Abstract
Kinesin-14 family proteins are minus-end directed motors that cross-link microtubules and play key roles during spindle assembly. We showed previously that the Xenopus Kinesin-14 XCTK2 is regulated by Ran via the association of a bipartite NLS in the tail of XCTK2 with importin α/β, which regulates its ability to cross-link microtubules during spindle formation. Here we show that mutation of the nuclear localization signal (NLS) of human Kinesin-14 HSET caused an accumulation of HSET in the cytoplasm, which resulted in strong microtubule bundling. HSET overexpression in HeLa cells resulted in longer spindles, similar to what was seen with NLS mutants of XCTK2 in extracts, suggesting that Kinesin-14 proteins play similar roles in extracts and in somatic cells. Conversely, HSET knockdown by RNAi resulted in shorter spindles but did not affect pole formation. The change in spindle length was not dependent on K-fibers, as elimination of the K-fiber by Nuf2 RNAi resulted in an increase in spindle length that was partially rescued by co-RNAi of HSET. However, these changes in spindle length did require microtubule sliding, as overexpression of an HSET mutant that had its sliding activity uncoupled from its ATPase activity resulted in cells with spindle lengths shorter than cells overexpressing wild-type HSET. Our results are consistent with a model in which Ran regulates the association of Kinesin-14s with importin α/β to prevent aberrant cross-linking and bundling of microtubules by sequestering Kinesin-14s in the nucleus during interphase. Kinesin-14s act during mitosis to cross-link and slide between parallel microtubules to regulate spindle length.
INTRODUCTION
During mitosis, eukaryotic cells form a microtubule (MT)-based bipolar spindle that exerts forces to congress the chromosomes to the metaphase plate and to segregate them to the two daughter cells. Although different cell types form spindles with varying morphologies, for a certain cell type the spindle morphology remains steady (Mitchison et al., 2005). It is therefore critical to understand how cells organize and maintain the spindle machinery of a certain size and shape so that we can understand how spindle organization affects chromosome behavior.
A bipolar spindle is composed of three classes of dynamic MTs: the astral MTs that extend from the centrosomes and project toward the cell cortex, the interpolar MTs that overlap in the spindle midzone in an antiparallel manner, and the kinetochore fibers (K-fibers), which are parallel bundles of 20–40 MTs that come from kinetochores and focus at the spindle poles (Compton, 2000; Walczak and Heald, 2008). The coordinated regulation of the dynamics and organization of these three classes of MTs is needed to form a morphologically normal spindle. Molecular perturbation of factors that regulate dynamics of MTs in the spindle or the addition of anti-MT drugs affects spindle organization and changes spindle length (Severin et al., 2001; Goshima et al., 2005b; Mitchison et al., 2005; Ohi et al., 2007). However, perturbation of molecular motors involved in MT sliding has given contradictory results on their roles in spindle length control (Hoyt et al., 1993; Sharp et al., 1999; Sharp et al., 2000a; Troxell et al., 2001; Goshima et al., 2005b; Burbank et al., 2007), raising the question of what are the principle mechanisms that control spindle length.
Two major models have been proposed for how spindles assemble during mitosis. In the search-and-capture model, MTs are nucleated around centrosomes with their dynamic plus ends growing toward the chromosomes until they are captured and stabilized by a kinetochore (Kirschner and Mitchison, 1986; McIntosh et al., 2002). In the chromatin-mediated spindle assembly model, MTs are nucleated in the vicinity of chromosomes and then become incorporated into the spindle by the action of molecular motors (McKim and Hawley, 1995; Heald et al., 1996; Walczak and Heald, 2008). It is thought that centrosome-mediated spindle formation is dominant in most mitotic cells that contain centrosomes. However, when centrosomes are laser-ablated in somatic cells spindle assembly still occurs, suggesting that chromatin-mediated spindle assembly also occurs in somatic cells (Khodjakov et al., 2000; Khodjakov and Rieder, 2001).
MT nucleation around chromosomes is mediated by a gradient of Ran-GTP that is highest near the chromosomes and diminishes away from the chromosomes (Carazo-Salas et al., 1999; Wilde and Zheng, 1999; Kalab et al., 2002, 2006; Caudron et al., 2005). Ran action in the spindle is mediated through nuclear localization signal (NLS)-containing spindle assembly factors that are kept in an inactive state by association with the nuclear import receptors importin α/β (Kalab and Heald, 2008; Walczak and Heald, 2008). The highest concentration of Ran-GTP is near the chromosomes where it binds to importin β, causing release of the spindle assembly factors that contribute to MT nucleation, stabilization, and organization (Caudron et al., 2005; Kalab and Heald, 2008).
One important Ran-regulated factor needed for proper spindle assembly is the Kinesin-14 family member XCTK2 (Walczak et al., 1997; Ems-McClung et al., 2004). Kinesin-14 family members are minus-end–directed MT motors (McDonald et al., 1990; Walker et al., 1990) that are needed for spindle assembly and pole organization in both mitotic and meiotic systems (Hatsumi and Endow, 1992a,b; Endow and Komma, 1996; Walczak et al., 1997; Matuliene et al., 1999; Mountain et al., 1999; Sharp et al., 1999; Goshima et al., 2005a). Structurally Kinesin-14s have a conserved kinesin-like motor at the C-terminus, a central coiled-coil stalk, and an N-terminal globular domain. The motor domain binds MTs in an ATP-sensitive manner, and the tail possesses a second ATP-insensitive MT binding site that enables Kinesin-14s to cross-link and slide MTs (Chandra et al., 1993; Walker, 1995; Walczak et al., 1997; Karabay and Walker, 1999; Ems-McClung et al., 2004). We showed previously that XCTK2 is regulated by Ran via the association of a bipartite NLS in the tail of XCTK2 with importin α/β (Ems-McClung et al., 2004). Binding of importin α/β to XCTK2 inhibits its association with MTs and blocks its ability to cross-link MTs. This data supports a model in which XCTK2 acts near chromosomes to cross-link and slide MTs during chromatin-mediated spindle formation. However, it is not clear whether Kinesin-14s are needed for spindle formation in somatic cells and how their MT sliding activity contributes to spindle organization.
MATERIALS AND METHODS
Constructs and Protein Purification
The HSET cDNA (Genbank ID: 68299768) was a generous gift from Duane Compton (Dartmouth Medical School, Hanover, NH). The cDNA encoding the nonmotor domain of HSET (amino acids 2–304) was subcloned into pGEX1* and p6HGFP-B vectors with the restriction enzymes BamHI and EcoRI to generate glutathione S-transferase (GST)- and 6-His-green fluorescent protein (GFP)–tagged fusion proteins, respectively. The HSET mutants HSET-NLSa (36KRR to AAA), HSET-NLSb (50KKR to AAA), and HSET N593K were made using the Quikchange site-directed mutagenesis system (Stratagene, La Jolla, CA). pEGFP-HSET and pEGFP-HSET N593K clones were generated by subcloning their cDNA into the pEGFPC1 vector by EcoRI/SalI. The XCTK2 constructs and mutants were described previously (Ems-McClung et al., 2004). All resulting clones were verified by sequencing.
GST-HSET(2–304) was expressed in Escherichia coli BL21(DE3) cells and affinity-purified on glutathione Sepharose 4B (Amersham Biosciences, Piscataway, NJ) as previously described (Ems-McClung et al., 2004), followed by the gel filtration on a Superose 12 column (GE Healthcare Life Sciences, Piscataway, NJ) equilibrated in column buffer (50 mM MOPS, pH 7.2, 50 mM NaCl, 0.1 mM EDTA, and 0.1 mM EGTA). Fractions were eluted, and sucrose was added to 10%. Fractions were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until use. His-GFP-HSET(2–304) was expressed, purified, and cross-linked to Affi-Gel 10 (Bio-Rad, Hercules, CA) as previously described (Walczak et al., 1997). Antibodies to the nonmotor domain of HSET were generated by immunizing a rabbit (Covance Research Products, Denver, PA) with GST-HSET(2–304) protein that was first treated with 0.5% glutaraldehyde for 45 min and then quenched with a trace of sodium borohydride for 5 min at room temperature (RT). HSET specific antibodies were then affinity-purified using a His-GFP-HSET(2–304) affinity column as described previously (Walczak et al., 1997).
For protein expression of the GFP-fusion proteins XCTK2, XCTK2 NLSa, XCTK2 NLSb, HSET and HSET-N593K, coding sequences were subcloned into pFastBac1-GFP using SacI and KpnI restriction enzymes, and baculovirus was produced using the Bac-to-Bac baculovirus system (Invitrogen, Carlsbad, CA). The GFP-tagged proteins were then expressed in Sf-9 insect cells and purified by conventional chromatography as previously described (Walczak et al., 1997). The proteins were stored in FPLC buffer (20 mM PIPES, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 50 μM Mg-ATP, 400 mM KCl, and 0.1 μg/ml LPC) with the addition of 10% sucrose, flash-frozen in liquid nitrogen, and stored at −80°C.
Nuclear Import and Spindle Assembly in Xenopus Egg Extracts
CSF extract was made from Xenopus laevis eggs as previously described (Murray, 1991). For the nuclear import assay, demembranated sperm nuclei were added at a concentration of 500 sperm/μl to CSF extract that was activated by the addition of 0.4 mM CaCl2 for 2 h until most of the sperm had formed a nuclear envelope and appeared round. At this time, a final concentration of 20 nM GFP-XCTK2 or the NLS mutants GFP-XCTK2 NLSa or GFP-XCTK2 NLSb were added to the extract with the preformed nuclei. Ten minutes after the addition of the recombinant proteins, samples (1 μl) were taken and squashed on a microscope slide with 3 μl of spindle fix (60% glycerol, 5 mM HEPES, pH 7.8, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10% formaldehyde, and 1 μg/ml Hoechst) and immediately imaged using fluorescence microscopy.
For spindle assembly assays, CSF extracts were supplemented with 200 nM X-Rhodamine-labeled tubulin, 500 sperm nuclei/μl, and the indicated recombinant proteins (GFP, GFP-XCTK2, GFP-XCTK2NLSa, GFP-XCTK2NLSb, GFP-HSET, and GFP-HSET N593K) at either a 2.5- or 5-fold molar excess over the endogenous XCTK2 as indicated in the text. The endogenous concentration of XCTK2 in CSF extract is ∼20 nM. All fusion proteins were diluted to a 10-fold concentrated stock in CSF-XB buffer, and then equivalent volumes were added to each spindle assembly reaction before spindle assembly for a final 1× concentration. For cycled extracts, CSF extracts were cycled into interphase with the addition of Ca2+ and then back into mitosis with fresh CSF extract as described (Sawin and Mitchison, 1991; Shamu and Murray, 1992). For SDS-PAGE, 5 μl of each extract reaction was diluted in 45 μl of sample buffer (0.125 M Tris, pH 6.8, 4% SDS, 20% glycerol, 4% β-mercaptoethanol, and a trace amount of bromophenol blue) and subjected to Western blot analysis to confirm that equivalent amounts of the proteins were added to the extract. Thirty minutes after initiation of the spindle assembly reaction, 30 μl of each extract sample was diluted in 1 ml of BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) containing 30% glycerol (vol/vol) and fixed by the addition of 1 ml of 30% glycerol, BRB80, and 4% formaldehyde for 20 min at RT. The fixed samples were layered on a 4-ml cushion of BRB80 containing 40% glycerol (vol/vol) and were centrifuged onto coverslips in a Beckman (Fullerton, CA) JS7.5 rotor at 6000 × g for 15 min. The coverslips were postfixed in −20°C methanol for 5 min and rehydrated in TBS-TX (10 mM Tris, pH 7.6, 150 mM NaCl and 0.1% Triton X-100). DNA was stained by incubation in 1 μg/ml Hoechst 33342 (Sigma-Aldrich, St. Louis, MO) diluted in TBS-TX, mounted in anti-fade (90% glycerol, 20 mM Tris-HCl, pH 8.8, and 0.5% p-phenylenediamine), and sealed with nail polish.
Cell Culture, RNA Interference, Gene Transfection, and Creation of Stable Cell Lines
HeLa cells were cultured at 37°C in Opti-MEM (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin (Invitrogen). For small interfering RNA (siRNA), HeLa cells at 4 × 104 cells/well were plated into each well of a six-well culture dish, arrested with 2 mM thymidine for 20 h, and then released into fresh media. Two hours after release from the thymidine block, 200 nM Luciferase RNA interference (RNAi) negative control no. 2 oligonucleotide (Dharmacon, Chicago, IL), 200 nM HSET RNAi oligonucleotide to the coding region (UCA GAA GCA GCC CUG UCA A), or 200 nM hNuf2 RNAi oligonucleotide (DeLuca et al., 2002) was transfected using Oligofectamine (Invitrogen). When comparing knockdown of multiple proteins, an equivalent amount of Luciferase negative control oligonucleotide was cotransfected with the specific oligonucleotide such that the total amount of oligonucleotide transfected was identical for each well. At 24 h after transfection, cells were blocked with 2 mM thymidine for 19 h to synchronize the cells and then released for 11–12 h to allow cells to progress to late G2. The cells were then processed for immunofluorescence, for live imaging, or for Western blot analysis.
To generate the stable cell lines, pEGFP-HSET or pEGFP-HSET N593K were individually introduced into HeLa cells using a calcium phosphate transfection in the presence of 2 mg/ml geneticin (Invitrogen; Rodriguez and Flemington, 1999). Transfected cells were replated at a density of 1 × 103 cells/plate in a 10-cm plate and grown for 7–10 d with daily changes in media until each colony had ∼200 cells. A single colony was isolated with a pipette tip by examining the colony under an inverted fluorescence microscope, transferred to a new dish, and maintained in Opti-MEM containing 1 mg/ml geneticin (Invitrogen). pEGFP-H2B and pmcherry-tubulin plasmids were sequentially introduced into HeLa cells to an established stable expressing cell line as described above. For transient transfections, pEGFP-HSET NLSa or pEGFP- HSET NLSb were introduced into HeLa cells using calcium phosphate transfection and fixed and processed for immunofluorescence 24 h after transfection.
Immunofluorescence and Western Blot Analysis
HeLa cells were fixed in −20°C methanol for 5 min at RT and rehydrated in TBS-TX for 2 min. Cells were blocked in Abdil (TBS-TX, 2% bovine serum albumin, and 0.1% sodium azide) for 1 h at RT or overnight at 4°C. Cells were stained with antibodies against HSET (2.5 μg/μl), Hec1 (1 μg/μl; Novus Biologicals, Littleton, CO), ACA (1:100; Antibodies Incorporated, Davis, CA), or α-tubulin DM1α (1:1000; Sigma-Aldrich) diluted in Abdil for 1 h at RT. Cells were subsequently stained with a 1:50 dilution of donkey anti-rabbit Alexa Fluor 488 (Invitrogen) or donkey anti-mouse TexasRed (Jackson ImmunoResearch Laboratories, West Grove, PA). To visualize DNA, fixed cells were stained with 2 μg/ml Hoechst (Sigma-Aldrich) in TBS-TX.
For Western blot analysis, HeLa cells were collected and lysed in sample buffer at 3000 cells/μl, and Xenopus extract was lysed by 1:10 dilution in sample buffer. 60,000 HeLa cells or 20 μl of extract sample were loaded on a 10% SDS-polyacrylamide gel and transferred to Protran nitrocellulose (Schleicher & Schuell, Waltham, MA). Blots were then probed with DM1α (1:10,000, Sigma-Aldrich), anti-HSET (2.5 μg/ml), anti-GFP (1.9 μg/ml), anti-importin β (1 μg/ml; Sigma-Aldrich), or anti-importin α (1:1000; from Mary Dasso, National Institutes of Health, Bethesda, MD), followed by sheep anti-mouse IgG HRP-linked whole antibody (1:10,000; Amersham, Piscataway, NJ) or donkey anti-rabbit IgG HRP-linked whole antibody (1:10,000; Amersham) and detected with SuperSignal West Pico enhanced chemiluminescence substrates according to the manufacturer's directions.
Immunoprecipitations
Immunoprecipitations from Xenopus egg extracts were performed as described previously (Ems-McClung et al., 2004). Anti-GFP and nonimmune rabbit IgG were covalently coupled to the Affi-prep protein A beads (Bio-Rad; Harlow and Lane, 1999). Where indicated, 200 nM purified GFP-XCTK2, GFP-XCTK2-NLSa, or GFP-XCTK2-NLSb was added to the immunoprecipitation reactions. Equivalent volumes of the eluted protein were electrophoresed on 10% SDS-PAGE gels and stained with Coomassie Blue G-250 or transferred to Protran. Western blots were probed as described above.
Immunoprecipitation in HeLa extract was performed as described (Luders et al., 2006) using anti-HSET antibodies cross-linked to Affi-prep protein A beads. The GFP-HSET overexpressing HeLa cells were plated in a 10-cm plate and treated with 100 ng/ml nocodazole overnight. Cells were trypsinized and diluted with ice-cold Opti-MEM. Cells were centrifuged at 1000 × g, resuspended in lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 1/1000 LPC) for 5 min on ice using a ratio of 1 ml buffer/10-cm plate. HeLa cell lysates were centrifuged in a TLS 55 rotor at 16,000 rpm for 10 min in a Beckman Optima L-90K Ultracentrifuge. For immunoprecipitations, 400 μl of supernatant were incubated with 50 μl of Affi-prep IgG beads or anti-GFP beads for 5 h at 4°C following the procedure described above for immunoprecipitations in egg extracts.
Cold Treatment for Analysis of Stable MTs
HeLa cells were transfected with the Luciferase control oligonucleotide or HSET RNAi oligonucleotide, and the GFP-HSET HeLa cell line was transfected with the Luciferase control oligonucleotide. At 24 h after transfection, cells were synchronized with 2 mM thymidine for 19 h and released in fresh medium for 11–12 h. The cells in dishes were put on an ice-water bath for 10 min to allow for MT depolymerization and then were fixed with −20°C methanol for 5 min and processed for immunofluorescence as described above.
Preparation of MT Substrates
Guanylyl-(α,β)-methylene-diphosphonate- (GMPCPP; Jena Scientific, Jena Germany) and paclitaxel-stabilized MTs were polymerized from cycled bovine tubulin as previously described (Desai and Walczak, 2001). Tubulin was clarified at 45,000 rpm for 5 min at 2°C in a TLA 100 rotor (Beckman Coulter) and then polymerized in the presence of 0.5 mM GMPCPP, BRB80, and 1 mM DTT for 30 min. Paclitaxel was added to 20 μM at 20 min after the start of polymerization. The MTs were pelleted at 37°C for 5 min in a TLA100 rotor and resuspended in BRB49 (49 mM PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA), 1 mM DTT, and 20 μM paclitaxel.
ELIPA ATPase Assays
ATPase assays were performed using the Enzyme Linked Inorganic Phosphate Assay (ELIPA) kit (Cytoskeleton, Denver, CO) to detect Pi release. For these experiments, 250 nM GFP-HSET or GFP-HSET N593K were incubated with 0.62 mM MgATP, 50 mM KCl, BRB80, and varying concentrations (0–3 μM) of GMPCPP- and paclitaxel-stabilized MTs (doubly stabilized). The rate of Pi released was measured using the SpectraMax190 (Molecular Devices, Sunnyvale, CA) with an absorbance of 360 nm at 30-s intervals for 30 min. Data were collected and analyzed from at least three independent experiments and fit to Equation 1 using Prism (GraphPad Software, San Diego, CA) to determine the kcat and Km of GFP-HSET or GFP-HSET N593K in the presence of MTs. The plots represent the average ± the SE of these experiments.
where kcat represents the maximum binding, MTt represents the total tubulin concentration, and Km represents the concentration of GFP-HSET to reach half-maximum binding.
HSET MT Cosedimentation Assays
The cosedimentation assays were performed similarly to Hertzer et al. (2006) using doubly stabilized MTs. Equal molar concentrations (0.35 μM) of purified protein (GFP-HSET or GFP-HSET N593K) were incubated in BRB20 (20 mM PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA), and 63 mM KCl with increasing concentrations of doubly stabilized MTs (0–0.7 μM). The reactions were pelleted at 45,000 rpm in a Beckman TLA 100 rotor. The supernatant was removed and mixed with an equal volume of 2× sample buffer. The pellet of each reaction was resuspended in a volume of 2× sample buffer equal to the supernatant and mixed with an equal volume of BRB80. Equal volumes of supernatant and pellet samples were analyzed by SDS-PAGE and stained with colloidal Coomassie Blue. The concentration of GFP-HSET partitioning between the supernatant and pellet fractions was quantified by densitometry of the gel using Image J (http://rsb.info.nih.gov/ij/; NIH, Bethesda, MD). Data collected and analyzed from at least three independent experiments were fit to a two-site quadratic binding equation (Equation 2; Wang and Jiang, 1996) using Prism (GraphPad Software) to determine the apparent Kd of GFP-HSET or GFP-HSET N593K for MTs. The plots represent the average ± the SE of these experiments:
where
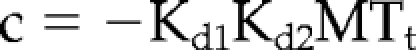 |
where Mt · E is the concentration of enzyme binding to MTs in nanomolar, E0 is the total amount of enzyme bound in nanomolar, E is the total enzyme concentration, MTt is the total tubulin concentration, Kd1 is the dissociation constant for site 1, and Kd2 is the dissociation constant for site 2.
Live Imaging
For HSET RNAi imaging, GFP-H2B/mcherry-tubulin HeLa cells were plated at 4000 cells/well for both control and HSET RNAi in 96-well BD Falcon imaging plates (Bedford, MA). HSET RNAi was performed using the same method described above except that the volumes of all reagents were normalized based on the smaller surface area of the plates. At 24 h after HSET RNAi, cells were blocked by 2 mM thymidine for 16 h and released in fresh medium for 9 h. Cell plates were placed in BD Pathway 855 and imaged at 20–40-ms exposure time at 5-min intervals ON. For imaging of HSET overexpressing cells, the GFP-HSET overexpressing cell line was plated at 8000 cells/well in a 96-well BD Falcon imaging plate and transiently transfected with p-mcherry-H2B plasmid using Oligofectamine 2000 (Invitrogen). At 8 h after transfection, cells were imaged using the same conditions as for the HSET RNAi cells. Mitotic progression was analyzed manually using the BD Attovision by recording spindle morphology from frame to frame. Mitotic progression was measured as the time from nuclear envelope breakdown (NEBD) to the de-condensation of daughter chromosomes.
Imaging and Statistical Analysis
All images were acquired on a Nikon Eclipse 90i (Melville, NY), using either a 20× (NA 0.5), 40× (NA 1.0), or 100× Plan Apo objective (NA 1.4) and a CoolSnap HQ CCD camera (Photometrics, Tucson, AZ). The camera and filters were controlled by Metamorph (Molecular Devices, Sunnyvale, CA). Image stacks were collected at 0.5-μm steps through the whole cell volume and then deconvolved using Autodeblur (Autoquant Imaging, Bethesda, MD) for 10–20 iterations. The extract samples were imaged using a 40× plan Apo objective (NA 1.0) and are single-plane images. All images were processed in Adobe Photoshop CS and assembled in Adobe Illustrator CS (San Jose, CA) equivalently for control and experimental samples. The mean spindle length and width were determined from three independent experiments and graphed with the SEM using Excel (Microsoft, Redmond, WA). The histogram of the length data were fit with Gaussian distribution by Prism (GraphPad). The curve was plotted in Excel (Microsoft) and superimposed with the histogram in Adobe Illustrator CS. To measure the fluorescence intensity of GFP-HSET and GFP-HSET N593K, Z series of images were taken with the 100× objective at 0.5-μm step intervals to cover the whole volume of the spindle. After 3D reconstruction in Metamorph, an equivalent 50 × 20-pixel box was drawn both in the spindle close to the pole and in the cytoplasm as the background. Background subtracted fluorescence intensity was correlated with the spindle length and presented in the graph. The interkinetochore distance was determined by measuring the center to center distance of ACA staining of sister kinetochores from 150 kinetochores in 30 cells from three independent experiments. Statistical significance was determined with a Student's t test performed in Excel.
RESULTS
HSET Plays a Role in Controlling Spindle Morphology
To study the role of HSET in controlling spindle organization, we generated a HeLa cell line that stably expresses GFP-HSET at >10-fold the endogenous level (data not shown). The spindles in the GFP-HSET–overexpressing cells had increased MT bundling and appeared longer than in control HeLa cells (Figure 1A). In some cases, the spindle poles even became curved at the periphery of the cell cortex (data not shown), suggesting that the two poles were pushed apart. We measured the pole-to-pole distance of these spindles using the centrosome staining of HSET as a marker and found a 16% increase in the pole-to-pole distance of the GFP-HSET cells compared with control HeLa cells (p < 0.01; Figure 1B). These spindles were not only longer, but were also 16% thinner based on a measurement of spindle width at the metaphase plate (p < 0.05; Figure 1B), suggesting that overexpression of HSET changes overall spindle morphology.
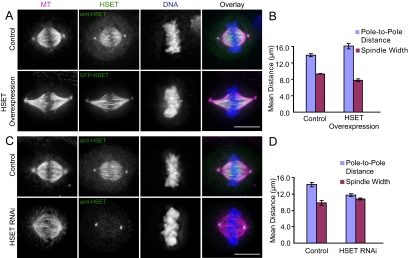
Figure 1.
HSET perturbation affects spindle morphology. (A) Control or GFP-HSET–overexpressing cells were processed for immunofluorescence to visualize MTs (magenta), HSET (green), and DNA (blue). Control cells were stained with anti-HSET, whereas transfected GFP-HSET was visualized directly by GFP fluorescence. (B) The average pole-to-pole distances and spindle widths of control and GFP-HSET–overexpressing cells are reported as mean ± SEM from three independent experiments. Average spindle lengths: control 13.9 ± 0.3 μm, overexpression 16.1 ± 0.6 μm; average spindle widths: control 9.34 ± 0.02 μm, overexpression 7.8 ± 0.4 μm (C) Cells transfected with either a control Luciferase or an HSET-specific siRNA were processed for immunofluorescence to visualize MTs (magenta), HSET (green), and DNA (blue). (D) The average pole-to-pole distances and spindle widths of control and HSET RNAi cells are reported as mean ± SEM from three independent experiments. Average spindle lengths: control 14.3 ± 0.5 μm, HSET RNAi 11.7 ± 0.4 μm; average spindle widths: control, 9.8 ± 0.6 μm; and HSET RNAi, 10.8 ± 0.3 μm. Scale bar, 10 μm.
If overexpression of HSET causes spindle elongation, then knocking down HSET by RNAi should cause a reduction in spindle length. HSET could be knocked down by greater than 80% as judged by Western blot (Supplementary Figure S1A) and by immunofluorescence (Figure 1C and Supplementary Figure S1B). The residual staining at spindle poles is due to nonspecific staining of another unknown centrosomal protein (data not shown). We found an 18% decrease in the pole-to-pole distance of HSET RNAi-treated cells compared with the spindles in control cells (p < 0.01; Figure 1, C and D) and a 10% increase in spindle width of HSET RNAi spindles (p = 0.05; Figure 1, C and D), indicating that spindles without HSET are broader and shorter than in control cells. This shortened spindle phenotype is distinct from the split pole phenotype seen with inhibition of Drosophila Kinesin-14 Ncd (Goshima et al., 2005a), because we did not see a significant increase in the percentage of multipolar spindles with HSET RNAi (Supplementary Figure S1C). Analysis of mitotic progression in HSET knockdown and HSET overexpression cells showed that neither perturbation significantly affected the average duration of mitosis (control: 102 ± 62 min; HSET RNAi: 99 ± 45 min; HSET overexpression: 118 ± 66 min; from ∼30 cells in each condition). The alignment of chromosomes to the metaphase plate was normal, and there was not an increased incidence of lagging chromosomes during anaphase (data not shown). Overall, these data suggest that HSET affects spindle morphology, perhaps by exerting an outward force on the spindle.
HSET Affects Spindle Morphology Independently of K-Fibers
It is possible that overexpressed HSET increases spindle length by sliding MTs within K-fibers along spindle MTs. To ask how K-fibers contribute to the morphological changes we observe, we treated cells with ice-cold buffer to depolymerize the highly dynamic spindle MTs and measured the length of the remaining stabilized MTs (Supplementary Figure 2). There was a reduction in the average length of the MTs after HSET RNAi (p = 0.05) and an increase in the average MT length in HSET overexpressing cells (p < 0.05; Supplementary Figure S2, A and B), suggesting that the length of the K-fiber may change upon HSET perturbation. We also measured the interkinetochore distance in HSET perturbed cells and found there was no difference in the interkinetochore distance in HSET RNAi cells (1.4 ± 0.2 μm) compared with control cells (1.6 ± 0.1 μm), whereas the HSET overexpression cells had a slight reduction in interkinetochore distance (1.2 ± 0.1 μm).
To ask whether the K-fibers are required for the change in spindle length after HSET perturbation, we knocked down hNuf2, one of the components of the Ndc80 complex, to inhibit formation of the K-fiber (Figure 2A). After hNuf2 RNAi, the spindle length was significantly increased in comparison to control cells (p < 0.01), consistent with previous findings (DeLuca et al., 2002; Figure 2B). When HSET was knocked down in the hNuf2 knockdown cells, the hNuf2-dependent increase in spindle length was rescued (p < 0.01), and the spindle length was slightly longer than control cells (p < 0.05), but was similar in length to cells overexpressing HSET (p = 0.32; Figure 2B). To ensure that there were no residual K-fibers contributing to the changes in spindle length, we only analyzed those mitotic cells with completely misaligned chromosomes. When K-fibers were diminished in GFP-HSET overexpressing cells, the spindle length was increased compared with hNuf2 RNAi spindles (p < 0.05; Figure 2, A and B). This increase is similar to that seen in HSET overexpression alone in normal spindles. Together these findings support the idea that K-fiber attachment to the kinetochores is not required for HSET-dependent changes in spindle length.
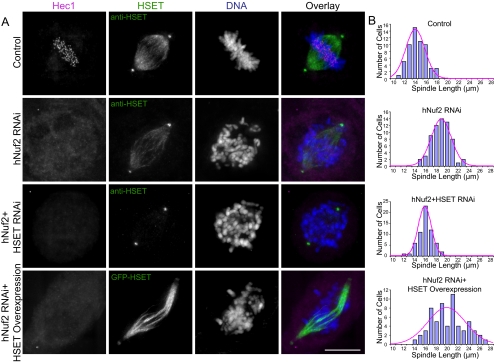
Figure 2.
HSET affects spindle morphology in spindles lacking K-fibers. (A) Control HeLa cells (top three rows) or GFP-HSET overexpressing HeLa cells (bottom row) were transfected with either control Luciferase, hNuf2, or hNuf2+HSET siRNAs, synchronized, released, and then processed for immunofluorescence. In each panel, the Hec1 (magenta), HSET (green,) and DNA (blue) staining are shown. Scale bar, 10 μm. (B) Histograms of pole-to-pole distances quantified from a total of 60 spindles in three independent experiments for each knockdown condition. The best fit Gaussian distribution is shown superimposed in magenta. Average spindle lengths: control, 14.0 ± 0.4 μm; hNuf2 RNAi, 18.3 ± 0.5 μm; hNuf2 + HSET RNAi, 15.5 ± 0.6 μm; and hNuf2 RNAi + HSET overexpression, 19.7 ± 0.4 μm.
HSET Cross-Links and Slides MTs to Regulate Spindle Length
Because Kinesin-14 proteins are known to cross-link and slide MTs, it is likely that the ability of HSET to slide MTs is needed to increase the spindle length. We took advantage of the observation that a mutation in Ncd (N600K) uncouples MT binding and MT-stimulated ATPase activity (Song and Endow, 1998), which results in a protein that should cross-link but not slide MTs. We made the corresponding mutation (N593K) in GFP-HSET and characterized the biochemical activity of the wild-type (wt) and N593K purified proteins. Although wt HSET had MT-stimulated ATPase activity, HSET N593K had no MT-stimulated ATPase activity (Supplementary Figure S3, A and B). The lack of ATPase activity of HSET N593K was not due to dead protein because it exhibited a similar apparent Kd for MTs compared with wt HSET (Supplementary Figure S3, C–E). This suggests that the HSET N593K mutant behaves as expected and uncouples MT binding and ATP hydrolysis.
To ask whether the N593K mutation affects spindle organization, we generated a stable cell line that overexpresses GFP-HSET N593K. The localization of GFP-HSET N593K in HeLa cells was the same as with endogenous HSET or as in cells expressing GFP-HSET, but the spindle MTs looked more bundled than in normal HeLa cells (Figure 3A). We measured the spindle length in both the GFP-HSET and GFP-HSET N593K cell lines and found that the pole-to-pole distance in the GFP-HSET N593K cell line was slightly longer than in normal control HeLa cells (p < 0.05), but shorter than in the GFP-HSET cell line (p < 0.01; Figures 1B and 3B), suggesting that the ability of HSET to elongate the spindle was reduced by the N593K mutation. One alternative possibility is that the decreased spindle length of GFP-HSET N593K cells relative to those expressing GFP-HSET alone is that the GFP-HSET was expressed to higher levels. To test this possibility, we measured the fluorescence intensity of the GFP signal as a function of the pole-to-pole distance in cells from each of the cell lines. Both GFP-HSET and GFP-HSET N593K–expressing cells had similar distributions of GFP fluorescence intensities, which had positive correlations with the pole-to-pole distances (Figure 3C), indicating that expression levels were not the root of the difference in the pole-to-pole distance. Because the GFP-HSET N593K expressing cells had a reduced slope compared with the GFP-HSET expressing cells, this suggests that the sliding ability of HSET is required for the elongation of the spindle.
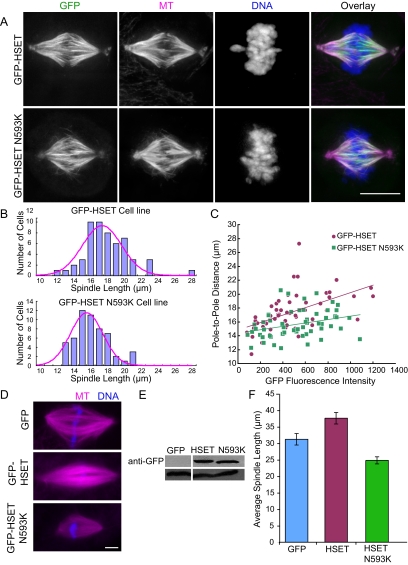
Figure 3.
Sliding activity is required for HSET/XCTK2-dependent spindle length change. (A) Cells expressing GFP-HSET or GFP-HSET N593K were synchronized, released, and then processed for immunofluorescence. In each panel, the MTs (magenta), HSET or HSET N593K mutant (green), and DNA (blue) staining are shown. (B) Histogram of spindle lengths quantified from a total of 60 spindles in three independent experiments for each expression condition. The Gaussian distribution is superimposed in magenta. Average spindle lengths: HSET, 17.5 ± 0.2 μm and HSET N593K, 15.7 ± 0.6 μm. (C) The fluorescence intensity of GFP-HSET (magenta) or GFP-HSET N593K (green) is plotted relative to the pole-to-pole distance from a total of ∼60 cells from three independent experiments. (D–F) GFP, GFP-HSET, or GFP-HSET N593K were added to Xenopus extracts at 2.5-fold the endogenous XCTK2 concentration before spindle assembly. (D) Representative images of spindles showing MTs (magenta) and DNA (blue). (E) Western blot of reactions probed with either anti-GFP antibodies (top panel) or anti-tubulin antibodies (bottom panel) to show that equivalent amounts of each protein were added to the extract. Note that 50 nM control GFP protein added to the extracts is below the detection limit of the anti-GFP antibody. (F) Quantification of the spindle length in extracts containing 2.5-fold excess GFP, GFP-HSET, or GFP-HSET N593K. A total of 60 structures were measured in three independent extracts, and the mean ± SEM is reported. Average spindle lengths: control GFP, 31.2 ± 1.7 μm; HSET, 37.6 ± 1.7 μm; and HSET 593K, 24.8 ± 1.1 μm. Scale bar, 10 μm.
We showed previously that addition of a molar excess of the Xenopus Kinesin-14 XCTK2 to spindle assembly reactions in egg extracts results in a stimulation of the rate and extent of bipolar spindle formation (Walczak et al., 1997), which could be mediated by MT sliding activity. To test the idea that HSET functions similar to XCTK2 in egg extracts and that MT sliding is involved in spindle assembly in egg extracts, we added a 2.5-fold molar excess of GFP-XCTK2, GFP-HSET, or GFP-HSET N593K to extracts and analyzed spindle formation at 30 min, where the majority of chromatin structures are composed of half spindles (Supplementary Figure S4). GFP-XCTK2 and GFP-HSET dramatically increased the percentage of bipolar spindles formed to similar extents, whereas GFP-HSET N593K failed to stimulate spindle assembly (Supplementary Figure S4F). This indicates that HSET functions similarly to XCTK2 in egg extracts and that sliding activity contributes to the stimulation of spindle assembly induced by XCTK2 and HSET. We also measured the lengths of spindles formed under these conditions to determine whether MT sliding also affected spindle length in egg extracts (Figure 3, D–F). Although addition of wt GFP-HSET caused a slight increase in spindle length compared with GFP control spindles (p <0.05), addition of GFP-HSET N593K caused a reduction in spindle length compared with GFP control spindles (p < 0.05) and to wt GFP-HSET addition (p < 0.05; Figure 3F), supporting the idea that the sliding activity of HSET/XCTK2 is required for the increase in spindle length in extracts and in cells.
The Spatial Distribution of HSET/XCTK2 during Interphase Contributes to MT Organization
We showed previously that Xenopus Kinesin-14 XCTK2 binds to importin α/β in a RanGTP-dependent manner to regulate the ability of XCTK2 to cross-link MTs in vitro (Walczak et al., 1997; Ems-McClung et al., 2004). Because the importin α/β complex transports cargo from the cytoplasm to the nucleus where RanGTP dissociates the complex to unload the cargo, we first asked whether full-length GFP-XCTK2 and GFP-HSET were also cargos of importin α/β in extracts and in cells. Our previous work with XCTK2 demonstrated that mutation of either end of the bipartite NLS disrupted the ability of the nonmotor domain of XCTK2 to be imported into nuclei in egg extracts (Ems-McClung et al., 2004; Figure 4A). Similar to mutation of the nonmotor domain of XCTK2, mutation of the NLS in full-length GFP-XCTK2 resulted in the failure of XCTK2 NLSa and XCTK2 NLSb to bind importin α/β in vitro (data not shown). Addition of wt GFP-XCTK2 or the NLS mutants to preformed nuclei in egg extracts showed that wt GFP-XCTK2 was effectively imported into the nucleus, whereas both of the NLS mutants were excluded from the nucleus and localized on the surrounding MTs (Figure 4B). In addition, wt GFP-XCTK2 associated with importin α/β in anti-GFP immunoprecipitations, whereas mutation of the NLS abrogated this association (Figure 4C). This indicates that full-length Xenopus Kinesin-14 is imported into the nucleus through an association with importin α/β to spatially regulate its distribution.
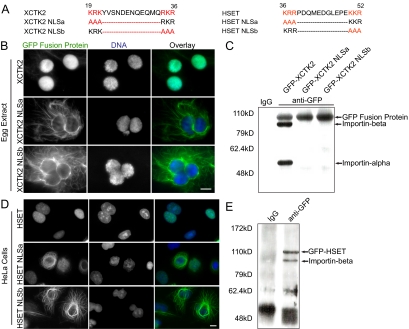
Figure 4.
The spatial distribution of HSET and XCTK2 is dependent on the NLS. (A) Bipartite NLS sequences within the tail domains of XCTK2 and HSET showing the canonical positively charged residues highlighted in red. The corresponding mutant sequences are shown below the wt sequence with the mutations highlighted in red. (B) GFP-XCTK2, GFP-XCTK2-NLSa, or GFP-XCTK2-NLSb, at 20 nM, was added to interphase Xenopus egg extracts and incubated for 60 min. Representative structures are shown with the GFP fusion protein (green) and DNA (blue). Scale bar, 10 μm. (C) Western blot of GFP fusion proteins (XCTK2 or the NLS mutants) from Xenopus egg extracts that were immunoprecipitated with a control IgG or an anti-GFP antibody and then probed sequentially with antibodies to GFP and importin β and α. (D) HeLa cells were transiently transfected with pEGFP-HSET, pEGFP-HSET NLSa, or pEGFP-HSET-NLSb and then processed for immunofluorescence to visualize the GFP fusion protein (green) and the DNA (blue). Scale bar, 10 μm. (E) Western blot of GFP-HSET that was immunoprecipitated from HeLa cells stably expressing GFP-HSET with either a control IgG or an anti-GFP antibody and then probed with antibodies to GFP and importin β.
To ask whether the importin α/β complex also spatially regulates HSET in human cells, we identified the bipartite NLS in HSET using PSORT II (http://psort.nibb.ac.jp) and mutated both ends of the bipartite NLS sequence in full-length GFP-HSET (Figure 4A). Transient expression of wt GFP-HSET and the GFP-HSET NLS mutants in HeLa cells showed a parallel distribution to that of XCTK2 in Xenopus nuclei (Figure 4D). Specifically, wt GFP-HSET was sequestered in the nucleus, and the NLSa and NLSb mutants were excluded. The nuclear excluded GFP-HSET NLS mutants also caused the MTs to become highly bundled in the cytosol. In addition, these two NLS mutants were highly toxic to cells in that most of the transfected cells died within 48 h after transfection, and we rarely found any mitotic cells in these transfections (data not shown). To test whether the import of HSET is through binding to importin α/β, we performed an immunoprecipitation with an anti-GFP antibody from extracts of GFP-HSET–overexpressing cells and found that importin β associated with GFP-HSET in this cell line (Figure 4E). Because the NLS mutants were not transported into the nucleus, we infer that it is because of a disrupted interaction with the importin α/β import machinery. Together, these data suggest that Kinesin-14s in higher eukaryotes are sequestered in the nucleus in interphase to prevent inadvertent disruption of the interphase cytoskeleton.
Association of Importin α/β to the NLS Regulates XCTK2 MT Cross-Linking and/or Sliding to Control Spindle Morphology
Because transport of HSET/XCTK2 in cells and extracts is regulated by importin α/β, we asked whether the function of these Kinesin-14s during mitosis was also regulated by the Ran system. Unfortunately, we were unable to generate stable cell lines expressing the GFP-HSET NLS mutants because of the toxicity induced by their overexpression. Therefore, we decided to mimic the cellular spindle assembly process by manipulation of Xenopus egg extracts. We used cycled spindle assembly reactions in Xenopus egg extracts and then added a 5-fold molar excess of GFP protein, wt GFP-XCTK2, or the NLS mutants at the time of the second CSF addition. Spindles were assembled for 80 min and then analyzed. Addition of excess wt GFP-XCTK2 caused bundling of the spindle MTs; however, the spindle length did not change significantly (Figure 5, A–C). In contrast, addition of either NLS mutant dramatically increased the spindle length (XCTK2 vs. NLSa, p < 0.01; XCTK2 vs. NLSb, p < 0.05) and caused excessive bundling of the spindle MTs. This effect was not limited to spindle assembly, as addition of the NLS mutants to preformed spindles also resulted in the formation of excessively long spindles (Supplementary Figure S5), suggesting that the importin α/β–mediated regulation of spindle length by XCTK2 is required both for spindle assembly and for spindle maintenance in extracts.
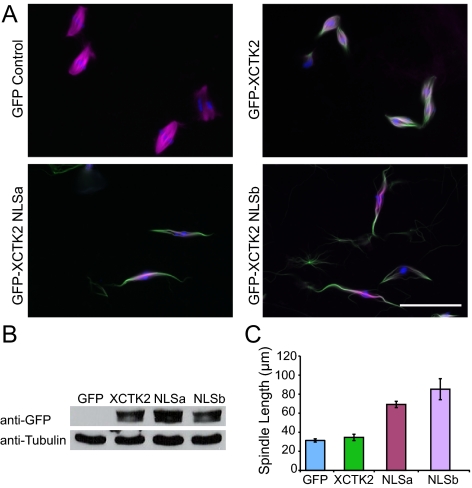
Figure 5.
Ran regulates the ability of XCTK2 to control spindle length. (A–C) GFP, GFP-XCTK2, GFP-XCTK2 NLSa, and GFP-XCTK2 NLSb proteins were added to cycled Xenopus egg extracts at a fivefold molar excess relative to the endogenous XCTK2 concentration at the time of the second CSF addition. (A) Representative fields of view showing MTs (magenta), the GFP fusion protein (green), and DNA (blue). (B) Western blot of samples from A that were probed with both anti-GFP antibodies (top panel) and anti-tubulin antibodies (bottom panel) to show that equivalent amounts of each protein were added to the extract. The GFP control concentration is below the detection limit of the antibody. (C) Quantification of the spindle length in extracts containing 10-fold excess exogenous GFP, GFP-XCTK2 (XCTK2), GFP-XCTK2 NLSa (NLSa), and GFP-XCTK2 NLSb (NLSb) proteins. A total of ∼80 structures were measured in each of three independent extracts, and the mean ± SEM is reported. Average spindle lengths: control GFP, 31.4 ± 1.7 μm; XCTK2, 34.6 ± 3.3 μm; NLSa, 69.2 ± 3.3 μm; and NLSb, 85.2 ± 11.0 μm. Scale bar, 50 μm.
DISCUSSION
In this study, we showed that sliding activity of HSET and XCTK2 on MTs is needed to control spindle length in both mitotic and meiotic spindles. HSET/XCTK2 is sequestered in the nucleus during interphase to prevent aberrant MT cross-linking and is released to the cytoplasm during mitosis to control MT cross-linking and sliding during mitosis. Our data suggest that Kinesin-14 function is conserved within vertebrates and is important to control spindle morphogenesis.
HSET Perturbation Does Not Affect Spindle Bipolarity in HeLa Cells
Previous studies have shown Kinesin-14 affects spindle bipolarity. Inhibition of Xenopus Kinesin-14 in egg extracts results in monopolar spindle formation (Walczak et al., 1997), but the fact that we and others do not find any defects in spindle bipolarity upon HSET perturbation supports the idea that the role of Kinesin-14 on spindle bipolarity is more dramatic in meiotic rather than mitotic systems. Perturbation of Drosophila Kinesin-14 Ncd splays the spindle poles and disrupts centrosome clustering (Endow et al., 1994a; Goshima et al., 2005a), causing multipolar spindles. For human Kinesin-14 HSET, its activity on spindle pole formation seems to be masked by centrosomes during mitosis, whereas in mouse oocytes, it causes unfocused poles with lost cohesion (Mountain et al., 1999). More recently, Kwon et al. (2008) showed that the requirement for HSET in spindle multipolarity is directly correlated to the number of centrosomes in mammalian cells. In cells with multiple centrosomes, HSET appears to be required to cluster those centrosomes; however, the clustering of centrosomes is not the predominant pathway for spindle bipolarity in HeLa cells. In support of our finding, Kwon et al. showed that perturbation of HSET did not significantly affect centrosome clustering in HeLa cells, which equates to our observation that there was not a substantial increase in spindle multipolarity after HSET knockdown.
Kinesin-14s Affect Spindle Length through MT Sliding
The spindle is a stable but very dynamic structure. Several conceptual models have been proposed to explain how spindles are organized to reach a steady-state size with a relatively constant morphology (Brinkley, 1985; Sharp et al., 2000b; Karsenti and Vernos, 2001; Chakravarty et al., 2004; Cytrynbaum et al., 2005; Goshima et al., 2005b; Rogers et al., 2005; Burbank et al., 2007). Of these models, it is clear that MT dynamics control MT length and affect spindle size; however, how MT sliding contributes to spindle structure is more complicated. Different models result from assumptions of different force production schemes and different MT organizations. One model predicted that MT dynamics play the major role, whereas MT sliding plays a minor role in the maintenance of spindle length, consistent with their experiments in Drosophila S2 cells in which knockdown or overexpression of the Kinesin-14 Ncd did not significantly change the spindle length (Goshima et al., 2005b); however, the model is inconsistent with data from Drosophila embryos, which showed that the Kinesin-5 Klp61F antagonizes Ncd to regulate the spindle pole-to-pole distance (Sharp et al., 1999). In contrast, recent computer simulations propose that the plus-end motors slide between anti-parallel MTs to generate the outward movement of MTs in the spindle, whereas the minus-end motors slide between parallel MTs and help cluster MT minus ends to form the spindle poles (Goshima et al., 2005a; Burbank et al., 2007). One possible link between these differing models would be if Kinesin-14s actually regulated spindle MT dynamics. In this scenario the increase in spindle length seen with HSET overexpression would be due to an increased stability of MTs. In support of this idea, the N-terminus of Ncd was shown to promote MT assembly and stabilize MTs against conditions that induce MT disassembly (Karabay and Walker, 1999). However, we do not favor this model, because we find no increase in the overall fluorescence intensity of tubulin in the spindles after XCTK2 addition (our unpublished results). Furthermore, several studies suggest that members of the Kinesin-14 family actually possess the ability to destabilize rather than to stabilize MTs (Endow et al., 1994b; Chu et al., 2005; Sproul et al., 2005).
Our data showed that in human cells and in Xenopus extracts Kinesin-14 proteins promote the outward movement of the spindle poles and increase half spindle length in monopolar spindles (data not shown). Thus our results are consistent with the model that Kinesin-14s slide between parallel MTs (Goshima et al., 2005a; Burbank et al., 2007), because a minus-end motor positioned between anti-parallel MTs would generate inward movement of the spindle poles and not outward movement as we demonstrated. HSET localization between parallel MTs as visualized by electron microscopy supports this theory (Mountain et al., 1999), as do studies showing that overexpression of CHO2 results in an increase in spindle length (Matuliene et al., 1999). We propose that one mechanism of action for the Kinesin-14 family is to cross-link and transport parallel MTs in a half spindle, not only to facilitate pole focusing, but also for spindle elongation (Figure 6). This model implies that the binding of Kinesin-14s between two MTs should have a certain orientation. Goshima et al. (2005a) propose that Ncd cross-links K-fiber MTs to spindle MTs by binding its motor domain to the astral/spindle MTs and its N-terminal domain to the K-fiber. Our data show that HSET can induce spindle elongation in the absence of K-fibers, suggesting that HSET cross-linking occurs through parallel spindle MTs within a half-spindle. Our data can be explained more readily by a recent single-molecule study showing that Ncd has different motility properties on single MTs versus bundles of MTs, which provides a possible mechanism for this biased binding (Furuta and Toyoshima, 2008).
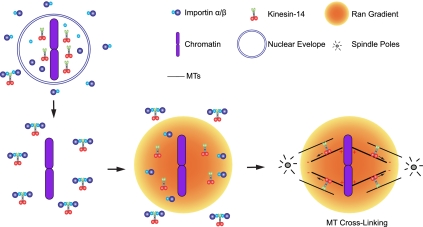
Figure 6.
Model for regulation of Kinesin-14 function in spindle organization. During interphase, Kinesin-14s are transported into the nucleus through association of the NLS in its tail with importin α/β, which prevents the bundling of cytoplasmic MTs. After nuclear envelope breakdown, a Ran-GTP gradient is formed in the vicinity of chromosomes, which results in the dissociation of Kinesin-14s from importin α/β. Once Kinesin-14s are activated by Ran, they likely cross-link MTs and transport MTs to the spindle pole through the minus-end movement of the motor domain. This activity is mediated through parallel MTs within a half spindle.
The Activity of Kinesin-14s in Spindle Morphology Control Is Regulated by Its NLS
Previously, we showed that Ran regulated the cross-linking activity of XCTK2 in vitro (Ems-McClung et al., 2004). Here we show that disruption of the association between XCTK2 and importin α/β through mutagenesis of NLS-impaired XCTK2 spatial distribution and spindle morphogenesis in extracts. This regulation is likely physiologically important for HSET regulation in cells because overexpression of the NLS mutants was lethal. Ncd contains a putative bipartite NLS in its tail domain, is similarly sequestered in the nucleus during interphase, and when artificially exported out of the nucleus by the addition of a nuclear export sequence, localizes to MTs and strongly bundles MTs similar to HSET and XCTK2 (Goshima et al., 2005c). This further supports the theory that Kinesin-14 function is conserved. Together these data demonstrate that the NLS is a critical region for the biological function of Kinesin-14s and is likely to be regulated by importin α/β, which is tightly controlled by Ran (Figure 6).
The Ran gradient controls spindle morphogenesis through the regulation of many spindle assembly factors, such as the chromatin-driven MT-nucleating proteins TPX2 (Gruss et al., 2001) and NuSAP (Ribbeck et al., 2006), the spindle pole organizing factor NuMA (Nachury et al., 2001; Wiese et al., 2001), the K-fiber–binding protein HURP (Koffa et al., 2006; Sillje et al., 2006), the mRNA export factor Rae1 (Blower et al., 2005), the tumor suppressor BRCA1 (Joukov et al., 2006), and many others. Because of the complexity of various Ran substrates, we were unable to establish a direct link between Ran regulation and Kinesin-14 function in spindle assembly in somatic cells. However, our data demonstrate that the function of Ran substrates must be tightly regulated, and disruption of this regulation pathway causes abnormal spindle organization in extracts. This supports the idea that the Ran gradient physiologically controls spindle formation and organization. Further investigations will be needed to clearly demonstrate this point.
Although our data suggest that the Ran system regulates the activity of Kinesin-14s in controlling spindle length, many questions remain unanswered. For example, how does the Ran system sense the size of the spindle and regulate the activity of Kinesin-14s? There are many Ran substrates involved in spindle assembly, but it is unclear how the Ran system coordinates the different factors and how Ran selectively regulates some factors without affecting others. Clearly Kinesin-14s function to focus poles, but how is the mechanism of pole focusing related spindle length control? The action of Kinesin-14s seem to be mediated by parallel MTs within a half-spindle and do not require either overlapping MTs or K-fibers. Perhaps other motors or MAPs also regulate spindle length through these other MT subsets. The answers to these interesting questions will help us unveil the detailed structure and organization of spindle.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all members of the Walczak lab for thoughtful ideas and suggestions on this work. S.C. is especially grateful to the Wood's Hole MBL course, which was instrumental in generating many ideas and helpful discussion. We are grateful to Duane Compton for sharing constructs and antibodies for HSET and to Mary Dasso for importin antibodies. This work was supported by National Institutes of Health (NIH) Grant GM59618 to C.E.W. and in part by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, The time-lapse imaging experiments were performed in the Indiana University Light Microscopy Imaging Center using a BD Pathway 855, supported by NIH Grant S10RR025033-01.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0971) on December 30, 2008.
REFERENCES
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Brinkley B. R. Microtubule organizing centers. Annu. Rev. Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- Burbank K. S., Mitchison T. J., Fisher D. S. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Chakravarty A., Howard L., Compton D. A. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian mitotic extract. Mol. Biol. Cell. 2004;15:2116–2132. doi: 10.1091/mbc.E03-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R., Salmon E. D., Erickson H. P., Lockhart A., Endow S. A. Structural and functional domains of the Drosophila ncd microtubule motor protein. J. Biol. Chem. 1993;268:9005–9013. [PubMed] [Google Scholar]
- Chu H. M., Yun M., Anderson D. E., Sage H., Park H. W., Endow S. A. Kar3 interaction with Cik1 alters motor structure and function. EMBO J. 2005;24:3214–3223. doi: 10.1038/sj.emboj.7600790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum E. N., Sommi P., Brust-Mascher I., Scholey J. M., Mogilner A. Early spindle assembly in Drosophila embryos: role of a force balance involving cytoskeletal dynamics and nuclear mechanics. Mol. Biol. Cell. 2005;16:4967–4981. doi: 10.1091/mbc.E05-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Moree B., Hickey J. M., Kilmartin J. V., Salmon E. D. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Walczak C. E. Assays for microtubule-destabilizing kinesins. Methods Mol. Biol. 2001;164:109–121. doi: 10.1385/1-59259-069-1:109. [DOI] [PubMed] [Google Scholar]
- Ems-McClung S. C., Zheng Y., Walczak C. E. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Chandra R., Komma D. J., Yamamoto A. H., Salmon E. D. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 1994a;107:859–867. doi: 10.1242/jcs.107.4.859. [DOI] [PubMed] [Google Scholar]
- Endow S. A., Kang S. J., Satterwhite L. L., Rose M. D., Skeen V. P., Salmon E. D. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994b;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Komma D. J. Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd-GFP fusion proteins. J. Cell Sci. 1996;109:2429–2442. doi: 10.1242/jcs.109.10.2429. [DOI] [PubMed] [Google Scholar]
- Furuta K., Toyoshima Y. Y. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 2008;18:152–157. doi: 10.1016/j.cub.2007.12.056. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R. D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005a;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J. M., Vale R. D. Length control of the metaphase spindle. Curr. Biol. 2005b;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005c;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. Immunoaffinity purification; pp. 321–325. [Google Scholar]
- Hatsumi M., Endow S. A. The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 1992a;103:1013–1020. doi: 10.1242/jcs.103.4.1013. [DOI] [PubMed] [Google Scholar]
- Hatsumi M., Endow S. A. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 1992b;101:547–559. doi: 10.1242/jcs.101.3.547. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hertzer K. M., Ems-McClung S. C., Kline-Smith S. L., Lipkin T. G., Gilbert S. P., Walczak C. E. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol. Biol. Cell. 2006;17:700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Totis L., Saunders W. S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics. 1993;135:35–44. doi: 10.1093/genetics/135.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kalab P., Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Karabay A., Walker R. A. The Ncd tail domain promotes microtubule assembly and stability. Biochem. Biophys. Res. Commun. 1999;258:39–43. doi: 10.1006/bbrc.1999.0572. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Cole R. W., Oakley B. R., Rieder C. L. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Kocher T., Wilm M., Mattaj I. W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., Thery M., Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J., Patel U. K., Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Matuliene J., Essner R., Ryu J., Hamaguchi Y., Baas P. W., Haraguchi T., Hiraoka Y., Kuriyama R. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J. Cell Sci. 1999;112:4041–4050. doi: 10.1242/jcs.112.22.4041. [DOI] [PubMed] [Google Scholar]
- McDonald H. B., Stewart R. J., Goldstein L. S. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., West R. R. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- McKim K. S., Hawley R. S. Chromosomal control of meiotic cell division. Science. 1995;270:1595–1601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E. D., Kapoor T. M. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D. A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Ohi R., Burbank K., Liu Q., Mitchison T. J. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 2006;17:2646–2660. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Flemington E. K. Transfection-mediated cell-cycle signaling: considerations for transient transfection-based cell-cycle studies. Anal. Biochem. 1999;272:171–181. doi: 10.1006/abio.1999.4156. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Sharp D. J. Spindle microtubules in flux. J. Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Mitchison T. J. Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 2001;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu C. E., Murray A. W. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Brown H. M., Kwon M., Rogers G. C., Holland G., Scholey J. M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Roles of motor proteins in building microtubule-based structures: a basic principle of cellular design. Biochim. Biophys. Acta. 2000b;1496:128–141. doi: 10.1016/s0167-4889(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Yu K. R., Sisson J. C., Sullivan W., Scholey J. M. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Sillje H. H., Nagel S., Korner R., Nigg E. A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Song H., Endow S. A. Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature. 1998;396:587–590. doi: 10.1038/25153. [DOI] [PubMed] [Google Scholar]
- Sproul L. R., Anderson D. J., Mackey A. T., Saunders W. S., Gilbert S. P. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr. Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell C. L., Sweezy M. A., West R. R., Reed K. D., Carson B. D., Pidoux A. L., Cande W. Z., McIntosh J. R. pkl1(+)and klp2(+): Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol. Biol. Cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Verma S., Mitchison T. J. XCTK 2, a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. A. ncd and kinesin motor domains interact with both alpha- and beta-tubulin. Proc. Natl. Acad. Sci. USA. 1995;92:5960–5964. doi: 10.1073/pnas.92.13.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. A., Salmon E. D., Endow S. A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Wang Z. X., Jiang R. F. A novel two-site binding equation presented in terms of the total ligand concentration. FEBS Lett. 1996;392:245–249. doi: 10.1016/0014-5793(96)00818-6. [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.