Abstract
The Ro autoantigen is a ring-shaped RNA-binding protein that binds misfolded RNAs in nuclei and is proposed to function in quality control. In the cytoplasm, Ro binds noncoding RNAs, called Y RNAs, that inhibit access of Ro to other RNAs. Ro also assists survival of mammalian cells and at least one bacterium after UV irradiation. In mammals, Ro undergoes dramatic localization changes after UV irradiation, changing from mostly cytoplasmic to predominantly nuclear. Here, we report that a second role of Y RNAs is to regulate the subcellular distribution of Ro. A mutant Ro protein that does not bind Y RNAs accumulates in nuclei. Ro also localizes to nuclei when Y RNAs are depleted. By assaying chimeric proteins in which portions of mouse Ro were replaced with bacterial Ro sequences, we show that nuclear accumulation of Ro after irradiation requires sequences that overlap the Y RNA binding site. Ro also accumulates in nuclei after oxidative stress, and similar sequences are required. Together, these data reveal that Ro contains a signal for nuclear accumulation that is masked by a bound Y RNA and suggest that Y RNA binding may be modulated during cell stress.
INTRODUCTION
It has become increasingly clear that eukaryotic cells contain an astounding variety of noncoding RNAs that function in diverse ways. Many of these RNAs function by base pairing with nucleic acid targets. RNAs that function in this way include the U small nuclear RNAs that function in pre-mRNA splicing, the many small nucleolar RNAs that guide the processing and modification of the ribosomal RNAs, and the telomerase RNA that provides the template for addition of repeat sequences to the ends of chromosomes (Hannon et al., 2006). More recently, a plethora of short (∼20–30 nucleotide [nt]) RNAs have been shown to base pair with complementary sites on RNA to influence mRNA translation and stability, to guide heterochromatin formation, and to prevent mobilization of transposable elements (Farazi et al., 2008).
In contrast, some noncoding RNAs function by binding proteins and in some cases inhibiting their activities. For example, the abundant nuclear 7SK RNA, through binding a protein called HEXIM1, sequesters the transcription elongation factor P-TEFb in an inactive complex. In the presence of stress, such as UV irradiation or treatment with the transcription inhibitor actinomycin B, 7SK dissociates, allowing increased transcription by RNA polymerase II (Michels and Bensaude, 2008). In heat-shocked mouse cells, B2 RNA binds RNA polymerase II to repress transcription, whereas in human cells, Alu RNAs carry out this function (Mariner et al., 2008). In contrast, the signal recognition particle (SRP) RNA acts as a scaffold for protein binding and also facilitates interaction between components of the SRP and its receptor (Neher et al., 2008). For each of these noncoding RNAs, both those that function by base pairing and those that function in other ways, identification of the function has resulted in a wealth of information about basic cellular processes.
Y RNAs are a class of noncoding RNAs whose function is beginning to be uncovered. These RNAs are ∼100 nt and are transcribed by RNA polymerase III (Chen and Wolin, 2004). The number of distinct Y RNAs varies from one to four depending on the species (Mosig et al., 2007; Perreault et al., 2007). Although the primary sequences differ, all Y RNAs fold into a structure consisting of a long stem formed by base pairing the 5′ and 3′ ends and a large internal loop (Chen and Wolin, 2004). Newly synthesized Y RNAs are initially bound by La, a nuclear protein that recognizes the UUUOH that is at the 3′ terminus of all nascent RNA polymerase III transcripts (Wolin and Cedervall, 2002). However, the majority of Y RNAs in animal cells are complexed in the cytoplasm to a ring-shaped protein known as the Ro autoantigen. Because Ro also associates with misfolded RNAs in some vertebrate nuclei, Ro is proposed to function in RNA quality control (O'Brien and Wolin, 1994; Chen et al., 2003). Both Ro and at least one Y RNA are also present in the radiation-resistant eubacterium Deinococcus radiodurans, where they function in 23S rRNA maturation (Chen et al., 2007). In both D. radiodurans and mammalian cells, Ro is important for cell survival after UV irradiation (Chen et al., 2000, 2003). In mammalian cells, Ro undergoes dramatic alterations in subcellular localization after UV irradiation, changing from mostly cytoplasmic to largely nuclear within 3 h (Chen et al., 2003).
Structural, biochemical, and genetic studies have shown that one role of Y RNAs is to regulate access of other RNAs to Ro. Crystallographic and biochemical analyses revealed that Ro contains two overlapping RNA binding sites. The 3′ ends of misfolded RNAs bind in the central cavity of the Ro ring, whereas helical portions of these RNAs bind on the basic surface. In contrast, Y RNAs bind on the outside of the ring to surfaces that partially overlap the misfolded RNA site (Stein et al., 2005; Fuchs et al., 2006). Evidence that Y RNAs prevent binding of other RNAs in vivo was provided in D. radiodurans. During normal growth of these bacteria, 23S rRNA maturation is inefficient, resulting in precursor accumulation. During growth at elevated temperature, maturation becomes efficient and requires Ro. Consistent with the hypothesis that Y RNAs inhibit Ro function, maturation is efficient at all temperatures in bacteria that either lack the major Y RNA or express a mutant Ro that does not bind Y RNAs (Chen et al., 2007).
In addition to regulating access to Ro, it is likely that Y RNAs have additional functions. First, because Ro binds Y RNAs in the cytoplasm and misfolded RNAs in nuclei (O'Brien and Wolin, 1994; Chen et al., 2003), Y RNA binding could influence the subcellular distribution of Ro. Second, although some organisms contain a single Y RNA, vertebrate cells contain between two and four distinct Y RNAs, raising the possibility that these RNAs have specialized roles. Consistent with this possibility, several proteins associate with only a subset of the four discrete Y RNAs in human cells (Bouffard et al., 2000; Fabini et al., 2001; Hogg and Collins, 2007). However, because Y RNAs are unstable in both worms and mouse cells lacking Ro (Labbe et al., 2000; Chen et al., 2003), any additional functions likely involve the Ro protein.
Here, we report that Y RNAs influence the subcellular distribution of Ro in mammalian cells. We show that a mutant Ro protein that does not bind Y RNAs accumulates in nuclei and that Ro increases in nuclei when Y RNAs are depleted. Because the D. radiodurans Ro, when expressed in mammalian cells, fails to accumulate in nuclei, we identified sequences important for nuclear accumulation by assaying chimeric proteins in which portions of mouse Ro were fused to the bacterial Ro. Accumulation of mouse Ro in nuclei after UV irradiation or oxidative stress requires sequences that overlap the Y RNA binding site. As these sequences are masked by Y RNA binding, our findings reveal a novel role for noncoding RNAs and suggest that Y RNA binding to Ro is modulated during cell stress.
MATERIALS AND METHODS
Cell Lines and Cell Culture
To prepare wild-type and Ro−/− mouse embryonic fibroblasts, 129/Sv × C57BL/6 Ro−/− mice (Xue et al., 2003) were backcrossed for six successive generations to C57BL/6 mice. Embryonic fibroblasts were prepared and immortalized by repeated passage (Todaro and Green, 1963). To generate FLAG3-Ro cells, three copies of the FLAG epitope were fused to a cDNA containing mouse Ro (Wang et al., 1996; a gift of E. Chan, University of Florida), and the resulting cDNA was cloned into the KpnI/BamHI sites of pUB6/V5-HisA (Invitrogen, Carlsbad, CA) to create pUB-3FRo. In this construct, FLAG3-Ro is expressed under control of the human ubiquitin C promoter. After transfection into immortalized Ro−/− fibroblasts, stable cell lines were selected with 5 μg/ml blasticidin S (Cayla, Toulouse, France). The FLAG3-Ro(H187S) and FLAG3-Ro(K170A R174A) mutants were generated from pUB-3FRo by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and transfected into Ro−/− fibroblasts. All fibroblast lines were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum, 100 mM β-mercaptoethanol, and 2 mM l-glutamine. For UV treatment, cells were washed with phosphate buffered saline (Invitrogen) and irradiated with UVC (253.7 nm; 10 J/m2) by using a germicidal lamp (Royal Philips Electronics, Amsterdam, The Netherlands). Fresh medium was added, and the plates were incubated for 24 h before fixation. To examine oxidative stress, cells were incubated with 50 μM hydrogen peroxide for 3 h. DBT mouse astrocytoma cells (Hirano et al., 1974) were cultured in minimal essential medium (Invitrogen) with 7% newborn calf serum and 10% tryptose phosphate broth.
To express D. radiodurans Ro in mouse cells, the coding region was amplified from genomic DNA and three copies of the FLAG epitope fused to the N terminus by using the polymerase chain reaction (PCR). The resulting DNA was cloned into the BamHI/XhoI sites of pUB6/V5-HisA. Chimeras between the mouse and D. radiodurans proteins were created using PCR.
Knockdown of Y RNAs
To generate short hairpin RNAs (shRNAs) targeting nt 48–67 of mY1, the oligonucleotides 5′-GATCCCCGTTACAGATTGAACTCCTGTTCAAGAGACAGGAGTTCAATCTGTAACTTTTTA-3′ and 5′-AGCTTAAAAAGTTACAGATTGAACTCCTGTCTCTTGAACAGGAGTTCAATCTGTAACGGG-3′ were annealed and cloned, by using the BglII/HindIII sites, behind the human H1 promoter in the enhanced green fluorescent protein (GFP)-expressing plasmid pG-SHIN2 (Kojima et al., 2004) (a gift of S. Kojima and G. Borisy, Northwestern University). To target nt 47-65 of mY3, the oligonucleotides 5′-GATCCCCGTTACAGATTTCTTTGTTCTTCAAGAGAGAACAAAGAAATCTGTAACTTTTTA-3′ and 5′-AGCTTAAAAAGTTACAGATTTCTTTGTTCTCTCTTGAAGAACAAAGAAATCTGTAACGGG-3′ were used. Sequences of chemically synthesized small interfering RNAs (siRNAs) (Dharmacon, Lafayette, CO) were mY1 sense, 5′-GUUACAGAUUGAACUCCUGUU-3′; mY1 antisense, 5′-CAGGAGUUCAAUCUGUAACUU-3′; mY3 sense, 5′-GUUACAGAUUUCUUUGUUCUU-3′; and mY3 antisense, 5′-GAACAAAGAAAUCUGUAACUU-3′. For efficient targeting, siRNAs were designed with symmetric 3′ UU overhangs (Elbashir et al., 2001). The control (NT) siRNA was siCONTROL NonTargeting siRNA #1 (Dharmacon).
The plasmid expressing shRNAs was transfected into cells using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) as described by the manufacturer. Briefly, cells were plated to 60% confluence in 10-cm culture dishes and rinsed once with Opti-MEM (Invitrogen) before transfection. Five micrograms of plasmid and 15 μl of FuGENE 6 were mixed in 500 μl of Opti-MEM, and the mixture was allowed to sit for 20 min at room temperature. After incubation, 500 μl of mixture was directly added to cells in 5 ml of Opti-MEM. After 4 h, the medium was replaced by the standard culture medium. At 2 d after transfection, GFP-positive cells were selected by fluorescence-activated cell sorting. To transfect DBT cells with chemically synthesized siRNAs, 400 pmol of each siRNA (40 nM) and 30 μl of Lipofectamine 2000 (Invitrogen) were each diluted with 1000 μl of Opti-MEM in separate tubes. After 5 min, the two mixtures were combined, incubated for 20 min, and added directly to 8 × 105 cells containing 10 ml of growth medium and plated in a 10-cm culture dish. After transfection, cells were incubated for 2 d before assaying.
Immunoprecipitations, Immunoblotting, and Immunofluorescence
Cells were harvested, washed in Tris-buffered saline (40 mM Tris-HCl, pH 7.5, and 150 mM NaCl), and sonicated in NET-2 (40 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% NP-40) containing 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail (Roche Diagnostics). Lysates were cleared by centrifugation at 100,000 × g in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA) for 20 min at 4°C and incubated with either anti-FLAG M2-conjugated agarose (Sigma-Aldrich, St. Louis, MO) or rabbit anti-mouse Ro antibodies (Chen et al., 2003) bound to protein A-Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) as described previously (Wolin and Steitz, 1984). RNAs in immunoprecipitates were labeled with [32P]pCp (England et al., 1980) and fractionated in 5% polyacrylamide/8 M urea gels, or they were subjected to Northern analysis.
Immunoblots and immunofluorescence were performed as described previously (Chen et al., 2003). Cells were photographed using an LSM 510 confocal laser scanning microscope equipped with a krypton/argon laser (Carl Zeiss, Thornwood, NY) or an Axioplan 2 microscope (Carl Zeiss) with a digital charge-coupled device camera (Hamamatsu, Bridgewater, NJ). A monoclonal anti-Ro antibody (Xue et al., 2003), which does not recognize native Ro, was used in the Western blot (Figure 1A), whereas a rabbit anti-mouse Ro antibody (Chen et al., 2003) was used for immunofluorescence. Other antibodies were mouse anti-FLAG M2 (Sigma-Aldrich), mouse anti-actin (Millipore Bioscience Research Reagents, Temecula, CA) and monoclonal anti-Sm (Y12; a gift of J. Steitz, Yale University, New Haven, CT).
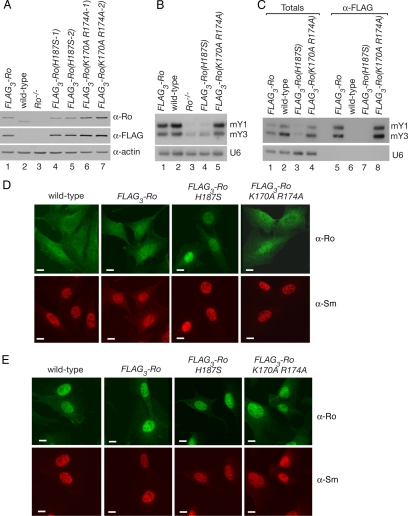
Figure 1.
A mutant Ro protein that does not bind Y RNAs accumulates in nuclei. (A) Lysates from wild-type (lane 2), Ro−/− (lane 3) and Ro−/− cell lines stably expressing either FLAG3-Ro (lane 1) or FLAG3-Ro containing the indicated mutations (lanes 4–7) were subjected to Western blotting with anti-Ro (top) and anti-FLAG antibodies (middle). To control for loading, the blot was reprobed to detect actin (bottom). (B) RNA extracted from the cell lines was subjected to Northern blotting to detect the two mouse Y RNAs, mY1 and mY3. The blot was reprobed to detect U6 snRNA (bottom). (C) Lysates from the cell lines were subjected to immunoprecipitation with anti-FLAG antibodies. RNAs from the immunoprecipitates (lanes 5–8) and equivalent amounts of the total extracts (lanes 1–4) were subjected to Northern blotting to detect mY1 and mY3. As a control, the blot was reprobed to detect the U6 snRNA. (D and E) The indicated cell lines were subjected to immunofluorescence with anti-Ro (top) and anti-Sm antibodies (bottom). Cells were either unirradiated (D) or irradiated with 10 J/m2 UVC and allowed to recover for 24 h (E) before staining. Bars, 10 μm.
Northern Blots
RNAs were fractionated in 5% polyacrylamide/8 M urea gels and transferred to ZetaProbe GT membranes (Bio-Rad, Hercules, CA) or Hybond-N (GE Healthcare). [γ-32P]ATP-labeled oligonucleotides were hybridized as described previously (Tarn et al., 1995). Oligonucleotide probes were mY1, 5′-AAGGGGGGAAAGTGTAGAACAGGA-3′; mY3, 5′-GAGCGGAGAAGGAACAAAGAAATCTG-3′; U6, 5′-ATGGAACGCTTCACGAATTTGCGAGTC-3′; U2, 5′-CAGATACTACACTTGATCTTAGCC-3′; and SRP RNA, 5′-TGCTCCGTTTCCGACCTGGGCCGGTTC-3′. The oligonucleotides 5′-CTCACTACCTTCGGACCAGCC-3′ and 5′-CCACTACTCTCGGACCAACC-3′ were used to detect the 5′ ends of mY1 and mY3, respectively.
RESULTS
A Mutant Ro Protein That Does Not Bind Y RNAs Accumulates in Nuclei
To examine whether RNA binding influences the subcellular distribution of Ro, we generated stable cell lines containing wild-type and mutant Ro proteins. Previous studies had revealed that mutating a conserved histidine in the Y RNA binding platform of X. laevis Ro to serine (H187S) decreased the affinity for Y RNAs by ∼30-fold in vitro, with a more modest decline (4-fold) in affinity for misfolded pre-5S rRNA. In contrast, mutation of two conserved basic residues in the central cavity (K170A R174A) decreased the affinity of Ro for misfolded pre-5S rRNA but did not affect Y RNA binding (Stein et al., 2005). To determine the effects in vivo, we introduced the mutations into cDNAs encoding mouse Ro, transfected the cDNAs into Ro−/− fibroblasts and selected for stable cell lines. To facilitate detection, three copies of the FLAG epitope were fused to the N terminus of each protein. In the resulting cells, all Ro present is the epitope-tagged version. Using Western blotting, we confirmed that each tagged protein was expressed at levels comparable to wild-type cells (Figure 1A).
Because Ro stabilizes bound Y RNAs (Labbe et al., 1999; Chen et al., 2003), the levels of Y RNAs in total cellular RNA provide a measure of the ability of mutant Ro proteins to bind these RNAs. In FLAG3-Ro cells, the levels of the two mouse Y RNAs (mY1 and mY3) were similar to wild-type cells (Figure 1B, lanes 1 and 2). However, the levels of Y RNAs in cells carrying the H187S mutation were similar to Ro−/− cells (Figure 1B, lanes 3 and 4), consistent with the vastly decreased binding observed for this mutant in vitro (Stein et al., 2005). As expected, mutation of central cavity residues K170 and R174 had no effect on Y RNA levels (Figure 1B, lane 5). By performing immunoprecipitations with anti-FLAG antibodies, we confirmed that the majority of Y RNAs in FLAG3-Ro and FLAG3-Ro(K170A R174A) cells were bound by Ro and that Y RNAs were undetectable in immunoprecipitates from the FLAG3-Ro(H187S) cells (Figure 1C, lanes 5, 7, and 8).
Immunofluorescence using anti-Ro antibodies revealed striking differences in the subcellular distribution of Ro between the cell lines. As reported previously (Chen et al., 2003), Ro is both cytoplasmic and nuclear in wild-type cells (Figure 1D). The distribution of Ro was similar in FLAG3-Ro cells, although slightly more Ro was detected in nuclei. However, in the presence of the H187S mutation, but not the central cavity K170A R174A mutations, Ro was significantly enhanced in nuclei (Figure 1D). After UV irradiation, Ro accumulated in nuclei in all cell lines (Figure 1E).
Increased Ro in Nuclei upon RNA Interference-mediated Knockdown of Y RNAs
The finding that FLAG3-Ro(H187S) accumulated strongly in nuclei was consistent with a model in which Y RNA binding influenced Ro's subcellular location. Alternatively, the H187S mutation could cause nuclear accumulation of Ro through a mechanism that is independent of Y RNAs, such as by interfering with binding of nuclear export receptor(s) to Ro. To determine whether Y RNA binding was important, we used RNA interference to deplete Y RNAs.
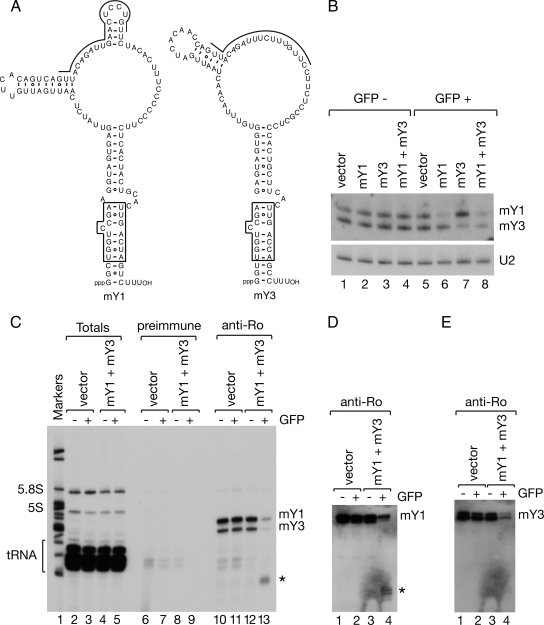
Although other noncoding RNAs have been depleted using RNA interference (Liang et al., 2003; Robb et al., 2005), we first confirmed that both mouse Y RNAs could be targeted by the RNA interference pathway. For our initial experiments, we expressed short hairpin RNAs under control of the human H1 promoter in a vector that also expresses GFP (Kojima et al., 2004). For the target sequences, we chose the large internal loops of mY1 and mY3 (Figure 2A), because the loops are single-stranded in native Ro ribonucleoproteins (RNPs) (Matera et al., 1995). The shRNA-containing plasmids were introduced into a mouse astrocytoma cell line DBT. We chose these cells because of their higher transfection efficiency and because slightly less Ro is detected in nuclei compared with fibroblasts, making small increases in nuclear accumulation upon siRNA knockdown more apparent (data not shown; but see Figure 3B). At 2 d after transfection, cells were sorted for GFP expression and RNA isolated and subjected to Northern analyses. In GFP-positive cells expressing either the mY1 or mY3 shRNA plasmid, the levels of mY1 or mY3 were reduced by 67 and 71%, respectively (Figure 2B, lanes 6 and 7). Moreover, when cells were transfected with both shRNA plasmids, both Y RNAs were reduced (Figure 2B, lane 8). Immunoprecipitation with anti-Ro antibodies, followed by end labeling of RNAs in the immunoprecipitates, confirmed that the levels of Ro-bound Y RNAs declined (Figure 2C, lane 13). Interestingly, immunoprecipitates from cells expressing the shRNA plasmids also contained a diffuse RNA band migrating at ∼60 nt (Figure 2C, lane 13, asterisk). Using cDNA cloning, we confirmed that this band contained Y RNA fragments (data not shown). In addition, Northern blotting of the immunoprecipitates revealed that 5′ fragments of the mY1 RNA stem remained bound to Ro after knockdown (Figure 2D). In contrast, less of the mY3 stem remained bound (Figure 2E).
Figure 2.
Y RNAs are efficient targets of the RNA interference pathway. (A) Proposed secondary structures of mY1 and mY3 RNAs are shown. The boxed region is a conserved helix that is critical for Ro recognition (Green et al., 1998). The sequences targeted by the shRNAs are indicated by lines. (B) Mouse astrocytes were transfected with plasmids expressing shRNAs against mY1, mY3, or both RNAs and sorted for GFP expression. Vector-transfected cells were used as a negative control. RNA extracted from GFP-negative cells (lanes 1–4) and GFP-positive cells (lanes 5–8) was subjected to Northern blotting to detect mY1 and mY3 RNA. As a loading control, the membrane was reprobed for the U2 small nuclear RNA (bottom). (C) Extracts from GFP-positive cells were subjected to immunoprecipitation with preimmune (lanes 6–9) or anti-Ro antibodies (lanes 10–13). RNAs in immunoprecipitates (lanes 6–13), and a small fraction of the total lysates (lanes 2–5) were labeled at the 3′ end with [32P]pCp. Lane 1, molecular size markers. Asterisk, a fragment of Y RNAs. (D and E) Unlabeled RNAs from the immunoprecipitates shown in C were subjected to Northern blotting to detect the 5′ half of the conserved helix of mY1 (D) and mY3 (E). Asterisk, a fragment corresponding to the 5′ end of mY1.
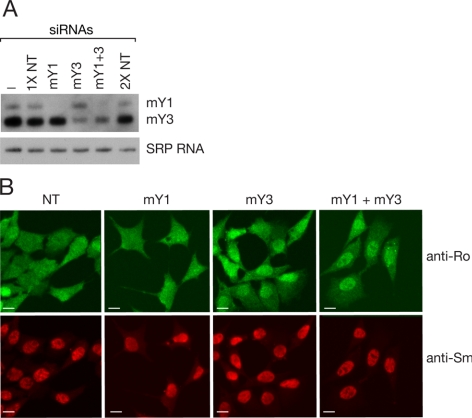
Figure 3.
Ro accumulates in nuclei after siRNA-mediated knockdown of Y RNAs. (A) siRNAs against the indicated Y RNAs were transfected into mouse astrocytes. After 2 d, the levels of Y RNAs were analyzed by Northern blotting. As a control, the blot was reprobed to detect SRP RNA. NT, nontarget control siRNAs. (B) At 2 d after transfection, the cells were subjected to immunofluorescence with anti-Ro (top) and anti-Sm antibodies (bottom). Bars, 10 μm.
We examined whether the subcellular distribution of Ro changed upon Y RNA depletion. Because attempts to use immunofluorescence to localize Ro in the GFP-positive cells were complicated by the strong GFP signal, we used siRNAs to reduce Y RNA levels (Figure 3A). Knockdown of mY1 RNA had little effect on Ro localization (Figure 3B), possibly because mY1 RNA fragments remain bound to Ro. In contrast, upon knockdown of mY3 RNA or both Y RNAs, Ro concentrated in nuclei (Figure 3B). We conclude that Y RNA binding influences Ro subcellular localization.
Sequences Required for Nuclear Accumulation of Ro Overlap the Y RNA Binding Site
Because Ro accumulates strongly in nuclei after UV irradiation (Chen et al., 2003), it seemed possible that this change in subcellular distribution was also modulated by Y RNA binding. To examine the mechanism, we defined the sequences in Ro that govern nuclear accumulation after UV treatment. Experiments in which we deleted parts of Ro or fused fragments to GFP failed to identify any sequences that conferred nuclear accumulation, suggesting the Ro ring is very sensitive to perturbations (data not shown). Consistent with this, previous attempts to use deletion analyses to identify Ro sequences involved in Y RNA binding were unsuccessful (Kenan et al., 1991; Pruijn et al., 1991).
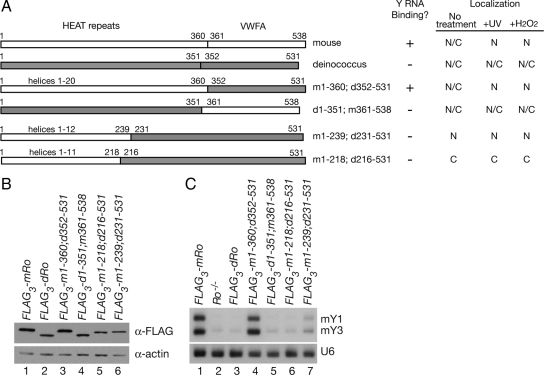
We examined whether chimeric Ro proteins, consisting of portions of mouse Ro fused to parts of the D. radiodurans Ro, could be used to identify sequences important for nuclear accumulation. First, to determine whether D. radiodurans Ro accumulates in mouse nuclei after UV, we expressed the FLAG-tagged protein under control of the UbC promoter in Ro−/− fibroblasts. After performing Western blotting, we chose cell lines in which the protein was expressed at roughly similar levels to the mouse FLAG3-Ro cells (Figure 4B). Northern blotting revealed that cells carrying D. radiodurans Ro (FLAG3-dRo) resembled Ro−/− cells in that mouse Y RNAs were nearly undetectable (Figure 4C, lanes 2 and 3). Finally, immunofluorescence revealed that although both proteins were present throughout the nucleus and cytoplasm in the majority of unirradiated cells, only mouse Ro accumulated strongly in nuclei after UV (Figure 5A, a–d, and B). Although some unirradiated cells expressing D. radiodurans Ro exhibited enhanced nuclear staining, the percentage did not increase after UV (Figure 5B). Thus, despite significant sequence conservation and structural homology between the vertebrate and bacterial proteins (Stein et al., 2005; Ramesh et al., 2007), the bacterial Ro does not bind the mouse Y RNAs in vivo and does not accumulate strongly in nuclei after UV.
Figure 4.
The HEAT repeat domain of mouse Ro is required for Y RNA binding. (A) Summary of the constructs assayed for Y RNA binding and nuclear accumulation after stress. Mouse sequences are shown in white, whereas deinococcal sequences are shaded. Numbers correspond to the amino acids of each Ro protein present within the construct. Because the mouse HEAT repeat domain of mouse Ro contains several insertions that are not present in the bacterial domain, chimeras containing mouse HEAT repeats contain slightly more amino acids than those containing bacterial HEAT repeats. Each construct also contains three copies of the FLAG epitope at the N terminus. (B) After transfection and selection of stable cell lines, Ro protein expression was examined by Western blotting by using an anti-FLAG antibody. To control for loading, the blot was reprobed to detect actin. (C) RNA extracted from the indicated cell lines was subjected to Northern analysis to detect mY1 and mY3 RNAs. As a control, the blot was reprobed to detect U6 snRNA.
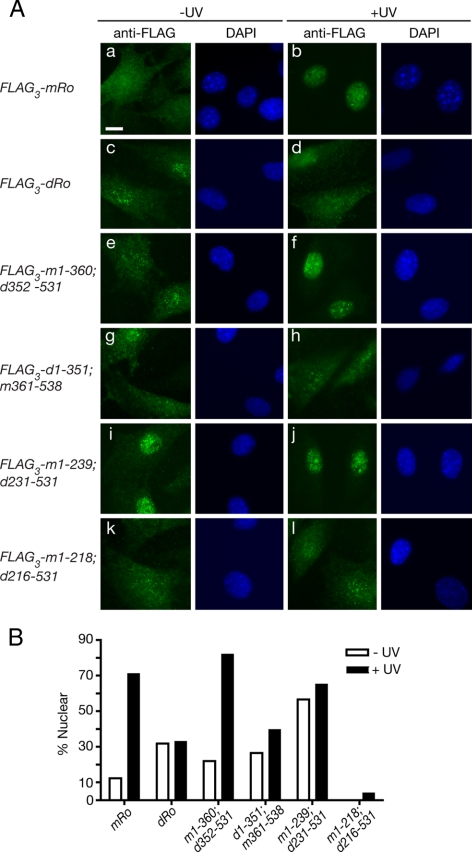
Figure 5.
Sequences for nuclear accumulation reside in the mouse HEAT repeat domain. (A) Ro−/− fibroblasts stably expressing either FLAG-tagged mouse Ro (a and b), D. radiodurans Ro (c and d), or the indicated chimeric proteins (e–l) were assayed for their location in unirradiated cells (a, c, e, g, i, and k) and 24 h after irradiation with 10 J/m2 UVC (b, d, f, h, j, and l). Left, immunofluorescence with anti-FLAG antibodies. Right, nuclei were visualized by staining with DAPI. Bar, 10 μm. (B) Histogram showing the percentage of cells in each cell line that exhibited predominantly nuclear staining before and after UV irradiation. For each measurement, at least 100 cells were counted. For FLAG3-m1-218; d216-531, no unirradiated cells with predominantly nuclear staining were detected.
To determine which domain of mouse Ro was important for nuclear accumulation, we constructed chimeric proteins. Ro consists of two domains (Figure 4A). One domain is made up of a series of α-helical HEAT repeats that form the ring. The ring is closed by the second domain, which resembles the von Willebrand Factor A (vWFA) domains found in integrins and some extracellular matrix proteins. These domains have been best characterized in integrins, in which they serve as ligand binding sites (Luo et al., 2007). By constructing FLAG-tagged proteins in which either the D. radiodurans or mouse HEAT repeat domain was fused to the vWFA domain of the other species, we found that only the chimera containing the mouse HEAT repeat domain fused to the bacterial vWFA domain (FLAG3-m1-360; d352-531) stabilized Y RNAs (Figure 4C, compare lanes 4 and 5) and accumulated in nuclei after UV (Figure 5A, e and f, and B). Thus, the mouse HEAT repeat domain is required for both Y RNA stabilization and nuclear accumulation. This result is consistent with the crystal structure of Xenopus laevis Ro, which revealed that the HEAT repeat domain contains the Y RNA binding site (Stein et al., 2005).
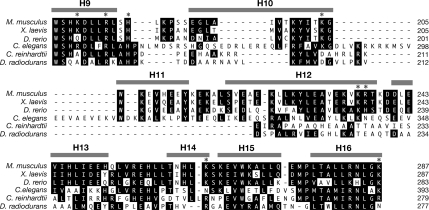
Because the mouse HEAT repeat domain contains 20 α-helices, we attempted to define the portion of Ro required by examining chimeras that contained varying numbers of mouse helices. In the presence of a chimera containing the first 12 mouse α-helices (FLAG3-m1-239; d231-531) Y RNA levels were only slightly elevated compared with Ro−/− cells (Figure 4C, compare lane 7 with lane 2). Although this could reflect a failure of the chimera to fold correctly, two residues outside helices 1–12, K264 (helix 14) and K287 (helix 16), have been shown to contribute to Y RNA binding in vitro (Fuchs et al., 2006). Interestingly, the chimeric protein was constitutively nuclear (Figure 5A, i and j). As this chimera exhibited significantly more nuclear accumulation in unstressed cells than D. radiodurans Ro (Figure 5Ac), a signal for nuclear accumulation resides within mouse helices 1–12. In Ro proteins that contain the intact mouse HEAT repeat domain, these sequences may be masked by Y RNA binding. Consistent with this hypothesis, a chimera containing only the first 11 α-helices (FLAG3-m1-218; d216-531) remained cytoplasmic (Figure 5A, k and l). Although misfolding of the chimeric protein could account for this result, an alternative explanation is that helix 12 is required for nuclear accumulation. In this regard, we note that helix 12 is poorly conserved between vertebrate and bacterial Ro proteins (Figure 6).
Figure 6.
Alignment of Ro proteins. Sequence alignment of Ro proteins from Mus musculus, X. laevis, Danio rerio, Caenorhabditis elegans, Chlamydomonas reinhardtii, and D. radiodurans. Residues that are identical or similar (L = V = I = M, F = Y = W, S = T, E = D, R = K = H) are shaded. α-Helices (gray bars) are assigned based on the X. laevis structure (Stein et al., 2005). Asterisks indicate amino acids shown by mutagenesis to contribute to Y RNA binding (Stein et al., 2005; Fuchs et al., 2006).
Attempts to further define the nuclear accumulation signal were unsuccessful. A derivative of the chimera containing mouse helices 1–12 in which mouse helix 1 was replaced by its bacterial counterpart (FLAG3-d1-52; m45-218; d216-531) was unstable (data not shown). Experiments in which helices 1–12 were fused to GFP revealed that these sequences did not confer nuclear accumulation, suggesting that the sequences do not fold correctly when removed from the context of the Ro ring (data not shown). Although the large size of the fused sequence may have contributed to misfolding, fusion of smaller portions to GFP (helices 3–12, 5–12, 7–12, 9–12, 11–12, and the isolated helix 12) also failed to confer nuclear accumulation (data not shown). A likely explanation for these data are that nuclear accumulation requires sequences, in addition to helix 12, that must be present in the context of the Ro ring to fold correctly.
Y RNA Binding Also Regulates Ro Nuclear Accumulation during Oxidative Stress
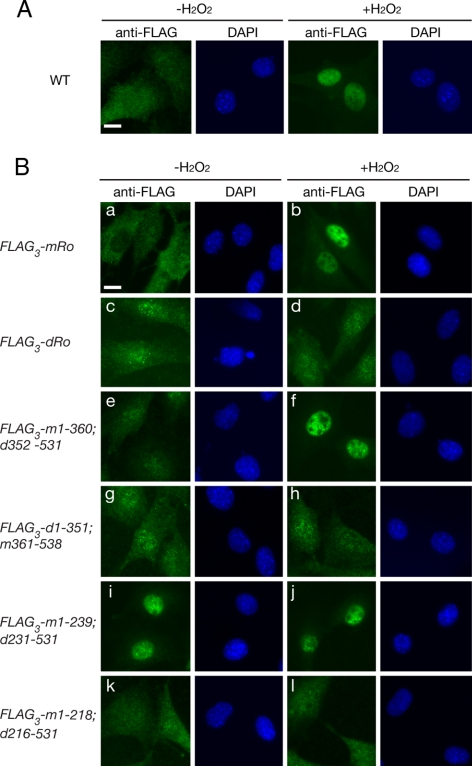
During experiments to determine whether other environmental stresses affect Ro subcellular distribution, we discovered that Ro also accumulates in nuclei during oxidative stress. Specifically, upon incubation of mouse fibroblasts with 50 μM hydrogen peroxide, Ro accumulated in nuclei within 3 h (Figure 7A). Examination of chimeric mouse-bacterial Ro proteins revealed that, as for UV, only chimeras containing the entire mouse HEAT repeat domain exhibited regulated nuclear accumulation (Figure 7B, e and f). Moreover, as observed for UV, the chimera containing only the first 12 mouse HEAT repeats was constitutively nuclear. These results suggest that sequences or structural features contained within helices 1–12 are important for nuclear accumulation after UV and oxidative stress, and that, in the absence of stress, these sequence(s) are masked by Y RNA binding.
Figure 7.
Nuclear accumulation of Ro during oxidative stress is regulated by Y RNA binding. (A) Wild-type mouse fibroblasts were subjected to immunofluorescence with anti-Ro antibodies before (left) and after incubation with 50 μM hydrogen peroxide for 3 h (right). Nuclei were detected by staining with DAPI. Bar, 10 μm. (B) Ro−/− fibroblasts expressing either FLAG-tagged mouse Ro (a and b), D. radiodurans Ro (c and d), or chimeric proteins (e–l) were assayed for their location in untreated cells (a, c, e, g, i, and k) and after treatment with 50 μM H2O2 for 3 h (b, d, f, h, j, and l). Left, immunofluorescence with anti-FLAG antibodies. Right, nuclei were visualized by staining with DAPI. Bar, 10 μm.
DISCUSSION
Although it was reported more than 25 years ago that most Y RNAs are found in the cytoplasm complexed to the Ro protein (Hendrick et al., 1981), the function of these RNAs has been enigmatic. We have shown that one function of these RNAs is to regulate the subcellular distribution of Ro. In the absence of bound Y RNAs, Ro accumulates in nuclei. Using chimeric proteins in which parts of the mouse Ro protein were fused to a portions of a bacterial orthologue, we determined that sequences important for nuclear accumulation after UV or oxidative stress reside within the HEAT repeat domain and overlap the Y RNA binding site. Our data are consistent with a model in which bound Y RNAs mask a signal for nuclear accumulation that becomes accessible after environmental stress. Importantly, these findings demonstrate one way that a noncoding RNA can regulate the subcellular distribution of a protein.
Together with recent results that Y RNAs inhibit binding of Ro to other RNAs (Chen et al., 2007), our experiments suggest that Y RNAs are members of a growing category of noncoding RNAs that function by modulating the activity of proteins. Members of this group include the prokaryotic 6S RNA, which binds RNA polymerase and inhibits transcription (Wassarman, 2007), the eukaryotic 7SK RNA, which binds HEXIM1 and converts it into an inhibitor of the transcription elongation factor P-TEFb, and the mouse B2 RNA and human Alu RNAs, which bind RNA polymerase II and inhibit transcription in response to heat shock (Mariner et al., 2008). However, although these other RNAs act by inhibiting transcription, Y RNAs are unique in modulating both the subcellular distribution of Ro and its binding to other RNAs.
We do not know whether the sole role of Y RNAs is to block a sequence required for Ro nuclear import or whether Y RNA binding also enhances export of Ro. Our finding that a chimera containing mouse helices 1–12 accumulates strongly in nuclei, whereas the D. radiodurans protein is both nuclear and cytoplasmic, indicates that the first 239 amino acids of mouse contains sequences required for Ro nuclear accumulation. Although helix 12 contains several lysines and arginines (Figure 6), the isolated helix was not sufficient to direct nuclear import of a reporter, suggesting it does not function as a classic importin-α–dependent nuclear localization sequence. Thus, sequences within helices 1–12 could be recognized by a different import receptor or be the binding site for an adaptor protein. In addition, if Ro is primarily exported as a Ro/Y RNA complex by exportin-5, as suggested by microinjection experiments in X. laevis oocytes (Simons et al., 1996; Rutjes et al., 2001; Gwizdek et al., 2003), a decrease in Y RNA association could impair exportin-5 binding, contributing to nuclear accumulation of Ro. However, the D. radiodurans Ro, which does not bind mouse Y RNAs, is present in both the nucleus and cytoplasm of unstressed cells and does not accumulate strongly in nuclei (Figures 4 and 5). Thus, a failure of Y RNA-free Ro to be exported is unlikely to be the primary cause of the observed nuclear accumulation.
Our data are consistent with a model in which sequences required for nuclear accumulation become accessible as a consequence of environmental stress. This change in accessibility could be accomplished either by dissociating bound Y RNAs from Ro or by a stress-induced rearrangement of Y RNA positioning that exposes the nuclear accumulation sequences. As is the case for 7SK RNA, B2 RNAs and Alu RNAs, all of which exhibit stress-regulated binding to their protein targets, the signals that modulate Y RNA binding to Ro are not yet known. Because human Ro was identified as a possible target of the DNA damage-activated ataxia telangiectasia-mutated (ATM) and ATM and Rad-3 related kinases (Matsuoka et al., 2007; Stokes et al., 2007), phosphorylation of Ro by stress-regulated kinases could influence Y RNA binding. In addition, purified human Ro has been reported to contain the reactive lipid oxidation product 4-hydroxy-2-nonenal (Scofield et al., 2005), raising the possibility that oxidative modifications contribute. Alternatively, stress-activated binding of other proteins or RNAs to Ro could modulate Y RNA binding, contribute to import, or both. It is also possible that a stress-regulated block of export contributes to the strong accumulation of Ro in nuclei.
Our finding that Y RNAs influence the subcellular distribution of Ro is consistent with some previous observations. First, upon manual enucleation of Xenopus oocytes, Ro/Y RNA complexes were only detected in the cytoplasm, whereas Ro/misfolded RNA complexes were nuclear (O'Brien and Wolin, 1994; Simons et al., 1994; Chen et al., 2003). Similarly, separation of mouse cells into cytoplasts and karyoplasts revealed that Y RNAs were primarily cytoplasmic, and only RNA-free Ro was detected in nuclei (O'Brien et al., 1993; Peek et al., 1993). However, using in situ hybridization, we found that mY3, like Ro, accumulates in nuclei after UV irradiation (Chen et al., 2003). Moreover, using Northern blotting, the levels of both mouse Y RNAs are unchanged after irradiation (unpublished data). Because a D. radiodurans Y RNA is made in excess and stabilized by Ro binding (Chen et al., 2007), the Y RNAs that accumulate in nuclei after irradiation could represent new Y RNA synthesis or stabilization of a normally unstable pool of Y RNAs. Alternatively, if stress results in a rearrangement of Y RNA positioning that exposes the nuclear accumulation signal, Y RNAs may remain bound to Ro during import.
In addition to inhibiting the binding of Ro to other RNAs and regulating the subcellular distribution of Ro, it is likely that Y RNAs contribute in other ways to Ro function. Specifically, because Ro binds a conserved helix in the stems of all Y RNAs (Stein et al., 2005), these two functions of Y RNAs may not involve the large internal loops that are a characteristic of these RNAs. Because several abundant RNA-binding proteins, including nucleolin, heterogeneous nuclear (hn)RNP I and ribosomal protein L5, have been reported to associate with the loops of one or more Y RNAs in human cells, binding of specific proteins to individual Y RNA loops may specialize Ro RNPs for distinct functions (Bouffard et al., 2000; Fabini et al., 2001; Hogg and Collins, 2007). However, a recent proposal that the human Ro recognizes misfolded pre-5S rRNAs only when Ro is bound to the Y5 RNA (Hogg and Collins, 2007) is difficult to reconcile with the fact that mice and certain other vertebrates lack a Y5 RNA (Mosig et al., 2007; Perreault et al., 2007) and with the observation that, in X. laevis oocytes, the Ro/pre-5S complex is located in nuclei, whereas the Ro/Y5 RNA is exclusively cytoplasmic (O'Brien and Wolin, 1994). Yet another possibility is that binding of other components involved in noncoding RNA quality control, such as helicases or nucleases, to the large internal loops of Y RNAs facilitates their association with the Ro RNP.
ACKNOWLEDGMENTS
We thank J. Lou for technical assistance; E. Chan, S. Kojima, and G. Borisy for gifts of plasmids; J. Steitz for the Y12 antibody; N. Conrad for helpful advice; and E. Wurtmann for comments on the manuscript. This work was supported by a postdoctoral fellowship from the Arthritis Foundation (to S. S.), a Yale-Howard Hughes Medical Institute Future Scientists Fellowship (to D.E.W.), a postdoctoral fellowship from the American Heart Association (to K. C.), and grants from the Ellison Medical Foundation and National Institutes of Health (GM-073863) (to S.L.W.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1094) on December 30, 2008.
REFERENCES
- Bouffard P., Barbar E., Briere F., Boire G. Interaction cloning and characterization of RoBPI, a novel protein binding to human Ro ribonucleoproteins. RNA. 2000;6:66–78. doi: 10.1017/s1355838200990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Quinn A. M., Wolin S. L. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–782. [PMC free article] [PubMed] [Google Scholar]
- Chen X., Smith J. D., Shi H., Yang D. D., Flavell R. A., Wolin S. L. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr. Biol. 2003;13:2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Chen X., Wolin S. L. The Ro 60 kDa autoantigen: insights into cellular function and role in autoimmunity. J. Mol. Med. 2004;82:232–239. doi: 10.1007/s00109-004-0529-0. [DOI] [PubMed] [Google Scholar]
- Chen X., Wurtmann E. J., Van Batavia J., Zybailov B., Washburn M. P., Wolin S. L. An orthologue of the Ro autoantigen functions in 23S rRNA in D. radiodurans. Genes Dev. 2007;21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Fabini G., Raijmakers R., Hayer S., Fouraux M. A., Pruijn G. J., Steiner G. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of Ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J. Biol. Chem. 2001;276:20711–20718. doi: 10.1074/jbc.M101360200. [DOI] [PubMed] [Google Scholar]
- Farazi T. A., Juranek S. A., Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Fuchs G., Stein A. J., Fu C., Reinisch K. M., Wolin S. L. Structural and biochemical basis for misfolded RNA recognition by the Ro protein. Nat. Struct. Mol. Biol. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- Green C. D., Long K. S., Shi H., Wolin S. L. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–765. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C., Ossareh-Nazari B., Brownawell A. M., Doglio A., Bertrand E., Macara I. G., Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 2003;278:5505–5508. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- Hannon G. J., Rivas F. V., Murchison E. P., Steitz J. A. The expanding universe of noncoding RNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Wolin S. L., Rinke J., Lerner M. R., Steitz J. A. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol. Cell. Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch. Gesamte Virusforschung. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Hogg J. R., Collins K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal quality control. Genes Dev. 2007;21:3067–3072. doi: 10.1101/gad.1603907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Kojima S., Vignjevic D., Borisy G. G. Improved silencing vector co-expressing GFP and small hairpin RNA. Biotechniques. 2004;36:74–79. doi: 10.2144/04361ST02. [DOI] [PubMed] [Google Scholar]
- Labbe J. C., Burgess J., Rokeach L. A., Hekimi S. ROP-1, an RNA quality-control pathway component, affects Caenorhabditis elegans dauer formation. Proc. Natl. Acad. Sci. USA. 2000;97:13233–13238. doi: 10.1073/pnas.230284297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe J. C., Hekimi S., Rokeach L. A. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics. 1999;151:143–150. doi: 10.1093/genetics/151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. H., Liu Q., Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl. Acad. Sci. USA. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.-H., Carman C. V., Springer T. A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner P. D., Walters R. D., Espinoza C. A., Drullinger L. F., Wagner S. D., Kugel J. F., Goodrich J. A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Frey M. R., Margelot K., Wolin S. L. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Michels A. A., Bensaude O. RNA-driven cyclin-dependent kinase regulation: when CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotechnology J. 2008;3:1022–1032. doi: 10.1002/biot.200800104. [DOI] [PubMed] [Google Scholar]
- Mosig A., Guofeng M., Stadler B.M.R., Stadler P. F. Evolution of the vertebrate Y RNA cluster. Theory Biosci. 2007:9–14. doi: 10.1007/s12064-007-0003-y. [DOI] [PubMed] [Google Scholar]
- Neher S. B., Bradshaw N., Floor S. N., Gross J. D., Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat. Struct. Mol. Biol. 2008;15:916–923. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien C. A., Margelot K., Wolin S. L. Xenopus Ro ribonucleoproteins: members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 1993;90:7250–7254. doi: 10.1073/pnas.90.15.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien C. A., Wolin S. L. A possible role for the 60 kd Ro autoantigen in a discard pathway for defective 5S ribosomal RNA precursors. Genes Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Peek R., Pruijn G.J.M., van der Kemp A., van Venrooij W. J. Subcellular distribution of Ro ribonucleoprotein complexes and their constituents. J. Cell Sci. 1993;106:929–935. doi: 10.1242/jcs.106.3.929. [DOI] [PubMed] [Google Scholar]
- Perreault J., Perreault J.-P., Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol. Biol. Evol. 2007;24:1678–1689. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- Pruijn G.J.M., Slobbe R. L., van Venrooij W. J. Analysis of protein-RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–5180. doi: 10.1093/nar/19.19.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A., Savva C. G., Holzenburg A., Sacchettini J. C. Crystal structure of Rsr, an ortholog of the antigenic Ro protein, links conformational flexibility to RNA binding activity. J. Biol. Chem. 2007;282:14960–14967. doi: 10.1074/jbc.M611163200. [DOI] [PubMed] [Google Scholar]
- Robb G. B., Brown K. M., Khurana J., Rana T. M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Rutjes S. A., Lund E., van der Heijden A., Grimm C., van Venrooij W. J., Pruijn G. J. Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs. RNA. 2001;7:741–752. doi: 10.1017/s1355838201002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield R. H., Kurien B. T., Ganick S., McClain M. T., Pye Q., James J. A., Schneider R. I., Broyles R. H., Bachmann M., Hensley K. Modification of lupus-associated 60-kDa Ro protein with the lipid oxidation product 4-hydroxy-2-nonenal increases antigenicity and facilitates epitope spreading. Free Radic. Biol. Med. 2005;38:719–928. doi: 10.1016/j.freeradbiomed.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Simons F.H.M., Pruijn G.J.M., van Venrooij W. J. Analysis of the intracellular localization and assembly of Ro ribonucleoprotein particles by microinjection into Xenopus laevis oocytes. J. Cell Biol. 1994;125:981–988. doi: 10.1083/jcb.125.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons F.H.M., Rutjes S. A., van Venrooij W. J., Pruijn G.J.M. The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA. 1996;2:264–273. [PMC free article] [PubMed] [Google Scholar]
- Stein A. J., Fuchs G., Fu C., Wolin S. L., Reinisch K. M. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes M. P., et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. USA. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.-Y., Yario T. A., Steitz J. A. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of intron. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- Todaro G., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Buyon J. P., Chan E. K. Cloning and expression of mouse 60 kDa ribonucleoprotein SS-A/Ro. Mol. Biol. Rep. 1996;23:205–210. doi: 10.1007/BF00351170. [DOI] [PubMed] [Google Scholar]
- Wassarman K. M. 6S RNA: a regulator of transcription. Mol. Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–402. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Steitz J. A. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc. Natl. Acad. Sci. USA. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., et al. A lupus-like syndrome develops in mice lacking the Ro 60 kDa protein, a major lupus autoantigen. Proc. Natl. Acad. Sci. USA. 2003;100:7503–7508. doi: 10.1073/pnas.0832411100. [DOI] [PMC free article] [PubMed] [Google Scholar]