Abstract
Organisms respond to circumstances threatening the cellular protein homeostasis by activation of heat-shock transcription factors (HSFs), which play important roles in stress resistance, development, and longevity. Of the four HSFs in vertebrates (HSF1-4), HSF1 is activated by stress, whereas HSF2 lacks intrinsic stress responsiveness. The mechanism by which HSF2 is recruited to stress-inducible promoters and how HSF2 is activated is not known. However, changes in the HSF2 expression occur, coinciding with the functions of HSF2 in development. Here, we demonstrate that HSF1 and HSF2 form heterotrimers when bound to satellite III DNA in nuclear stress bodies, subnuclear structures in which HSF1 induces transcription. By depleting HSF2, we show that HSF1-HSF2 heterotrimerization is a mechanism regulating transcription. Upon stress, HSF2 DNA binding is HSF1 dependent. Intriguingly, when the elevated expression of HSF2 during development is mimicked, HSF2 binds to DNA and becomes transcriptionally competent. HSF2 activation leads to activation of also HSF1, revealing a functional interdependency that is mediated through the conserved trimerization domains of these factors. We propose that heterotrimerization of HSF1 and HSF2 integrates transcriptional activation in response to distinct stress and developmental stimuli.
INTRODUCTION
Cells react to stressful conditions by activation of heat-shock factors (HSFs), of which there are three mammalian isoforms: HSF1, HSF2, and HSF4 (Pirkkala et al., 2001). Activated HSFs bind to heat-shock elements (HSEs) within the promoters of their target genes and induce synthesis of protective molecular chaperones called heat-shock proteins (Hsps). Hsps prevent protein misfolding and are required for stress resistance and healthy cell growth, development, and aging (Bukau et al., 2006, Morimoto, 2008). They also protect against metabolic, neurodegenerative and cardiovascular disorders (Balch et al., 2008). In addition to Hsps, HSFs are associated with expression of noncoding satellite III (sat III) RNA in nuclear stress bodies (nSBs) (Jolly et al., 2004, Rizzi et al., 2004), to which HSF1 and HSF2 translocate upon stress (Jolly et al., 1997, Alastalo et al., 2003). The nSBs form on the locus 9q12 consisting of pericentromeric heterochromatin, and the sat III transcripts provide scaffolds for docking of other nSB components, as shown for the splicing factor ASF/SF2 (Chiodi et al., 2004, Metz et al., 2004). Besides ASF/SF2, the RNA processing factors heterogeneous nuclear ribonucleoprotein (hnRNP) A1-associated protein (HAP), hnRNPM, Sam68, and SRp30c localize to the nSBs (Weighardt et al., 1999, Denegri et al., 2001).
HSF1 is activated by classical stresses such as heat shock and heavy metals and responds to elevated temperatures in vitro (Ahn et al., 2001, Anckar and Sistonen, 2007). HSF1 is also involved in development and has critical roles in longevity and cancer (Xiao et al., 1999, Hsu et al., 2003, Morley and Morimoto, 2004, Dai et al., 2007). Unlike HSF1, HSF2 lacks intrinsic stress responsiveness (Ahn et al., 2001). Another major difference between these factors is that while HSF1 is evenly expressed, the levels of HSF2 fluctuate. These changes in expression coincide temporally with HSF2 DNA binding activity during developmental processes (Rallu et al., 1997, Min et al., 2000). The function of HSF2 in development was revealed by hsf2−/− mice, which display neurological and reproductive abnormalities in both genders (Kallio et al., 2002, Wang et al., 2003, Chang et al., 2006, Åkerfelt et al., 2008). In addition, the stress-induced expression of hsps in hsf2−/− cells is altered (Östling et al., 2007). However, the mechanism by which HSF2 is recruited to stress-inducible promoters is not known. How HSF2 is activated, and the functional relationship between HSF1 and HSF2 also remain to be elucidated.
In this study, we use the nSBs as a model system to show that HSF1 and HSF2 interact as DNA-bound heterotrimers. When HSF1-HSF2 heterotrimerization is inhibited by depletion of HSF2, the transcription of sat III DNA is enhanced, indicating that HSF1-HSF2 heterotrimerization regulates transcription. We mimic the elevated HSF2 concentration during development and demonstrate that increased HSF2 expression induces transcription of sat III DNA and localization of both HSF1 and HSF2 to the nSBs. In testis, where HSF2 is abundantly expressed and plays a role in spermatogenesis (Sarge et al., 1994, Fiorenza et al., 1995, Kallio et al., 2002, Wang et al., 2003, Åkerfelt et al., 2008), we show interaction between HSF1 and HSF2. Increased HSF2 expression also induces transcription of the classical HSF target hsp70, suggesting that HSF2 is activated by its elevated expression. Importantly, although the stress-induced DNA binding of HSF2 is dependent on HSF1 activity, induced HSF2 expression converts HSF1 to a transcriptionally competent state. We propose that heterotrimerization is a transcriptional switch at the interface of activation by either HSF1 or HSF2.
MATERIALS AND METHODS
Cell Culture, Treatments, Plasmids, and Transfections
HeLa and human embryonic kidney (HEK) 293T cells were cultured in Dulbecco′s modified Eagle's medium and K562 cells in RPMI 1640 medium in 5% CO2 at 37°C. The media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, and penicillin and streptomycin. Heat shocks were performed at 42°C in a water bath for the indicated times. HeLa and HEK293T cells were transfected by electroporation (975 μF, 220 V; Gene Pulser; Bio-Rad, Hercules, CA). Plasmid DNA and 5 × 106 cells in 0.4 ml of Opti-MEM (Invitrogen, Carlsbad, CA) were added to a 0.4-cm-gap electroporation cuvette (BTX, San Diego, CA) and subjected to a single electric pulse. The mouse HSF1-yellow fluorescent protein (YFP) was generated as the mouse HSF1-cyan fluorescent protein (CFP), described in Jolly et al., 2002. The human HSF2-YFP, containing amino acids 1-214, was constructed by polymerase chain reaction (PCR) and cloned into the BamHI and XhoI sites of pEYFP-N1 (Clontech, Mountain View, CA). The tandem CFP-YFP construct was a kind gift of Richard I. Morimoto (Northwestern University, Evanston, IL). The mHSF2α and mHSF2β plasmids used for over-expression of HSF2 were described in Alastalo et al., 2003. All constructs were sequenced.
RNA Interference (RNAi)
Transient down-regulation of HSF1 was performed by electroporation of Scramble and HSF1 RNAi plasmids in HEK293T (Östling et al., 2007). The cells were harvested after 72-h incubation. The stable scrambled and HSF1–down-regulating cell lines were generated by transfection of the pSuper shRNA Scrambled and HSF1 RNAi plasmids (Östling et al., 2007) to HeLa cells, and single clones were established after selection with neomycin. For down-regulation of HSF2, small interfering RNA (siRNA) against HSF2 or AllStars negative control siRNA was transfected using HIPerFect Transfection reagent (all from QIAGEN, Hilden, Germany).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as in Östling et al., 2007. K562 cells were cross-linked with 1% formaldehyde. Chromatin was sonicated and immunoprecipitated with antibodies against HSF1 (SPA-901; Nventa Biopharmaceuticals, San Diego, CA), HSF2 SFI58 (Östling et al., 2007), and normal rabbit serum (NS; Jackson ImmunoResearch Laboratories, West Grove, PA). The following primers were used for ChIP: sat III (based on clone17 in Valgardsdottir et al., 2005), For 5′-AAT GAA CCC GAT GCA AT-3′, Rev 5′-CCA TTC TTG TTG AAT CCA TT-3′; and β-actin, For 5′-AAC TCT CCC TCC TCC TCT TCC TC-3′, Rev 5′-GAG CCA TAA AAG GCA ACT TTC GG-3′.
Immunofluorescence
For immunofluorescence analysis, HeLa cells cultured on coverslips were fixed with −20°C methanol for 6 min or with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min. After three washes with PBS-0.5% Tween 20, the cells were incubated in blocking solution (1% bovine serum albumin PBS-0.5% Tween 20) for 1 h. Rabbit anti-HSF1 (Holmberg et al., 2000), rat anti-HSF1 (NeoMarkers, Fremont, CA), rabbit anti-HSF2 (Sarge et al., 1993), or rat anti-HSF2 (NeoMarkers) antibodies were diluted in blocking solution and added for 1 h. Secondary antibodies, anti-rabbit Alexa 488 and anti-rat Alexa 568 (Invitrogen), were incubated for 1 h. The coverslips were mounted and DNA was visualized using VECTASHIELD mounting medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). The cells were analyzed with an LSM510-Meta scanning confocal microscope (Carl Zeiss, Jena, Germany) equipped with the SP2 (version 3.2) software. The images were acquired using a Plan-Apochromat 63×/1.4 oil differential interference contrast objective and further processed using Adobe Photoshop (Adobe Systems, Mountain View, CA) and CorelDRAW software.
Western Blot Analysis
Soluble cell extracts were prepared and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by transfer to nitrocellulose membrane (Protran nitrocellulose; Whatman Schleicher and Schuell, Dassel, Germany). HSF1 was detected by polyclonal anti-HSF1 antibodies (Sarge et al., 1993; Holmberg et al., 2000), HSF2 by polyclonal anti-HSF2 antibodies (Sarge et al., 1993; Östling et al., 2007) and Hsc70 by SPA-815 (Nventa Biopharmaceuticals). Secondary antibodies were horseradish peroxidase conjugated and purchased from Promega (Madison, WI) or GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). The immunoblots were developed with an enhanced chemiluminescence method (ECL kit; GE Healthcare).
Immunoprecipitation
Male hybrid mice of the B6129SF2/J strain were used in coimmunoprecipitation experiments. HSF2 knockout mice were obtained by matings of heterozygous mice that have been described previously (Kallio et al., 2002) and were maintained in a C57BL/6N background. The pathogen-free mice were housed under controlled environmental conditions and fed with complete pellet chow and allowed tap water. The mice were killed by CO2 asphyxiation. All mice were handled in accordance with the institutional animal care policies of the Åbo Akademi University (Turku, Finland). For coimmunoprecipitation experiments, testes were isolated from 60- to 80-d-old mice and lysed in 2 ml of lysis buffer (Alastalo et al., 2003). The precleared cellular lysate was incubated with anti-HSF1 (NeoMarkers), anti-HSF2 (NeoMarkers), or anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO) antibodies at 4°C for 1 h under rotation, after which 40 μl of a 50% slurry of protein-G/Sepharose was added to the reaction mixture and incubated for 1 h at 4°C under rotation. After centrifugation, the Sepharose beads were washed with supplemented TEG buffer, and the immunoprecipitated proteins were run on 8% SDS-PAGE and transferred to nitrocellulose filter for immunoblotting as described above.
Semiquantitative Reverse Transcription (RT)-PCR and Real-Time RT-PCR
RNA was isolated with the RNAeasy kit (QIAGEN). Contaminating genomic DNA was removed with two DNase I treatments according to the RNAeasy protocol (QIAGEN). Of each sample, 1 μg of RNA was subjected to reverse transcription using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). For semiquantitative RT-PCR, ABsolute Rox mix (Advanced Biotechnologies, Epsom, United Kingdom) was used and the PCR was run 40 cycles. The same sat III primers as for ChIP were used. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were GAPDH For 5′-ACC CAC TCC TCC ACC TTT GA-3′, GAPDH Rev 5′-TTG CTG TAG CCA AAT TCG TTG T-3′. Real-time RT-PCR analyses were performed with ABsolute cybrgreen mix (Advanced Biotechnologies) and the ABI Prism 5700 and 7900 (Applied Biosystems). Relative RNA quantities were normalized to GAPDH. For real-time RT-PCR, the following primers and probes were used: sat III For 5′-AAT GGA ATG CAA TGG AAT GG-3′, sat III Rev 5′-CCT GTA CTC GGG TTG ATT CC-3′, GAPDH For 5′-ACC CAC TCC TCC ACC TTT GA-3′, and GAPDH Rev 5′-CTG TTG CTG TAG CCA AAT TCG T-3′ (Shumaker et al., 2006); and hsp70 Probe 5′-FAM TTACACACCTGCTCCAGCTCCTTCCTCTT TAMRA-3′, hsp70 For 5′-GCCGAGAAGGACGAGTTTGA-3′, hsp70 Rev 5′-CCTGGTACAGTCCGCTGATGA-3′, GAPDH Probe 5′-FAM ACCAGGCGCCCAATACGACCAA TAMRA-3′, GAPDH For 5′-GTTCGACAGTCAGCCGCATC-3′, and GAPDH Rev 5′-GGAATTTGCCATGGGTGGA-3′.
Structural Modeling
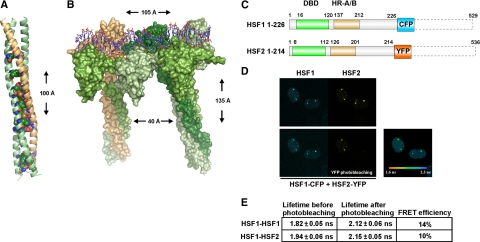
The structural model of the human HSF heterotrimer of two HSF1 (amino acids [aa] 16-205) molecules and one HSF2 (aa 8-194) was done in three steps. First, a template of the DNA binding domain of six Kluyveromyces lactis HSF monomers bound to a 32-base pair DNA was generated using SYBYL 7.3 (Tripos, St. Louis, MO) by aligning three dimers of the crystal structure of K. lactis HSF bound to DNA next to each other as suggested by Littlefield and Nelson (1999). Second, the HR-A domain was aligned against the Escherichia coli Lpp-56 x-ray structure (Shu et al., 2000), and the HR-B domain was aligned against the mH38-P1 GCN4 Leucine Zipper x-ray structure (Shu et al., 1999), resulting in the template structure for the HR-A/B trimerization domain. The alignments were done according to the characteristic heptad repeat sequence (abcdefg)n seen in coiled coil structures (Supplemental Figure 3). Third, the final template used for modeling the heterotrimer of the DNA binding and HR-A/B domain was generated by linking the two domains by using the x-ray structure of human GABPα protein (Batchelor et al., 1998). In the final model of the heterodimer, HSF2 makes both head-to-head and tail-to-tail contacts with HSF1. For sequence alignments, MALIGN and MALFORM (Johnson and Overington, 1993) were used within the Bodil visualization and modeling package (Lehtonen et al., 2004). Ten models were generated with Modeler (Sali and Blundell, 1993), and the model with the lowest objective function was chosen. Sequence alignment in Supplemental Figure 3 was done with ALSCRIPT (Barton, 1993), and Figure 2, A and B, were created with the PYMOL Molecular Graphics System (Delano Scientific, Palo Alto, CA).
Figure 2.
HSF1 and HSF2 interact as DNA-bound heterotrimers. (A) Structural model of a heterotrimer formed by the HR-A/B domains of two HSF1 molecules (green) and one HSF2 molecule (beige). The conserved amino acids found at positions a and d in the heptad repeat (see Supplemental Figure 3) are shown as spheres. (B) Surface representation of the structural model of an HSF1-HSF2-HSF1 heterotrimer and an HSF1 homotrimer bound to DNA. HSF2 is colored beige and the different HSF1 molecules are colored in different shades of green. The HSF trimers are separated on DNA by two nucleotides as in a consensus HSE. The height and width of the complex as well as the distance between the two coiled coils are indicated. (C) Schematic presentation of the HSF1 and HSF2 C-terminal deletion constructs used for FRET. The position of amino acids is shown, and the DNA-binding domain (DBD) and trimerization domains HR-A/B are indicated. The deleted C-terminal regions are illustrated by dashed lines. (D) Interaction between HSF1 and HSF2 on DNA in the nSBs was detected with FLIM-FRET on live cells. HeLa cells were transfected with C-terminal HSF1 and HSF2 deletions fused to CFP and YFP, and the fluorescence lifetime of the donor (HSF1-CFP) was measured before or after photobleaching of the acceptor (HSF2-YFP). The fluorescence lifetime after photobleaching of the acceptor is indicated by a color scale bar. (E) A mean FLIM-FRET efficiency was calculated for the FRET pairs HSF1-CFP − HSF1-YFP and HSF1-CFP − HSF2-YFP. The data for each experimental condition represents measurements from at least five cells, and the SD is indicated.
Confocal Microscopy and Two-Photon Fluorescence Lifetime Imaging
The two-photon and confocal microscopy on HeLa cells was performed with an inverted two-photon laser scanning microscope Axiovert 200M (LSM510 NLO META; Carl Zeiss). During the experiment, cells were maintained at 37°C in a humidified atmosphere containing 5% CO2 by using an on-stage incubator (PeCon, Frankfurt, Germany). All measurements were performed using a 63×/1.4 oil immersion plan-apochromat objective. In the fluorescence lifetime imaging (FLIM) experiments, the fluorescence decays were measured by the time-correlated single photon counting technique. Fluorescence decays were fitted using a biexponential model and the corresponding mean decay time in each pixel was color coded to obtain FLIM images (SPCImage software; Becker & Hickl, Berlin, Germany). Fluorescence resonance energy transfer (FRET) was identified by the shorter lifetime of donor (CFP) in the presence of acceptor (τDA) as compared with that (τD) in the control donor-only cells. The FLIM/FRET efficiency was calculated as EFILM/FRET = 1 − τDA/τD.
Additional acceptor photobleaching experiments were carried out on the same cell and completed with FLIM measurements to confirm FRET. At least five cells were measured for each experimental condition.
RESULTS
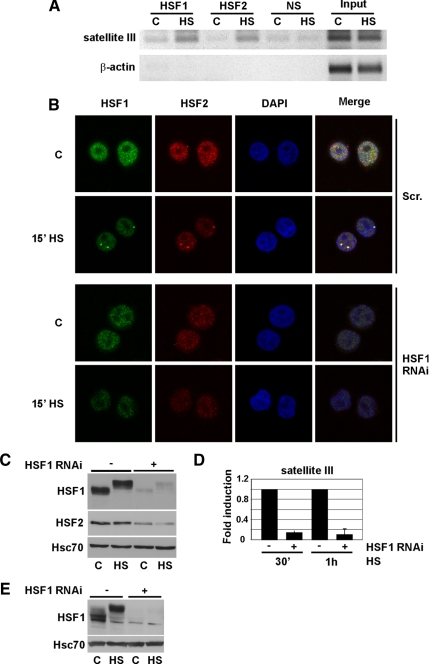
Stress-induced Translocation of HSF2 in the nSBs Is HSF1 Dependent
The colocalization of HSF1 and HSF2 into the nSBs (Alastalo et al., 2003) prompted us to further investigate their functional relationship. HSF1 binds to DNA in the nSBs (Jolly et al., 2002), and we performed ChIP in K562 cells to examine the binding of HSF2 to sat III DNA. Heat shock-induced binding of HSF1 and HSF2 was detected (Figure 1A), indicating that both HSFs occupy the same sat III DNA sequences (also see Supplemental Figure 1). To investigate how HSF1 affects the localization of HSF2, we generated a HeLa cell line stably down-regulating HSF1 by using vector-based RNAi (Figure 1C). In HSF1-depleted cells, the localization of both HSF1 and HSF2 into nSBs was abrogated (Figure 1B; for additional times, see Supplemental Figure 2A), inferring that stress-induced HSF2 DNA binding activity is dependent on HSF1. This result is supported by the finding that transient down-regulation of HSF1 by shRNAs in HEK293T cells (Figure 1E) abolishes sat III transcription (Figure 1D and Supplemental Figure 2B).
Figure 1.
Stress-induced localization of HSF2 into the nSBs is HSF1 dependent. (A) HSF1 and HSF2 binding to sat III in untreated (C) and 1-h heat-shocked (HS) K562 cells was analyzed with ChIP. β-actin was used as a control promoter. Input represents 1% of the total material and a nonspecific antibody (NS) was used as a negative control. The ChIP assay on sat III was performed on four biological samples. (B) HSF1 was down-regulated in HeLa cells (HSF1 RNAi) and the nSB formation followed by staining of HSF1 (green) and HSF2 (red). As a control, a scrambled cell line (Scr.) was used. Note that the settings used for image acquisition of HSF1 and HSF2 in Scr. cells were reused for HSF1 RNAi cells. For staining, polyclonal and monoclonal antibodies were used against HSF1 and HSF2, respectively. The nucleus is shown in blue by staining of DNA with DAPI. Heat shock and control are indicated as HS and C, respectively. For additional time points, see Supplemental Figure 2A. (C) Western blot analysis of HSF1 and HSF2 in the stable Scr. (−) and HSF1-RNAi (+) cell lines. The retarded mobility of HSF1 in heat-shocked (HS) samples is due to hyperphosphorylation (Sarge et al., 1993). Hsc70 serves as a loading control. (D) Real-time RT-PCR analysis of sat III transcription in Scr. (−) and HSF1-RNAi (+) cells. The results are shown as fold induction upon 30 min and 1 h of HS. Fold induction was calculated by comparing the induction in HSF1 RNAi samples to the induction in scrambled samples, which were arbitrarily set to 1. The data represent three biological samples, and relative quantities of sat III RNA were normalized to GAPDH. Error bars indicate SD. (E) Western blot analysis of HSF1 down-regulation in HEK293T cells. HS indicates a 1-h heat shock.
HSF1 and HSF2 Interact as DNA-bound Heterotrimers
The dependence of HSF2 on HSF1 activity for localization to the nSBs (Figure 1B) raised the question of the underlying mechanism. Upon activation, both HSF1 and HSF2 form homotrimers through their trimerization domain consisting of the heptad repeats A and B (HR-A/B) (Sarge et al., 1993). Because HSF2 interacts with HSF1 via the HR-A/B domain (Alastalo et al., 2003), we studied whether HSF1 and HSF2 can form heterotrimers. We aligned the HR-A/B sequences of HSF1 and HSF2 and found that the amino acids involved in trimerization are conserved, especially well within the midsection of the HR-A/B domains of HSF1 and HSF2 (Supplemental Figure 3). Using two HSF1 HR-A/B helices and one HSF2 HR-A/B, we made a structural model of the left-handed coiled coil that constitutes the trimer interface (Figure 2A). All buried polar residues, which have been suggested to play a role in partner verification, are conserved in the HSF1-HSF2 coiled coil structure (Figure 2A). Based on the crystal structure of DNA-bound K. lactis (KLUELA) HSF (Littlefield et al., 1999), we generated a model of a human HSF1-HSF2 heterotrimer. The heterotrimer is bound to DNA and composed of the DNA binding and HR-A/B domains of HSF1 and HSF2 (Figure 2B). Adjacent to the HSF1-HSF2 heterotrimer, an HSF1 homotrimer is shown. The trimers are bound to a 32-base pair DNA double-stranded helix, with two nucleotide spacers as in a canonical HSE. We measured a distance of ∼40 Å between the HR-A/B coiled coils of two HSF trimers (Figure 2B). Noncovalent contacts require proximities of <4 Å (Laberge, 1998), and at a distance of 40 Å, electrostatic interactions are unlikely to occur (Creighton, 1993), excluding interactions between separate trimers. This suggests that interactions between HSF1 and HSF2 on DNA are mediated through heterotrimerization.
FLIM-FRET was used to investigate whether HSF1 and HSF2 interact on DNA within the nSBs. We could not detect FRET when fusing CFP and YFP to the C termini of full-length HSF1 and HSF2, presumably because the C termini of HSF1 and HSF2 are highly mobile, preventing FRET. Therefore, to facilitate the proper positioning of the CFP and YFP moieties, we used HSF1 and HSF2 C-terminal deletion constructs (Figure 2C), that translocate into nSBs spontaneously (Supplemental Figure 4A; Jolly et al., 2004). These C-terminal deletion constructs contain the DNA binding and HR-A/B domains of HSF1 and HSF2 and should be apt for the study of interaction between the proteins as HSF1 and HSF2 interact through the HR-A/B domains (Alastalo et al., 2003). Upon transfection of HeLa cells with the HSF1 and HSF2 constructs, the fluorescence lifetime of the donor (HSF1-CFP) was shorter in the presence of acceptor (HSF2-YFP) compared with that in cells with only HSF1-CFP (Figure 2D), indicating that FRET occurs. The FRET signal was predominantly localized to the nSBs with a mean FRET efficiency of 10%, a significant number considering that the FRET efficiency of an HSF1-CFP/HSF1-YFP pair representing HSF1 homotrimers was 14% (Figure 2E). Additional FACS-FRET (Supplemental Figure 4B) and acceptor photobleaching experiments (data not shown) confirmed the FRET results, showing that HSF1 and HSF2 interact as DNA-bound heterotrimers.
Stress-induced HSF Activity Is Regulated through HSF1-HSF2 Heterotrimerization
To investigate the impact of HSF1-HSF2 heterotrimerization on stress-induced transcription, we abrogated heterotrimer formation by depleting HSF2. HSF2-specific siRNAs were transfected to HEK293T cells (Figure 3B), and the transcription of sat III DNA was measured by real-time RT-PCR. A robust increase in the accumulation of sat III transcripts was evident when HSF2 was depleted (Figure 3A), demonstrating that HSF1-HSF2 heterotrimerization regulates HSF-mediated transcription. As expected, HSF2 knockdown did not alter the stress-induced relocalization of HSF1 to the nSBs (Figure 3C).
Figure 3.
Stress-induced HSF activity is regulated through HSF1-HSF2 heterotrimerization. (A) Real-time RT-PCR was used to assess the impact of HSF2 down-regulation on the transcriptional activity during heat shock (HS) in the nSBs. The fold induction was calculated by comparing the induction in HSF2 RNAi (+) samples to the induction in Scr. (−) samples, which were arbitrarily set to 1. The data represent three biological samples, and relative quantities of sat III RNA were normalized to GAPDH. Error bars indicate SD. (B) Western blot analysis of Scr. (−) and HSF2 RNAi (+) HEK293T cells. HS indicates a 1-h heat shock. (C) The localization of HSF1 (green) and HSF2 (red) to nSBs in Scrambled (Scr.) or HSF2–down-regulating cells (HSF2 RNAi) was followed by staining. The settings used for acquiring images of HSF1 and HSF2 in Scr. cells were reused for HSF2 RNAi cells. For staining, polyclonal and monoclonal antibodies were used against HSF1 and HSF2, respectively. The nucleus is shown in blue by staining of DNA with DAPI. Heat shock and control are indicated by HS and C, respectively.
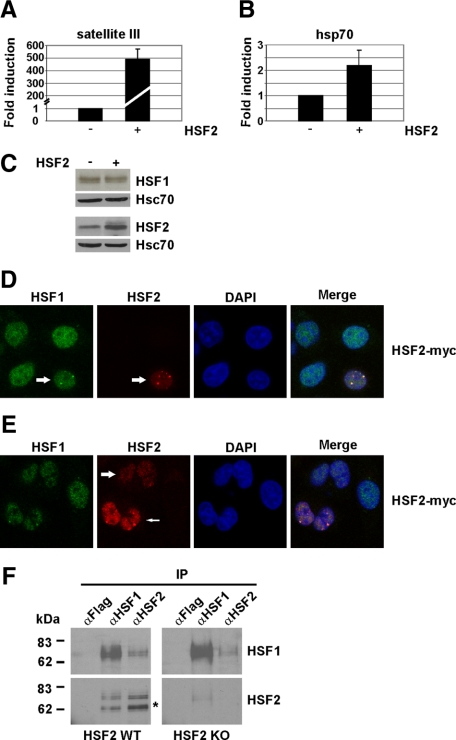
Elevated HSF2 Expression Induces HSF1-HSF2 Heterotrimerization and Activates Transcription
HSF2 is involved in development, and several reports show a correlation between increased HSF2 expression and DNA binding activity (Murphy et al., 1994, Rallu et al., 1997, Min et al., 2000, Chang et al., 2006, Åkerfelt et al., 2008). Abrogation of HSF2 DNA binding in the nSBs was associated with reduced HSF2 protein levels (Figure 1C), indicating that HSF2 is regulated in a concentration-dependent manner. We investigated the impact of elevated HSF2 expression on sat III transcription in HeLa and HEK293T cells by real-time RT-PCR, and we found that increased HSF2 concentration (Figure 4C and Supplemental Figure 5C) led to a prominent induction (∼500-fold in HeLa cells; ∼150-fold in HEK293T cells) of sat III transcription in the absence of stress (Figure 4A and Supplemental Figure 5A). To extend the study to other HSF targets, we measured the impact of HSF2 overexpression on the transcription of hsp70. In HeLa cells, transcription of also hsp70 was induced approximately twofold (Figure 4B), suggesting that HSF2 is activated when abundantly expressed. No similar induction of hsp70 was detected in HEK293T cells (Supplemental Figure 5B), which is probably due to the constitutive HSF activity in these cells, caused by the adenoviral transactivator E1A (Phillips et al., 1991).
Figure 4.
Elevated HSF2 expression activates transcription and induces HSF1-HSF2 heterotrimerization. (A) Real-time RT-PCR analysis of sat III transcription upon HSF2 over-expression (+) in untreated HeLa cells. To exclude the possibility of unspecific activation of the heat shock response, GFP was over-expressed as a control (−). (B) hsp70 transcription upon HSF2 overexpression (+) was measured in untreated HeLa cells by real-time RT-PCR. GFP was over-expressed as a control (−). In A and B, fold induction was calculated by comparing transcription in the HSF2-over-expressing cells to the control cells, in which transcription was arbitrarily set to 1. The data represent three (A) and five (B) biological samples, and the relative quantities of RNA were normalized to GAPDH. Error bars indicate SD. (C) Western blot analysis of HSF1 (top) and HSF2 (bottom) in control (−) and HSF2-over-expressing (+) HeLa cells. (D) HSF2 was over-expressed in HeLa cells and the localization of HSF1 (green) and HSF2 (red) was monitored in the absence of stress. The arrow marks a cell over-expressing HSF2. (E) HSF1 and HSF2 localization in untreated HSF2-over-expressing HeLa cells. Moderate and robust HSF2 expression is indicated by a thick and thin arrow, respectively. In D and E, monoclonal anti-myc and polyclonal anti-HSF1 antibodies were used for staining. The nucleus is shown in blue by staining of DNA with DAPI. (F) Coimmunoprecipitation of endogenous HSF1 and HSF2 in mouse wild-type testis (HSF2 WT). To show interaction during spermatogenesis, antibodies against HSF1 and HSF2 were used for immunoprecipitation. In HSF2 knockout testis (HSF2 KO), no interaction could be detected. As a negative control, anti-FLAG antibodies were used. The asterisk indicates small molecular weight isoforms of HSF2, as described in Alastalo et al. (1998).
The effect of elevated HSF2 expression on the localization of HSF1 was studied with immunofluorescence in HeLa and HEK293T cells. When abundantly expressed, HSF2 translocated to the nSBs (Figure 4D and Supplemental Figure 5D), whereas it remained dispersed in the nucleoplasm of moderately over-expressing HeLa cells (Figure 4E), further suggesting that HSF2 is regulated by its concentration. Intriguingly, HSF2 recruited also HSF1 to the nSBs (Figure 4D), implying that the increased expression of HSF2 seen during development leads to activation of HSF1.
To provide more evidence for HSF1-HSF2 heterotrimerization as a regulatory mechanism of transcription during developmental processes, we chose to investigate interaction between HSF1 and HSF2 in mouse testis, a tissue undergoing active differentiation. Moreover, HSF2 is abundantly expressed in testis and has been shown to be active during spermatogenesis (Sarge et al., 1994, Fiorenza et al., 1995, Alastalo et al., 1998, Kallio et al., 2002, Wang et al., 2003, Åkerfelt et al., 2008). We performed coimmunoprecipitation of HSF1 and HSF2 in both wild-type and HSF2 knockout testis, and we found that HSF2 could be coimmunopreciptiated with HSF1 antibodies and vice versa (Figure 4F). These results indicate that HSF1 and HSF2 form heterotrimers during spermatogenesis.
DISCUSSION
Integration of HSF Activity in Response to Stress and Developmental Stimuli
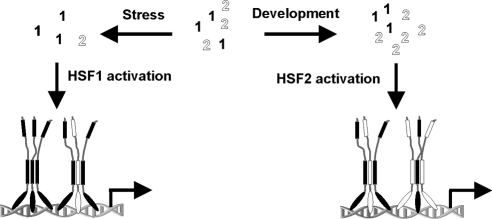
Trimerization is a crucial step in the activation process of HSFs that greatly increases the affinity for DNA (Xiao et al., 1991). In this study, we show that HSF1 and HSF2 form heterotrimers and propose that HSF1-HSF2 heterotrimerization provides a switch that integrates transcriptional activation in response to stress and developmental stimuli (Figure 5). HSF1 is known to be activated by stress and responds to elevated temperatures in vivo and in vitro. We suggest that when activated, HSF1 does not only form homotrimers but also trimerizes with HSF2, which itself is inert to stress. This model explains why the translocation of HSF2 into the nSBs is abrogated when HSF1 is depleted and also the previously suggested dependence of HSF2 on HSF1 for stress-induced DNA binding activity (Östling et al., 2007). Moreover, we show that elevated expression of HSF2 leads to its activation, suggesting that concentration regulates HSF2 during development (Figure 5). When activated, HSF2 incorporates HSF1 into a transcriptionally competent heterotrimer, illustrating the capacity of HSFs to initiate heterotrimerization. This shows, for the first time, an interdependent cooperativity between HSF1 and HSF2 that is mediated through the conserved HR-A/B domains. We propose that HSF1 needs cooperation from HSF2 in responding to developmental stimuli and that some of the functions during development earlier ascribed to HSF1 are in fact a consequence of HSF1-HSF2 heterotrimer activity.
Figure 5.
Schematic illustration of HSF1-HSF2 heterotrimerization as a mechanism integrating HSF activity. HSF1 and HSF2 are indicated in black and white, respectively. Upon stress, HSF1 is activated, leading to formation of HSF1-HSF2 heterotrimers. Stress-induced HSF activity is regulated through HSF1-HSF2 heterotrimerization, a mechanism that probably provides also temporal regulation as heat shock diminishes HSF2 levels, thereby restricting heterotrimerization through limited availability of HSF2. During development, HSF2 levels are increased at certain stages and in a tissue-specific manner, leading to activation of HSF2. Elevated HSF2 expression in turn induces HSF1-HSF2 heterotrimerization, illustrating the integrating role for HSF1-HSF2 heterotrimerization in response to distinct stimuli.
Implications of HSF1-HSF2 Heterotrimerization
Our results show that HSF1-HSF2 heterotrimerization regulates stress-induced transcription, as demonstrated by increased expression of sat III transcripts when HSF2 is depleted. Also, the positive impact on hsp70 and hsp25, and the negative impact on hsp40 and hsp110 transcription seen in hsf2−/− cells (Östling et al., 2007), can now be explained by HSF1-HSF2 heterotrimerization. HSF1 and HSF2 prefer architecturally different HSEs (Kroeger et al., 1993). This suggests that HSF1-HSF2 heterotrimers bind DNA differently than homotrimers and that the distinct regulation of HSF target genes could arise from binding of HSF1-HSF2 heterotrimers to specific sites. Another possibility is that HSF1-HSF2 heterotrimers compete with homotrimers for common binding sites. For example, the clusterin promoter contains an HSE which binds only one trimer, and on which HSF1-HSF2 heterotrimerization has been proposed to occur (Loison et al., 2006). However, no formal proof for HSF1-HSF2 heterotrimerization on the clusterin promoter has yet been provided.
Another example of a protein family using heterotrimerization is the matrilins. The matrilin heterotrimers have a variable stoichiometry that has been suggested to be determined by the concentration of the individual monomers (Frank et al., 2002). Our model does not exclude variations in the stoichiometry of the HSF1-HSF2 heterotrimers, which may change similarly to that of the matrilin analogs. Because the expression of HSF2 varies between different cell types and tissues (Rallu et al., 1997, Fiorenza et al., 1995), the HSF1-HSF2 heterotrimer stoichiometry could be modified accordingly, allowing tissue-specific regulation of HSF-mediated transcription.
Interestingly, HSF2 levels are reduced when HSF1 is down-regulated (Figure 1C), a phenomenon that has been noted also by others (Rossi et al., 2006) and is not due to unspecific down-regulation by RNAi (Supplemental Figure 6). Besides HSF1 down-regulation, heat shock reduces the levels of HSF2 (Figure 3B). It is plausible that the HSF-mediated stress-induced transcription upon prolonged stress is determined by the receding amounts of HSF2 available for heterotrimerization with HSF1 and that HSF1-HSF2 heterotrimerization regulates transcription in a temporal manner.
The nSBs: Versatile Centers for Regulation of Gene Expression?
The localization of HSF1 and HSF2 to the locus 9q12 is followed by expression of sat III transcripts and formation of nSBs. Transcription of sat III DNA is a general response to stress (Valgardsdottir et al., 2008) and the sat III transcripts are noncoding and heterogenous in size (Jolly et al., 2004, Rizzi et al., 2004). The transcripts bind a subset of mRNA-processing factors that localize to the nSBs (Weighardt et al., 1999, Denegri et al., 2001). Because the ratio between splicing factors determines the choice of splicing site, the nSBs are thought to induce alternative splicing upon stress (Jolly and Lakhotia, 2006). However, the nSBs may have multiple functions. The sat III transcripts could be involved in genomic silencing by incorporation into the RNAi system (Biamonti, 2004). Possibly, the sat III transcripts may play a role in the regulation of gene expression. In Drosophila, noncoding RNAs (ncRNAs) can activate transcription (Sanchez-Elsner et al., 2006). Interestingly, this regulation is mediated via binding of Ash1 to the ncRNA molecules, which bind to the same sequences from which they are transcribed, a feature they share with the sat III transcripts in the nSBs. Moreover, it has been proposed that the expression of human and chick coding mRNAs containing α-like sequences in their untranslated regions is controlled by small and developmentally expressed ncRNAs derived from α-satellite DNA (Li and Kirby, 2003). In human genes, segments of sat III DNA have been detected in the flanking regions and introns (Borstnik et al., 1994). It is tempting to speculate that an analogous control system of gene expression, involving the sat III transcripts and regulated by HSF1 and HSF2 in response to distinct stimuli, is used in humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Helena Saarento, Pia Roos-Mattjus, Mia Blomqvist, Gunilla Högnäs, Jung Hue Ryung, Marianne Suominen, and John Eriksson for valuable contributions and critical review of the manuscript. Perttu Terho, Jouko Sandholm, and Oso Rissanen (Turku Centre for Biotechnology Cell Imaging Core and DNA-chips Facility) are acknowledged for excellent technical assistance. This work was supported by the Academy of Finland (L. S., T.A.S.), the Sigrid Jusélius Foundation (L. S., T.A.S.), the Finnish Cancer Organizations, the Åbo Akademi University (L. S.), the Turku Graduate School of Biomedical Sciences (A. S., J.K.B., M. Å.), and the EpiPro (CLARA/INCa) and ARECA (ARC) programs (C. V., C. J., A. G.).
Abbreviations used:
- HSF
heat-shock factor
- Hsp
heat-shock protein
- nSB
nuclear stress body
- sat III
satellite III.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0864) on January 7, 2009.
REFERENCES
- Ahn S. G., Liu P. C., Klyachko K., Morimoto R. I., Thiele D. J. The loop domain of heat shock transcription factor 1 dictates DNA-binding specificity and responses to heat stress. Genes Dev. 2001;15:2134–2145. doi: 10.1101/gad.894801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfelt M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., Sistonen L. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl. Acad. Sci. USA. 2008;105:11224–11229. doi: 10.1073/pnas.0800620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alastalo T-P., Hellesuo M., Sandqvist A., Hietakangas V., Kallio M., Sistonen L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 2003;116:3557–3570. doi: 10.1242/jcs.00671. [DOI] [PubMed] [Google Scholar]
- Alastalo T.-P., Lönnström M., Leppä S., Kaarniranta K., Pelto-Huikko M., Sistonen L., Parvinen M. Stage-specific expression and cellular localization of the heat shock factor 2 isoforms in the rat seminiferous epithelium. Exp. Cell Res. 1998;240:16–27. doi: 10.1006/excr.1997.3926. [DOI] [PubMed] [Google Scholar]
- Anckar J., Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv. Exp. Med. Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Barton G. J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Batchelor A. H., Piper D. E., de la Brousse F. C., McKnight S. L., Wolberger C. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Biamonti G. Nuclear stress bodies: a heterochromatin affair? Nat. Rev. Mol. Cell Biol. 2004;5:493–498. doi: 10.1038/nrm1405. [DOI] [PubMed] [Google Scholar]
- Borstnik B., Pumpernik D., Lukman D., Ugarkovic D., Plohl M. Tandemly repeated pentanucleotides in DNA sequences of eucaryotes. Nucleic Acids Res. 1994;22:3412–3417. doi: 10.1093/nar/22.16.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chang Y., et al. Role of heat-shock factor 2 in cerebral cortex formation and as a regulator of p35 expression. Genes Dev. 2006;20:836–847. doi: 10.1101/gad.366906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I., Corioni M., Giordano M., Valgardsdottir R., Ghigna C., Cobianchi F., Xu R. M., Riva S., Biamonti G. RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies. Nucleic Acids Res. 2004;32:4127–4136. doi: 10.1093/nar/gkh759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton E. T. Proteins: Structures and Molecular Properties. New York: W. H. Freeman and Company; 1993. [Google Scholar]
- Dai C., Whitesell L., Rogers A. B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denegri M., Chiodi I., Corioni M., Cobianchi F., Riva S., Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza M. T., Farkas T., Dissing M., Kolding D., Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Schulthess T., Landwehr R., Lustig A., Mini T., Jeno P., Engel J., Kammerer R. A. Characterization of the matrilin coiled-coil domains reveals seven novel isoforms. J. Biol. Chem. 2002;277:19071–19079. doi: 10.1074/jbc.M202146200. [DOI] [PubMed] [Google Scholar]
- Holmberg C. I., Illman S. A., Kallio M., Mikhailov A., Sistonen L. Formation of nuclear HSF1 granules varies depending on stress stimuli. Cell Stress Chap. 2000;5:219–228. doi: 10.1379/1466-1268(2000)005<0219:fonhgv>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A. L., Murphy C. T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Johnson M. S., Overington J. P. A structural basis for sequence comparisons. An evaluation of scoring methodologies. J. Mol. Biol. 1993;233:716–738. doi: 10.1006/jmbi.1993.1548. [DOI] [PubMed] [Google Scholar]
- Jolly C., Konecny L., Grady D. L., Kutskova Y. A., Cotto J. J., Morimoto R. I., Vourc'h C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 2002;156:775–781. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Lakhotia S. C. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34:5508–5514. doi: 10.1093/nar/gkl711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Metz A., Govin J., Vigneron M., Turner B. M., Khochbin S., Vourc'h C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Morimoto R., Robert-Nicoud M., Vourc'h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci. 1997;110:2935. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- Kallio M., et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger P. E., Sarge K. D., Morimoto R. I. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol. Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge M. Intrinsic protein electric fields: basic non-covalent interactions and relationship to protein-induced Stark effects. Biochim. Biophys. Acta. 1998;1386:305–330. doi: 10.1016/s0167-4838(98)00100-9. [DOI] [PubMed] [Google Scholar]
- Lehtonen J. V., et al. BODIL: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 2004;18:401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- Li Y. X., Kirby M. L. Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev. Dyn. 2003;228:72–81. doi: 10.1002/dvdy.10355. [DOI] [PubMed] [Google Scholar]
- Littlefield O., Nelson H.C.M. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat. Struct. Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- Loison F., Debure L., Nizard P., Le Goff P., Michel D., Le Drean Y. Up-regulation of the clusterin gene after proteotoxic stress. Implication of HSF1/HSF2 heterocomplexes. Biochem. J. 2006;395:223–231. doi: 10.1042/BJ20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz A., Soret J., Vourc'h C., Tazi J., Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 2004;117:4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- Min J. N., Han M. Y., Lee S. S., Kim K. J., Park Y. M. Regulation of rat heat shock factor 2 expression during the early organogenic phase of embryogenesis. Biochim. Biophys. Acta. 2000;1494:256–262. doi: 10.1016/s0167-4781(00)00053-1. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J. F., Morimoto R. I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. P., Gorzowski J. J., Sarge K. D., Phillips B. Characterization of constitutive HSF2 DNA-binding activity in mouse embryonal carcinoma cells. Mol. Cell Biol. 1994;14:5309–5317. doi: 10.1128/mcb.14.8.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östling P., Björk J. K., Roos-Mattjus P., Mezger V., Sistonen L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J. Biol. Chem. 2007;282:7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- Phillips B., Abravaya K., Morimoto R. I. Analysis of the specificity and mechanism of transcriptional activation of the human hsp70 gene during infection by DNA viruses. J. Virol. 1991;65:5680–5692. doi: 10.1128/jvi.65.11.5680-5692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L., Nykänen P., Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rallu M., Loones M., Lallemand Y., Morimoto R., Morange M., Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Ciafre S., Balsamo M., Pierimarchi P., Santoro M. G. Targeting the heat shock factor 1 by RNA interference: a potent tool to enhance hyperthermochemotherapy efficacy in cervical cancer. Cancer Res. 2006;66:7678–7685. doi: 10.1158/0008-5472.CAN-05-4282. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T., Gou D., Kremmer E., Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Murphy S. P., Morimoto R. I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K. D., Park-Sarge O-K., Kirby J. D., Mayo K. E., Morimoto R. I. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol. Reprod. 1994;50:1334–13343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- Shu W., Ji H., Lu M. Trimerization specificity in HIV-1 gp 41, analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- Shu W., Liu J., Ji H., Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J. Mol. Biol. 2000;299:1101–1112. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- Shumaker D. K., et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Rossi A., Bazzini S., Ghigna C., Riva S., Biamonti G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang J., Moskophidis D., Mivechi N. F. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200. [DOI] [PubMed] [Google Scholar]
- Weighardt F., Cobianchi F., Cartegni L., Chiodi I., Villa A., Riva S., Biamonti G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]
- Xiao H., Perisic O., Lis J. T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- Xiao X., Zuo X., Davis A. A., McMillan D. R., Curry B. B., Richardson J. A., Benjamin I. J. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.