Abstract
Background
Although microRNAs (miRNAs) have been found to play an important role in many biological and metabolic processes, their functions in animal response to environmental toxicant exposure are largely unknown.
Objectives
We used hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a common environmental contaminant, as a toxicant stressor to investigate toxicant-induced changes in miRNA expression in B6C3F1 mice and the potential mechanism of RDX-induced toxic action.
Methods
B6C3F1 mice were fed diets with or without 5 mg/kg RDX for 28 days. After the feeding trials, we isolated RNAs from both brain and liver tissues and analyzed the expression profiles of 567 known mouse miRNAs using microarray and quantitative real-time polymerase chain reaction technologies.
Results
RDX exposure induced significant changes in miRNA expression profiles. A total of 113 miRNAs, belonging to 75 families, showed significantly altered expression patterns after RDX exposure. Of the 113 miRNAs, 10 were significantly up-regulated and 3 were significantly down-regulated (p < 0.01) in both mouse brain and liver. Many miRNAs had tissue-specific responses to RDX exposure. Specifically, expression of seven miRNAs was up-regulated in the brain but down-regulated in the liver or up-regulated in the liver but down-regulated in the brain (p < 0.01). Many aberrantly expressed miRNAs were related to various cancers, toxicant-metabolizing enzymes, and neurotoxicity. We found a significant up-regulation of oncogenic miRNAs and a significant down-regulation of tumor-suppressing miRNAs, which included let-7, miR-17-92, miR-10b, miR-15, miR-16, miR-26, and miR-181.
Conclusions
Environmental toxicant exposure alters the expression of a suite of miRNAs.
Keywords: carcinogenesis, gene regulation, microarray, microRNA, miRNA, qRT-PCR, RDX, toxicant, toxicity, toxicogenomics
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX, also known as hexogen or cyclonite) is a common environmental pollutant resulting from military and civil activities. According to the U.S. Department of Defense, an estimated 12,000 sites across the United States have been contaminated with explosives, including RDX. Concentrations of RDX in soils exceed thousands of milligrams per kilogram (Jenkins et al. 2006). RDX and its metabolites were also identified in water sources, including groundwater (Beller and Tiemeier 2002).
Environmental contamination by RDX and its N-nitroso metabolites has raised health concerns about human and environmental exposure (Zhang et al. 2008). It has long been known that RDX exposure causes neurotoxicity, immunotoxicity, and an increased likelihood of cancers. A causal relationship has been established between seizures and acute occupational RDX exposure in humans, with results confirmed in laboratory animals (Burdette et al. 1988; Pan et al. 2007; Testud et al. 1996). One early study demonstrated that RDX exposure elevated the incidence of tumors in B6C3F1 mice (Lish et al. 1984). Based on these findings, RDX has been classified as a class C potential human carcinogen by the U.S. Environmental Protection Agency (U.S. EPA 1988). Despite evidence supporting the role of RDX as a cytotoxic agent and potential chemical carcinogen, the molecular mechanism of RDX-induced neurotoxicity and potential carcinogenesis remains unknown.
Recently identified microRNAs (miRNAs) may play an important role in RDX exposure and in the process of RDX-induced tumorigenesis and neurotoxicity. miRNAs are a group of small non-protein-coding endogenous RNAs that posttranscriptionally regulate the expression of > 30% of human protein-coding genes (Lewis et al. 2005). miRNAs negatively regulate gene expression through translation inhibition, mRNA cleavage, or deadenylation/decap-mediated mRNA decay (Giraldez et al. 2006; Wu et al. 2006). These mechanisms are likely governed by the degree of complementarity between mRNAs and their targeting miRNAs (Zhang et al. 2007b). In animals, most miRNAs bind to their target mRNAs imperfectly for repressing protein translation at multiple complementary sites within the 3k untranslated regions (UTRs) (Ambros 2001), with a few exemptions at the open reading frames or 5k UTRs (Lytle et al. 2007). Many miRNAs are evolutionarily conserved in animals, from worms to humans (Pasquinelli et al. 2000), suggesting that miRNA functions are conserved from species to species.
miRNAs play an important role in almost all fundamental biological and metabolic processes in eukaryotic organisms (Zhang et al. 2007b). Many recent studies have demonstrated that miRNAs regulate cancer development, including cancer invasiveness and metastasis. Most commonly occurring cancers are associated with the aberrant expression of at least one miRNA (Zhang et al. 2007a). Moreover, it has been established that specific cancers have their unique miRNA expression profiles (Lu et al. 2005), suggesting that miRNAs respond to different cancers in different ways. Thus, miRNA profiles are useful for classifying and identifying cancers. Changes in miRNA expression can regulate the cancer development cascade. A recent in vitro study showed that transient overexpression of the miRNA let-7 in A549 lung adenocarcinoma cell lines inhibited lung cancer cell proliferation (Takamizawa et al. 2004). Another study found that changes in the expression levels of a single miRNA (miR-10b) could initiate tumor invasion and metastasis (Ma et al. 2007).
It is well known that many chemical toxicants and biological toxins cause different types of cancers. Although various environmental carcinogens cause DNA and chromosome damage as well as the aberrant expressions of cancer-related genes and toxicant-metabolizing enzymes, the pathogenic mechanisms of toxicant/toxin-induced cancers are unclear (de Kok et al. 2005; Myllynen et al. 2007). Because miRNAs play important roles in cancer development, and the aberrant expression of miRNAs has been observed in different cancers, we hypothesized that the exposure to a specific environmental procarcinogen, such as RDX, would induce alterations in miRNA expression, and that the altered miRNA expression contributes to carcinogenesis. To test this hypothesis, we exposed B6C3F1 mice to RDX and investigated the effect of RDX exposure on the global expression profile of miRNAs, particularly on the oncogenic, tumor-suppressing, and disease-related miRNAs. We assayed RDX-induced changes in miRNAs using microarray and quantitative real-time polymerase chain reaction (qRT-PCR) technologies. Given that miRNAs are highly conserved between mice and humans, the results would help us better understand the molecular mechanisms of RDX-related diseases, including potential carcinogenic and neurologic damages.
Materials and Methods
Animal treatment
We purchased female, virgin, B6C3F1 mice (Mus muscaris), at 9–11 weeks of age, from Charles River Laboratories, Inc. (Wilmington, MA). Mice were acclimated to the laboratory environment for 5 days before administering dietary RDX. Twenty-one mice were randomly assigned to treatment or control groups. Each cage housed three mice, and all cages were located in an animal room with temperature ranging from 68°F to 72°F and 25–75% relative humidity with 16/8-hr light/dark cycle. RDX-spiked food was used to feed mice and tap water was provided ad libitum, and their behavior and the weight of food and drinking water were carefully monitored twice per day. Animal use and handling protocols complied with Texas Tech University Animal Use and Care Committee guidelines. We treated all mice humanely and with regard for alleviation of suffering.
The mice in treatment groups were fed using finely ground Purina Certified Rodent Chow No. 5002 (Purina Mills, St. Louis, MO) containing 5 mg/kg RDX. RDX was dissolved in acetone and then sprayed onto the rodent chow to produce a dosage of 5 mg/kg. For the control group, we sprayed the same amount of acetone used in the RDX food preparation onto the rodent chow as a vehicle control. We thoroughly mixed the RDX-treated mouse food for at least 30 min after spraying with the acetone-solubilized solution and then spread it in a well-ventilated area to allow the acetone to evaporate for 4 days before use. The actual amount of RDX in the rodent chow was measured using pressurized liquid extraction followed by gas chromatography with electron-capture detection (Pan et al. 2005).
Mouse euthanasia and organ collection
After 28 days of exposure, we euthanized both the RDX-fed and control mice by CO2 asphyxiation. The brain and liver tissues were immediately removed, weighed, and transferred into a 2-mL microcentrifuge tube and stored in liquid nitrogen during necropsy. We then transferred the frozen tissue samples into a −80°C freezer until miRNA analysis.
RNA isolation
We extracted total RNA from each sample (brain and liver) using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer’s instructions. Briefly, 0.03–0.05 g tissues were weighed and placed in a new 2-mL micro-centrifuge tube, followed by adding 300 μL lysis/binding buffer. Then, the tissues were thoroughly disrupted and homogenized using a Sonic Dismembrator (model 100, Fisher Scientific, Atlanta, GA). After homogenization, we added 30 μL miRNA homogenate additive to each tissue lysate, vortexed it for 10 sec, and then incubated it on ice for 10 min. After washing, we eluted the total RNAs using 100 μL elution buffer provided in the miRNA isolation kit. We performed all these operations on ice. The extracted RNA was quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE), aliquoted, and immediately stored at −80°C until analysis.
miRNA microarray
The miRNA micro-array analysis was performed by LC Sciences (Houston, TX). Briefly, the assay started with approximately 6 μg total RNA. After the total RNAs were fractionated by size using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA), poly(A) tails were added to RNA sequences with lengths less than 300 nucleotides using poly(A) polymerase. Then, an oligonucleotide tag was ligated to the poly(A) tail for later fluorescent dye staining. RNA samples from liver and brain extracts were hybridized overnight using two different tags on a μParaflo microfluidic chip using a microcirculation pump developed by Atactic Technologies (Houston, TX) (Gao et al. 2004). One of the major advantages of the μParaflo microfluidic technology is that it allows efficient parallel synthesis of a large number of different oligonucleotide molecules (Sun et al. 2008). Each micro-fluidic chip contained the following probes: a) detection probes, which consisted of all chemically modified nucleotide sequences complementary to all 567 mouse miRNAs listed in the Sanger miRNA miRBase database (Release 10.1: December 2007; http://www.microrna.sanger.ac.uk/sequences/); b) a total of 49 positive and negative control probes designed by LC Sciences to ensure uniformity of sample labeling and assay conditions; and c) a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. LC Sciences made the probes in situ using the photogenerated reagent chemistry. The melting temperatures of hybridization were balanced by chemical modifications of the probes, and hybridization reactions were performed in 100 μL 6× SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide at 34°C. After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated to control and treatment samples, respectively, for dye staining. Each analyzed miRNA was repeated six times and the controls were repeated for 4–16 times. A GenePix 4000B (Molecular Devices, Union City, CA) laser scanner was used to collect the fluorescence images, which were digitized using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD).
miRNA microarray data analysis
We analyzed miRNA microarray data by subtracting the background and then normalizing the signals using a LOWESS filter (locally weighted regression), as described in a previous report (Bolstad et al. 2003). We listed a miRNA as a detectable miRNA when it met at least three criteria: a) signal intensity greater than three times background standard deviation; b) spot coefficient of variance (CV) < 0.5, in which we calculated CV as (standard deviation)/(signal intensity); and c) at least three of the six repeats have signal greater than three times background standard deviation. The ratio of the two sets (control and treatment) of detected signals (log2 transformed, balanced) and p-values of the t-test were calculated. Differentially detected signals were those with p-values less than 0.01.
The detailed method for microarray data analysis and normalization is described in the Supplemental Material (http://www.ehponline.org/members/2008/11841/suppl.pdf (Bolstad et al. 2003).
RT-PCR and qRT-PCR
We selected miRNAs with aberrant expression in micro-array analysis and then validated them using qRT-PCR on an ABI7300 system (Applied Biosystems, Foster City, CA). We used TaqMan miRNA assays to detect and quantify mouse miRNAs using stem-loop RT-PCR according to the manufacturer’s instructions. We provide a detailed description of the method in the Supplemental Material (http://www.ehponline.org/members/2008/11841/suppl.pdf).
Prediction of miRNA targets
We used two different computational programs, TargetScan (Lewis et al. 2003) and PicTar (Krek et al. 2005), to predict miRNA targets. Each predicted gene was assigned a score from 1 to n according to the gene’s original score generated by each computational program. Then, the two scores from both programs were added up to give a total score for each predicted gene. The lower the total score a predicted gene has, the more likely it is the real target gene of a specific miRNA. We identified the 50 most promising candidate genes based on the additive scores. For details, see the Supplemental Material (http://www.ehponline.org/members/2008/11841/suppl.pdf).
Results
miRNA expression in mouse brain and liver tissues
We observed differentially expressed patterns of miRNAs in mouse brain and liver. In this study, 369 miRNAs were detected in mouse brain and/or liver out of the 567 currently known mouse miRNAs (Figure 1). Of the 369 detected miRNAs, 284 were detected in the liver and 326 in the brain. Among detected miRNAs, 20 miRNAs were highly expressed in the liver, and 53 miRNAs were highly expressed in the brain [see Supplemental Material, Tables 1–3 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. Fifteen miRNAs were highly expressed in both liver and brain: miR-709, let-7a, let-7f, let-7c, let-7d, miR-26a, let-7b, let-7g, miR-26b, miR-29a, miR-126-3p, miR-23b, miR-30c, miR-16, and miR-23a.
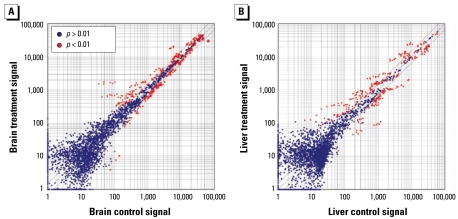
Figure 1.
miRNA microarray signal distribution between control samples and RDX-exposed samples in mouse brain tissues (A) and liver tissues (B).
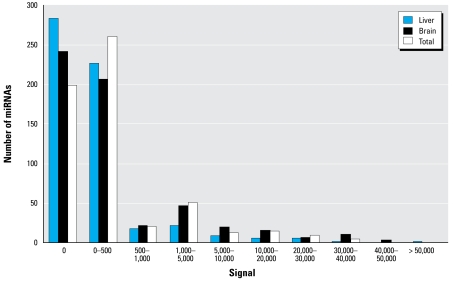
The expression levels were low for most of the detected miRNAs. Of the 369 detected miRNAs, 269 gave signals lower than 500 (Figures 1 and 2). However, 89 miRNAs were expressed with signals greater than 1,000; some of them with signals greater than 40,000. This suggests that the expression levels of miRNAs in mice vary greatly, with some produced at only a few copies per cell and others present in hundreds of copies. This observation is similar to that of a previous report (Lagos-Quintana et al. 2002).
Figure 2.
Signal distribution of all analyzed miRNAs in mouse liver and brain tissues by microarray assay.
miRNA expression patterns significantly differ between mouse liver and brain. Some miRNAs were highly expressed in the brain but not in the liver, and vice versa [see Supplemental Material, Tables 1–3 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. In this study, 85 miRNAs were expressed in the brain only. Some of these miRNAs, for example, miR-9, miR-9*, miR-218, miR-204, and miR-129-3p, are highly expressed in the brain with signals higher than 40,000. However, we did not detect these miRNAs in the liver, suggesting that these miRNAs are specific to brain relative to liver. Except for the brain-specific miRNAs, the expression levels of 68 miRNAs in the brain were at least 10-fold higher than those in the liver. On the contrary, 43 miRNAs were expressed in the liver but not in the brain, suggesting that these miRNAs are liver specific compared with brain. In addition to the 43 liver-specific miRNAs, five miRNAs (miR-148a, miR-192, miR-194, miR-122, and miR-21) were highly expressed in the liver but at low levels in the brain; four of them (miR-148a, miR-192, miR-194, miR-122) were expressed at levels at least 10-fold greater in the liver than in the brain. miR-10a and the miR-689 were also expressed in the liver at levels 10-fold higher than those in the brain[see Supplemental Material, Table 2 (http://www.ehponline.org/members/2008/11841/suppl.pdf)].
RDX exposure altered miRNA expression profiles
A comparison of miRNA expression levels between control and RDX-treated mice revealed that RDX exposure significantly altered miRNA expression profiles [Figure 3; see Supplemental Material, Table 1–3 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. RDX exposure resulted in the expression of a greater number of miRNAs in brain tissues from RDX-treated mice compared with controls. By comparison, the total number of miRNAs expressed in liver tissues from RDX-treated mice was lower than that in controls [see Supplemental Material, Table 1 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. In brain tissues, we detected 58 miRNAs in treated mice but not in controls, whereas we detected only four miRNAs (miR-10a, miR-10b, miR-712*, and miR-715) in control brain tissues but not in treated samples [see Supplemental Material, Table 3 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. Although many miRNAs were aberrantly expressed after RDX exposure, almost all detected miRNAs, either in control or in treated mice but not in both, gave low microarray signals, suggesting these miRNAs were expressed at low levels. Interestingly, of the total 191 miRNAs detected in one sample (either control or treatment) but not in both, one-third were miRNA* sequences. Usually, miRNA* sequences were quickly degraded during the final stage of miRNA biogenesis (Schwarz et al. 2003). What caused these miRNA* sequences to become detectable after exposure to RDX is unclear.
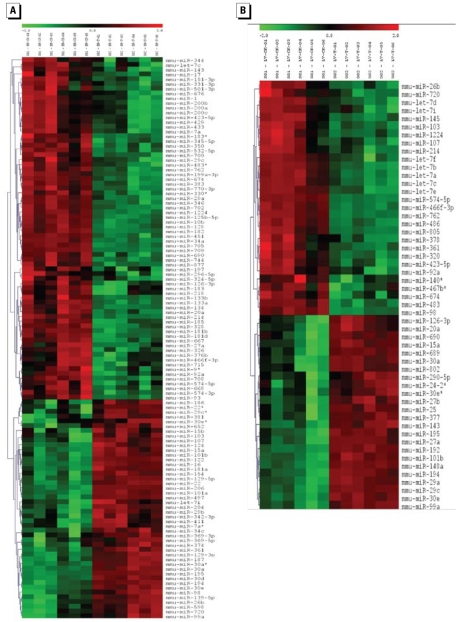
Figure 3.
Hierarchical clustering of the differentially expressed miRNAs in control and RDX-treated mouse brain (A) and liver (B) samples after exposure to 5 mg/kg RDX for 28 days. Each row represents one miRNA with significantly differential expressions between control and treatment (p < 0.01). Each column represents a biological replicates; in each panel, the left six columns are for controls and the right six for RDX treatments. Colors represent expression levels of each individual miRNA: red, up-regulation; green, down-regulation.
RDX exposure affected not only the total number of detectable miRNAs in mouse brain and liver, but also the expression levels of miRNAs (p < 0.01; Figure 3, Tables 1 and 2). The expression levels of 84 miRNAs were significantly altered in mouse brain after RDX exposure (Table 1, Figures 3, 4A). Of the 84 miRNAs, 38 were up-regulated and 46 were down-regulated. However, the extent of changes varied among miRNAs. For example, the expression levels of miR-206 and miR-497 were increased significantly by 26- and 9-fold, respectively, whereas the expression of miR-10b was decreased significantly by 14-fold (Table 1). Similarly, the changes in miRNA expression of liver tissues varied among miRNAs (Table 2, Figures 3, 4B). RDX exposure significantly affected the expression of 56 miRNAs in mouse liver (Table 2, Figures 3, 4B). Of these 56 miRNAs, 31 were up-regulated and 15 were down-regulated. The most up-regulated miRNAs were miR-689, miR-802, miR-29c, and miR-30e, and their expression levels were increased at least 3-fold. In contrast, the expressions of some miRNAs were significantly inhibited by RDX exposure; the miRNAs with at least 4-fold inhibition were miR-574-5p, miR-466f-3p, and let-7e.
Table 1.
Eighty-four miRNAs with significant expression levels in mouse brain tissues after exposure to 5 mg/kg RDX for 28 days (p < 0.01).
| miRNAs | Control (Ck) signal | Treatment (T) signal | Ln (T/Ck) | Fold change | miRNAs | Control (Ck) signal | Treatment (T) signal | Ln (T/Ck) | Fold change |
|---|---|---|---|---|---|---|---|---|---|
| Up-regulated (n = 38) | miR-29c | 18243.37 | 15988.93 | −0.22 | 0.86 | ||||
| miR-206 | 62.19 | 1603.19 | 4.73 | 26.53 | miR-218 | 19242.65 | 16257.02 | −0.22 | 0.86 |
| miR-497 | 78.32 | 683.40 | 3.18 | 9.04 | miR-222 | 7927.73 | 6898.47 | −0.22 | 0.86 |
| miR-195 | 5420.90 | 18586.65 | 1.85 | 3.60 | miR-191 | 12497.48 | 10563.47 | −0.24 | 0.85 |
| miR-101a | 414.29 | 1384.95 | 1.82 | 3.52 | miR-137 | 7815.85 | 6503.00 | −0.25 | 0.84 |
| miR-194 | 197.96 | 672.73 | 1.80 | 3.49 | miR-29a | 35778.41 | 30011.86 | −0.25 | 0.84 |
| miR-101b | 145.54 | 378.38 | 1.43 | 2.70 | miR-126-3p | 14784.18 | 12600.23 | −0.27 | 0.83 |
| miR-30a* | 301.75 | 823.11 | 1.40 | 2.64 | miR-183 | 11607.92 | 9357.58 | −0.27 | 0.83 |
| miR-129-5p | 218.09 | 576.45 | 1.39 | 2.63 | miR-128 | 44764.78 | 36552.18 | −0.31 | 0.81 |
| miR-598 | 392.37 | 957.26 | 1.22 | 2.34 | miR-125b-5p | 43002.58 | 33354.35 | −0.35 | 0.78 |
| miR-122 | 741.33 | 1683.98 | 1.20 | 2.30 | miR-27a | 4920.33 | 3500.16 | −0.36 | 0.78 |
| miR-98 | 1676.79 | 3775.53 | 1.19 | 2.28 | miR-335-5p | 5043.28 | 3912.21 | −0.40 | 0.76 |
| miR-30e* | 250.04 | 540.47 | 1.16 | 2.23 | miR-429 | 12415.46 | 9289.49 | −0.42 | 0.75 |
| miR-7a* | 219.35 | 429.92 | 1.03 | 2.04 | miR-200b | 19737.09 | 14444.82 | −0.44 | 0.74 |
| miR-187 | 349.58 | 687.98 | 0.89 | 1.85 | miR-200a | 9521.66 | 6901.67 | −0.45 | 0.73 |
| miR-146b | 362.94 | 611.32 | 0.82 | 1.77 | miR-690 | 5227.26 | 3879.89 | −0.45 | 0.73 |
| miR-28 | 276.74 | 463.37 | 0.80 | 1.74 | miR-34a | 1717.74 | 1251.73 | −0.45 | 0.73 |
| miR-411 | 287.46 | 486.04 | 0.71 | 1.64 | miR-185 | 4515.15 | 3236.95 | −0.46 | 0.72 |
| miR-16 | 15325.33 | 23835.23 | 0.65 | 1.57 | miR-200c | 10497.13 | 7489.56 | −0.47 | 0.72 |
| miR-30d | 6457.60 | 10059.60 | 0.64 | 1.56 | miR-383 | 1408.73 | 991.23 | −0.50 | 0.71 |
| miR-26b | 8889.40 | 13087.02 | 0.61 | 1.52 | miR-674 | 1832.33 | 1267.67 | −0.54 | 0.69 |
| miR-30e | 2673.12 | 4166.48 | 0.60 | 1.52 | miR-376b | 1191.48 | 807.15 | −0.55 | 0.69 |
| miR-15a | 1416.54 | 2199.71 | 0.57 | 1.48 | miR-433 | 4293.85 | 2893.60 | −0.55 | 0.68 |
| miR-22 | 986.42 | 1449.72 | 0.53 | 1.45 | miR-328 | 2162.73 | 1441.62 | −0.61 | 0.66 |
| miR-124 | 24353.80 | 34061.86 | 0.53 | 1.44 | miR-744 | 1334.35 | 888.29 | −0.62 | 0.65 |
| miR-204 | 6615.88 | 9502.37 | 0.52 | 1.44 | miR-182 | 9376.05 | 6027.11 | −0.63 | 0.65 |
| miR-99a | 2198.46 | 3034.79 | 0.51 | 1.43 | miR-330* | 820.65 | 497.40 | −0.64 | 0.64 |
| miR-129-3p | 5616.21 | 7610.62 | 0.43 | 1.35 | miR-181d | 970.01 | 600.68 | −0.65 | 0.64 |
| miR-30a | 8398.62 | 11447.01 | 0.42 | 1.34 | miR-181b | 2470.77 | 1563.50 | −0.67 | 0.63 |
| miR-139-5p | 8644.69 | 11096.41 | 0.40 | 1.32 | miR-451 | 2271.21 | 1403.72 | −0.71 | 0.61 |
| let-7i | 19829.87 | 24407.14 | 0.38 | 1.30 | miR-705 | 1915.43 | 1168.91 | −0.74 | 0.60 |
| miR-361 | 5810.54 | 7223.99 | 0.34 | 1.26 | miR-762 | 1214.54 | 727.44 | −0.76 | 0.59 |
| miR-99b | 3474.25 | 4415.96 | 0.33 | 1.26 | miR-7a | 6007.06 | 3500.84 | −0.78 | 0.58 |
| miR-181a | 8172.97 | 9590.44 | 0.24 | 1.18 | miR-423-5p | 1247.31 | 741.65 | −0.80 | 0.57 |
| miR-29b | 6102.84 | 7130.04 | 0.23 | 1.17 | miR-1224 | 1215.84 | 660.03 | −0.84 | 0.56 |
| let-7g | 28189.65 | 32541.92 | 0.21 | 1.15 | miR-667 | 2752.98 | 1448.71 | −0.88 | 0.54 |
| let-7e | 24340.51 | 27969.73 | 0.18 | 1.13 | miR-134 | 439.35 | 224.72 | −0.91 | 0.53 |
| miR-125a-5p | 30571.05 | 33774.05 | 0.14 | 1.10 | miR-770-3p | 566.45 | 307.74 | −0.98 | 0.51 |
| let-7f | 35296.63 | 39682.61 | 0.14 | 1.10 | miR-709 | 65443.45 | 30144.97 | −1.10 | 0.47 |
| Down-regulated (n = 46) | miR-199a-3p | 306.18 | 144.31 | −1.17 | 0.44 | ||||
| miR-9* | 24490.19 | 21896.30 | −0.16 | 0.89 | miR-214 | 258.41 | 108.21 | −1.21 | 0.43 |
| miR-23b | 20805.98 | 18196.75 | −0.20 | 0.87 | miR-1 | 462.98 | 194.98 | −1.22 | 0.43 |
| miR-23a | 17809.02 | 15248.49 | −0.22 | 0.86 | miR-10b | 137.98 | 10.11 | −3.80 | 0.07 |
Table 2.
Fifty-six miRNAs with significant expression levels in mouse liver tissues after exposure to 5 mg/kg RDX for 28 days (p < 0.01).
| miRNAs | Control (Ck) Signal | Treatment (T) Signal | Ln (T/Ck) | Fold change |
|---|---|---|---|---|
| Up-regulated (n = 31) | ||||
| miR-689 | 53.64 | 321.49 | 2.69 | 6.47 |
| miR-802 | 102.06 | 452.31 | 2.25 | 4.76 |
| miR-29c | 650.72 | 2353.21 | 1.87 | 3.66 |
| miR-30e | 241.16 | 872.41 | 1.85 | 3.62 |
| miR-148a | 2798.83 | 8285.51 | 1.53 | 2.88 |
| miR-192 | 8975.90 | 23801.68 | 1.49 | 2.82 |
| miR-99a | 62.76 | 182.85 | 1.48 | 2.79 |
| miR-101b | 271.45 | 691.85 | 1.32 | 2.49 |
| miR-152 | 138.25 | 411.63 | 1.31 | 2.48 |
| miR-29a | 5693.53 | 13089.25 | 1.24 | 2.36 |
| miR-30a | 1231.17 | 2988.00 | 1.23 | 2.34 |
| miR-93 | 85.17 | 186.80 | 1.10 | 2.14 |
| miR-15a | 493.75 | 978.07 | 1.04 | 2.06 |
| miR-139-5p | 129.23 | 295.12 | 1.03 | 2.05 |
| miR-194 | 6147.52 | 12165.48 | 1.00 | 1.99 |
| miR-27a | 806.70 | 1598.30 | 0.99 | 1.98 |
| miR-690 | 827.18 | 1477.09 | 0.86 | 1.82 |
| miR-20a | 344.54 | 589.10 | 0.80 | 1.75 |
| miR-195 | 375.51 | 619.41 | 0.79 | 1.73 |
| miR-22 | 900.37 | 1538.86 | 0.79 | 1.73 |
| miR-451 | 885.93 | 1482.35 | 0.74 | 1.67 |
| miR-30d | 1156.83 | 1791.33 | 0.60 | 1.51 |
| miR-126-3p | 7922.97 | 11017.61 | 0.54 | 1.45 |
| miR-25 | 557.96 | 796.59 | 0.51 | 1.42 |
| miR-24 | 1582.59 | 2283.18 | 0.50 | 1.41 |
| miR-27b | 2272.75 | 3102.84 | 0.48 | 1.39 |
| miR-143 | 667.41 | 1005.61 | 0.48 | 1.39 |
| miR-23b | 7607.74 | 9496.82 | 0.35 | 1.27 |
| miR-26a | 19218.42 | 23515.59 | 0.34 | 1.27 |
| miR-23a | 5561.59 | 6882.01 | 0.30 | 1.23 |
| miR-30c | 6790.49 | 8329.45 | 0.29 | 1.22 |
| Down-regulated (n = 25) | ||||
| miR-26b | 12561.71 | 9299.89 | −0.38 | 0.77 |
| let-7i | 3777.32 | 2842.92 | −0.42 | 0.75 |
| miR-125a-5p | 1505.59 | 1067.46 | −0.47 | 0.72 |
| miR-92a | 3658.17 | 2635.02 | −0.47 | 0.72 |
| let-7a | 31984.07 | 20972.51 | −0.54 | 0.69 |
| let-7c | 29197.98 | 18108.06 | −0.60 | 0.66 |
| miR-151-5p | 1583.62 | 967.48 | −0.61 | 0.66 |
| let-7d | 27906.40 | 17206.77 | −0.62 | 0.65 |
| let-7f | 31965.56 | 20730.37 | −0.63 | 0.65 |
| miR-361 | 1316.31 | 813.85 | −0.66 | 0.63 |
| miR-185 | 766.30 | 489.99 | −0.67 | 0.63 |
| miR-15b | 1419.61 | 831.88 | −0.75 | 0.60 |
| miR-107 | 1044.56 | 607.84 | −0.80 | 0.57 |
| let-7b | 24395.46 | 13082.76 | −0.80 | 0.57 |
| miR-103 | 1112.64 | 590.07 | −0.83 | 0.56 |
| miR-145 | 861.14 | 456.09 | −0.88 | 0.54 |
| miR-423-5p | 640.50 | 338.97 | −0.91 | 0.53 |
| miR-320 | 1031.88 | 524.74 | −0.95 | 0.52 |
| miR-762 | 2142.91 | 1069.24 | −0.98 | 0.51 |
| miR-486 | 866.92 | 347.48 | −1.27 | 0.42 |
| miR-98 | 304.26 | 119.05 | −1.32 | 0.40 |
| miR-805 | 3046.89 | 1115.89 | −1.42 | 0.37 |
| let-7e | 6778.31 | 1657.50 | −2.01 | 0.25 |
| miR-466f-3p | 241.33 | 55.51 | −2.17 | 0.22 |
| miR-574-5p | 1012.75 | 94.86 | −3.37 | 0.10 |
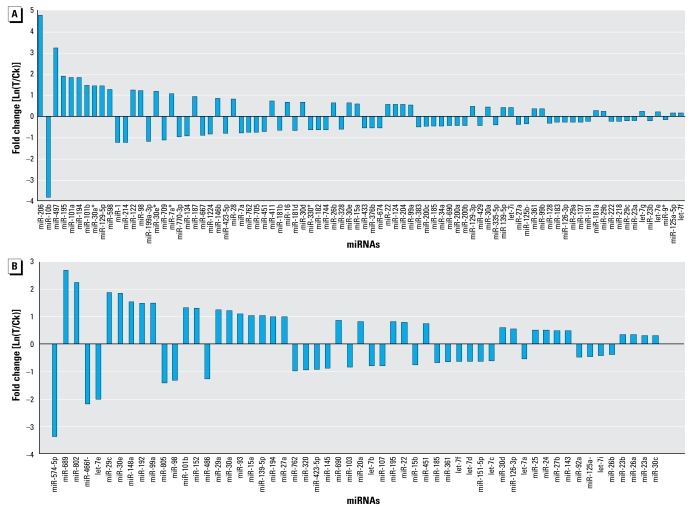
Figure 4.
Fold changes of miRNAs whose expression levels were significantly altered in brain tissues (A) and liver tissues (B) of mice exposed to 5 mg/kg RDX for 28 days.
Two major evidences suggest that brain was more sensitive to RDX exposure than liver. First, the number of miRNAs exhibiting expression alterations after RDX exposure is larger in brain tissues than in liver tissues. Eighty-four miRNAs have significant expression alterations in mouse brain, whereas only 56 miRNAs have significant expression alterations in the liver. Second, the changes in miRNA expression level were much higher and wider in the brain than in the liver. The fold change miRNA expression ranged from 14-fold down (miR-10b) to 26.5-fold up (miR-206) in the brain, compared with only 10-fold down (miR-574 -5p) to 6.5-fold up (miR-689) in the liver (Tables 1 and 2).
Of the 113 miRNAs with significantly aberrant expressions after RDX exposure, the expression levels of 10 miRNAs were significantly increased in both mouse liver and brain (p < 0.01): miR-99a, miR-30a, miR-30d, miR-30e, miR-22, miR-194, miR-195, miR-15a, miR-139-5p, and miR-101b. Three miRNAs (miR-762, miR-423-5p, and miR-185) had expression levels significantly decreased in both brain and liver (p < 0.01). For most miRNAs, their responses to RDX exposure were not consistent between brain and liver (Table 3). Seven miRNAs (let-7e, miR-98, miR-361, miR-26b, miR-125a-5p, let-7i, and let-7f) were significantly up-regulated in the brain but down-regulated in the liver after RDX exposure. In contrast, another seven miRNAs (miR-126-3p, miR-23b, miR-27a, miR-29a, miR-29c, miR-451, and miR-690) were significantly up-regulated in the liver but down-regulated in the brain. Although 37 miRNAs had significant expression changes in both brain and liver, 57 miRNAs had significant expression changes in the brain but not in the liver and 29 miRNAs had significant expression changes in the liver but not in the brain (Table 3).
Table 3.
Comparison of miRNA expression profiles between mouse liver tissues and brain tissues after exposure to 5 mg/kg RDX for 28 days (p < 0.01).
| miRNAs
|
||
|---|---|---|
| miRNA category | No. | Name |
| Up-regulated in both brain and liver | 10 | miR-99a, miR-30e, miR-30d, miR-30a, miR-22, miR-195, miR-194, miR-15a, miR-139-5p, miR-101b |
| Down-regulated in both brain and liver | 3 | miR-762, miR-423-5p, miR-185 |
| Up-regulated in brain but down-regulated in liver | 7 | let-7e, miR-98, miR-361, miR-26b, miR-125a-5p, let-7i, let-7f |
| Up-regulated in liver but down-regulated in brain | 7 | miR-126-3p, miR-23b, miR-27a, miR-29a, miR-29c, miR-451, miR-690 |
| Significant expression changes in brain but not in liver | 57 | miR-99b, miR-1, miR-101a, miR-10b, miR-122, miR-1224, miR-124, miR-125b-5p, miR-128, miR-129-3p, miR-129-5p, miR-134, miR-137, miR-146b, miR-16, miR-181a, miR-181b, miR-181d, miR-182, miR-183, miR-187, miR-191, miR-199a-3p, miR-200a, miR-200b, miR-200c, miR-204, miR-206, miR-214, miR-218, miR-222, miR-23a, miR-28, miR-29b, miR-30a*, miR-30e*, miR-328, miR-330*, miR-335-5p, miR-34a, miR-376b, miR-383, miR-411, miR-429, miR-433, miR-497, miR-598, miR-667, miR-674, miR-705, miR-709, miR-744, miR-770-3p, miR-7a, miR-7a*, miR-9*, let-7g |
| Significant expression changes in liver but not in brain | 29 | let-7a, let-7b, let-7c, let-7d, miR-103, miR-107, miR-143, miR-145, miR-148a, miR-151-5p, miR-152, miR-15b, miR-192, miR-20a, miR-23a, miR-24, miR-25, miR-26a, miR-27b, miR-30c, miR-320, miR-466f-3p, miR-486, miR-574-5p, miR-689, miR-802, miR-805, miR-92a, miR-93 |
Confirmatory studies on differentially expressed miRNAs by qRT-PCR
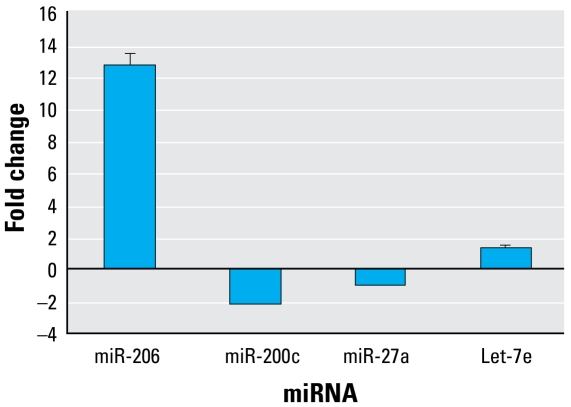
To validate the microarray data, we assayed expression levels of four miRNAs (miR-206, miR-200c, miR-27a, and let-7e) by qRT-PCR and compared the results from the microarray and qRT-PCR. Of the miRNAs selected for comparison, two miRNAs (miR-206 and let-7e) were up-regulated whereas two miRNAs (miR-200c and miR-27a) were down-regulated based on the results of microarray analysis. The expression data obtained by qRT-PCR analysis are comparable with the microarray analysis data (Figure 5).
Figure 5.
Confirmatory studies of selected miRNAs by TaqMan RT-PCR. The fold changes refer to the expression fold changes of the selected miRNAs in RDX-treated mice comparing with control mice. Values represent the mean ± SD of three independent samples, each run in triplicate.
Prediction of the target genes of miR-206
miRNAs function by targeting mRNAs for mRNA cleavage or translation repression. Thus, miRNA function studies depend heavily on the identification of miRNA targets. Currently, the major strategy to identify miRNA targets is based on various computational programs (Zhang et al. 2006). Almost all currently available computational programs overpredict miRNA targets. Several hundreds of genes have been predicted to be the targets of one single miRNA; only a few of them have been validated by experimental approaches, and most of these predicted targets are likely to be false targets (Zhang et al. 2006). One good approach to circumvent this problem is to employ two different computational programs to predict miRNA targets independently and then compare the two lists of targeted genes predicted by each program. The genes predicted by both programs are more likely to be the real targets of the miRNA.
In both microarray and qRT-PCR analyses, miR-206 was significantly up-regulated in mouse brain after exposure to RDX. The change of miR-206 expression was higher than all other miRNAs detected in this study. This suggests that miR-206 may play an important function in animal response to RDX exposure. To explore the potential functions of miR-206, we employed two computational programs (TargetScan and PicTar) to predict the targets of miR-206. After systematically analyzing two lists of miR-206–targeted genes predicted by TargetScan and PicTar, we identified the 50 most promising potential targeted genes [see Supplemental Material, Table 4 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. These candidate targets include several categories. First, these candidate targets contained several transcriptional factors (e.g., the zinc finger protein, the cAMP-responsive element binding protein, and the paired box gene). It is well known that miRNAs target transcriptional factors for regulating animal development. Second, stress-associated genes were predicted as targets of miR-206. Third, miR-206 may target brain-derived neurotrophic factors (BDNFs), which may cause neurologic disorders. Fourth, miR-206 may directly target cancer-related genes, for example, met proto-oncogene (NM_000245, GenBank) and v-ets erythroblastosis virus E26 oncogene homolog 1 (NM_005238, GenBank). This suggests that miR-206 may regulate multiple biological and metabolic processes in response to RDX exposure. Further investigating the functions of miR-206 on these potential targeted genes will help us better understand the mechanism of RDX-induced neurotoxicity and potential carcinogenesis.
Discussion
miRNA response to chemical exposure
We found the expressions of 113 miRNAs, belonging to 75 families, to be affected by RDX exposure in mice. Although a number of the RDX-responsive miRNAs (let-7, miR-34, miR-146, and miR-222) found in this study have been reported previously to have aberrant expressions in response to different chemical exposures in human cell lines, most of the RDX-responsive miRNAs reported in this study have not been shown to respond to chemical treatment previously (Blower et al. 2008; Marsit et al. 2006; Moffat et al. 2007; Pogribny et al. 2007; Rossi et al. 2007; Saito et al. 2006; Sun et al. 2008). These results suggest that either the spectrum of miRNAs that respond to chemical treatment is unique for specific groups of chemicals or that there are methodologic differences in detection of chemically responsive miRNAs. The investigation of the effect of different chemicals on miRNA expression profiles will allow a better understanding of the response spectrum of miRNAs in relation to chemical-induced toxicity and diseases. A particularly interesting potential application of these results is the use of miRNA response profile to develop a set of sensitive biomarkers for monitoring and assessing the health effects of environmental toxicants.
The research reported here possibly marks the first time that the effects of an environmental toxicant on the expression of miRNAs has been examined in vivo. Consequently, we are now in a position to compare these results with similar studies that have measured the response of miRNAs to pharmaceutical agents, which allows us to determine whether there are common miRNA response pathways for xenobiotic agents or whether the collective miRNA response is a function of specific chemical exposure. We will also be able to use the results of this research to understand the effects of environmental toxicants on environmental and human health.
This is also the first comparison study of miRNA expression in different tissues of mice exposed to an environmental toxicant. The contrasting expression of a same miRNA in brain relative to liver (some miRNAs were up-regulated in brain but down-regulated in liver and vice versa) indicates that there is tissue-specific variation in response to the same chemical and that miRNAs may play different roles in different tissues. We also determined that the response of brain miRNAs to RDX exposure was more pronounced than that of the liver, which is indicative that the toxic effects of RDX exposure may be more severe in the brain than in the liver (Figure 4). This further suggests that miRNAs respond to chemical exposure in a tissue-specific manner and that the toxicity may be mediated, at least in part, through miRNAs.
RDX-induced regulation of miRNAs and their targets involved in neurotoxicity
It has been well known for some time that RDX exposure causes adverse central nervous system (CNS) syndromes, including convulsion, epileptic seizure, and loss of reflexes in human and experimental animals (Burdette et al. 1988; Goldberg et al. 1992; Levine et al. 1981; Pan et al. 2007; Testud et al. 1996). However, the molecular mechanism of RDX-induced neurotoxicity is unknown. In this study, we documented significant changes in miRNA expression in the brains of RDX-treated animals relative to their untreated controls. Of particular interest, miR-206 exhibited the most significant up-regulation (26-fold) in RDX-exposed mouse brain relative to controls. This result is notable because the BDNF gene is among the potential miR-206 targets. BDNF is one of the most important members of the neurotrophin family, and it is broadly expressed in mammalian (including human) CNS and peripheral nervous system and is thought to play important roles in supporting neuronal survival and differentiation, neurite outgrowth, and synaptic plasticity (Murer et al. 2001). Neurodegenerative diseases, including Alzheimer’s disease and Parkinson disease, have been associated with reduced BDNF expression (Murer et al. 2001). One recent study demonstrated that the environmental toxicant polybrominated diphenyl ethers reduced BDNF proteins in hippocampus of mice (Viberg et al. 2008), which suggests that neurotoxic environmental toxicants may exert their toxic effects through their activity on BDNF expression. In this present study, six transcript variants of BDNF genes are uniformly predicted to be the targets of miR-206 by both PicTar and TargetScan programs [see Supplemental Material, Table 4 (http://www.ehponline.org/members/2008/11841/suppl.pdf)] underscoring the likelihood that RDX-induced increases in expression of miR-206 may contribute to the neurotoxicity of RDX in the brain through its reduction of BDNF gene expression.
Although the regulation of the BDNF gene by various factors has been extensively investigated at the mRNA and protein levels (Murer et al. 2001), the role of miRNAs in BDNF gene regulation was only recently reported in a study that demonstrated that multiple differentially expressed miRNAs act as inhibitors of BDNF in human prefrontal cortex (Mellios et al. 2008). In that study, luciferase assays confirmed that BDNF was targeted by two miRNAs, miR-30a-5p and miR-195 (Mellios et al. 2008). Interestingly, we also found these two confirmed BDNF-targeting miRNAs (miR-30a and miR-195) were significantly up-regulated in mouse brain and liver after RDX exposure (Tables 1 and 2, Figure 4). In addition, two other members of the miR-30 family (miR-30d and miR-30e) that target BDNF (Mellios et al. 2008) were also overexpressed (Tables 1 and 2, Figure 4). This supports the role of BDNF as a potentially significant target gene of multiple miRNAs. Mellios et al. (2008) failed to detect miR-206 expression in human parietal cortex tissues using microarray analysis, although they predicted computationally that a target site for miR-206 exists in BDNF gene. However, both our microarray and qRT-PCR findings indicate that miR-206 was strongly up-regulated in brain tissues of mice exposed to RDX. These results suggests several alternative but not mutually exclusive hypotheses, including a regulatory interaction between miR-206 and BDNF that responds significantly to RDX treatment in brain tissues, or that miR-206 may be a stress response miRNA that only activates in response to specific toxicant exposure, including RDX.
RDX-induced regulation of miRNAs and their targets involved in carcinogenesis
U.S. EPA has classified RDX as a class C potential human carcinogen (U.S. EPA 1988) based on a laboratory study in which RDX exposure elevated tumor incidence in B6C3F1 mice (Lish et al. 1984). However, the molecular mechanism of the RDX-induced carcinogenesis remains unknown. An interesting finding of this study is that a number of miRNAs with aberrant expressions after RDX exposure are also frequently deregulated in a wide range of cancers. Since the initial report linking miRNAs and cancer development (Calin et al. 2002), almost all types of cancers have been found to be associated with the aberrant expression of at least one miRNA, in which miRNAs function as oncogenes or tumor suppressor genes (Wiemer 2007; Zhang et al. 2007a). Importantly, over- or underexpression of one single miRNA resulted in tumor cell growth inhibition (Takamizawa et al. 2004) or tumor invasion and metastasis (Ma et al. 2007). For example, overexpression of miRNA let-7 significantly inhibited A549 lung cancer cell growth in vitro (Takamizawa et al. 2004), suggesting a potential novel approach for clinical cancer gene therapy by controlling the expression of a single miRNA (Zhang and Farwell 2008). In this study, we found that many cancer-related miRNAs, such as let-7, miR-17-92, miR-10b, 125b, miR-146, miR-15, miR-200, and miR-16, were significantly affected by RDX exposure (Table 4). Expression profiles of these miRNAs were significantly altered in many types of cancers, such as breast, lung, ovarian, liver, and prostate cancer, colorectal neoplasia, hepatocellular carcinoma, and chronic lymphocytic leukemia (Zhang et al. 2007a). This suggests that miRNAs may be involved in RDX-induced carcinogenesis.
Table 4.
Identification of currently known cancer-related miRNAs, whose expression levels were significantly altered after exposure to 5 mg/kg RDX for 28 days (p < 0.01).
| miRNAs with significantly altered expression levels after exposure to RDX (p < 0.01)
|
||
|---|---|---|
| Brain | Liver | Cancersa |
| let-7↑ | let-7↓ | Lung cancer, prostate cancer, colorectal neoplasia |
| miR-103↓ | Stomach cancer | |
| miR-10b↓ | Breast cancer, colorectal neoplasia | |
| miR-125a↑, miR-125b↓ | miR-125a↓ | Breast cancer, ovarian cancer, HCC |
| miR-128↓ | Brain cancer (glioblastoma), prostate cancer | |
| miR-143↑ | Colorectal neoplasia | |
| miR-145↓ | Breast cancer, ovarian cancer, colorectal neoplasia | |
| miR-146b↑ | Breast cancer, papillary thyroid carcinoma | |
| miR-148a↑ | Pancreatic cancer | |
| miR-15a↑ | miR-15a↑, miR-15b↓ | CLL, pituitary adenomas |
| miR-152↑ | Pituitary adenomas | |
| miR-16↑ | CLL, pituitary adenomas | |
| miR-181a↑, miR-181b,d↓ | Brain cancer (glioblastoma), papillary thyroid carcinoma, pancreatic cancer | |
| miR-183↓ | Colorectal neoplasia | |
| miR-191↓ | Pituitary adenomas | |
| miR-192↑ | Pituitary adenomas | |
| miR-195↑ | miR-195↑ | Prostate cancer, HCC |
| miR-199a3p↓ | Ovarian cancer | |
| miR-200a,b,c↓ | HCC, ovarian cancer, cholangiocarcinoma | |
| miR-20a↑ | Colorectal neoplasia | |
| miR-218↓ | Stomach cancer | |
| miR-222↓ | Papillary thyroid carcinoma | |
| miR-24↑ | Pituitary adenomas | |
| miR-26b↑ | miR-26a↑, miR-26b↓ | Pituitary adenomas |
| miR-98↑ | miR-98↓ | Pituitary adenomas |
↑, up-regulation; ↓, down-regulation.
Abbreviations: CLL, chronic lymphocytic leukemia; HCC, hepatocellular carcinoma.
In addition to the possibility that RDX-induced miRNAs are involved in carcinogenesis, we found that several potential miRNA-targeted genes are involved in cancer development. One example is the TNKS-2 gene cluster (tankyrase 2, TRF1-interacting ankyrin-related ADP-ribose polymerase 2; NM_025235, GenBank), which is predicted to be a potential target of miR-206 [see Supplemental Material, Table 4 (http://www.ehponline.org/members/2008/11841/suppl.pdf)]. TNKS-2 and its close homolog TNKS-1 are positive regulators of telomere elongation, which is important for the perpetual growth of cancer cells (Cook et al. 2002). TNKS functions by interacting with telomeric repeat binding factor 1 (TRF1), which inhibits the telomerase activity to terminate telomere elongation. Binding of TRF1 to the telomere can be inhibited by ADP ribosylation of TRF1 by TNKS-1 and TNKS-2 (Cook et al. 2002; Smith et al. 1998). Aberrant expression of TNKS has been found in cancers and neoplasms, including multiple myeloma, plasma cell leukemia (Xu et al. 2001), bladder and colon cancer (Shebzukhov et al. 2008), and breast cancer (Sidorova et al. 2006). Knockdown of TNKS-1 by small interfering RNA blocked mitosis due to sister chromatids remaining associated in telomeres (Dynek and Smith 2004). The inhibition of tankyrase and telomerase caused telomere shortening and apoptosis, which has been proposed as a potential cancer therapy (Seimiya 2006). Despite the association among TRF-1 and TNKS and cell growth control, there have been no reports concerning miRNA-mediated regulation of TNKS gene clusters. We observed, using combined PicTar and TargetScan analyses, that TNKS-2 was a target of miR-206, which implies that the strong up-regulation of miR-206 may target TNKS-2, which in turn may be a protective mechanism against the potential carcinogenic effects of RDX by inducing telomere elongation termination and apoptosis. This provides a new insight to elucidate carcinogenesis potentials of RDX.
Analyses of differentially expressed miRNAs and their targets in brain and liver tissues of mice exposed to RDX indicate that several miRNAs and their potential targets are involved in a complex regulatory network that encompasses a host of biological and metabolic processes. The RDX-induced miRNA-mediated changes in these processes have the potential to contribute to the RDX-induced heath effects, including neurotoxicity and potential carcinogenesis, two major concerns related to RDX exposure. miRNAs may directly and/or indirectly (through regulating other genes) affect the action of RDX on tumor pathogenesis and neurotoxicity (Figure 6). Recent studies on miRNA functions in carcinogenesis, cancer metastasis, and CNS disorders provide further support for this hypothesis. Further investigating the roles of miR-206 in regulation of BDNF and TNKS-2 may lead to a novel miRNA-related gene therapy for the treatment of diseases associated with BDNF and TNKS gene expression. Also, the study of stress response miRNAs will facilitate the development of sensitive miRNA biomarkers for monitoring the health effects of environmental toxicants.
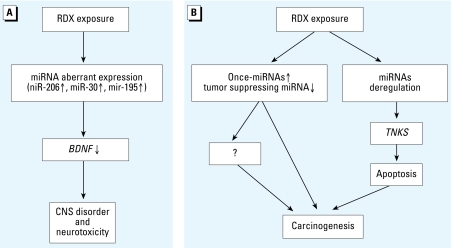
Figure 6.
A proposed model showing the mechanisms of RDX-induced CNS disorder (A) and carcinogenesis (B). In this model, continuous lines show steps confirmed in this study or elsewhere; the dashed lines represent processes that have not been proved by direct evidence. RDX exposure caused aberrant expressions of many miRNAs. Some of them, for example, miR-106, miR-30, and miR-195, target BDNF, one of the most important members in the neurotrophin family that is widely expressed in mammalian CNS and peripheral nervous system (Murer et al. 2001; Wetmore et al. 1991). RDX-induced overexpression of miRNAs (miR-206, miR-30, and miR-195) inhibits the expression of BDNF gene, which contributes to neurotoxicity and CNS disorders. RDX exposure also induced aberrant expressions of onco-miRNAs and tumor-suppressing miRNAs that would directly or indirectly regulate tumor pathogenesis or target genes related to cell cycle (e.g., TNKS) to regulate cell apoptosis and cancer development. These proposed molecular changes, together with others, eventually cause RDX-induced carcinogenesis and CNS disorders. However, more studies need to be performed for testing this model.
Footnotes
Supplemental Material is available online at http://www.ehponline.org/members/2008/11841/suppl.pdf
We performed the animal treatment experiments at Texas Tech University (TTU) with the support of the Strategic Environmental Research and Development Program. We greatly appreciated the help and support of G. Cobb at the Institute of Environmental and Human Health and the staff at the TTU Animal facility. We also thank S. McConnell at Western Illinois University (WIU) and E. Stellwag and A. Spuches at East Carolina University (ECU) for reviewing and editing the manuscript.
This work was supported by ECU New Faculty Research Startup Funds Program, WIU New Faculty Research Startup Funds Program, and WIU University Research Council.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Beller HR, Tiemeier K. Use of liquid chromatography/tandem mass spectrometry to detect distinctive indicators of in situ RDX transformation in contaminated groundwater. Environ Sci Technol. 2002;36:2060–2066. doi: 10.1021/es0157696. [DOI] [PubMed] [Google Scholar]
- Blower PE, Chung JH, Verducci JS, Lin SL, Park JK, Dai ZY, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Burdette LJ, Cook LL, Dyer RS. Convulsant properties of cyclotrimethylenetrinitramine (RDX): spontaneous audiogenic, and amygdaloid kindled seizure activity. Toxicol Appl Pharmacol. 1988;92:436–444. doi: 10.1016/0041-008x(88)90183-4. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok TM, Hogervorst JG, Briede JJ, van Herwijnen MH, Maas LM, Moonen EJ, et al. Genotoxicity and physicochemical characteristics of traffic-related ambient particulate matter. Environ Mol Mutagen. 2005;46:71–80. doi: 10.1002/em.20133. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- Gao XL, Gulari E, Zhou XC. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–596. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goldberg DJ, Green ST, Nathwani D, McMenamin J, Hamlet N, Kennedy DH. RDX intoxication causing seizures and a widespread petechial rash mimicking meningococcemia. J R Soc Med. 1992;85:181–181. [PMC free article] [PubMed] [Google Scholar]
- Jenkins TF, Hewitt AD, Grant CL, Thiboutot S, Ampleman G, Walsh ME, et al. Identity and distribution of residues of energetic compounds at Army live-fire training ranges. Chemosphere. 2006;63:1280–1290. doi: 10.1016/j.chemosphere.2005.09.066. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Levine BS, Furedi EM, Gordon DE, Burns JM, Lish PM. 13 week toxicity study of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in Fischer 344 rats. Toxicol Lett. 1981;8:241–245. doi: 10.1016/0378-4274(81)90108-9. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lish PM, Levine BS, Furedi-Machacek EM, Sagartz EM, Rac VS. Chicago: U.S. Army Medical Research and Development Command; 1984. Determination of the Chronic Mammalian Toxicological Effects of RDX: Twenty-Four Month Chronic Toxicity/Carcinogenicity Study of RDX in the B6C3F1 Hybrid Mouse. Document No. AD A 160774. [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Mellios N, Huang H-S, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17(19):3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat ID, Boutros PC, Celius T, Linden J, Pohjanvirta R, Okey AB. MicroRNAs in adult rodent liver are refractory to dioxin treatment. Toxicol Sci. 2007;99:470–487. doi: 10.1093/toxsci/kfm189. [DOI] [PubMed] [Google Scholar]
- Murer MG, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol. 2001;63:71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Myllynen P, Kurttila T, Vaskivuo L, Vahakangas K. DNA damage caused by benzo(a)pyrene in MCF-7 cells is increased by verapamil, probenecid and PSC833. Toxicol Lett. 2007;169:3–12. doi: 10.1016/j.toxlet.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Pan XP, San Francisco MJ, Lee C, Ochoa KM, Xu XZ, Liu J, et al. Examination of the mutagenicity of RDX and its N-nitroso metabolites using the Salmonella reverse mutation assay. Mutat Res. 2007;629:64–69. doi: 10.1016/j.mrgentox.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Pan XP, Zhang BH, Cobb GP. Extraction and analysis of trace amounts of cyclonite (RDX) and its nitroso-metabolites in animal liver tissue using gas chromatography with electron capture detection (GC-ECD) Talanta. 2005;67:816–823. doi: 10.1016/j.talanta.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Tryndyak VP, Boyko A, Rodriguez-Juarez R, Beland FA, Kovalchuk O. Induction of microRNAome deregulation in rat liver by long-term tamoxifen exposure. Mutat Res. 2007;619:30–37. doi: 10.1016/j.mrfmmm.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248–253. doi: 10.1016/j.phrs.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu ZS, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seimiya H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer. 2006;94:341–345. doi: 10.1038/sj.bjc.6602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebzukhov YV, Lavrik IN, Karbach J, Khlgatian SV, Koroleva EP, Belousov PV, et al. Human tankyrases are aberrantly expressed in colon tumors and contain multiple epitopes that induce humoral and cellular immune responses in cancer patients. Cancer Immunol Immunother. 2008;57:871–881. doi: 10.1007/s00262-007-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova N, Zavalishina L, Kurchashova S, Korsakova N, Nazhimov V, Frank G, et al. Immunohistochemical detection of tankyrase 2 in human breast tumors and normal renal tissue. Cell Tissue Res. 2006;323:137–145. doi: 10.1007/s00441-005-0053-8. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Testud F, Glanclaude JM, Descotes J. Acute hexogen poisoning after occupational exposure. J Toxicol Clin Toxicol. 1996;34:109–111. doi: 10.3109/15563659609020244. [DOI] [PubMed] [Google Scholar]
- U.S. EPA. Health Advisory for RDX. Washington, DC: U.S. Environmental Protection Agency; 1988. [Google Scholar]
- Viberg H, Mundy W, Eriksson P. Neonatal exposure to decabrominated diphenyl ether (PBDE 209) results in changes in BDNF, CaMKII and GAP-43, biochemical substrates of neuronal survival, growth, and synaptogenesis. Neurotoxicology. 2008;29:152–159. doi: 10.1016/j.neuro.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Wetmore C, Cao YH, Pettersson RF, Olson L. Brain-derived neurotrophic factor—subcellular compartmentalization and interneuronal transfer as visualized with antipeptide antibodies. Proc Natl Acad Sci USA. 1991;88:9843–9847. doi: 10.1073/pnas.88.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wu LG, Fan JH, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zheng C, Bergenbrant S, Holm G, Bjorkholm M, Yi Q, et al. Telomerase activity in plasma cell dyscrasias. Br J Cancer. 2001;84:621–625. doi: 10.1054/bjoc.2000.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Cox SB, McMurry ST, Jackson WA, Cobb GP, Anderson TA. Effect of two major N-nitroso hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) metabolites on earthworm reproductive success. Environ Pollut. 2008;153:658–667. doi: 10.1016/j.envpol.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Farwell MA. MicroRNAs: a new emerging class of players for disease diagnostics and gene therapy. J Cell Mol Med. 2008;12:3–21. doi: 10.1111/j.1582-4934.2007.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007a;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Computational identification of microRNAs and their targets. Comput Biol Chem. 2006;30:395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Wang QL, Pan XP. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007b;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]