Abstract
Cohesin is an essential protein complex required for sister chromatid cohesion. Cohesin associates with chromosomes and establishes sister chromatid cohesion during interphase. During metaphase, a small amount of cohesin remains at the chromosome-pairing domain, mainly at the centromeres, whereas the majority of cohesin resides in the cytoplasm, where its functions remain unclear. We describe the mitosis-specific recruitment of cohesin to the spindle poles through its association with centrosomes and interaction with nuclear mitotic apparatus protein (NuMA). Overexpression of NuMA enhances cohesin accumulation at spindle poles. Although transient cohesin depletion does not lead to visible impairment of normal spindle formation, recovery from nocodazole-induced spindle disruption was significantly impaired. Importantly, selective blocking of cohesin localization to centromeres, which disrupts centromeric sister chromatid cohesion, had no effect on this spindle reassembly process, clearly separating the roles of cohesin at kinetochores and spindle poles. In vitro, chromosome-independent spindle assembly using mitotic extracts was compromised by cohesin depletion, and it was rescued by addition of cohesin that was isolated from mitotic, but not S phase, cells. The combined results identify a novel spindle-associated role for human cohesin during mitosis, in addition to its function at the centromere/kinetochore regions.
INTRODUCTION
Mitotic spindle formation is critical for proper chromosome congression, alignment, and segregation during cell division (Compton, 2000; Scholey et al., 2003; Kline-Smith and Walczak, 2004). Microtubules (MTs) undergo drastic changes in their dynamics and organization as the cell enters mitosis to form the bipolar spindle apparatus. As the nuclear membrane breaks down at the G2/M transition, interphase cytoskeletal MTs are destabilized, and mitotic spindle MTs nucleate from two opposing centrosomes/spindle poles with increased growth and turnover rates (Saxton et al., 1984; Zhai et al., 1996). This mitosis-specific change of microtubule behavior indicates the presence of cell cycle-specific regulators of spindles. Indeed, studies identified many important microtubule-associated proteins (MAPs) that regulate different aspects of mitotic spindle dynamics, such as the efficiency of MT nucleation and minus-end focusing at spindle poles, MT plus- and minus-end dynamics, and overall spindle stability (Kline-Smith and Walczak, 2004). However, the intricate regulatory mechanisms for spindle organization are not completely understood, and there are most likely additional factors that contribute to proper spindle regulation during mitosis.
Cohesin is a conserved and essential multiprotein complex required for sister chromatid cohesion (Losada et al., 1998; Uhlmann and Nasmyth, 1998; Toth et al., 1999). Genetic studies in both yeast and metazoans revealed that inhibition of cohesin function leads to premature separation of sister chromatids, chromosome misalignment, and meloteric attachment of spindles to kinetochores, which all result in missegregation of chromosomes, indicating the crucial role of cohesin in mitosis (Tanaka et al., 2000; Hauf et al., 2001; Sonoda et al., 2001; Hoque and Ishikawa, 2002; Vass et al., 2003). Cohesin is composed of two structural maintenance of chromosomes family proteins, SMC1 and SMC3, which form a stable heterodimer that associates with the two non-SMC components Rad21 (Scc1/Mcd1) and SA protein (Scc3) (Toth et al., 1999; Losada et al., 2000; Sumara et al., 2000; Tomonaga et al., 2000). In vertebrates, two mitotic SA protein homologues, SA1 and SA2, were found to form distinct cohesin complexes (Losada et al., 2000).
In metazoans, cohesin is loaded onto chromosomes in interphase to establish sister chromatid cohesion. During G2/prophase, the majority of cohesin dissociates from chromosomes as an intact complex in a Polo-like kinase-regulated manner (Losada et al., 2000; Sumara et al., 2000; Gregson et al., 2001). Only a small amount of cohesin (∼5%) remains at the chromosome-pairing domain to mediate sister chromatid cohesion, which is eventually terminated by the cleavage of Rad21 at the onset of anaphase leading to chromosome segregation (Waizenegger et al., 2000; Hoque and Ishikawa, 2001; Gimenez-Abian et al., 2004). However, the functional significance of the abundant cytoplasmic cohesin remained an enigma.
Previously, we showed that microinjection of anti-human (h)SMC1 antibody into human metaphase cells led to immediate disorganization of the metaphase plate and cell cycle arrest (Schmiesing et al., 1998). Because this particular antibody detects hSMC1 in the cytoplasm, but not at the pairing domain of metaphase chromosomes (likely due to steric hindrance), the notion was raised that cytoplasmic cohesin may play an important role in mitosis. Subsequently, we found that a subpopulation of cytoplasmic cohesin localizes to the mitotic spindle poles and interacts with the nuclear mitotic apparatus protein (NuMA) (Gregson et al., 2001). NuMA is a factor that localizes to the spindle poles in a mitosis-specific manner and plays an important role in mitotic spindle assembly at the spindle poles (Gaglio et al., 1995; Merdes et al., 1996). We also demonstrated that cohesin is required for mitotic spindle aster assembly in vitro, suggesting that cytoplasmic cohesin plays a role in mitotic spindle organization (Gregson et al., 2001). However, the in vivo significance of this observation was unclear. Here, we report that cohesin localizes to spindle poles and functions in mitotic spindle formation in vivo and in vitro. Our results indicate a mitosis-specific and chromosome-independent role for cohesin in spindle function in vertebrates.
MATERIALS AND METHODS
Cell Lines and Cell Synchronization
HeLa cells were grown in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum, l-glutamate, and penicillin/streptomycin. Wild-type, CENP-H or Scc1/Rad21 conditional knockout DT40 cells were cultured as described previously (Fukagawa et al., 2001, 2004; Sonoda et al., 2001). HeLa cells were synchronized to S phase by using double-thymidine block and to M phase by an additional nocodazole treatment as described previously (Schmiesing et al., 2000; Gregson et al., 2001).
Antibodies
Rabbit polyclonal antibody was raised against the N terminus of NuMA (amino acids 1-307) expressed in Escherichia coli. Antigen-affinity purified antibodies against hSMC1, SA-1, and hRAD21 were described previously (Gregson et al., 2001, 2002). Mouse monoclonal antibody (mAb) specific for CENP-E and rabbit polycloncal antibody specific for Tripin/hSgo2 were published previously (Yen et al., 1991; Huang et al., 2007). Polyclonal antibodies against Aurora A (rabbit; Novus Biologicals, Littleton, CO) and SA2 (goat; Bethyl Laboratories, Montgomery, TX) as well as mouse monoclonal antibodies specific for NuMA (Calbiochem, San Diego, CA), Polo-like kinase 1 (Plk1) (Abcam, Cambridge, MA), SUV39H1 (Millipore, Billerica, MA), CENP-A (Abcam), and α-tubulin, β-tubulin, and γ-tubulin (Sigma-Aldrich) were used. Secondary antibodies conjugated with alkaline phosphatase or horseradish peroxidase (Promega, Madison, WI) for Western blot, and with Cy3 (The Jackson Laboratory, Bar Harbor, ME) and fluorescein (Vector Laboratories, Burlingame, CA) for immunostaining, were also used.
Plasmids and Transfection
NuMA deletion mutants were generated by polymerase chain reaction (PCR) by using NuMA cDNA (kindly provided by Dr. D. Compton, Dartmouth Medical School) as the template and cloned into the Tet-Off vector. Cells were transfected using Effectene (QIAGEN, Valencia, CA) according to the manufacturer's instructions. The SMC1 gene, with green fluorescent protein (GFP) or FLAG peptide fused in-frame at its C-terminal end, was cloned into the pIRESneo3 vector (Clontech, Mountain View, CA) for eukaryotic gene expression. G418-resistant stable cell lines, expressing either hSMC1-GFP or hSMC1-FLAG (hSMC1-flg), were generated and used for further experiments.
Cell Extraction, Immunofluorescent Staining, and Image Analysis
In situ cell extraction was performed essentially as described to remove the majority of cytoplasmic cohesin to visualize cohesin associated at the spindle poles (Schmiesing et al., 2000; Gregson et al., 2001). Briefly, cells were extracted using the cytoskeleton (CSK) buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 7.0, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2] with 0.5% Triton X for 5 min at 4°C to remove soluble cytoplasmic proteins, and then they were fixed with 4% paraformaldehyde at 4°C for 10 min. For metaphase chromosome spreads, mitotic cells were enriched by the addition of 0.05 μg/ml nocadazole for 4–6 h. Mitotic cells were collected by shaking, washed with phosphate-buffered saline (PBS), and resuspended in 75 mM KCl. Cells were spun onto polylysine-coated slides by using Cytospin at 1900 rpm for 2 min. Cells were fixed with 3.7% formaldehyde for 10 min at 4°C. Slides were rinsed and stored in SNBP buffer (0.02% saponin, 0.05% NaN3, and 1% bovine serum albumin in PBS) for subsequent immunostaining. Immunofluorescent staining of cells was carried out as described previously (Schmiesing et al., 2000; Gregson et al., 2001). Immunofluorescent image analysis was performed using an IX70 with the MagnaFire digital charge-coupled device camera system (Olympus, Tokyo, Japan). Quantity One software (Bio-Rad, Hercules, CA) was used for quantification of the immunofluorescent images.
Coimmunoprecipitation, Silver Staining, and Western Analysis
Coimmunoprecipitation was performed as described previously (Schmiesing et al., 2000; Gregson et al., 2001). The immunoprecipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis and silver staining or transferred to nitrocellulose membrane for Western blotting as described previously (Schmiesing et al., 2000; Gregson et al., 2001). Western blots were developed by either colorimetric reaction (Promega) or enhanced chemiluminescence (ECL kit; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Centrosome Purification
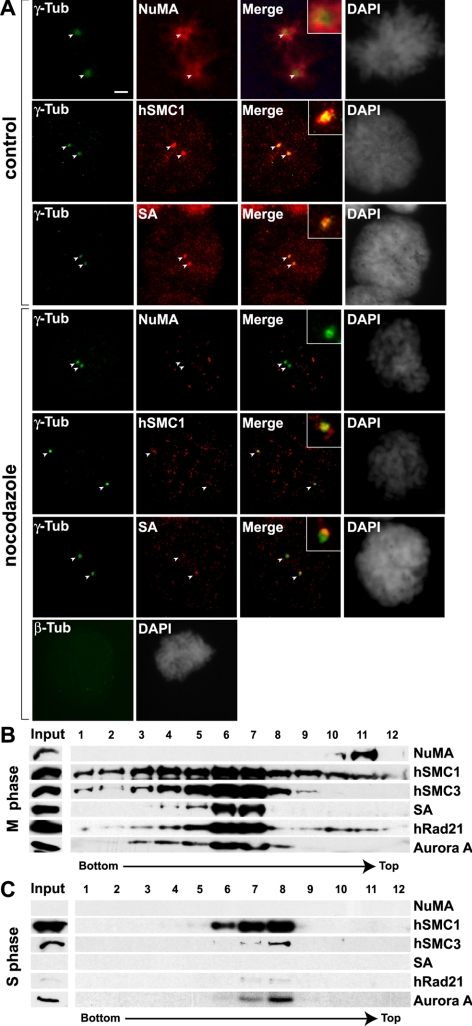
Isolation of human centrosomes from HeLa cells was carried out following protocols described previously (Moudjou and Bornens, 1998). Briefly, HeLa cells were synchronized to S phase and M phase, and treated with nocodazole (1 μg/ml) and cytochalasin B (5 μg/ml) for 1 h before harvest. Cells were then collected and washed with PBS and 8% sucrose. The pellet was resuspended in buffer containing 1 mM Tris-HCl and 0.1% β-mercaptoethanol (βME), spun down at 3K for 5 min, and resuspended in the same buffer containing 1% NP-40. After spinning down the pellet at 13K for 30 s, a 1/50 volume of 50× PE (10 mM PIPES and 1 mM EDTA, pH 7.2) was added to the supernatant. The supernatant was then applied to a 70% sucrose cushion at 10,000 × g for 30 min. Most of the supernatant was aspirated, and the sucrose interface (∼150 mg of extract) was applied to a continuous sucrose gradient (40–70%) at 100,000 × g for 16 h in a SW28 rotor. Twelve fractions were collected from the bottom. Western blot analysis with anti-Aurora A antibody was used to identify the fractions containing centrosomes.
Cohesin Purification
Crude extracts of the HeLa stable cell line expressing hSMC1-flg synchronized to M phase or S phase were incubated with anti-FLAG M2 affinity gel for 3 h and then washed sequentially with 0.1 M HNAD (25 mM HEPES, pH 7.6, 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1 M KCl, 0.1% NP-40, 1 mM dithiothreitol [DTT], and 0.2 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride [AEBSF]), 1 M HAD (25 mM HEPES, pH 7.6, 0.1 mM EDTA, 12.5 mM MgCl2, 1 M KCl, 10% glycerol, 1 mM DTT, and 0.2 M AEBSF) and KHM buffer (78 mM KCl, 50 mM HEPES, pH 7.0, 4 mM MgCl2, and 2 mM EGTA) with DTT and AEBSF. The purified cohesin containing hSMC1-flg (i.e., flg-cohesin) was eluted with 100 μg/ml FLAG peptide (Sigma-Aldrich) in KHM buffer.
In Vitro Mitotic Spindle Aster Assembly
The in vitro aster assembly assay was performed as described previously (Gregson et al., 2001), by using the protocol of Gaglio et al. (1995, 1996, 1997)). Briefly, HeLa cells were synchronized to mitosis and were collected by shake-off and incubated with 20 μg/ml cytochalasin B. Cells were then washed with phosphate-buffered saline and resuspended in KHM buffer containing cytochalasin B at a concentration of ∼3 × 107 cells/ml. Cells were Dounce homogenized, and the crude extract was subjected to ultracentrifugation at 100,000 × g for 15 min. at 4°C. The supernatant was collected, and a fraction of it was subjected to immunodepletion. Approximately 10–20 μg of either preimmune immunoglobulin (IgG), anti-hSMC1, anti-Rad21, or anti-NuMA antibody was coupled to protein A beads and incubated with 20 μl of mitotic extracts for 45 min at 4°C. The beads were spun down, the supernatants were collected, and the depletion process was repeated. The final supernatants were passed through empty spin columns to remove any remaining beads. In add-back experiments, flg-cohesin immunopurified from S phase or M phase extracts was added to 20 μl of each depleted extract. The supernatants were incubated for one hour at 30°C in the presence of 2.5 mM ATP and 10 μM Taxol for the in vitro aster assembly. After the reaction, a small portion of each sample of equal volume was dropped onto a coverslip, fixed with methanol at −20°C for 20 min, and subjected to immunofluorescent staining with an antibody specific for β-tubulin (Sigma-Aldrich) or costained with anti-NuMA antibody. The criteria for asters used in the experiments are as follows: 1) distinct clustering of β-tubulin with colocalization of NuMA at the center, and 2) diameter of the β-tubulin/NuMA clustering must be >1 μm. The number of such asters was counted in 40 randomly chosen areas on the coverslip under the microscope with the 100× objective. The number of asters in the preimmune-depleted extracts was considered to be 100%.
Small Interfering RNA (siRNA) Transfection
The 21-nucleotide siRNA duplexes were designed and synthesized by QIAGEN against hSMC1 (5′-CAC CAT CAC ACT TTA ATT CCA-3′), hRad21 (5′-CTG GGA GTA GTT CGA ATC TAT-3′), SA1 (5′-CAC GTA GAA TCA GAT GTT CTA-3′), SA2 (5′-TCG GTG GTA GAT GAT TGG ATA-3′), and CENP-E (5′-AAC ACG GAT GCT GGT GAC CTC-3′) (Tanudji et al., 2004). The target sequence for SUV39H1 is as described previously (Ait-Si-Ali et al., 2004). The control siRNA consists of a random sequence (QIAGEN). Transfection of siRNA into HeLa cells was performed twice using RNAiFect (QIAGEN) following the manufacturer's instructions. Samples were analyzed 24 h after the second siRNA transfection by immunofluorescence and Western blot analyses.
Spindle Recovery and Taxol Experiments In Vivo
For the spindle recovery experiments, cells were treated with 0.5 μg/ml nocodazole for 30 min at 24 h after the second siRNA transfection. Cells were then washed three times and were incubated in fresh medium for 20 min. Alternatively, cells were treated with 10 μM Taxol for 5 min after the nocodazole treatment to induce centrosome-independent spindle aster formation. Experiments were repeated three times, and an average of 50 cells was examined per sample.
Spindle Assembly Analysis in Chicken DT40 Cells
Wild-type or Rad21 knockout DT40 cells were treated with doxycycline (Dox) for 24 h before the 1-h 0.1 μg/ml nocodazole treatment for the disruption of spindles. Cells were then released from nocodazole treatment by being washed three times and further incubated in nocodazole-free media. The experiments were done in the continuous presence of Dox. At different time points after the nocodazole release, cells were harvested by centrifugation (2000 rpm; 5 min; GS-6R tabletop refrigerated centrifuge; Beckman Coulter, Fullerton, CA), and they were resuspended to a final concentration of 0.4–1.5 × 106 cells/ml. Next, 50–100 μl of each cell suspension was spun onto poly-l-lysine–coated slides by Cytospin and fixed with 3.5% formaldehyde (in PBS, at room temperature for 10 min). Alternatively, wild-type, Rad21, or CENP-H knockout cells were treated with Dox for 27 h, and spindle morphology was analyzed without any nocodazole treatment.
RESULTS
Mitosis-specific Association of Cohesin with Spindle Poles
In previous work, we found that hSMC1, hSMC3, and SA (SA1 or SA2) localize to mitotic spindle poles in human cells by immunofluorescent staining of the endogenous proteins by using antibodies specific for each cohesin component (Gregson et al., 2001). Human Rad21 localization at spindle poles was also reported previously (Hoque and Ishikawa, 2001). These results were further confirmed by the localization of the recombinant hSMC1 fused to GFP at mitotic spindle poles in a stable HeLa cell line expressing hSMC1-GFP (Supplemental Figure S1A). Together, these results demonstrate unequivocally that a subpopulation of cohesin localizes to the spindle poles.
To distinguish whether cohesin associates with the centrosomes or localizes to the pericentrosomal area in a spindle-dependent manner similar to NuMA (Kallajoki et al., 1991), we examined cohesin localization after disruption of spindle MTs. After nocodazole treatment, no β-tubulin was observed, and NuMA no longer clustered to the spindle poles (Figure 1A). In contrast, hSMC1 and SA remained at the spindle poles, although at lower levels (∼20–30% compared with the control cells). This suggests that a subpopulation of cohesin associates with centrosomes independently of spindle MTs. To further confirm this, centrosomes from M and S phase cells were purified by sequential sucrose gradient centrifugation and probed for the presence of cohesin by Western blot (Figure 1, B and C). In this method, spindle MTs were removed by nocodazole treatment, which also removes NuMA from mitotic centrosome fractions (Figure 1B). Consistent with the immunolocalization results (Figure 1A), all four cohesin components cofractionated with centrosomes purified from mitotic cells as marked by the presence of Aurora A (Figure 1B) (Stenoien et al., 2003). In contrast, the cohesin complex was not detected in centrosomes purified from S phase cells (Figure 1C). Weak signals of hSMC3 and hRAD21 were detected, but at levels significantly lower than those in mitosis. This is consistent with the fact that the majority of cohesin localizes in the nucleus during interphase (Schmiesing et al., 1998; Losada et al., 2000; Sumara et al., 2000). A complete absence of NuMA in the S phase centrosome fractions further ensures that there is no contamination of nuclear proteins into this preparation. Interestingly, however, a significant population of hSMC1 was found to cofractionate with Aurora A (Figure 1C). Consistent with this, hSMC1, but not SA, was detected at the centrosomes by immunofluorescent staining of interphase cells (Supplemental Figure S1B). This indicates that hSMC1 alone is capable of interacting with interphase centrosomes, providing the first example of SMC1 behaving differently from either the SMC1-SMC3 heterodimer or the holo-cohesin complex. Together, these results demonstrate mitosis-specific association of cohesin with centrosomes, and indicate that one mechanism of cohesin recruitment to the mitotic spindle poles is through cell cycle-specific interaction with centrosomes.
Figure 1.
Cohesin association with centrosomes. (A) Microtubule-independent recruitment of cohesin to spindle poles. HeLa cells were treated with 0.1 μg/ml nocodazole for 1 h before preextraction and fixation. Cells were then stained with antibodies specific for γ-tubulin, β-tubulin, NuMA, SMC1, and SA antibodies as indicated. Untreated control mitotic cells are also shown. Bar, 5 μm. The exposure time for all images was identical, with the exception of NuMA in the control cell, which was decreased to one-fifth due to the strong signal. (B) Western blot analysis of the 40–70% sucrose gradient fractions of purified centrosomes from HeLa cells synchronized to mitotic (M) phase. Centrosome peak fractions were detected by the presence of Aurora A. (C) Same as in B, except for S phase-synchronized cells.
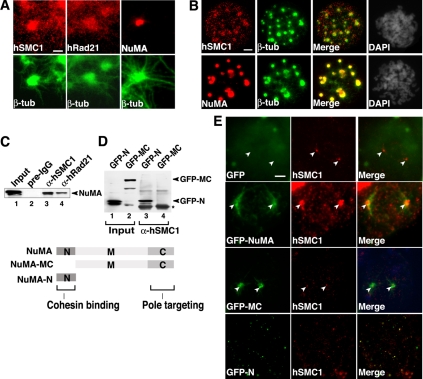
We next tested whether cohesin also localizes to the spindle poles in a centrosome-independent manner. We found that cohesin localizes to in vitro assembled mitotic asters, distributing diffusely along astral MTs with NuMA clustering at the center (Figure 2A). Furthermore, we found that both cohesin and NuMA cluster to the Taxol-induced centrosome-independent asters in vivo (Figure 2B). These results indicate that cohesin localizes to the mitotic spindle pole by two different mechanisms, both centrosome dependent and independent.
Figure 2.
NuMA recruits cohesin to the spindle poles through its N-terminus domain. (A) Cohesin localization to in vitro assembled mitotic spindle asters. Immunofluorescent staining of in vitro assembled mitotic spindle asters with mAb specific for β-tubulin (β-tub) and polyclonal antibodies specific for SMC1, Rad21, or NuMA as indicated. Bar, 2 μm. (B) Cohesin localization to Taxol-induced centrosome-independent asters in vivo. HeLa cells were treated with nocodazole and Taxol sequentially followed by preextraction and fixation. Cells were then stained with antibodies specific for β-tubulin, SMC1, and NuMA as indicated. Bar, 5 μm. (C) Coimmunoprecipitation-Western analysis of the interaction between endogenous cohesin and GFP-NuMA in vivo. Extracts from 293T cells expressing the full-length NuMA fused to GFP (lanes 1–4) were used for coimmunoprecipitation with antibody against SMC1 (αSMC1; lane 3), Rad21 (αRad21; lanes 4), or preimmune IgG (pre-IgG; lane 2), and eluted with 2 M guanidine-HCl after a 1 M KCl wash. Anti-GFP antibody was used as the primary antibody for Western analysis. (D) Coimmunoprecipitation-Western analysis of the interaction between GFP-NuMA deletion mutants and endogenous cohesin. The experiments were carried out in a manner similar to C by using anti-hSMC1 antibody for immunoprecipitation and anti-GFP antibody for Western analysis. An asterisk indicates the IgG heavy chain. A schematic diagram of NuMA deletion mutants is shown. The C-terminal spindle pole-targeting domain of NuMA is indicated. (E) The effects of overexpression of the full-length and deletion mutants of GFP-NuMA on cohesin targeting to the spindle poles in vivo. A GFP-only control is also shown. Cells were preextracted before fixation and immunostained with anti-SMC1 antibody. Bar, 5 μm.
The N Terminus of NuMA Is Important for Mitotic Cohesin Targeting to Spindle Poles
Because cohesin interacts and colocalizes with NuMA at the spindle poles (Figure 2, A and B) (Gregson et al., 2001), NuMA may play a role in recruiting cohesin to the spindle poles. To address this, we first mapped the domain of NuMA that is involved in the cohesin interaction. Antibodies against hSMC1 and hRad21, which precipitate the entire cohesin complex (Gregson et al., 2001), both coprecipitated GFP-full-length NuMA, confirming the in vivo interaction between cohesin and NuMA (Figure 2C). We next tested deletion mutants of NuMA and found that cohesin interacts with the N-terminal region, but not with either the middle or C-terminal regions, of NuMA (Figure 2D).
Based on the above-mentioned results, we examined the possible dominant-negative effects of overexpression of the full-length and deletion mutants of NuMA on cohesin localization in vivo. GFP-full-length NuMA localized to the spindle poles and significantly enhanced the cohesin clustering at the spindle poles (Figure 2E). In contrast, the N-terminal deletion mutant GFP-NuMA-MC that retains the spindle pole-targeting domain (Compton and Cleveland, 1993) but lacks hSMC1-binding site localized to the spindle poles with no positive effect on cohesin localization at spindle poles. The N-terminal fragment of NuMA that interacts with hSMC1, but lacks the pole-targeting domain failed to cluster at spindle poles and inhibited cohesin accumulation there. These results indicate that the interaction of cohesin with the N terminus of NuMA is important for efficient cohesin targeting to mitotic spindle poles. Together, the results suggest that cohesin is capable of associating with the mitotic centrosomes in a NuMA-independent manner, but spindle pole clustering of cohesin is further enhanced by cohesin's interaction with NuMA in the pericentrosomal region.
Cohesin Is Required for Mitotic Spindle Formation in Chicken DT40 Cells
Although a role for cohesin in mitotic spindle assembly was suggested in vitro in human mitotic extracts (Gregson et al., 2001), its in vivo relevance was undefined. Thus, we tested the effect of cohesin depletion using Dox-inducible Rad21 knockout chicken DT40 cells (Figure 3). Cohesin components also localize to mitotic spindle poles in these cells, demonstrating that cohesin localization at spindle poles is conserved in at least two vertebrates (Supplemental Figure S1C).
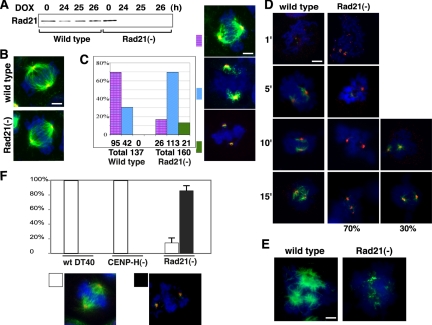
Figure 3.
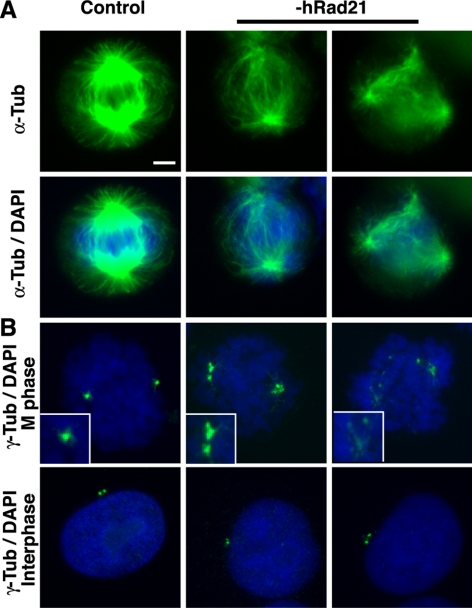
Effects of Rad21 depletion on mitotic spindle organization in DT40 cells. (A) Western analysis of Rad21 conditional knockout [Rad21(−)] chicken DT40 cells after Dox treatment. The wild-type and Rad21(−) cells were treated with Dox for the duration of time indicated at top. (B) Spindle morphology of the wild-type and mutant cells after 24-h Dox treatment. Dox-treated wild-type and Rad21(−) mitotic cells were fixed and stained with antibody specific for α-tubulin (green) and NuMA (red) as well as 4,6-diamidino-2-phenylindole (DAPI; blue). The merged images are shown. Bar, 5 μm. (C) Rad21 depletion affects the efficiency of spindle reassembly. Wild-type or Rad21(−) cells were treated with nocodazole for 1 h after 24-h incubation with Dox. Cells were then released from nocodazole treatment and incubated for 1 h to restore the spindle. Immunostaining is same as described in B. The percentage of normal and abnormal spindles is shown in the graph. The total numbers of mitotic cells examined are shown at the bottom. Representative spindle morphology for each category (specifically, “normal spindles,” “abnormal short spindles,” and “poles with no visible asters”) is shown. Bar, 5 μm. (D) Time course analysis of spindle reassembly in Rad21(−) DT40 cells after nocodazole release. Spindle morphology in mitotic cells was analyzed at 1, 5, 10, and 15 min after nocodazole release as indicated. Representative morphology of spindles is shown. Green, α-tubulin; red, NuMA; and blue, DAPI. Almost no spindle was observed within the first 15 min of reassembly in 70% of the Rad21-depleted mitotic cells examined, in contrast to the efficient recovery of spindles in the wild-type cells. The other 30% of mutant mitotic cells exhibited shorter spindles compared with wild type cells. Bar, 5 μm. (E) Rad21 depletion inhibits Taxol-induced nonspecific aster assembly in vivo. After the nocodazole treatment as described in C, wild-type or Rad21(−) cells were further incubated with Taxol. Cells were then fixed and stained with anti-α-tubulin antibody and DAPI. The wild-type cells exhibited centrosome-independent aster formation upon Taxol treatment, whereas Rad21-depleted cells failed to form asters. Bar, 5 μm. (F) Prolonged Rad21 depletion abolishes mitotic spindle formation. Rad21(−) cells were treated with Dox for 27 h, and mitotic spindle morphology was compared with Dox-treated wild type cells and CENP-H(−) cells. Approximately 40 mitotic cells for each cell line were analyzed in one experiment and the experiments were repeated three times to obtain the SE of the mean. The percentage of mitotic cells with and without spindles is shown in the graph. Representative images (“spindle-positive” and “poles with no visible asters”) (staining is the same as described in B) is shown below.
After 24 h of Dox treatment, there was no detectable Rad21 in these cells by Western blotting (Figure 3A). At this time point, there was no significant difference in mitotic spindle morphology between the wild-type and Rad21 knockout cells (Figure 3B), which is consistent with published observations (Sonoda et al., 2001). However, when we briefly treated these cells with nocodazole to disrupt the spindles and examined their subsequent reassembly, there were significant differences in the efficiency of spindle recovery between the wild-type and mutant cells (Figure 3C). Although ∼70% of the wild-type mitotic cells reassembled normal mitotic spindles, an abnormal morphology with short spindles was observed in ∼70% of the mutant mitotic cells. In addition, some of the mutant cells failed to assemble any visible asters. Thus, Rad21 depletion in chicken cells compromised efficient spindle reassembly.
One possible explanation for the observed spindle defect is that it is an indirect result of destabilization of spindles at the kinetochores by centromere dysfunction (loss of tension) caused by cohesin depletion at the centromere pairing domain. To address this, a time course analysis of spindle reassembly was carried out. If the spindles are destabilized indirectly due to kinetochore defects, we should first observe extension of spindles to kinetochore regions before degradation after nocodazole release. However, the results show that spindles that emanate from the poles remain short and never reach the chromosomes, suggesting that the defect is independent of kinetochores (Figure 3D).
To further substantiate these results, these cells were further treated with Taxol immediately after nocodazole treatment to examine the induction of centrosome- and chromosome-independent nonspecific multiaster formation (Figure 3E). Taxol-induced aster formation was significantly compromised in the mutant cells. Thus, the efficiency of spindle aster formation seems to be decreased in cohesin-depleted cells.
Cohesin at spindle poles seems to be stable and relatively resistant to depletion because there was still a small amount of residual Rad21 at the spindle poles in some of the mutant cells treated with Dox for 24 h (Supplemental Figure S1C, Rad21). Therefore, the Dox treatment was further extended to 27 h, and the mitotic spindle morphology was examined. At this time point, without nocodazole treatment, the majority of Rad21-depleted mitotic cells exhibited very weak, if any, spindle formation (Figure 3F). This is not due simply to spindle checkpoint activation, because similar conditional depletion of CENP-H by Dox treatment, which was shown to activate the spindle checkpoint resulting in mitotic arrest (Fukagawa et al., 2001), did not result in loss of spindle formation (Figure 3F). After 27 h of Dox treatment, Rad21 and CENP-H depletion caused the increase of the mitotic indices to 30 and 20%, respectively (Supplemental Figure S2). The mitotic spindles in CENP-H–depleted cells are as dense as those in the control cells, and there was no loss of spindles as observed in the Rad21-depleted cells. Similarly, the normal spindles were observed in CENP-H–depleted mitotic cells at 35 h of Dox treatment, at which time the mitotic index was 30% (Supplemental Figure S2). In fact, a previous study reported that mitotic spindles were still observed after 48 h of Dox treatment, with a mitotic index of 85% (Fukagawa et al., 2001). Thus, the observed Rad21 depletion phenotype is specific, strongly suggesting that cohesin is required for mitotic spindle formation in vivo.
Cohesin Depletion Affects the Efficiency of Spindle Assembly in Human Cells
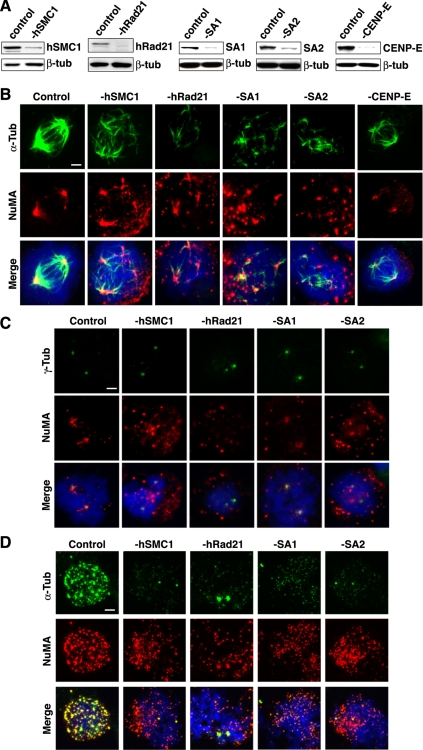
To further substantiate the observations in chicken cells, we treated HeLa cells with siRNAs targeting both SMC and non-SMC components of cohesin (i.e., hSMC1, hRad21, SA1, and SA2). Two sequential transfections of siRNAs resulted in >80% depletion of each component (Figure 4A).
Figure 4.
Cohesin depletion inhibits mitotic spindle recovery in vivo. (A) Western blot analysis of siRNA-transfected cell lysates. Cell lysates were prepared 24 h after the second transfection with siRNA for hSMC1, Rad21, SA1, SA2, or CENP-E. Control cells were transfected with a randomized siRNA. β-Tubulin serves as a loading control. (B) Cohesin depletion inhibits the spindle recovery. Cells transfected with siRNA were treated with nocodazole and incubated in fresh medium for 20 min before fixation. Mitotic cells were stained with antibodies specific for α-tubulin (green) and NuMA (red). DNA is visualized by DAPI staining (blue). The depleted proteins are indicated at top. Bar, 5 μm. (C) Multiple NuMA signals are not due to centrosome amplification. In a similar experiment as described in B, cells were stained with γ-tubulin (green) to visualize centrosomes and colocalization with NuMA was analyzed. Bar, 5 μm. (D) Cohesin depletion inhibits Taxol-induced spindle asters in vivo. Cohesin-depleted cells and control cells were treated with Taxol for 5 min as in Figure 2B. Bar, 5 μm.
Twenty-four hours after the second siRNA transfection, no significant defect of mitotic spindle morphology was observed (Supplemental Figure S3). However, similar to what was observed in Rad21-depleted chicken DT40 cells, after transient disruption of spindles by nocodazole, reassembly of spindles was significantly abrogated in cohesin-depleted mitotic cells (Figure 4B). In mitotic cells treated with a negative control siRNA of randomized sequence, almost complete recovery of spindles was observed at 20 min after the removal of nocodazole, accompanied with efficient reclustering of NuMA at the spindle poles. In contrast, the mitotic cells depleted of any tested cohesin component exhibited similar defects in spindle recovery. Specifically, multiple short disorganized spindle asters were formed in the mitotic cytoplasm colocalizing with some of the NuMA foci, which failed to cluster to the spindle poles. Consistent with the absence of holo-cohesin at interphase centrosomes (Figure 1C and Supplemental Figure S1B), hRad21 depletion had no effect on microtubule regrowth in interphase cells (Supplemental Figure S4). Abnormal mitotic spindle aster formation in cohesin-depleted cells was not due to abnormal centrosome numbers because only two γ-tubulin foci were present in these cells (Figure 4C).
Cohesin defects were shown to lead to merotelic attachment of spindles to kinetochores, indicating that cohesin is important for proper kinetochore–spindle attachment (Tanaka et al., 2000; Hoque and Ishikawa, 2002; Deehan Kenney and Heald, 2006). To address whether the observed spindle abnormality is due to kinetochore dysfunction, CENP-E was depleted by siRNA (Figure 4A). CENP-E depletion was shown to impair bipolar spindle attachment to kinetochores in human cells (Schaar et al., 1997; Tanudji et al., 2004). Furthermore, CENP-E depletion leads to spindle checkpoint activation (Schaar et al., 1997; Yao et al., 2000; McEwen et al., 2001; Tanudji et al., 2004). In contrast to the cohesin depletion, however, no spindle assembly defect was observed in CENP-E–depleted cells in mitosis (Figure 4B). The results suggest that the defect associated with cohesin depletion is not due to kinetochore dysfunction and checkpoint activation. This is in agreement with the clear phenotypic differences between Rad21 and CENP-H depletion in chicken DT40 cells (Figure 3F). Furthermore, although Rad21 depletion increased the mitotic index to 20–50% under this condition, the mitotic indices after hSMC1 and SA depletion were comparable with that of the control siRNA-treated cells (4–5%), indicating that the cohesin depletion phenotype is not due simply to the prolonged mitotic arrest. In addition, cells expressing GFP-NuMA-N, which abrogates cohesin localization at spindle poles (Figure 2E), also exhibited similar spindle reassembly defect in mitosis (Supplemental Figure S5), further supporting the notion that the cohesion–NuMA interaction and cohesin localization at mitotic spindle poles are functionally significant.
To further assess the efficiency of spindle assembly in cohesin-depleted cells, cells were treated with Taxol. Immediately after nocodazole release, the Taxol treatment induced multiple spindle asters in the control mitotic cells (Figure 4D). In contrast, in cohesin-depleted mitotic cells, no significant aster formation was observed, except to a minimal extent at the two spindle poles, which is similar to the observation in cohesin-depleted chicken DT40 cells (compare Figures 3E and 4D). Thus, the efficiency of spindle aster nucleation is decreased in cohesin-depleted cells. The limited aster formation at the spindle poles is likely due to residual cohesin (see below; Supplemental Figure S7).
When further depletion was achieved after three rounds of hRad21 siRNA transfections, ∼40% of mitotic cells exhibited spindle abnormality with a weaker β-tubulin signal accompanied by a multispindle pole phenotype (Figure 5A). Similar multipole phenotype by hRad21 (Scc1) depletion was also reported previously (Losada et al., 2005). γ-Tubulin staining also revealed that centrosomes were fragmented in a mitosis-specific manner, but not in interphase cells, suggesting a structural defect of mitotic spindle poles (Figure 5B). Together, these results demonstrate that cohesin is important for spindle pole function as well as centrosome integrity in human cells.
Figure 5.
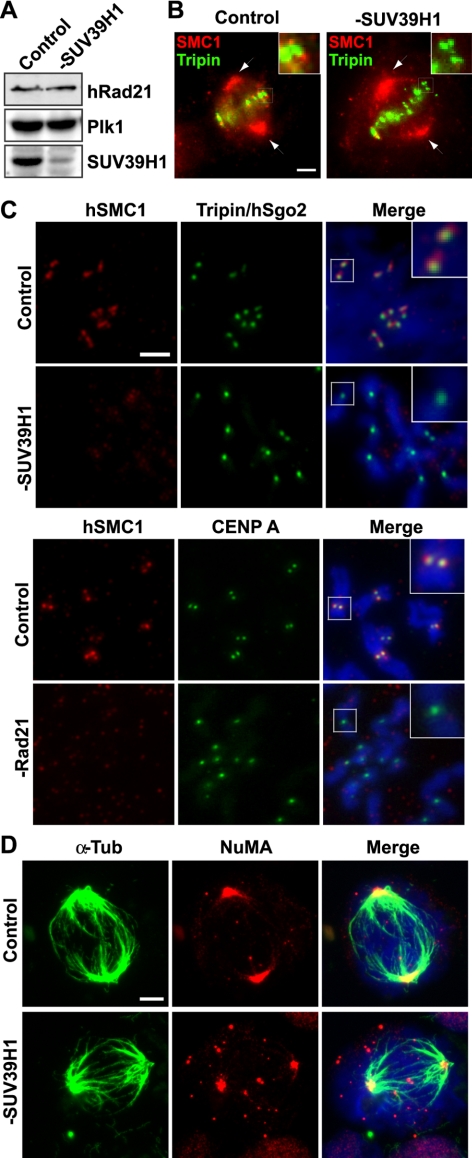
hRad21 depletion induces spindle abnormality and centrosome fragmentation. (A) Spindle morphology of control- and hRad21-siRNA transfected cells. For prolonged Rad21 depletion, siRNA transfection was repeated three times at 24-h intervals. At 12 h after the third siRNA transfection, hRad21 depletion led to spindle abnormality in ∼40% of mitotic cells. Top, α-tubulin only. Bottom, α-tubulin and DAPI overlay. Bar, 5 μm. (B) γ-Tubulin and DAPI staining of mitotic and interphase cells. M phase centrosomes in high magnification are shown. Magnification is identical to that described in A.
The Effect of Cohesin Depletion on Spindle Assembly Is Distinct from the Effect on Centromeric Sister Chromatid Cohesion
To further distinguish cohesin's function at spindle poles from cohesin's role in centromeric sister chromatid cohesion, SUV39H1 was depleted by siRNA (Figure 6). SUV39H1 is a major histone H3 lysine 9 (H3K9) methyltransferase (HMTase) important for H3K9 methylation (H3K9me) at the pericentromeric heterochromatin in mammalian cells (Aagaard et al., 1999, 2000). Enrichment of H3K9me at the pericentromeric heterochromatin was shown to recruit the heterochromatin binding protein HP1 (Swi6 in S. pombe), which further recruits cohesin in Schizosaccharomyces pombe (Bernard et al., 2001; Nonaka et al., 2002). Pericentromeric heterochromatin-dependent cohesin recruitment is necessary for centromeric sister chromatid cohesion during mitosis in S. pombe (Bernard et al., 2001; Nonaka et al., 2002). A similar requirement for H3K9me and HP1 for cohesin recruitment to the centromeres and subsequent centromeric sister chromatid cohesion was demonstrated in chicken DT40 cells (Fukagawa et al., 2004). Consistent with these observations, we found that SUV39H1 depletion in HeLa cells results in displacement of cohesin from centromeres and disruption of centromeric sister chromatid cohesion (Figure 6, B and C). We found that cohesin localization at the spindle poles is not affected by SUV39H1 depletion, thus separating the cohesin function at centromeres and spindle poles (Figure 6B). Consistent with the intact cohesin localization at the spindle poles, we found that spindle assembly and recovery after nocodazole-mediated disruption at the poles is intact (Figure 6D). It should be noted that despite the clear sister chromatid cohesion loss observed in metaphase chromosome spreads after hypotonic treatment (Figure 6C), centromeres are still relatively clustered in both cohesin- and SUV39H1-depleted mitotic cells after CSK extraction (Supplemental Figure S6A and B). Interestingly, however, after disruption and reassembly of spindles by transient nocodazole treatment, centromeres were severely dispersed in cohesin-depleted cells with lack of proper spindle reassembly, whereas they remained relatively clustered in the SUV39H1-depleted cells with efficient spindle recovery (Supplemental Figure S6C). The results further indicate that the spindle defect caused by cohesin depletion additionally compromised chromosome positioning, further accentuating the cohesion defect. Together, the results clearly separate the function of cohesin at centromeres from that at spindle poles and indicate that the spindle abnormality caused by cohesin depletion is not due to depletion of cohesin at the centromere regions of chromosomes.
Figure 6.
Depletion of SUV39H1 selectively inhibits cohesin function at centromeres but not at spindle poles. (A) Western blot analysis of control- and SUV39H1-siRNA–treated cells. There is no effect on the cohesin subunit hRad21. Plk1 serves as a loading control. (B) Cohesin localization at kinetochores, but not spindle poles, is affected by SUV39H1 depletion. Cells growing on glass coverslips were extracted with MTSB buffer (MTSB: 4 M glycerol, 100 mM PIPES, pH 6.8, 1 mM EGTA, and 5 mM MgCl2) containing 0.5% Triton X-100 for 2 min at room temperature followed by rinsing in MTSB. They were then fixed in −20°C methanol for 10 min. Cohesin is detected by antibody specific for hSMC1 (red). Antibody specific for Tripin/hSgo2 (green) was used as a centromere marker. The kinetochore regions in higher magnification are shown at the top right corners. Spindle poles are indicated by arrows. Bar, 5 μm. (C) Metaphase chromosome spreads of control- and SUV39H1-siRNA–treated cells. Immunofluorescent staining was carried out using antibodies specific for hSMC1 (red) and Tripin/Sgo2 (green). For comparison, metaphase chromosome spreads of control- and Rad21-depleted cells are also shown using antibodies specific for hSMC1 (red) and CENP-A (green). DNA is visualized by DAPI staining (blue). Bar, 2 μm. The kinetochore regions in higher magnification are shown at the top right corners. (D) SUV39H1 depletion does not affect spindle recovery. Cells transfected with siRNA as indicated were treated with nocodazole and incubated in fresh medium for 20 min before fixation similar to the experiments in Figure 4B. Mitotic cells were stained with antibodies specific for α-tubulin (green) and NuMA (red) as indicated at top. Bar, 5 μm.
Purified M Phase Cohesin, but not S Phase Nuclear Cohesin, Functions in Mitotic Aster Assembly In Vitro
Because cohesin targeting to the spindle poles is specific to mitosis as shown above, we next addressed whether cohesin's ability to support spindle assembly is also specific to mitosis. As observed previously, depletion of cohesin from HeLa mitotic extracts by using anti-hRad21 antibody resulted in abrogation of spindle aster assembly in vitro (Figure 7A) (Gregson et al., 2001). Because this assay does not include any chromosomes, the effect of cohesin on spindle assembly is likely independent of its function in sister chromatid cohesion. The depleted extracts were then complemented with human cohesin immunopurified from a stable cell line expressing FLAG-tagged hSMC1 (hSMC1-flg). The bound cohesin holocomplex was stringently washed (i.e., 1 M salt) before elution with FLAG peptide, and the purity of both mitotic and S phase cohesin was verified by silver stain (Figure 7B). When cohesin purified from mitotic extracts was added back to the cohesin-depleted extracts, spindle aster assembly was partially restored (Figure 7A, lane 3). However, adding back a comparable amount of S phase nuclear cohesin failed to restore any aster assembly (Figure 7A, lane 4, and B). Thus, cohesin's ability to support spindle aster assembly is specifically associated with mitotic cohesin, and not with S phase nuclear cohesin, providing further evidence that cohesin's role in spindle assembly is distinct from that in sister chromatid cohesion.
Figure 7.
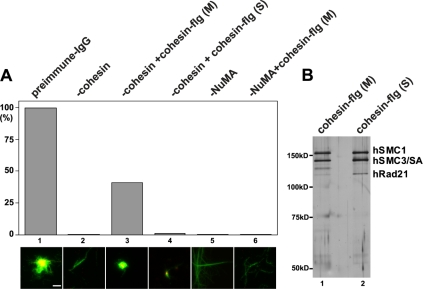
M phase-, but not S phase-, cohesin complements mitotic spindle aster assembly in vitro. Bar, 2 μm. (A) Cohesin add-back to cohesin- and NuMA-depleted extracts. Cohesin was depleted from HeLa mitotic aster assembly extracts using anti-Rad21 antibody (-cohesin, lanes 2–4). Aster assembly was compared with those of the preimmune IgG-depleted extracts (lane 1), and the extracts depleted with anti-NuMA antibody (-NuMA, lanes 5 and 6). Lane 3, FLAG-purified cohesin (cohesin-flg) from M phase extracts was added to the cohesin-depleted extracts before aster assembly. Lane 4, a comparable amount of cohesin-flg purified from S phase-synchronized extracts was added to the cohesin-depleted extracts. Lane 6, M phase cohesin-flg was added to the NuMA-depleted extracts before aster assembly. Typical spindle morphology observed in each depletion (and add-back) is shown below. Green, β-tubulin. Red, NuMA. Yellow, overlap between β-tubulin and NuMA. (B) A silver stain of cohesin-flg from S phase and M phase. Cohesin components are indicated.
The spindle phenotype of cohesin depletion is distinct from that of NuMA depletion (Figure 7A). Although loss of either NuMA or cohesin drastically decrease the numbers of asters, a few short asters are observed in the cohesin-depleted extracts with residual NuMA at the center (Figure 7A, lane 2) (Gregson et al., 2001). In contrast, long and unfocused microtubule fibers with no central clustering were observed in the NuMA-depleted extracts (Figure 7A, lanes 5 and 6). This is consistent with the previous observations that NuMA is important for both proper focusing of spindle asters and formation of spindles (Gordon et al., 2001; Levesque et al., 2003). In addition, purified mitotic cohesin failed to reestablish spindle assembly activity in the NuMA-depleted extracts (Figure 7A, lane 6), further supporting the notion that the functions of cohesin and NuMA in spindle assembly are distinct.
DISCUSSION
Two Distinct Functions of Cohesin in Mitosis
Our results suggest that cohesin promotes proper chromosome segregation in two different ways. First, there is its well-characterized function of sister chromatid cohesion through interaction with chromatin. The second mode involves localization at spindle poles to function in spindle organization as described in this study. Although it is unusual for a chromatin-associated factor to play a role in spindle organization, it is not unprecedented. For example, origin recognition complex protein 2 (ORC2) and ORC6 have, in addition to their activities at the replication origins, spindle-related functions at the centrosomes and kinetochores, respectively (Prasanth et al., 2002, 2004). DNA repair factors such as BRCA1 were also shown to localize to the centrosomes and play a role in centrosome duplication (Xu et al., 1999; Fukasawa, 2005).
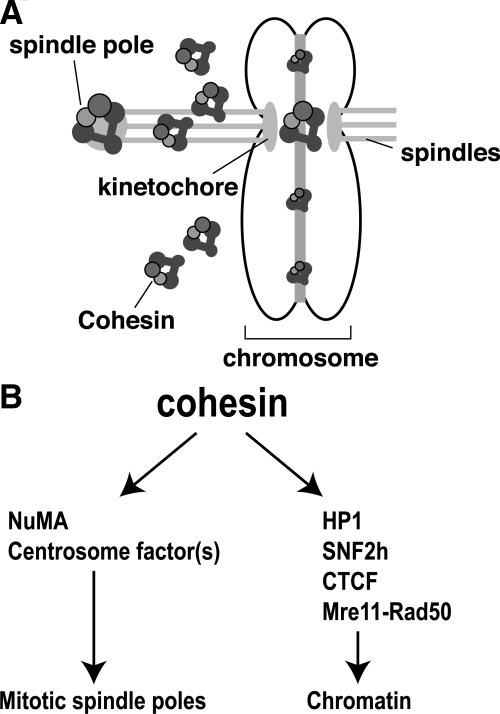
We hypothesize that specific interactions with other cellular factors determine subcellular localization and functional specificity of cohesin (Figure 8). Several factors have been identified to be required for specific targeting of cohesin. For example, Dicer and HP1 are required for pericentromeric heterochromatin recruitment of cohesin in S. pombe and vertebrates, whereas kinetochore components are involved in centromere targeting of cohesin in Saccharomyces cerevisiae (Megee and Koshland, 1999; Megee et al., 1999; Tanaka et al., 2000; Bernard et al., 2001; Nonaka et al., 2002; Hall et al., 2003; Fukagawa et al., 2004). Furthermore, hSNF2h is important for human cohesin recruitment to a type of Alu repeat sequence, whereas Mre11-Rad50 recruits cohesin to DNA damage sites (Hakimi et al., 2002; Kim et al., 2002). CCCTC-binding factor (CTCF) was also shown to target cohesin to many of the CTCF-binding sites in mammalian cells (Parelho et al., 2008; Stedman et al., 2008; Wendt et al., 2008). Our results reveal that cohesin is recruited to the spindle poles in a mitosis-specific manner through two mechanisms: interaction with centrosomes or with NuMA. Therefore, we propose that NuMA and centrosomal protein(s) are critical for recruiting, and thus determining the functional specificity of, cohesin in spindle organization (Figure 8). This is consistent with our observation that depletion of SUV39H1, which is required for proper localization of HP1 and subsequent recruitment of cohesin to the centromeres, had no effect on cohesin localization at spindle poles and spindle assembly in mitosis. Interestingly, a recent study reported that SUV39h1 and SUV39h2 double knockout had no effect on centromeric localization of cohesin in mouse embryonic fibroblasts despite the significant loss of H3K9me and HP1 (Koch et al., 2008). The discrepancy between this study and our current study in human cells, as well as the previous study demonstrating loss of H3K9me, HP1, and cohesin in Dicer-depleted chicken cells (Fukagawa et al., 2004), is currently unclear. It is possible that in mouse embryonic fibroblasts, other HMTases may compensate for the lack of Suv39h1/h2. The residual H3K9me may thus be sufficient to recruit some HP1 and cohesin to the centromeres, which may also explain the increased distance between sister centromeres, but not the total loss of centromeric cohesion, in these cells (Koch et al., 2008).
Figure 8.
A model for two distinct roles of cohesin in mitotic chromosome organization and function. (A) During mitosis, cohesin has two distinct functions. By associating with chromosomes, cohesin mediates the pairing of sister chromatids. By associating with mitotic spindle poles, cohesin plays a role in mitotic spindle dynamics. (B) The functional specificities of cohesin at different subcellular locations may be determined by the interactions with factors specific for each role. For example, HP1, SNF2h, CTCF, and Mre11-Rad50 were shown to function in chromosome loading of cohesin, whereas NuMA and an unknown centrosome component(s) are required for its spindle pole localization in vertebrate cells.
Mitosis-specific Activity of Cohesin at the Spindle Poles
It was shown that cohesin dissociates from chromosomes before metaphase, which requires the Plk1 (Sumara et al., 2002). A recent study revealed that Rad21/Scc1 and SA2/Scc3 are the major targets of Plk1 (Hauf et al., 2005). It is possible that cell cycle-specific covalent modification such as phosphorylation of cohesin subunit(s) and/or its interacting protein(s) at centrosomes is involved in mitosis-specific centrosome targeting of cohesin. Our observation that only mitotic cohesin, but not S phase-derived cohesin, is able to restore spindle aster formation in vitro further suggests that the interphase nuclear cohesin and mitotic cytoplasmic cohesin are qualitatively different.
Because a subpopulation of interphase nuclear cohesin interacts with NuMA (Gregson et al., 2001), it is possible that this NuMA-interacting cohesin clusters to the mitotic spindle poles by piggy-backing onto NuMA, which accumulates at the spindle poles in a mitosis-specific manner. This NuMA clustering is dynein dependent (Merdes et al., 2000). Consistent with this, a weak but specific interaction of cohesin with dynein was observed previously (Gregson et al., 2001). Interestingly, the mRNA export factor Rae1, which also binds to NuMA and is required for mitotic spindle assembly (Blower et al., 2005; Wong et al., 2006), was recently found to interact with hSMC1 at the mitotic spindle pole (Wong and Blobel, 2008), further supporting the notion that cohesin functionally localizes to the spindle poles. In addition, hSMC1 found at the interphase centrosomes may serve as a core to assemble the holo-cohesin complex at spindle poles for mitosis. Further study is necessary to address the role of hSMC1 at interphase centrosomes.
Role of Cohesin in Spindle Formation
Our results reveal a novel function of cohesin in mitotic spindle assembly at the spindle poles. This function of cohesin in human and chicken cells may also be conserved in other organisms. Localization of Rad21 (DRAD21) at spindle poles was reported previously in Drosophila (Warren et al., 2000). siRNA against DRAD21 led to abnormal spindle morphology lacking astral MTs (Vass et al., 2003). The abnormal spindle morphology was originally interpreted as a result of destabilization of kinetochore MTs due to the lack of tension caused by the defective centromeric cohesion. However, our results with SUV39H1-depleted cells strongly argue that the observed spindle defects caused by cohesin depletion are not caused by loss of cohesin at centromeres. Thus, the lack of astral MTs observed in Drosophila could also be explained by dysfunction of cohesin at spindle poles. Based on the Western blot analysis, the amount of cohesin associated with centrosomes in a microtubule-independent manner is estimated to be <1% of the cytoplasmic cohesin (Figure 1B; data not shown). However, based on the immunofluorescent signals (compare Figures 1A and 6B), up to 10-fold more cohesin was estimated to cluster to the intact spindle poles, including the pericentrosomal area (which is facilitated by NuMA), suggesting that cohesin's role at spindles poles is an important function of the cytoplasmic cohesin in mitosis.
Although cohesin depletion by siRNA in human cells affected the efficiency of mitotic spindle assembly, it did not lead to the complete abolishment of mitotic spindle formation in our study. This is in contrast to the in vitro mitotic spindle aster assembly assay using human cell extracts, in which immunodepletion of cohesin led to almost complete abrogation of spindle assembly (Figure 7A). This discrepancy can be explained by the incomplete depletion of cohesin by transient siRNA transfection. We found that cohesin remains at the spindle poles in mitotic cells even after the majority of cytoplasmic cohesin is depleted by siRNAs (Supplemental Figure S7). Consistent with this, a prolonged depletion in chicken DT40 cells did lead to the loss of mitotic spindles. Although a secondary effect due to long-term depletion of cohesin cannot be excluded, the results agree with the in vitro depletion experiments.
It remains unclear as to how cohesin influences centrosome/spindle pole activity. One possibility is that loss of cohesin from centrosomes disrupts the association of essential centrosome components, thus affecting centrosome integrity and function. Alternatively, but not mutually exclusively, cohesin may directly participate in microtubule nucleation/assembly. The fact that purified cohesin (resistant to a 1 M salt wash) has the ability to restore spindle assembly activity in vitro strongly suggests that cohesin itself, and not necessarily an associated factor, is functionally important. For example, although NuMA interacts with cohesin, their depletion phenotypes are different and adding back the purified cohesin to NuMA-depleted extracts failed to even partially restore spindle assembly, indicating that the observed effect is not due to cohesin-associated NuMA. According to the model of cohesin ring formation (Gruber et al., 2003), one provocative idea is that cohesin may bundle spindle fibers instead of chromatin fibers to stabilize the spindles. The importance of cohesin for the maintenance of centrosome integrity is supported by the observation that Rad21 depletion leads to centrosome fragmentation. However, the fact that impairment of spindle reassembly can be observed without centrosome fragmentation suggests that there may be two different aspects to cohesin's function at spindle poles. Although further study is necessary to dissect this novel function of cohesin, our results demonstrate that cohesin functions in mitotic spindle formation in vivo, which is distinct from its role in sister chromatid cohesion. The spindle function of cohesin may contribute to its chromatid cohesion function to ensure proper chromosome organization and segregation in mitosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Duane Compton for the NuMA cDNA. We also thank Drs. Duane Compton and Jonathan Scholey for critical reading of the manuscript. This work was supported in part by the Yanagida Center of Excellence grant for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japanese Government (to S. T.); the National Institutes of Health (GM86877 and GM44762), a core grant (CA06927), the Leukemia and Lymphoma Society, and an appropriation from the Commonwealth of Pennsylvania (to T.J.Y.); grant GM-59150 from the National Institutes of Health and research grant 1-FY02-207 from the March of Dimes Birth Defects Foundation (to K. Y.). K. Y. was a Leukemia and Lymphoma Society Scholar.
Abbreviations used:
- H3K9me
histone H3 lysine 9 methylation
- HMTase
histone methyltransferase
- MTs
microtubules
- NuMA
nuclear mitotic apparatus protein
- plk1
polo-like kinase 1
- SMC
structural maintenance of chromosomes.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0419) on January 7, 2009.
REFERENCES
- Aagaard L., et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L., Schmid M., Warburton P., Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- Ait-Si-Ali S., Guasconi V., Fritsch L., Yahi H., Sekhri R., Naguibneva I., Robin P., Cabon F., Polesskaya A., Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure J.-F., Partridge J. F., Genier S., Javerzat J.-P., Allshire R. C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Compton D. A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Cleveland D. W. NuMA is required for the proper completion of mitosis. J. Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deehan Kenney R., Heald R. Essential roles for cohesin in kinetochore and spindle function in Xenopus egg extracts. J. Cell Sci. 2006;119:5057–5066. doi: 10.1242/jcs.03277. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Dionnne M. A., Compton D. A. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J. B., Hasbani M. J., Gill S. R., Schroer T. A., Compton D. A. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Compton D. A. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Abian J. F., Sumara I., Hirota T., Hauf S., Gerlich D., De La Torre C., Ellenberg J., Peters J. M. Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Gordon M. B., Howard L., Compton D. A. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 2001;152:425–434. doi: 10.1083/jcb.152.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson H. C., Schmiesing J. A., Kim J.-S., Kobayashi T., Zhou S., Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- Gregson H. C., Van Hooser A. A., Ball J., A R, Brinkley B. R., Yokomori K. Localization of human SMC1 protein at kinetochores. Chrom. Res. 2002;10:267–277. doi: 10.1023/a:1016563523208. [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C. H., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Hakimi M. A., Bochar D. A., Schmiesing J. A., Dong Y., Barak O. G., Speicher D. W., Yokomori K., Shiekhattar R. A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- Hall I. M., Noma K., Grewal S. I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., Peters J. M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Waizenegger I. C., Peters J.-M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Hoque M. D., Ishikawa F. Cohesin defects lead to premature sister chromatid separation, kinetochore dysfunction and spindle-assembly checkpoint activation. J. Biol. Chem. 2002;277:42306–42314. doi: 10.1074/jbc.M206836200. [DOI] [PubMed] [Google Scholar]
- Hoque M. T., Ishikawa F. Human chromatid cohesin component hRad21 is phosphorylated in M phase and associated with metaphase centromeres. J. Biol. Chem. 2001;276:5059–5067. doi: 10.1074/jbc.M007809200. [DOI] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J. B., Liu S. T., Kao G. D., Muschel R., Chan G. K., Yen T. J. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki M., Weber K., Osborn M. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO J. 1991;10:3351–3362. doi: 10.1002/j.1460-2075.1991.tb04899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Krasieva T. B., LaMorte V. J., Taylor A.M.R., Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Koch B., Kueng S., Ruckenbauer C., Wendt K. S., Peters J. M. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- Levesque A. A., Howard L., Gordon M. B., Compton D. A. A functional relationship between NuMA and kid is involved in both spindle organization and chromosome alignment in vertebrate cells. Mol. Biol. Cell. 2003;14:3541–3552. doi: 10.1091/mbc.E03-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Yokochi T., Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Losada A., Yokochi T., Kobayashi R., Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B. F., Chan G. K., Zubrowski B., Savoian M. S., Sauer M. T., Yen T. J. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P. C., Koshland D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- Megee P. C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W. C., Cleveland D. W. Formation of spindle poles by dynein/dynactin-dependent transport. J. Cell Biol. 2000;149:851–861. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J. D., Cleveland D. W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Moudjou M., Bornens M. Method of centrosome isolation from cultured animal cells. In: Celis J. E., editor. Cell Biology: A Laboratory Handbook. Vol. 2. London, United Kingdom: Academic Press; 1998. pp. 111–119. [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S. S., Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Parelho V., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Saxton W. M., Stemple D. L., Leslie R. J., Salmon E. D., Zavortink M., McIntosh J. R. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B. T., Chan G.K.T., Maddox P., Salmon E. D., Yen T. J. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing J. A., Ball A. R., Gregson H. C., Alderton J., Zhou S., Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc. Natl. Acad. Sci. USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing J. A., Gregson H. C., Zhou S., Yokomori K. A human condensin complex containing hCAP-C/hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M., Brust-Mascher I., Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Sonoda E., et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- Stedman W., Kang H., Lin S., Kissil J. L., Bartolomei M. S., Lieberman P. M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D. L., Sen S., Mancini M. A., Brinkley B. R. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton. 2003;55:134–146. doi: 10.1002/cm.10120. [DOI] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Gieffers C., Peters B. H., Peters J.-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Stukenberg P. T., Kelm O., Redemann N., Nigg E. A., Peters J.-M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanudji M., Shoemaker J., L'Italien L., Russell L., Chin G., Schebye X. M. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Vass S., Cotterill S., Valdeolmillos A. M., Barbero J. L., Lin E., Warren W. D., Heck1 M.M.S. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr. Biol. 2003;13:208–218. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Waizenegger I. C., Hauf S., Meinke A., Peters J.-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Warren W., et al. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Wendt K. S., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wong R. W., Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl. Acad. Sci. USA. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R. W., Blobel G., Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. USA. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Weaver Z., Linke S. P., Li C., Gotay J., Wang X. W., Harris C. C., Ried T., Deng C. X. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yen T. J., Compton D. A., Wise D., Zinkowski R. P., Brinkley B. R., Earnshaw W. C., Cleveland D. W. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P. J., Simon P. M., Borisy G. G. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J. Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.