared with baseline (difference = −0.61 %5mC; p = 0.02).
Conclusions
We observed changes in global and gene specific methylation that should be further characterized in future investigations on the effects of PM.
Keywords: DNA methylation, epigenetics, etiology, interspersed repetitive sequences, nitric oxide synthase, particulate matter
Foundry work has been associated in several early investigations with adverse health outcomes, including cardiovascular and respiratory disease as well as increased risk of lung cancer [Andjelkovich et al. 1990; International Agency for research on Cancer (IARC) 1987; Kuo et al. 1999; Xu et al. 1996]. Exposures responsible for the excess in risk have not been clearly identified (IARC 1987). In modern foundry facilities, exposures to chemicals are remarkably lower than in the past (Bergamaschi et al. 2005), but particulate matter (PM) levels are still well above the concentrations found in ambient outdoor air. Ambient PM has also been associated with increased hospitalization and mortality due to cardiorespiratory disease and lung cancer (Brook et al. 2004; Peters 2005; Samet et al. 2000; Vineis and Husgafvel-Pursiainen 2005). Epidemiologic (Brook et al. 2004; Peters 2005; Schulz et al. 2005) and in vivo studies (Chang et al. 2005; Chen and Hwang 2005; Corey et al. 2006) suggest that the transition metal components of PM may be responsible for such effects.
The mechanisms linking PM inhalation to adverse health outcomes have not been completely clarified. Inhaled particulate pollutants have been shown to produce systemic changes in gene expression, which can be detected in peripheral blood of exposed individuals (Wang et al. 2005). Gene expression of human genes is controlled by DNA methylation, which, in mammals, involves the postreplication addition of methyl groups to the 5′ position of cytosine ring within the context of CpG dinucleotides to form 5-methylcytosine (5mC). Initial observations of in vitro and animal models have shown that air particles, or air particle components such as toxic metals, can induce changes in DNA methylation (Belinsky et al. 2002; Takiguchi et al. 2003). Whether DNA methylation changes occur in human subjects exposed to PM has never been determined.
Reduced genomic methylation content in blood DNA has been observed in subjects with cardiovascular disease, as well as in cancer subjects (Robertson 2005). Genomic DNA hypomethylation is likely to result from demethylation in transposable repetitive elements, which plays a crucial role in gene regulation and genomic stability. More than 90% of all genomic 5-methylcytosines lies within CpG islands located in transposable repetitive elements, including Alu and long interspersed nuclear element-1 (LINE-1) sequences, which are those most common and well characterized. Measurements of Alu and LINE-1 methylation have been used to estimate global genomic DNA methylation content (Yang et al. 2004). In vitro studies have shown that reactive oxygen species (ROS), which are considered one of the main cellular stressors generated by PM exposure (Borm et al. 2007), may produce genomic hypomethylation (Valinluck et al. 2004). Conditions associated with reduced global DNA methylation content, such as specific dietary and genetic variations (Friso and Choi 2002; Friso et al. 2002), have been shown to interact with ambient PM exposure to produce health-related outcomes (Baccarelli et al. 2008).
Elevated expression of the inducible nitric oxide synthase gene (iNOS, also known as NOS2, Genbank accession number AF017634) has been observed in animal experiments of exposure to PM or PM components in the lung and across other different tissues (Folkmann et al. 2007; Thomson et al. 2007; Ulrich et al. 2002), including blood leukocytes (Blackford et al. 1994). Specific studies on iNOS have shown that lower DNA methylation in the gene promoter is associated with increased expression (Chan et al. 2005). iNOS expression and activity are increased in the presence of ROS (Zhen et al. 2008) and other factors, such as cigarette smoke (Anazawa et al. 2004; Chyu et al. 1999; Wright et al. 1999), associated with cardiorespiratory outcomes.
In the present work, we investigated short- and long-term effects of particle exposure on DNA methylation in peripheral blood DNA from workers with well-characterized exposure to a wide range of PM levels in an electric steel furnace plant. We measured global genomic DNA methylation content, estimated in Alu and LINE-1 repetitive elements, and promoter methylation of iNOS.
Material and Methods
Study subjects
We recruited 63 healthy, male workers (mean age 44 years; range between 27 and 55 years) free of cardiovascular and pulmonary disease in a steel production plant in Brescia, Northern Italy. All participants had been working in the present job position for at least 1 year. Thirty-seven subjects had a rotating weekly schedule based on four consecutive working days of 8 hr each, followed by 2 days of rest. The remaining 26 subjects worked Monday through Friday, also in 8-hr shifts. Twenty-five subjects (40%) were current smokers, who reported a mean (±SD) number of 13.0 ± 7.2 cigarettes smoked every day. The average body mass index of the study participants was 26.5 ± 2.7 kg/m2.
A self-administered questionnaire was used to collect detailed information on lifestyle, drug use, recent medical conditions, and residential history. Records from the factory administrative and clinical files were used to abstract information on occupational and past medical history.
In order to discriminate short- and long-term effects of PM, we obtained blood samples for DNA methylation analysis at two different times. Sample 1 was collected in the morning of the first day of work (after 2 days off work) before the beginning of any work activity. Sample 2 was collected at the same hour on the fourth day of work, after 3 consecutive days of work. Individual written informed consent and approval from the local Institutional Review Board were obtained before the study.
Exposure assessment
Measures of PM with aerodynamic diameters < 10 μm (PM10) obtained in each of the 11 work areas of the steel production plant were used to estimate individual exposures. PM10 was measured during the days between sample 1 and sample 2 collection using a GRIMM 1100 light-scattering dust analyzer (Grimm Technologies, Inc. Douglasville, GA, USA).
During the 3 working days between blood samples 1 and 2, each of the study subjects recorded in a personal log the time he spent in each of the work areas. Individual exposure was calculated as the average of PM10 weighted by the time spent in each area. PM10 levels in each of the work areas have shown very little variability over time, as measures repeated over 1 year showed very high correlation between PM10 concentrations (r2 > 0.90). Because all the study subjects reported in the questionnaire to have performed their standard work routine during the 3 days of the study, the time-weighted PM10 represented, in addition to the exposure during the week of the study, a measure of the usual exposure of the study subjects. Therefore, we considered our study subjects to be usually exposed to the levels of PM10 measured during the week of the examination. In the statistical analysis, as specified below, we evaluated the association of PM10 levels with a) DNA methylation measured at the end of the work week (sample 1), which, together with short-term changes between sample 1 and sample 2, were taken as measures of short term effects; b) DNA methylation measured at the beginning (sample 1) and at the end of the work week (sample 2), which were analyzed jointly in repeated measure analysis and taken as a measure of long-term effects.
DNA methylation analysis
We used EDTA tubes to collect 7 mL whole blood that was promptly centrifuged on site at 2,500 rpm for 15 min. The buffy coat (400 μL) was transferred in a cryovial, immediately frozen in vapor phase of liquid nitrogen, and shipped in nitrogen dry shippers to the laboratory. DNA was extracted using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) following the manufacturer’s instructions.
We performed DNA methylation analyses on bisulfite-treated DNA using highly quantitative analysis based on PCR pyrosequencing; 1 μg DNA (concentration 50 ng/μL) was treated using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. Final elution was performed with 30 μL M-Elution Buffer.
To estimate global DNA methylation content, we performed DNA methylation analyses of Alu and LINE-1 repeated sequences, which allow for the amplification of a representative pool of repetitive elements, as previously described (Bollati et al. 2007). Measures of Alu and LINE-1 methylation have been shown to be highly correlated with 5-methylcytosine content measured through high performance liquid chromatography and are commonly used as a surrogate of global methylation (Weisenberger et al. 2005; Yang et al. 2004).
We developed the assay for iNOS methylation by locating the iNOS promoter using the Genomatix Software (Genomatix Software Inc, Ann Arbor, MI, USA) on chromosome 17 (start = 23149861, end = 23150461), and amplified the sequence between 23149872 and 23149990. A 50-μL PCR was carried out in 25 μL GoTaq Green Master mix (Promega), 10 pmol forward primer, 10 pmol reverse primer, 50 ng bisulfite-treated genomic DNA, and water. PCR cycling conditions were 95°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec for 40 cycles. PCR products were purified and sequenced by pyrosequencing as previously described (Bollati et al. 2007) using 0.3 μM sequencing primer.
Primers for Alu, LINE-1, and iNOS assay are shown in Table 1. For all assays we used built-in controls to verify bisulfite conversion efficiency. Compared with other common methods of DNA methylation analysis, pyrosequencing-based assays have the advantage of producing individual measures of methylation at more than one CpG dinucleotide, thus reflecting more accurately DNA methylation in the region. In the Alu or LINE-1 assays, we measured the percentage of 5mC (%5mC) at each of three CpG dinucleotide positions that are repeated over the human genome with the sequence of interest.
Table 1.
Primers for DNA methylation analysis.
| Sequence ID | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Sequencing primer (5′ to 3′) | Sequence analyzeda |
|---|---|---|---|---|
| Global methylation analysis | ||||
| Alu | Biotin-TTTTTATTAAAAATATAAAAATT | CCCAAACTAAAATACAATAA | AATAACTAAAATTACAAAC | G/AC/TG/AC/TG/ACCACCA |
| LINE-1 | TTTTGAGTTAGGTGTGGGATATA | Biotin-AAAATCAAAAAATTCCCTTTC | AGTTAGGTGTGGGATATAGT | TTC/TGTGGTGC/TGTC/TG |
| Gene-specific methylation analysis | ||||
| iNOS | AATGAGAGTTGTTGTTGGGAAGTGTTT | Biotin-CCACCAAACCCAACCAAACT | TAAAGGTATTTTTGTTTTAA | C/TGATTTTC/TGGGTTTTTTTTTATTTTG |
Nucleotides at which DNA methylation was measured are underlined
In the iNOS promoter assay, we measured %5mC at each of two individual CpG dinucleotides within a CpG island located in the gene promoter.
The within-sample coefficients of variation were 0.7% for LINE-1, 1.6% for Alu, and 0.7% for iNOS. The between-sample coefficients of variation in this study population were 1.7% for LINE-1, 3.3% for Alu, and 5.3% for iNOS. Every sample was tested two times for each assay to confirm reproducibility. The resulting data were analyzed using mixed models, as described in the statistical analysis section below.
Statistical analysis
In each blood sample, the pyrosequencing-based analysis of DNA methylation produced six values each for Alu or LINE-1 (methylation at three CpG dinucleotide positions replicated in two measurements) and four values for iNOS (methylation at two individual CpG dinucleotide positions replicated in two measurements). Each subject was tested twice [at the beginning of the work week (sample 1) and after 3 days of work (sample 2)]. To account for the data structure, we used mixed effects models, as described below.
Analysis of short-term effects of PM exposure on DNA methylation
We first evaluated differences between sample 1 and sample 2 in two-way crossed random effects models:
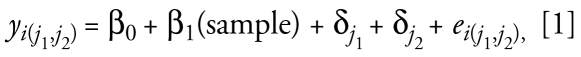 |
where β0 is the overall intercept; β1 is the regression coefficient for the difference between Samples 1 and 2; j1 represents the subject; j2 represents the CpG dinucleotides position; δj1 is the random effect for subject j1; δj2 is the random effect for CpG dinucleotides position; and ei(j1,j2) is the residual error term. Likelihood ratio tests were used to test for the significance of β1.
We then evaluated whether DNA methylation measured after 3 days of work (Sample 2) was associated with the PM10 exposure level estimated during the previous 3 days, using two-way crossed random effects models:
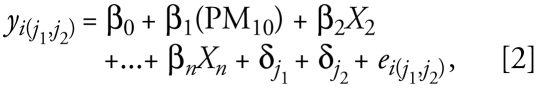 |
where β0 is the overall intercept; β1 is the regression coefficient for PM10 exposure; β2… βn are the regression coefficients for the covariates included in multivariate models; j1 represents the subject; j2 represents the CpG dinucleotides position; δj1 is the random effect for subject j1; δj2 is the random effect for CpG dinucleotides position, and ei(j1,j2) is the residual error term. Covariates for multivariate models included the following potential confounders that were chosen a priori and included in the analysis: age, body mass index, smoking, and number of cigarettes/day. These variables were not significantly associated in univariate analysis with methylation of Alu (β = 0.00, SE = 0.01, p = 0.99 for age; β = 0.01, SE = 0.03, p = 0.83 for BMI; β = 0.11, SE = 0.15, p = 0.46 for smoking; β = −0.01, SE = 0.01, p = 0.73 for cigarettes/day); LINE-1 (β = 0.00, SE = 0.02, p = 0.94 for age; β = −0.06, SE = 0.05, p = 0.19 for BMI; β = −0.15, SE = 0.27, p = 0.57 for smoking; β = −0.02, SE = 0.03, p = 0.41 for cigarettes/day) or iNOS (β = −0.03, SE = 0.06, p = 0.57 for age; β = −0.19, SE = 0.16, p = 0.23 for BMI; β = 0.47, SE = 0.89, p = 0.59 for smoking; β = −0.02, SE = 0.09, p = 0.82 for cigarettes/day).
Analysis of long-term effects of PM exposure on DNA methylation
As noted in the exposure assessment section, PM10 exposure levels estimated during the study also represented a measure of the usual exposure of the study subjects. To estimate long-term effects of PM10 on DNA methylation, we evaluated the level of individual exposure to PM10 in relation to all the measures of DNA methylation performed in the study, regardless of whether they were measured on samples taken on the first day of work (sample 1), or after 3 consecutive days of exposure to PM10 (sample 2), thus assuming that PM10 effects operating over an extended time frame produced similar modifications at the two time points.
For DNA methylation measures that did not show changes in the analysis of short-term effects, we fit two-way error-components models, as described in the formula [2] above. If a significant difference between samples 1 and 2 was found in the analysis of short-term effects, we fit a three-way error-components model, as described in the following notation:
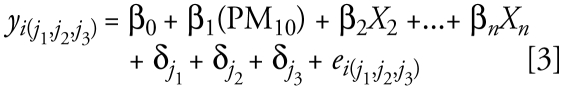 |
where β0 is the overall intercept; β1 represents the mean PM10 effect; β2… βn are the regression coefficients for the covariates included in multivariate models; j1 represents the subject; j2 represents the CpG dinucleotide; and j3 represents the blood sample (sample 1 or 2); δj1 is the random effect for subject j1; δj2 is the random effect for the CpG dinucleotide position j2; and δj3 is the random effect for blood sample j3; ei(j1,j2,j3) is the residual error term.
Covariates for multivariate models included the same variables as in the analysis of short-term effects (age, body mass index, smoking, and number of cigarettes/day).
Results
Distribution of DNA methylation data
DNA methylation showed changes among different blood DNA samples that were relatively small compared with the mean methylation. DNA methylation in Alu repeated elements ranged between 24.3 and 28.9 %5mC, with a mean of 25.8 %5mC (SD = 0.83). DNA methylation in LINE-1 repeated elements ranged between 75.9 and 86.1 %5mC, with a mean of 78.8 %5mC (SD = 1.22). DNA methylation in iNOS ranged between 56.2 and 75.6 %5mC, with a mean of 67.8 %5mC (SD = 3.52).
Short-term effects of PM10 exposure on DNA methylation
Individual PM10 average levels estimated for each subject during the 3 work days between the first and the second DNA methylation measurement (sample 1 and sample 2) ranged between 73.4 and 1220.2 (μg/m3) (average 233.4 μg/m3). As shown in Table 2, DNA methylation of Alu and LINE-1 repeated elements did not show any change after 3 days of work (sample 2) compared with the baseline measurements taken at the beginning of the first day of work (sample 1) (mean difference = 0.00 %5mC, SE = 0.08, p = 0.99 for Alu; mean difference = 0.02 %5mC, SE = 0.11, p = 0.89 for LINE-1). DNA methylation in the iNOS promoter was significantly decreased after 3 days of work compared with the baseline measurement (mean difference = −0.61 %5mC; SE = 0.26, p = 0.02) (Table 2). The average level of individual exposure to PM10 during the 3 days of work showed negative correlations with DNA methylation of Alu, LINE-1, and iNOS measured in sample 2 (Table 3), with associations that were not statistically significant in unadjusted analysis, as well as in models adjusted for age, body mass index, smoking, and number of cigarettes/day.
Table 2.
Change in methylation of Alu, LINE-1, and iNOS, after 3 days of work (sample 2) compared with measures on the first day of work (sample 1).
| Mean DNA methylation
|
Difference in DNA methylation (Sample 2 – Sample 1)
|
|||||
|---|---|---|---|---|---|---|
| No.of subjects | Sample 1 | Sample 2 | Mean | SE | p-Value | |
| Global methylation analysis | ||||||
| Alu (%5mC) | 61 | 25.8 (0.7) | 25.8 (0.6) | 0.00 | 0.08 | 0.99 |
| LINE-1 (%5mC) | 61 | 78.8 (1.0) | 78.8 (1.5) | 0.02 | 0.11 | 0.89 |
| Gene-specific methylation analysis | ||||||
| iNOS (%5mC) | 60 | 68.8 (3.5) | 68.2 (3.7) | −0.61 | 0.26 | 0.02 |
Table 3.
Association of PM10 average exposure with methylation of Alu, LINE-1, and iNOS measured after 3 consecutive work days of exposure.
| Unadjusted regression
|
Adjusted regressiona |
|||||
|---|---|---|---|---|---|---|
| βb | SE | p-Value | βb | SE | p-Value | |
| Global methylation analysis | ||||||
| Alu | −0.18 | 0.10 | 0.08 | −0.18 | 0.10 | 0.071 |
| LINE-1 | −0.25 | 0.25 | 0.31 | −0.28 | 0.25 | 0.26 |
| Gene-specific methylation analysis | ||||||
| iNOS | −0.27 | 0.63 | 0.66 | −0.39 | 0.62 | 0.53 |
Multivariable regression models adjusted for age, body mass index, smoking, number of cigarettes/day.
β for an increment equal to the difference between the 90th and 10th percentile of PM10.
Long-term effects of PM10 exposure on DNA methylation
To identify possible long-term effects of PM10 exposure, we evaluated the level of individual exposure to PM10, taken as a measure of usual exposure to particles, in relation to all the measures of DNA methylation performed in the study, regardless of whether they were measured on samples taken on the first day of work (i.e., after 2 days off, sample 1), or after 3 consecutive days of work (sample 2) (Table 4). In the models, the two samples collected at different times are exchangeable, thus assuming that PM10 effects operating over an extended time frame produced similar modifications at the two time points.
Table 4.
Association of PM10 average level with all measures of DNA methylation in blood samples taken from exposed workers.a
| Unadjusted regression | Adjusted regressionb | |||||
|---|---|---|---|---|---|---|
| βc | SE | p-Value | βc | SE | p-Value | |
| Global methylation analysis | ||||||
| Alu | −0.18 | 0.09 | 0.04 | −0.19 | 0.09 | 0.04 |
| LINE-1 | −0.30 | 0.17 | 0.07 | −0.34 | 0.17 | 0.04 |
| Gene-specific methylation analysis | ||||||
| iNOS | −0.48 | 0.58 | 0.41 | −0.55 | 0.58 | 0.34 |
To estimate long-term effects of PM10, the level of individual exposure to PM10, taken as a measure of usual exposure to particles, was examined in relation to all the measures of DNA methylation performed in the study, regardless of whether they were measured on samples taken on the first day of work (i.e., after 2 days off) or after 3 consecutive days of exposure to PM10 in the plant. In the models, the two samples collected at different times are exchangeable, thus assuming that PM10 effects operating over an extended time frame produced similar modifications at the two time points.
Multi-variable mixed models adjusted for age, body mass index, smoking, number of cigarettes/day.
β for an increment equal to the difference between the 90th and 10th percentile of PM10.
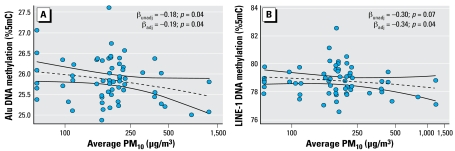
In unadjusted models, the average PM10 levels were significantly associated with decreased Alu methylation (β = −0.18, SE = 0.09; p = 0.04). A negative, nonsignificant association was also observed for LINE-1 methylation (β = −0.30, SE = 0.17, p = 0.07). In multivariable regression analysis adjusting for age, body mass index, smoking, and number of cigarettes, the average PM10 levels were significantly and negatively associated with both Alu (β = −0.19, SE = 0.09, p = 0.04) and LINE-1 (β = −0.34, SE = 0.17, p = 0.04) methylation. Scatter plots representing the association of average PM10 level with Alu and LINE-1 methylation are shown in Figure 1.
Figure 1.
Long-term effects of PM10 on blood DNA methylation in Alu (A) and LINE-1 (B) repeated elements. PM10 concentrations, taken as a measure of usual exposure to particles, was examined in relation to all the measures of DNA methylation performed in the study, regardless of whether they were measured on samples taken on the first day of work (i.e., after 2 days off) or after 3 consecutive days of exposure to PM10 in the plant. Abbreviations: adj, adjusted; nonadj, nonadjusted. Data points represent the average of DNA.
iNOS methylation showed no association with average PM10 level in both non-adjusted (β = −0.48, SE = 0.58, p = 0.41) and multivariable analyses (β = −0.55, SE = 0.58, p= 0.34) (Table 4).
In addition, we also evaluated the association of PM10 level with DNA methylation measured on blood DNA collected on the first day of the work week (Sample 2), as long-term effects of the exposure would likely be reflected also on samples collected after 2 days off work. The associations found between average PM10 levels and DNA methylation measured at the beginning of the first week were in the same directions as those in the primary analysis reported in Table 4 both in the unadjusted (β = −0.18 SE = 0.12, p = 0.14 for Alu; β = −0.34, SE = 0.16, p = 0.04 for LINE-1; β = −0.66, SE = 0.60 p = 0.24 for iNOS) and multivariable analysis (β = −0.19, SE = 0.12, p = 0.11 for Alu; β = −0.39, SE = 0.15, p = 0.01 for LINE-1; β = −0.67, SE = 0.59, p = 0.25 for iNOS).
Discussion
In the present study of workers in an electric furnace steel plant with well-characterized measures of exposure to a wide range of PM10 levels, global DNA methylation estimated in Alu and LINE-1 repeated elements were negatively associated with individual PM10 exposure, without changes related to short-term exposure during the week of the study. We observed short-term changes in iNOS promoter methylation, which decreased after 3 consecutive days of work in the plant.
Decreases in global DNA methylation content have been associated with widespread alterations in gene expression and chromatin packaging control, as well as with higher genomic instability (Dean et al. 2005). The decrease we observed in our study in association with PM10 exposure may represent an initial step reproducing decreases in global DNA methylation content that are eventually observed in cardiovascular disease and cancer (Baccarelli et al. 2007; Robertson 2005). In our study, the association between PM10 level and decreased methylation in Alu and LINE-1 was significant only when the two measurements of methylation taken before and after 3 consecutive work days, which showed no differences in Alu and LINE-1 methylation, were both included in repeated-measure models. In our analyses, the use in the same models of both methylation measures taken at the beginning and at the end of the work week was meant to evaluate long-term effects of PM10, which would have similar effects on measures of DNA methylation taken at the two different time points, and also provided our statistical analysis with added power to detect the PM10 effects. These results suggest that PM10 operated on genomic DNA methylation content over an extended time frame, possibly causing a persistent suppression of methylation levels that were not reset to baseline over the 2 days off between consecutive work weeks.
Air particle exposure has been shown to cause increased iNOS expression in animal models (Folkmann et al. 2007; Ulrich et al. 2002). In our study, iNOS methylation, which has been previously shown to keep iNOS expression suppressed (Chan et al. 2005), was significantly decreased after 3 days of work, compared with measures taken before the first day of work of the same week. However, we did not find any association with levels of PM10 exposure; thus, whether iNOS promoter methylation is modified by short-term exposure to PM remains uncertain. In vitro studies have shown that methylation of individual genes undergoes rapid changes in response to environmental factors (Bruniquel and Schwartz 2003; Takiguchi et al. 2003), and iNOS expression has been found to respond rapidly to different stimuli, including immunostimulatory cytokines, bacterial products, or infection (Alderton et al. 2001). Increased iNOS expression has been found in disease conditions that have also been associated with PM exposure, such as cardiovascular disease and lung cancer (Comini et al. 1999; Liu et al. 1998). Whether iNOS expression is increased after PM exposure due to iNOS promoter demethylation should be clarified in subjects exposed acutely to particles after an extended washout period, as well as in larger populations of exposed subjects. Future investigations should also aim at clarifying whether changes in iNOS promoter methylation modify its expression, as iNOS expression was not measured in this study.
Although several other genes might have been included in our study, we selected iNOS for DNA methylation analysis because the increase of expression after exposure to PM or PM components has been well substantiated in previous studies conducted on several tissues (Anazawa et al. 2004; Blackford et al. 1997; Castranova 2004; Folkmann et al. 2007; Porter et al. 2006) including blood leukocytes (Blackford et al. 1994), which were the source of DNA for our study. Further research is warranted to evaluate PM-related changes in DNA methylation of iNOS-related genes, as well as in other independent pathways.
Our study was based on quantitative analysis of DNA methylation using pyrosequencing, which is highly reproducible and accurate at measuring small changes in DNA methylation (Bollati et al. 2007; Yang et al. 2004). DNA methylation analysis measured multiple individual CpG dinucleotide positions for each marker and was repeated twice on each sample to minimize the assay variability. We used multilevel mixed models to fully represent the structure of the data and take advantage of the multiple measurements while also adjusting for potential confounders. Our data comprised each of the methylation markers investigated (Alu, LINE-1, and iNOS) of a matrix of six or four measures (3 × 2 or 2 × 2), including data from three (for Alu and LINE-1) or two (for iNOS) CpG dinucleotides and from two replicates. Commonly used statistical methods for the analysis of such data would include linear regression and analysis of variance, using as the outcome the mean computed from the multiple CpG dinucleotides and replicates from each sample. However, because the data in the methylation matrix obtained on an individual sample were not independent, the use of standard methods would not adequately represent the correlation existing within the matrix. We therefore elected to use multilevel mixed models that allowed us to fully utilize the information from all the measurements in our data and maximize statistical power by distinguishing between the different sources of variance in the data.
We investigated a population with well-characterized PM10 exposure that allowed for contrasting subjects over a wide range of different exposure levels. Our study was based on subjects working in several work areas of the same factory but did not include a different population of subjects without a specific condition of exposure to PM. However, the lowest level of PM10 observed in our study (74 μg/m3) was relatively low, particularly if compared with the highest level found in our population (1,220 μg/m3), and only marginally higher than ambient PM10 levels measured in the geographic area in which the plant is located [average annual ambient PM10 levels between 41 and 57 μg/m3 were recorded in the year of the study by different ambient monitoring stations in the Brescia area (Anselmi and Patelli 2006)]. In addition, limiting our investigation to individuals who have all been working in the same work facility avoided potential concerns related to the selection of external referents who might have differed from the exposed population in terms of socioeconomic factors and other characteristics determining hiring into the plant (Pearce et al. 2007).
In addition to PM, workers in foundries may have additional exposures, including heat, polycyclic aromatic hydrocarbons (Mirer 1998; Sorahan et al. 1994), carbon monoxide (Lewis et al. 1992; Park 2001), and non-ionizing radiations (Gomes et al. 2002). Although study subjects in our study were in a modern facility with state-of-the-art systems for exposure reduction, we cannot exclude that exposures other than PM might have contributed to the observed effects.
Our results showed alterations in blood DNA methylation in a population of foundry workers, including changes in global methylation estimated in Alu and LINE-1 repetitive elements and gene-specific methylation of the iNOS promoter. Further studies are required to determine the role of such alterations in mediating the effects of particles on human health.
Footnotes
This work was supported by research grants from the CARIPLO Foundation (2007-5469) and the National Institute of Environmental Health Sciences (ES015172-01) and by additional funding from CARIPLO Foundation and Lombardy Region Research Contracts UniMi 8614/2006 and UniMi 9167/2007.
References
- Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anazawa T, Dimayuga PC, Li H, Tani S, Bradfield J, Chyu KY, et al. Effect of exposure to cigarette smoke on carotid artery intimal thickening: the role of inducible NO synthase. Arterioscler Thromb Vasc Biol. 2004;24:1652–1658. doi: 10.1161/01.ATV.0000139925.84444.ad. [DOI] [PubMed] [Google Scholar]
- Andjelkovich DA, Mathew RM, Richardson RB, Levine RJ. Mortality of iron foundry workers. I. Overall findings. J Occup Med. 1990;32:529–540. doi: 10.1097/00043764-199006000-00010. [DOI] [PubMed] [Google Scholar]
- Anselmi U, Patelli R. Rapporto sulla qualità dell’aria di Brescia e provincia [in Italian] Milan: ARPA Lombardia; 2006. [Google Scholar]
- Baccarelli A, Cassano PA, Litonjua A, Park SK, Suh H, Sparrow D, et al. Cardiac autonomic dysfunction: effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation. 2008;117:1802–1809. doi: 10.1161/CIRCULATIONAHA.107.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Zanobetti A, Martinelli I, Grillo P, Hou L, Lanzani G, et al. Air pollution, smoking, and plasma homocysteine. Environ Health Perspect. 2007;115:176–181. doi: 10.1289/ehp.9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky SA, Snow SS, Nikula KJ, Finch GL, Tellez CS, Palmisano WA. Aberrant CpG island methylation of the p16(INK4a) and estrogen receptor genes in rat lung tumors induced by particulate carcinogens. Carcinogenesis. 2002;23:335–339. doi: 10.1093/carcin/23.2.335. [DOI] [PubMed] [Google Scholar]
- Bergamaschi E, Catalani S, Folesani G, Venco P, Bodini E, Guidetti F, et al. Environmental and biological monitoring of exposure to polycyclic aromatic hydrocarbons in workers of an electric steel foundry [in Italian] Med Lav. 2005;96:390–402. [PubMed] [Google Scholar]
- Blackford JA, Jr, Antonini JM, Castranova V, Dey RD. Intratracheal instillation of silica up-regulates inducible nitric oxide synthase gene expression and increases nitric oxide production in alveolar macrophages and neutrophils. Am J Respir Cell Mol Biol. 1994;11:426–431. doi: 10.1165/ajrcmb.11.4.7522485. [DOI] [PubMed] [Google Scholar]
- Blackford JA, Jr, Jones W, Dey RD, Castranova V. Comparison of inducible nitric oxide synthase gene expression and lung inflammation following intratracheal instillation of silica, coal, carbonyl iron, or titanium dioxide in rats. J Toxicol Environ Health. 1997;51:203–218. doi: 10.1080/00984109708984022. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Borm PJ, Kelly F, Kunzli N, Schins RP, Donaldson K. Oxidant generation by particulate matter: from biologically effective dose to a promising, novel metric. Occup Environ Med. 2007;64:73–74. doi: 10.1136/oem.2006.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Franklin B, Cascio W, Hong Y, Howard G, Lipsett M, et al. Air pollution and cardiovascular disease: a statement for healthcare professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation. 2004;109:2655–2671. doi: 10.1161/01.CIR.0000128587.30041.C8. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nature Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Castranova V. Role of nitric oxide in the progression of pneumoconiosis. Biochemistry. 2004;69:32–37. doi: 10.1023/b:biry.0000016348.34175.53. [DOI] [PubMed] [Google Scholar]
- Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- Chang CC, Hwang JS, Chan CC, Wang PY, Hu TH, Cheng TJ. Effects of concentrated ambient particles on heart rate variability in spontaneously hypertensive rats. J Occup Health. 2005;47:471–480. doi: 10.1539/joh.47.471. [DOI] [PubMed] [Google Scholar]
- Chen LC, Hwang JS. Effects of subchronic exposures to concentrated ambient particles (CAPs) in mice. IV. Characterization of acute and chronic effects of ambient air fine particulate matter exposures on heart-rate variability. Inhal Toxicol. 2005;17:209–216. doi: 10.1080/08958370590912789. [DOI] [PubMed] [Google Scholar]
- Chyu KY, Dimayuga P, Zhu J, Nilsson J, Kaul S, Shah PK, et al. Decreased neointimal thickening after arterial wall injury in inducible nitric oxide synthase knockout mice. Circ Res. 1999;85:1192–1198. doi: 10.1161/01.res.85.12.1192. [DOI] [PubMed] [Google Scholar]
- Comini L, Bachetti T, Agnoletti L, Gaia G, Curello S, Milanesi B, et al. Induction of functional inducible nitric oxide synthase in monocytes of patients with congestive heart failure. Link with tumour necrosis factor-alpha. Eur Heart J. 1999;20:1503–1513. doi: 10.1053/euhj.1999.1580. [DOI] [PubMed] [Google Scholar]
- Corey LM, Baker C, Luchtel DL. Heart-rate variability in the apolipoprotein E knockout transgenic mouse following exposure to Seattle particulate matter. J Toxicol Environ Health. 2006;69:953–965. doi: 10.1080/15287390500362105. [DOI] [PubMed] [Google Scholar]
- Dean W, Lucifero D, Santos F. DNA methylation in mammalian development and disease. Birth Defects Res C Embryo Today. 2005;75:98–111. doi: 10.1002/bdrc.20037. [DOI] [PubMed] [Google Scholar]
- Folkmann JK, Risom L, Hansen CS, Loft S, Moller P. Oxidatively damaged DNA and inflammation in the liver of dyslipidemic ApoE−/− mice exposed to diesel exhaust particles. Toxicology. 2007;237:134–144. doi: 10.1016/j.tox.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. J Nutr. 2002;132:2382S–2387S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methyl-enetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J, Lloyd O, Norman N. The health of the workers in a rapidly developing country: effects of occupational exposure to noise and heat. Occup Med (Oxford, England) 2002;52:121–128. doi: 10.1093/occmed/52.3.121. [DOI] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer) Overall Evaluations of Carcinogenicity: An Updating of IARC Volumes 1–42. IARC Monogr Eval Carcinog Risks Hum Suppl. 1987;7 [PubMed] [Google Scholar]
- Kuo HW, Chang CL, Liang WM, Chung BC. Respiratory abnormalities among male foundry workers in central Taiwan. Occup Med (Oxford, England) 1999;49:499–505. doi: 10.1093/occmed/49.8.499. [DOI] [PubMed] [Google Scholar]
- Lewis S, Mason C, Srna J. Carbon monoxide exposure in blast furnace workers. Aust J Public Health. 1992;16:262–268. doi: 10.1111/j.1753-6405.1992.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Liu CY, Wang CH, Chen TC, Lin HC, Yu CT, Kuo HP. Increased level of exhaled nitric oxide and up-regulation of inducible nitric oxide synthase in patients with primary lung cancer. Br J Cancer. 1998;78:534–541. doi: 10.1038/bjc.1998.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirer FE. Foundries. In: Stellman JM, editor. Encyclopedia of Occupational Health and Safety. Geneva: International Labor Office; 1998. pp. 82.81–82.56. [Google Scholar]
- Park RM. Mortality at an automotive engine foundry and machining complex. J Occup Environ Med. 2001;43:483–493. doi: 10.1097/00043764-200105000-00009. [DOI] [PubMed] [Google Scholar]
- Pearce N, Checkoway H, Kriebel D. Bias in occupational epidemiology studies. Occup Environ Med. 2007;64:562–568. doi: 10.1136/oem.2006.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. Particulate matter and heart disease: evidence from epidemiological studies. Toxicol Appl Pharmacol. 2005;207(suppl):477–482. doi: 10.1016/j.taap.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Porter DW, Millecchia LL, Willard P, Robinson VA, Ramsey D, McLaurin J, et al. Nitric oxide and reactive oxygen species production causes progressive damage in rats after cessation of silica inhalation. Toxicol Sci. 2006;90:188–197. doi: 10.1093/toxsci/kfj075. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. Fine particulate air pollution and mortality in 20 U.S. cities, 1987–1994. N Engl J Med. 2000;343:1742–1749. doi: 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- Schulz H, Harder V, Ibald-Mulli A, Khandoga A, Koenig W, Krombach F, et al. Cardiovascular effects of fine and ultrafine particles. J Aerosol Med. 2005;18:1–22. doi: 10.1089/jam.2005.18.1. [DOI] [PubMed] [Google Scholar]
- Sorahan T, Faux AM, Cooke MA. Mortality among a cohort of United Kingdom steel foundry workers with special reference to cancers of the stomach and lung, 1946–90. Occup Environ Med. 1994;51:316–322. doi: 10.1136/oem.51.5.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286:355–365. doi: 10.1016/s0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
- Thomson EM, Kumarathasan P, Calderon-Garciduenas L, Vincent R. Air pollution alters brain and pituitary endothelin-1 and inducible nitric oxide synthase gene expression. Environ Res. 2007;105:224–233. doi: 10.1016/j.envres.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Ulrich MM, Alink GM, Kumarathasan P, Vincent R, Boere AJ, Cassee FR. Health effects and time course of particulate matter on the cardiopulmonary system in rats with lung inflammation. J Toxicol Environ Health. 2002;65:1571–1595. doi: 10.1080/00984100290071676. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Husgafvel-Pursiainen K. Air pollution and cancer: biomarker studies in human populations. Carcinogenesis. 2005;26:1846–1855. doi: 10.1093/carcin/bgi216. [DOI] [PubMed] [Google Scholar]
- Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, et al. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113:233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JL, Dai J, Zay K, Price K, Gilks CB, Churg A. Effects of cigarette smoke on nitric oxide synthase expression in the rat lung. Lab Invest. 1999;7:975–983. [PubMed] [Google Scholar]
- Xu Z, Brown LM, Pan GW, Liu TF, Gao GS, Stone BJ, et al. Cancer risks among iron and steel workers in Anshan, China, Part II: Case-control studies of lung and stomach cancer. Am J Ind Med. 1996;30:7–15. doi: 10.1002/(SICI)1097-0274(199607)30:1<7::AID-AJIM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen J, Lu H, Wang XQ, Vaziri ND, Zhou XJ. Upregulation of endothelial and inducible nitric oxide synthase expression by reactive oxygen species. Am J Hypertens. 2008;21:28–34. doi: 10.1038/ajh.2007.14. [DOI] [PubMed] [Google Scholar]