Abstract
Cooperative communications between the central spindle and the contractile ring are critical for the spatial and temporal regulation of cytokinesis. Here we report that MyoGEF, a guanine nucleotide exchange factor that localizes to the central spindle and cleavage furrow, interacts with centrosome/spindle pole-associated protein (CSPP), which is concentrated at the spindle pole and central spindle during mitosis and cytokinesis. Both in vitro and in vivo pulldown assays show that MyoGEF interacts with CSPP. The C-terminus of MyoGEF and N-terminus of CSPP are required for their interaction. Immunofluorescence analysis indicates that MyoGEF and CSPP colocalize at the central spindle. Depletion of CSPP or MyoGEF by RNA-interference (RNAi) not only causes defects in mitosis and cytokinesis, such as metaphase arrest and furrow regression, but also mislocalization of nonmuscle myosin II with a phosphorylated myosin regulatory light chain (p-MRLC). Importantly, CSPP depletion by RNAi interferes with MyoGEF localization at the central spindle. Finally, MyoGEF interacts with ECT2, and RNAi-mediated depletion of MyoGEF leads to mislocalization of ECT2 and RhoA during cytokinesis. Therefore, we propose that CSPP interacts with and recruits MyoGEF to the central spindle, where MyoGEF contributes to the spatiotemporal regulation of cytokinesis.
INTRODUCTION
The small GTPase proteins, including RhoA, Rac1, and Cdc42, have been implicated in regulating a variety of biological processes such as cell migration, tissue morphogenesis, gene expression, and cytokinesis (Burridge and Wennerberg, 2004; Jaffe and Hall, 2005). The small GTPase proteins can cycle between a GDP-bound, inactive form and a GTP-bound, active form. This switch is largely regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs catalyze the exchange of bound GDP for GTP, thereby activating the small GTPase proteins, whereas GAPs increase the low intrinsic GTPase activity, leading to the inactivation of the small GTPase proteins (Mackay and Hall, 1998). RhoA can activate Rho-kinase, which in turn inhibits myosin phosphatase and directly phosphorylates myosin regulatory light chains, resulting in an increase in myosin contractile activity (Kimura et al., 1996; Matsui et al., 1996). RhoA activation at the cleavage furrow is critical for myosin contractile ring assembly and furrow ingression (Matsumura et al., 2001; Yoshizaki et al., 2004; Bement et al., 2005; Kamijo et al., 2006).
Lines of evidence suggest that small GTPase signaling can transduce signals from the central spindle to the myosin contractile ring (Burgess and Chang, 2005; Glotzer, 2005; Eggert et al., 2006). Pebble, a guanine nucleotide exchange factor that genetically interacts with Rho1, but not with Rac1 or Cdc42, localizes to the cleavage furrow and is required for cytokinesis in Drosophila (Prokopenko et al., 1999). More importantly, Pebble, RacGAP50C, and Pavarotti form a trimolecular complex that, in turn, positions the myosin contractile ring to the site of furrowing during cytokinesis (Somers and Saint, 2003). ECT2, the human homologue of pebble, localizes to the central spindle, and it is required for furrow formation and ingression as well as the completion of cytokinesis in mammalian cells (Tatsumoto et al., 1999; Kamijo et al., 2006; Yuce et al., 2005; Zhao and Fang, 2005). Further studies demonstrate that the human homologues of Pavarotti and RacGAP50C, i.e., mitotic kinesin-like protein 1 (Mklp1) and Male Germ Cell RacGAP (MgcRacGAP), are responsible for the localization of ECT2 to the central spindle. In turn, ECT2 is required for equatorial RhoA activation and contractile ring assembly (Kamijo et al., 2006; Yuce et al., 2005; Nishimura and Yonemura, 2006). A recent study demonstrates that another GEF, GEF-H1, can activate RhoA at the cleavage furrow during late stages of cytokinesis and plays a critical role in the regulation of furrow ingression (Birkenfeld et al., 2007). We also reported previously that MyoGEF (myosin II-interacting GEF), which localizes to the central spindle and cleavage furrow during cytokinesis, binds to nonmuscle myosin II and activates the small GTPase protein RhoA. Disruption of MyoGEF by RNA interference (RNAi) results in the formation of binucleated/multinucleated cells (Wu et al., 2006). Clearly, further work is required to elucidate how each of three GEFs contributes to the regulation of cytokinesis and whether they have distinct or overlapping functions in cytokinesis.
To understand how MyoGEF regulates cytokinesis, we have carried out a yeast two-hybrid screening to identify MyoGEF-interacting partners, and we have identified centrosome/spindle pole-associated protein (CSPP) as one of MyoGEF-interacting proteins. CSPP localizes to the centrosome, spindle pole, and central spindle (Patzke et al., 2005, 2006). Here we present evidence demonstrating that CSPP interacts with MyoGEF and that this interaction plays an important role in the regulation of mitotic progression and cytokinesis. Time-lapse microscopy reveals that depletion of CSPP by RNAi leads to cell cycle arrest at metaphase as well as cytokinesis defects. More importantly, CSPP depletion interferes with MyoGEF localization at the central spindle, leading to mislocalization of p-MRLC during cytokinesis. Further, we found that MyoGEF depletion by RNAi results in mislocalization of two critical cytokinesis regulators, ECT2 and RhoA, during cytokinesis. Our results suggest that MyoGEF-CSPP interaction contributes to the spatiotemporal regulation of cytokinesis.
MATERIALS AND METHODS
Yeast Two-Hybrid Screening
The bait plasmid was generated by subcloning the full-length human MyoGEF cDNA into pAS2-1 vector (Clontech, Palo Alto, CA), in which MyoGEF was fused in frame with the GAL4 DNA-binding domain. The bait plasmid was cotransformed into yeast strain Y187 with a mouse 11-d embryo matchmaker cDNA library (Clontech). The positive yeast colonies that were able to grow in SD synthetic medium lacking leucine, tryptophan, and histidine were further confirmed by performing a filter lift assay for β-galactosidase activity. The prey plasmids were then recovered from the positive yeast colonies and subjected to DNA sequencing. The mouse CSPP cDNAs were amplified by using the following primer pair: 5′-GAGCTCATGGCAGATAGCTTGGATGAA-3′ (forward primer; the underlined nucleotide sequence is the recognition site for SacI) and 5′-CCGCGGTTAAGCATGTGCAGCAGAGAG-3′ (reverse primer; the underlined nucleotide sequence is the recognition site for SacII).
Plasmids
The full-length MyoGEF was cloned into pCS3+MT vector as previously described (Wu et al., 2006). Four polypeptides corresponding to amino acids 71-388, 71-565, 392-565, and 392-780 in human MyoGEF were cloned into BamHI/XhoI sites of pCMV-3Tag2B (Stratagene, La Jolla, CA) and pGEX-5X-1 (GE Healthcare, Waukesha, WI) vectors. mCSPP-1, -2, and -3 were cloned into SacI/SacII sites of pEGFP-C3 (Clontech) to generate pEGFP-C3-mCSPP-1, -2, and -3. To generate Myc-tagged mCSPP-2, the full-length mCSPP-2 was amplified from pEGFP-C3-mCSPP-2 by PCR and then subcloned into BglII/XbaI sites of pCS3+MT vector. Green fluorescent protein (GFP)-tagged human CSPP was provided by Dr. Hans-Christian Aasheim (Norwegian Radium Hospital, Oslo, Norway). A plasmid encoding H2B-GFP was from Addgene (Cambridge, MA; plasmid 11680; Kanda et al., 1998). YFP-ceRhoA plasmid was provided by Dr. Michael Glotzer (University of Chicago, Chicago, IL).
Antibody Generation
We generated a peptide polyclonal antibody using the N-terminal 17 amino acids of mCSPP-1 and mCSPP-3 as antigen (Supplemental Figure S1; amino acid residues in red). This antibody recognized both mCSPP-1 and -3 as well as human CSPP-L.
Cell Culture and Transfection
HeLa Tet-ON cells (Clontech) were grown in DMEM supplemented with 10% fetal bovine serum. We will refer to HeLa Tet-ON cells as HeLa cells in this article. Transfection of plasmids or small interfering RNA (siRNA) into HeLa cells were done by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For transfection with plasmids, the transfected cells were analyzed ∼24 h after transfection. For transfection with siRNA or siRNA plus plasmids, the transfected cells were analyzed ∼48-72 h after transfection. The nucleotide sequence for hCSPP-L siRNA is as follows: 5′-AGUUCAAUCCAGUUCAUUACCACCC-3′. MyoGEF siRNA has been described previously (Wu et al., 2006).
Cell Synchronization
Cell cycle synchronization was performed by double thymidine block. Briefly, the transfected HeLa cells were treated with 2 mM thymidine for 24 h, cultured in thymidine-free medium for 10 h, and then treated with 2 mM thymidine for 16 h, followed by “release” to progress through the cell cycle in thymidine-free medium for 0 or 12 h.
Protein Expression and In Vitro Translation
Glutathione S-transferase (GST)-fused MyoGEF polypeptides were expressed in a bacterial expression system. BL21 bacterial cells expressing GST-MyoGEF polypeptides were homogenized by sonication and lysed in PBS containing 1% Triton X-100 for 1 h at 4°C. The GST-fusion proteins were purified by using glutathione-conjugated agarose beads, eluted with 100 mM Tris-HCl (pH 7.5) and 5 mM glutathione and dialyzed against 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl. In vitro–translated Myc-tagged mCSPP-2 protein was synthesized using the TNT SP6 quick-coupled transcription/translation system (Promega, Madison, WI) according to the manufacturer's instructions.
Immunoprecipitation and GST Pulldown Assays
Immunoprecipitation and GST pulldown assays were carried out as described previously (Wei, 2005; Wu et al., 2006). Briefly, transfected cells were lysed in RIPA (radioimmune precipitation assay) lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.25% deoxycholate, 1% NP-40, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF with Protease inhibitor mixture) for 10 min on ice. Cell extracts were collected and precleared with protein A/G agarose beads. The precleared lysate was incubated with agarose-conjugated anti-Myc or anti-GFP antibody overnight at 4°C. After washing three times with RIPA lysis buffer, the bound proteins were eluted with SDS loading buffer. For GST pulldown experiments, the immobilized GST-MyoGEF polypeptides were incubated with in vitro–translated Myc-mCSPP-2 protein overnight at 4°C. After washing four times with binding buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.05% Triton-X-100, 10% glycerol, 0.2 mM EDTA, and 1 mM DTT), the beads were resuspended in SDS loading buffer to elute the bound proteins.
Immunoblotting
Cell lysates and immunoprecipitated and GST pulldown proteins were separated by 7% SDS-PAGE gel, transferred to an Immobilon-P transfer membrane (Millipore, Bedford, MA), blocked in 5% nonfat milk, and incubated with primary antibodies as indicated. The following primary antibodies were used: mouse anti-Myc (9E10, 1:1000; Santa Cruz), rabbit anti-GFP (1:1000; Santa Cruz), and rabbit anti-CSPP (1:2000). The blots were washed and incubated with horseradish peroxide–conjugated secondary antibodies (1:5000; Santa Cruz) for 1 h at 23°C. The blots were visualized by SuperSignal West Pico Luminol/Enhancer solution (Pierce, Rockford, IL).
Immunofluorescence Staining and Time-Lapse Microscopy
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde for 12 min and permeabilized in 0.5% Triton X-100 for 10 min at 23°C. For RhoA staining, cells were fixed with 10% trichloroacetic acid (TCA) for 10 min at 4°C. For MyoGEF and CSPP staining, cells were fixed with methanol/acetone (1:1) for 10 min at −20°C. After blocking with 1% bovine serum albumin for 1 h at 23°C, the transfected cells were incubated with primary antibodies as indicated for 3 h at 23°C or overnight at 4°C, followed by incubation with secondary antibodies for 40 min at 23°C. The primary antibodies used for immunofluorescence were as follows: monoclonal anti-β-tubulin antibody (Sigma; 1:1000); rabbit polyclonal p-MRLC antibody (Cell Signaling, Beverly, MA; 1:500); rabbit polyclonal aurora B antibody (Invitrogen; 1:100); rabbit polyclonal CSPP antibody (1:200); rabbit polyclonal MyoGEF antibody (1:100); rabbit polyclonal ECT2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA; 1:100). The secondary antibodies rhodamine goat anti-mouse IgG (1:500) and rhodamine goat anti-rabbit IgG (1:500) were purchased from Invitrogen. The nuclei were visualized by DAPI (Sigma, St. Louis, MO). The coverslips were mounted using a Prolong antifade kit (Invitrogen). Images were taken using a Leica DMI 6000 B microscope (Leica, Deerfield, IL) and processed by deconvolution. For time-lapse microscopy, the cells grown on coverglass chamber were transfected with plasmids or siRNA as indicated. The transfected cells were cultured in Leibovitz's L-15 Medium (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum. Images of the live transfected cells were taken every 20 s using a Leica DMI 6000 B microscope.
RESULTS
Depletion of MyoGEF Caused Defects in Furrow Ingression
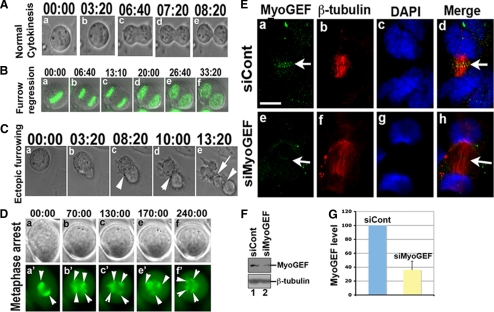
We previously reported that RNAi-mediated depletion of MyoGEF resulted in the formation of binucleated/multinucleated cells (Wu et al., 2006). To better understand the role of MyoGEF in regulating cytokinesis, we have used time-lapse microscopy to image HeLa cells that underwent mitosis and cytokinesis after RNAi-mediated depletion of MyoGEF. Plasmids encoding GFP-H2B or GFP-tubulin were cotransfected into HeLa cells with control or MyoGEF siRNA. We started imaging the transfected cells at 48-72 h after transfection. Our results indicated that there were mainly three kinds of defects resulting from RNAi-mediated depletion of MyoGEF in HeLa cells: 1) Furrow regression (n = 6/50 cells; in control siRNA-treated cells, n = 0/50 cells; see Table 1). As shown in Figure 1B, the cleavage furrow began to ingress but then regressed immediately, generating a binucleated cell. 2) Ectopic furrowing (n = 7/50 cells; in control siRNA-treated cells, n = 0/50 cells; see Table 1): As shown in Figure 1C, both major (arrowheads) and ectopic (arrow) furrows were formed during cytokinesis. However, the ectopic furrow eventually regressed and the major furrow continued to ingress until the completion of cytokinesis (not shown). 3) Metaphase arrest (n = 14/50 cells; in control siRNA-treated cells, n = 4/50 cells; see Table 1): As shown in Figure 1D, the bipolar spindle was formed at the beginning of metaphase (arrowheads in a′). After ∼60 min, the original bipolar spindle was converted into multiple-polar spindles (arrowheads in b′–f′). These results suggest that depletion of MyoGEF may compromise the integrity of the mitotic spindle and cause metaphase arrest. The dual effects of MyoGEF depletion on spindle assembly, as well as on cytokinesis are consistent with our observations that MyoGEF is localized at the spindle pole and central spindle (Wu et al., 2006; see Figures 3F and 5).
Table 1.
Summary of cell phenotypes resulting from MyoGEF or CSPP depletion in HeLa cells
| Cell phenotypes | Control siRNA | MyoGEF siRNA | hCSPP-L siRNA |
|---|---|---|---|
| Normal cytokinesis | 46 | 23 | 35 |
| Furrow regression | 0 | 6 | 8 |
| Ectopic furrowing | 0 | 7 | 0 |
| Metaphase arrest | 4 | 14 | 12 |
| Total cells (n) | 50 | 50 | 55 |
Figure 1.
Depletion of MyoGEF caused defects in furrow ingression. (A) Normal cytokinesis. Live cell images show normal cytokinesis in HeLa cells transfected with control siRNA. (B) Furrow regression. Time-lapse images show that furrow does not successfully ingress in HeLa cells transfected with MyoGEF siRNA and a plasmid encoding H2B-GFP (green). (C) Ectopic furrowing. Time-lapse images show ectopic furrow formation (arrow in e) in a HeLa cell transfected with MyoGEF siRNA. (D) Metaphase arrest. Time-lapse images show that a HeLa cell transfected with MyoGEF siRNA and a plasmid encoding GFP-α-tubulin (green) was arrested at metaphase. Arrowheads indicate the spindles. (E) Depletion of MyoGEF did not affect central spindle formation. HeLa cells treated with siCont (a–d) or siMyoGEF (e–h) were fixed with methanol/acetone and subjected to immunofluorescence with antibodies specific for MyoGEF (green) and β-tubulin (red). The nuclei were stained with DAPI (blue). Bar, 20 μm. (F) HeLa cells treated with control siRNA (siCont; lane 1) or MyoGEF siRNA (siMyoGEF; lane 2) were subjected to immunoblot analysis with antibodies specific for MyoGEF and β-tubulin. (G) The immunoblot images in F were quantified using the NIH Image program. The numbers in A–D indicate the elapsed time (min:sec).
Figure 3.
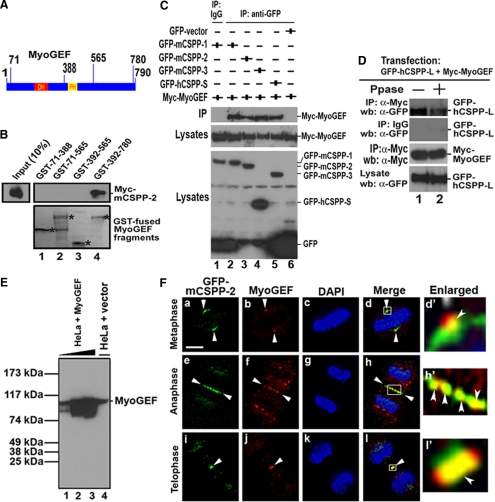
Interaction between MyoGEF and CSPP. (A) Schematic diagram of MyoGEF. The numbers indicate amino acids. DH and PH, Dbl-homology domain and pleckstrin homology domain, respectively. (B) An in vitro GST pulldown assay using GST-MyoGEF fragments and in vitro–translated mCSPP-2. GST-MyoGEF fragments are indicated by asterisks. (C) Myc-MyoGEF interacted with GFP-CSPP in vivo. HeLa cells were transfected with plasmids encoding Myc-MyoGEF and GFP-mCSPP-1, GFP-mCSPP-2, GFP-mCSPP-3, GFP-hCSPP-S, or GFP alone. The transfected cells were subjected to coimmunoprecipitation with anti-GFP antibody (α-GFP) followed by immunoblot analysis with anti-Myc antibody. (D) Myc-MyoGEF interacted with GFP-hCSPP-L in vivo. HeLa cells transfected with plasmids encoding GFP-hCSPP-L and Myc-MyoGEF were subjected to immunoprecipitation with anti-Myc antibody, followed by immunoblot with anti-GFP antibody. Note that treatment with λ-phosphatase (Ppase) decreases the interaction between Myc-MyoGEF and GFP-hCSPP-L (cf. lane 1 with lane 2 in top panel). (E) Characterization of MyoGEF antibody. HeLa cells were transfected with an empty vector (lane 4) or a MyoGEF plasmid (without any tags; lanes 1-3). Increasing amount of HeLa cell lysates from cells exogenously expressing MyoGEF was used (lanes 1–3). Same amount of cell lysates was used in lanes 3 and 4. (F) Colocalization of GFP-mCSPP-2 and endogenous MyoGEF to the central spindle and midbody. HeLa cells expressing GFP-mCSPP-2 were fixed with methanol/acetone and subjected to immunofluorescence with anti-MyoGEF antibody (red). The nuclei were stained with DAPI (blue). (d′, h′, and l′) Enlarged images from panels d, h, and l, respectively. Bar, 20 μm.
Figure 5.
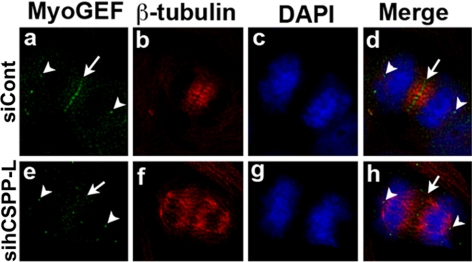
Depletion of CSPP affected MyoGEF localization. HeLa cells were transfected with control siRNA (siCont; a–d) or hCSPP-L siRNA (sihCSPP-L; e–l). The transfected HeLa cells were fixed with methanol/acetone and subjected to immunofluorescence with antibodies specific for MyoGEF (green) and β-tubulin (red). The nuclei were stained with DAPI (blue). Bar, 20 μm.
We then asked whether there were abnormalities in central spindle formation in cells depleted of MyoGEF. As shown in Figure 1E, depletion of MyoGEF did not affect central spindle formation (n = 50/50; cf. a–d with e–h), suggesting that furrow regression resulting from MyoGEF knockdown is not secondary to central spindle defects. In addition, we did not observe obvious DAPI-staining signals at the central spindle in cells depleted of MyoGEF (n = 50/50; Figure 1E; cf. a–d with e–h). Further, immunofluorescence with anti-lamin antibody showed no lamin-staining signals at the cleavage furrow in cells depleted of MyoGEF (data not shown). These results suggest that cytokinesis defects resulting from MyoGEF depletion were not secondary to chromosome bridging/lagging.
Identification of Mouse CSPP
To further understand the functions of MyoGEF, we set out to identify the proteins that can interact with MyoGEF. We carried out a yeast two-hybrid screening with full-length human MyoGEF as bait to screen a mouse 11-d embryo matchmaker cDNA library (Clontech). This screen resulted in the identification of a 286-amino acid polypeptide (Supplemental Figure S1; the underlined amino acid residues), which belongs to a protein family termed CSPP (Patzke et al., 2005, 2006). Two alternative spliced isoforms of human CSPP, i.e., hCSPP (shorter isoform that lacks 294 amino acids at the N-terminal end; we will refer to this isoform as hCSPP-S in this article) and hCSPP-L have been described previously (Patzke et al., 2005, 2006). One mouse CSPP isoform (mCSPP-1; NM_026493) has also been deposited in the database.
We have cloned the cDNAs for three alternative spliced isoforms of mCSPP: mCSPP-1, mCSPP-2, and mCSPP-3 (Figure 2A and Supplemental Figure S1). Analysis of the amino acid sequence alignment between mouse CSPP and human CSPP shows that 76% of amino acids in mCSPP-1 and 74% of amino acids in mCSPP-2 are identical to those in human CSPP-L. In addition, mCSPP-1, mCSPP-2, and hCSPP-L contain the similar number of amino acid residues, i.e., these three proteins contain 1197, 1142, and 1221 amino acid residues, respectively. Further, 92% of the amino acids in mCSPP-1 are identical to those in mCSPP-2. Thus, both mCSPP-1 and -2 are likely the mouse orthologues of human CSPP-L.
Figure 2.
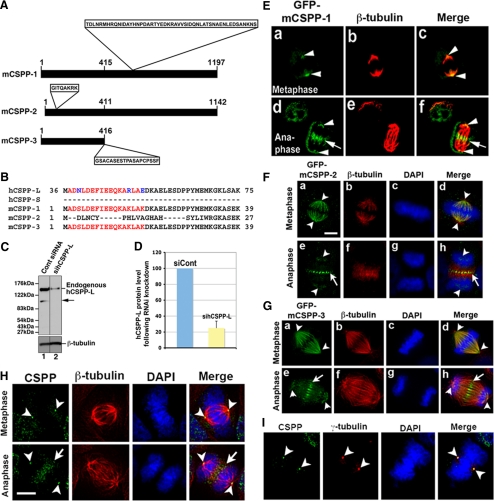
Identification of mCSPP. (A) Schematic diagram of three isoforms of mCSPP proteins. The major differences among mCSPP-1, -2, and -3 are shown: a 51-amino acid insert in the middle region of mCSPP-1; an 8-amino acid insert in the N-terminal region of mCSPP-2; 19 amino acids that are present in mCSPP-3 at the C-terminus, but not in mCSPP-1 and mCSPP-2. The number indicates the amino acids. (B) Alignments of the N-terminal amino acid sequences of human CSPP and mouse CSPP isoforms. The amino acids in red were used as antigen to raise an antibody specific for CSPP. (C) The polyclonal antibody specific for CSPP recognizes endogenous hCSPP-L. Treatment with hCSPP-L siRNA decreases the expression of hCSPP-L (cf. lane 1 with lane 2). (D) The immunoblot images in C were quantified using the NIH Image program. (E–G) GFP-mCSPP-1 localizes to the spindle pole and central spindle (E), whereas GFP-mCSPP-2 (F) and -3 (G) localize to the spindle microtubules and central spindle. The nuclei were stained with DAPI (blue). Bar, 20 μm. (H and I) Localization of endogenous CSPP during cytokinesis. HeLa cells were fixed and subjected to immunofluorescence with antibodies specific for CSPP (green) and β-tubulin (red) or γ-tubulin (red). Endogenous CSPP localizes to the spindle pole and central spindle (H) Endogenous CSPP also colocalizes with γ-tubulin at the spindle pole (I). The nuclei were stained with DAPI (blue). Bar, 20 μm. Note that cells in E–G were fixed with 4% paraformaldehyde, whereas cells in H and I were fixed with methanol/acetone (1:1).
To characterize the endogenous CSPP, we generated a polyclonal antibody using amino acid residues corresponding to amino acids 2-18 in mCSPP-1 and mCSPP-3 (Figure 2B; amino acids in red). This polyclonal antibody could recognize endogenous hCSPP-L (Figure 2C; lane 1). Further, an hCSPP-L siRNA (sihCSPP-L) that specifically targets the 3′ untranslated region (UTR) of hCSPP-L could silence the expression of hCSPP-L (Figure 2C; cf. lane 1 with lane 2 in the top panel). A 100-kDa protein could also be depleted by sihCSPP-L (arrow in Figures 2C). It is possible that this 100-kDa protein is a degraded product or an unidentified hCSPP isoform.
mCSPP Localized to the Spindle Pole and Central Spindle
To determine the cellular localization of mouse CSPP isoforms, we fused the cDNAs of three mCSPP isoforms to the 3′-end of GFP. HeLa cells transfected with plasmids encoding GFP-tagged mCSPP-1, -2, or -3 were stained with an antibody specific for β-tubulin (Figure 2, E–G, red). GFP-mCSPP-1 localized to the spindle pole (arrowheads in Figure 2E) and central spindle (Figure 2E; arrows in d and f). GFP-mCSPP-2 and -3 also localized to the central spindle (arrows in Figure 2, F and G) in anaphase. However, GFP-mCSPP-2 and -3 predominantly localized to the spindle microtubules, but did not concentrate at the spindle pole, in metaphase (Figure 2, F, arrowheads in a and d, and G, arrowheads in a and d). It should be noted that mCSPP-3 contains only the N-terminal region of mCSPP-1 (Figure 2A). Yet, it still predominantly localized to the spindle microtubules and central spindle (Figure 2G, a, d, e, and h), suggesting that the N-terminal region of mCSPP is critical for localization of mCSPP at the spindle microtubules and central spindle during cytokinesis.
To understand the dynamic localization of mouse CSPP during mitosis and cytokinesis, HeLa cells were transfected with plasmids encoding GFP-mCSPP-1, GFP-mCSPP-2, or GFP-mCSPP-3, and time-lapse microscopy was performed to monitor the cell cycle progression in the transfected HeLa cells. Consistent with GFP signals in fixed HeLa cells expressing GFP-mCSPP-1, GFP-mCSPP-3, and mCSPP-3 (Figure 2, E–G), time-lapse microscopy showed that mCSPP-1 localized to the spindle pole and central spindle (Supplemental Movie S1), whereas GFP-mCSPP-2 and GFP-mCSPP-3 localized to the spindle microtubules and central spindle (Supplemental Movies S2b and S3). Although GFP-mCSPP-1 showed a more diffuse distribution, careful analysis of the movie or immunofluorescence images indicated that some GFP-mCSPP-1 signals were consistently concentrated at the spindle pole and central spindle (Supplemental Movie S1 and Figure 2E). HeLa cells expressing GFP-mCSPP-1 (Supplemental Movie S1) or GFP-mCSPP-3 (Supplemental Movie S3) exhibited normal cell cycle progression. In contrast, ∼70% of HeLa cells expressing GFP-mCSPP-2 were arrested at metaphase (Supplemental Movie S2a; n = 29/40 cells). Nevertheless, in the GFP-mCSPP-2–expressing HeLa cells that could successfully complete cytokinesis, GFP-mCSPP-2 was concentrated at the spindle microtubules and central spindle (Supplemental Movie S2b).
It has not been reported whether endogenous CSPP also localizes to the spindle pole and central spindle. Therefore, we carried out immunofluorescence staining of HeLa cells with CSPP antibody to examine the localization of endogenous human CSPP during cytokinesis. HeLa cells were fixed with methanol/acetone (1:1) and processed for immunofluorescence with antibodies specific for CSPP and β-tubulin (Figure 2H). CSPP antibody could recognize the spindle pole/centrosome (arrowheads in Figure 2H) as well as the central spindle (arrows in Figure 2H). Immunofluorescence staining patterns of CSPP with CSPP antibody are comparable to GFP signals of GFP-mCSPP-1 in transfected cells (cf. Figure 2, E with H). It should be noted that our CSPP antibody only recognizes mCSPP-1, mCSPP-3, and hCSPP-L, but not mCSPP-2 and hCSPP-S (data not shown). We then asked whether CSPP colocalized with γ-tubulin at the spindle pole. HeLa cells fixed with methanol/acetone (1:1) were stained with antibodies specific for CSPP and γ-tubulin. As shown in Figure 2I, CSPP and γ-tubulin colocalized at the spindle pole during mitosis (arrowheads). These results demonstrate that endogenous human CSPP localizes to the spindle pole and central spindle during mitosis and cytokinesis.
In Vivo and In Vitro Interaction between MyoGEF and CSPP
To determine which regions in MyoGEF are responsible for its interaction with CSPP, we carried out in vitro GST pulldown assays using four overlapping MyoGEF fragments and in vitro–translated mCSPP-2. The four overlapping MyoGEF fragments were expressed as GST-fusion polypeptides, whereas mCSPP-2 was expressed as a Myc-tagged protein. As shown in Figure 3B, MyoGEF fragments 392-780, but not 392-565, 71-565, or 71-388, interacted with Myc-mCSPP-2 (Figure 3B; cf. lane 4 with lanes 1-3), suggesting that the C-terminal region (amino acids 565-780) of MyoGEF is required for its interaction with mCSPP-2. These results also support a direct interaction between CSPP and MyoGEF. To determine whether CSPP and MyoGEF interact in vivo, a plasmid encoding Myc-MyoGEF was cotransfected into HeLa cells with plasmids encoding GFP-mCSPP-1, mCSPP-2, mCSPP-3, GFP-hCSPP-S, or GFP alone. Twenty-four hours after transfection, the transfected cell lysates were subjected to immunoprecipitation with anti-GFP antibody, followed by immunoblot analysis with anti-Myc antibody. As shown in Figure 3C, Myc-MyoGEF could be coimmunoprecipitated with GFP-tagged mCSPP-1, -2, -3, or hCSPP-S in the transfected HeLa cells. However, anti-GFP antibody could not precipitate Myc-MyoGEF from HeLa cells transfected with GFP-vector and a plasmid encoding Myc-MyoGEF (lane 6 in Figure 3C). Myc-MyoGEF was also coprecipitated with GFP-hCSPP-L in transfected HeLa cells (Figure 3D). Treatment with λ-phosphatase decreased MyoGEF- hCSPP-L interaction (Figure 3D, cf. lane 1 with lane 2 in the top panel), suggesting that the MyoGEF-hCSPP-L interaction can be enhanced by phosphorylation.
Colocalization of CSPP and MyoGEF during Cytokinesis
To further demonstrate the correlation between CSPP and MyoGEF, HeLa cells transfected with GFP-mCSPP-2 were processed for immunofluorescence with an antibody specific for MyoGEF. HeLa cells expressing GFP-mCSPP-2 retained most of the GFP signals at the spindle microtubules and central spindle after fixation with methanol/acetone. In contrast, HeLa cells expressing GFP-mCSPP-1 and -3 lost most of the GFP signals after fixation with methanol/acetone. Therefore, we used HeLa cells transfected with GFP-mCSPP-2 plasmid for these experiments. We also developed a new polyclonal antibody against MyoGEF using full-length MyoGEF as antigen. This MyoGEF antibody could recognize exogenously expressed MyoGEF (Figure 3E, lanes 1-3) as well as endogenous protein (Figure 3E, lane 4). As shown in Figure 3F, endogenous MyoGEF localized to the spindle pole (arrowheads in b and d), central spindle (arrowheads in panels f and h), and midbody (arrowheads in j and l). GFP-mCSPP-2 localized to the spindle microtubules near the centrosome (arrowheads in panels a and d), central spindle (arrowheads in e and h), and midbody (arrowheads in i and l), although some GFP signals at the spindle microtubules were lost upon methanol/acetone fixation (cf. Figure 3F with Figure 2F). Importantly, endogenous MyoGEF colocalized with GFP-mCSPP-2 at the central spindle (Figure 3F, arrowheads in h and h′) and midbody (Figure 3F, arrowheads in l and l′). In addition, endogenous MyoGEF also partially overlapped with GFP-mCSPP-2 at the spindle pole (Figure 3F, arrowheads in d and d′). It should be noted that different patterns of GFP-mCSPP-2 in Figures 2Fa and 3Fa are due to differences in fixation (cells in Figures 2Fa and 3Fa were fixed with paraformaldehyde and methanol/acetone, respectively).
Depletion of CSPP Caused Defects in Cytokinesis
The previous flow cytometry analysis shows that RNAi-mediated depletion of CSPP leads to cell cycle arrest at S phase, but not at mitotic phase (Patzke et al., 2005). However, localization of CSPP at the spindle pole and central spindle strongly suggests that it may have a role in regulating mitotic progression and/or cytokinesis. Thus, we used time-lapse microscopy to examine whether depletion of CSPP by RNAi has an effect on mitosis and cytokinesis.
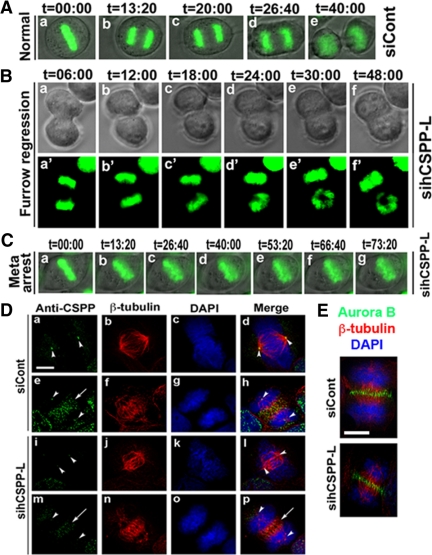
HeLa cells were transfected with a plasmid encoding GFP-H2B (histone 2B) and control siRNA or sihCSPP-L. Forty-eight to 72 h after transfection, the transfected cells that underwent mitosis were monitored using time-lapse microscopy. As shown in Figure 4B, we observed cytokinesis defects in sihCSPP-L-treated HeLa cells. Approximately 15% of cells (n = 8/55 cells; Table 1) that underwent mitosis showed furrow regression during cytokinesis, i.e., furrow successfully ingressed (a–c) and then regressed (d–f; cf. a with f). We did not observe furrow regression in control siRNA-treated cells (Figure 4A; n = 0/50 cells; Table 1). In addition, time-lapse microscopy also showed that ∼20% of hCSPP-L siRNA-treated HeLa cells (n = 12/55 cells; in control siRNA-treated cells, n = 4/50; Table 1) were arrested at metaphase (Figure 4C).
Figure 4.
Depletion of CSPP led to defects in mitosis and cytokinesis. (A) Normal cytokinesis in HeLa cells transfected with control siRNA and a plasmid encoding H2B-GFP (green). (B) Furrow regression in HeLa cells transfected with hCSPP-L siRNA and a plasmid encoding H2B-GFP (green; a′–f′). (C) Metaphase arrest in HeLa cells transfected with hCSPP-L siRNA and a plasmid encoding H2B-GFP (green). (D) Depletion of hCSPP-L did not affect central spindle formation. HeLa cells were transfected with control siRNA (siCont; a–h) or hCSPP-L siRNA (sihCSPP-L; i–p). The transfected HeLa cells were fixed with methanol/acetone and subjected to immunofluorescence with antibodies specific for CSPP (green) and β-tubulin (red). The nuclei were stained with DAPI (blue). (E) Cells treated with siCont or sihCSPP-L were subjected to immunofluorescence with antibodies specific for aurora B (green) and β-tubulin (red). Bar, (D) 20 μm; (E) 10 μm. Meta, metaphase. The numbers in A–C indicate the elapsed time (min:sec).
To determine whether furrow regression resulting from CSPP depletion was secondary to central spindle defects or chromosome lagging, we transfected HeLa cells with control siRNA (siCont) or hCSPP-L siRNAi (sihCSPP-L). Forty-eight to 72 h after transfection, the transfected cells were processed for immunofluorescence with antibodies against CSPP and β-tubulin. As shown in Figure 4D, knockdown of CSPP did not affect central spindle formation (cf. e–h with m–p; sihCSPP-L–treated cells: n = 65/65; siCont-treated cells: n = 50/50), suggesting that furrow regression after CSPP knockdown is not secondary to defects in central spindle formation. This was also confirmed by the finding that depletion of CSPP did not affect the localization of aurora B at the central spindle (Figure 4E; sihCSPP-L-treated cells: n = 79/79; siCont-treated cells: n = 50/50). In addition, we did not observe obvious DAPI-staining signals at the central spindle (Figure 4D, cf. e–h with m–p; sihCSPP-L-treated cells: n = 65/65; siCont-treated cells: n = 50/50). Further, immunofluorescence with anti-lamin antibody showed no lamin-staining signals at the cleavage furrow after CSPP depletion (data not shown). These results suggest that chromosome bridging/lagging are unlikely to be the major causes of furrow regression resulting from CSPP depletion. However, the spindle poles in cells treated with human CSPP siRNA (sihCSPP-L) were not completely focused compared with those in cells transfected with control siRNA (n = 14/68; Figure 4D, cf. b and d with j and l).
Depletion of CSPP Disrupted MyoGEF Localization at the Central Spindle
To further demonstrate the functional correlation between MyoGEF and CSPP, we asked whether depletion of hCSPP-L by RNAi had an impact on MyoGEF localization during cytokinesis. As shown in Figure 5, MyoGEF localized to the central spindle in cells transfected with siCont (n = 50; arrows in panels a and d). In contrast, MyoGEF did not concentrate at the central spindle in cells depleted of hCSPP-L (n = 17/89; Figure 5, cf. a and d with e and h). These results suggest that CSPP plays a role in regulating MyoGEF localization to the central spindle. However, depletion of CSPP-L did not affect the localization of MyoGEF to the spindle pole (Figure 5; cf. a and e).
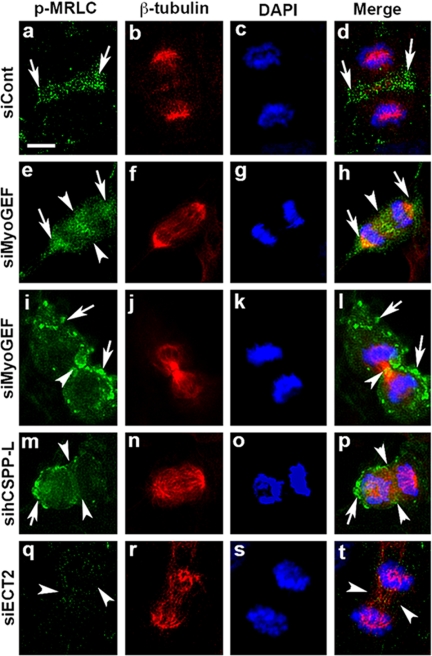
Depletion of MyoGEF or CSPP Resulted in Mislocalization of p-MRLC during Cytokinesis
Phosphorylation of MRLC is the hallmark of myosin II activation, and it is critically important for the initiation and progression of cytokinesis (Yamakita et al., 1994; Kosako et al., 2000; Matsumura, 2005). We have thus examined whether depletion of MyoGEF or CSPP alters localization of p-MRLC during cytokinesis. In control siRNA-transfected HeLa cells, p-MRLC was concentrated at the cleavage furrow zone (Figure 6; arrows in a and d). In anaphase cells transfected with MyoGEF siRNA (Figure 6, e–h), p-MRLC showed diffuse distribution with a high level of p-MRLC at the poles (arrows in e and h), but was not concentrated at the cleavage furrow (arrowheads in e and h; cf. h with d). During telophase in cells transfected with MyoGEF siRNA (Figure 6, i–l), p-MRLC was concentrated at the midbody (arrowheads in i and l). However, there was also a high level of p-MRLC that showed punctate distribution at other locations (arrows in i and l). During anaphase in cells transfected with hCSPP-L siRNA (Figure 6, m–p), p-MRLC was not concentrated at the cleavage furrow (arrowheads in m and p). Instead, p-MRLC showed diffuse distribution including at the poles (arrows in m and p). The mislocalization of p-MRLC contrasts to the effect of ECT2 depletion on p-MRLC: As shown in Figure 6, q and t, depletion of ECT2 almost completely eliminated p-MRLC at the cleavage furrow, the observation of which is consistent with previous reports (Zhao and Fang, 2005; Kamijo et al., 2006).
Figure 6.
Depletion of MyoGEF or CSPP resulted in p-MRLC mislocalization during cytokinesis. HeLa cells were transfected with control siRNA (a–d), MyoGEF siRNA (e–l), hCSPP-L siRNA (m–p), or ECT2 siRNA (q–t). Seventy-two hours after transfection, the transfected cells were fixed with 4% paraformaldehyde and stained with antibodies specific for β-tubulin (red) and p-MRLC (green). The chromosomes were stained with DAPI (blue). Bar, 20 μm.
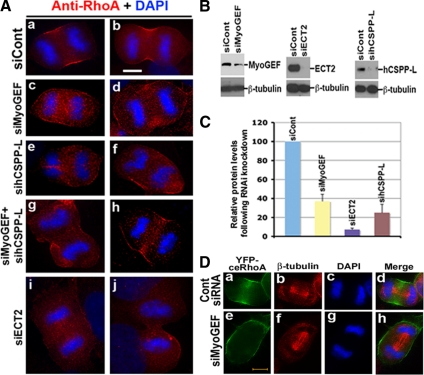
Depletion of MyoGEF or CSPP Resulted in Mislocalization of Active RhoA during Cytokinesis
Accumulation of active RhoA at the cleavage furrow is essential for cytokinesis (Yoshizaki et al., 2004; Yuce et al., 2005; Niiya et al., 2006; Nishimura and Yonemura, 2006; Petronczki et al., 2007). To gain an insight into how MyoGEF regulates cytokinesis, we investigated the effects of MyoGEF or CSPP depletion on RhoA localization during cytokinesis. We used RNAi to deplete MyoGEF, CSPP-L, or ECT2 in U2OS cells. The siRNA-treated cells were fixed with TCA and analyzed for the localization of active RhoA at the cleavage furrow. It has been shown that TCA fixation can retain active RhoA staining at the equatorial cortex (Nishimura and Yonemura, 2006). We selected U2OS cells for these experiments, because U2OS cells express higher levels of MyoGEF but lower levels of ECT2 than HeLa cells (data not shown). In cells treated with control siRNA (siCont), RhoA staining was restricted to the cleavage furrow (n = 28/30; 93%; Figure 7A, a and b). However, in cells depleted of MyoGEF, RhoA staining was not restricted to equatorial cortical regions (n = 27/58; 47%; Figure 7A, c and d). Instead, RhoA staining could be observed at the whole cortical region of the anaphase cells (Figure 7A, c and d). Similar RhoA staining patterns were also found in cells depleted of hCSPP-L (n = 7/20; 35%; Figure 7A, e and f) or both MyoGEF and hCSPP-L (n = 9/17; 53%; Figure 7A, g and h). Consistent with previous reports (Yuce et al., 2005; Nishimura and Yonemura, 2006), we also found that depletion of ECT2 resulted in dramatic reduction of RhoA staining at the equatorial cortex (n = 11/15; 73%; Figure 7A, i and j). In contrast, a recent study using the FRET-based RhoA biosensor reveals that active RhoA is mislocalized (but is not decreased) during cytokinesis in ECT2-depleted cells (Birkenfeld et al., 2007). These inconsistent results may be due to different methods that are used to measure RhoA activation at the cleavage furrow (immunostaining vs. FRET). Nevertheless, our results indicate that CSPP and MyoGEF play a role in restricting active RhoA to the cleavage furrow.
Figure 7.
Depletion of MyoGEF or CSPP affected the distribution of active RhoA during cytokinesis. (A) U2OS cells treated with siRNAs as indicated were fixed with TCA and subjected to immunofluorescence with an antibody specific for RhoA (red). The nuclei were stained with DAPI (blue). Bar, 20 μm. (B) U2OS cells treated with siRNAs as indicated were subjected to immunoblot analysis with antibodies as indicated. (C) The immunoblot images in B were quantified using the NIH Image program. (D) Depletion of MyoGEF interfered with YFP-ceRhoA distribution during cytokinesis. HeLa cells were transfected with a YFP-ceRhoA–expressing plasmid and control siRNA (a–d) or siMyoGEF (e–h). Forty-eight hours after transfection, the transfected cells were fixed with 4% paraformaldehyde and stained with an antibody specific for β-tubulin (red). The nuclei were stained with DAPI (blue). Bar, 20 μm.
We then asked whether MyoGEF depletion also affected YFP-ceRhoA localization at the cleavage furrow during cytokinesis. YFP-ceRhoA has been successfully used in monitoring the active RhoA localization during cytokinesis, i.e., active ceRhoA localizes to the cortical region of the cleavage furrow (Yuce et al., 2005). We transfected HeLa cells with YFP-ceRhoA plasmid and control siRNA or siMyoGEF. Forty-eight hours after transfection, the transfected cells were processed for immunofluorescence with β-tubulin antibody. We focused on the cells expressing a low level of YFP-ceRhoA. As shown in Figure 7D, YFP-ceRhoA localized to the equator of anaphase cells treated with control siRNA (a–d). In contrast, YFP-ceRhoA localized to the whole cortical region of the anaphase cell treated with siMyoGEF (Figure 7D, e–h). These results further confirm that MyoGEF is important for the localization of active RhoA at the equatorial cortex during cytokinesis.
Interaction between MyoGEF and ECT2
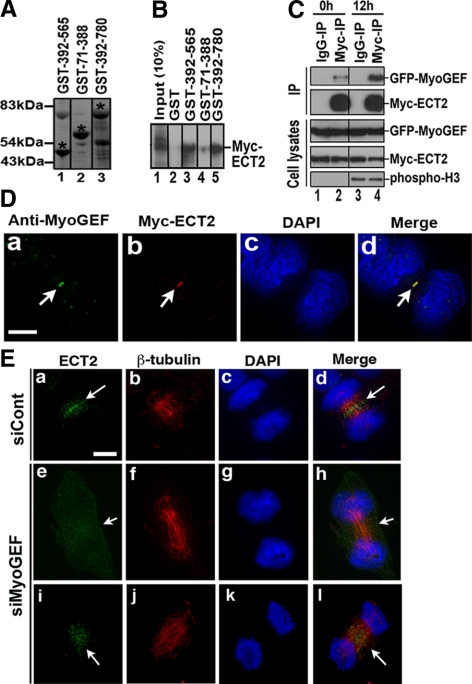
Depletion of MyoGEF led to mislocalization of p-MRLC and active RhoA at the cleavage furrow (Figures 6 and 7), suggesting that a significant amount of active RhoA is still generated in cells depleted of MyoGEF during cytokinesis. Another GEF, ECT2, also localizes to the central spindle, and it is critical for equatorial RhoA activation (Yuce et al., 2005; Zhao and Fang, 2005). Therefore, we asked whether MyoGEF could interact with ECT2 and whether depletion of MyoGEF could affect ECT2 localization at the central spindle.
Three overlapping GST-tagged MyoGEF fragments (Figure 8A) were used in the GST pulldown assay to assess whether MyoGEF could interact with ECT2 in vitro. As shown in Figure 8B, MyoGEF fragments 392-565 and 392-780, but not 71-388, could bind to the in vitro–translated ECT2, suggesting that amino acids 392-565 of MyoGEF can directly bind to ECT2. To confirm the in vivo interaction between MyoGEF and ECT2, HeLa cells were transfected with plasmids encoding GFP-MyoGEF and Myc-ECT2. The transfected cells were enriched at the G1/S border (0 h after release from thymidine block) or mitosis (12 h after release from thymidine block) as described in Materials and Methods. The transfected cells were then subjected to immunoprecipitation with anti-Myc antibody. As shown in Figure 8C, GFP-MyoGEF could be coimmunoprecipitated with Myc-ECT2 from the transfected cell lysates, suggesting that MyoGEF also interacts with ECT2 in vivo. Immunoblot analysis with anti-phospho-histone 3 antibody confirmed that the transfected cells were enriched at mitosis at 12 h after release from thymidine block (Figure 8C, bottom panel).
Figure 8.
Depletion of MyoGEF affected ECT2 localization at the central spindle. (A) GST-tagged MyoGEF fragments (indicated by asterisks). (B) The GST pulldown assay using GST-tagged MyoGEF fragments and in vitro–translated Myc-ECT2. (C) HeLa cells were transfected with plasmids encoding GFP-MyoGEF and Myc-ECT2. At 0 or 12 h after release from thymidine block, the transfected cells were subjected to coimmunoprecipitation with anti-Myc antibody, followed by immunoblot analysis with antibodies as indicated. (D) Colocalization of endogenous MyoGEF and Myc-ECT2 to the midbody. (E) HeLa cells were treated with control siRNA (siCont; a–d) or MyoGEF siRNA (siMyoGEF; e–l). Seventy-two hours after transfection, the transfected cells were processed for immunofluorescence with antibodies specific for β-tubulin (red) and ECT2 (green). The chromosomes were stained with DAPI (blue). Note that cells in D were fixed with methanol/acetone (1:1), whereas cells in E were fixed with 4% paraformaldehyde. Bar, 20 μm.
We then asked whether MyoGEF colocalized with ECT2 during cytokinesis. HeLa cells transfected with Myc-ECT2 plasmid were fixed with methanol/acetone and subjected to immunofluorescence with antibodies specific for MyoGEF and Myc. As shown in Figure 8D, MyoGEF colocalized with Myc-ECT2 to the midbody. We also used RNAi to examine whether depletion of MyoGEF affected ECT2 localization during cytokinesis. In control siRNA-treated cells, ECT2 was concentrated at the central spindle (n = 15; Figure 8E, a–d). In contrast, ECT2 showed a more diffuse distribution in siMyoGEF-treated cells that underwent cytokinesis (n = 6/17; Figure 8E, e–l). These results suggest that MyoGEF can bind ECT2 and promote its localization to the central spindle.
DISCUSSION
In this article, we have identified CSPP as an interacting partner of MyoGEF. Both in vitro and in vivo pulldown assays confirm that CSPP interacts with MyoGEF. Immunofluorescence analysis shows that CSPP and MyoGEF colocalize at the central spindle. Depletion of MyoGEF or CSPP by RNAi leads to cytokinesis defects as well as mislocalization of nonmuscle myosin II with a p-MRLC during cytokinesis. More importantly, CSPP depletion interferes with MyoGEF localization at the central spindle. Further, depletion of MyoGEF causes mislocalization of ECT2 and RhoA during cytokinesis. These findings suggest a role for CSPP-MyoGEF interaction in regulating cytokinesis.
A Role for CSPP in Both Cytokinesis and Cell Cycle Progression
A previous report shows that depletion of CSPP leads to cell cycle arrest in S phase, but not in mitotic phase by flow cytometry (Patzke et al., 2005). This appears to be inconsistent with our finding that RNAi depletion of CSPP caused cell cycle arrest at metaphase (∼20%) as well as defects in cytokinesis (∼15%). One explanation is that the small fraction of abnormal mitotic cells could be detected by time-lapse microscopy, but not by flow cytometry. Another possibility is that CSPP has cell type–specific effects on cytokinesis. The previous study used a different cell line HEK293T, and cell type–specific regulation of cytokinesis has been suggested (Yoshizaki et al., 2004). We believe that, in addition to its role in regulating cell cycle progression from G1/G0 through S-phase, CSPP also has a role in the regulation of mitosis and cytokinesis (Figure 4).
Consistent with this idea, CSPP not only localizes to the spindle pole and central spindle, but also localizes to the centrosome (Patzke et al., 2005, 2006). It has been well established that the centrosome, which is implicated in microtubule nucleation, plays a central role in both cytokinesis and cell cycle progression from G1/G0 through S-phase. Elimination of the centrosome with a microneedle or by laser microsurgery caused defects in cytokinesis (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Piel et al., 2001). In addition, these cells fail to enter S phase and arrest at G0/G1 phase. Components of the centrosome have also been shown to be important for cytokinesis and cell cycle progression from G1/G0 through S phase (Doxsey, 2001; Kaiser et al., 2002; Mailand et al., 2002). For instance, RNAi-mediated depletion of centriolin, a critical component of the centrosome, results in cell cycle arrest in G0/G1 phase as well as cytokinesis defects (Gromley et al., 2003). Time-lapse microscopy further indicates that a GFP-labeled mother centriole transiently migrates to the intracellular bridge near the midbody and then moves back to the center of the cell before abscission, suggesting a role for the centrosome in regulating abscission (Piel et al., 2001; Doxsey et al., 2005).
Cytokinesis Defects in MyoGEF- and CSPP-depleted Cells Are Not Likely Secondary to Central Spindle Defects or Mitotic Abnormalities
The spindle defect in MyoGEF-depleted cells (Figure 1) raised a question as to whether cytokinesis defects induced by siMyoGEF treatment were secondary to abnormalities in spindle apparatus. This is unlikely because of the following observations. First, in some of siMyoGEF-treated cells that underwent furrow regression, cytokinesis was almost completed before the furrow started to regress (data not shown), suggesting that furrow formation and ingression are not affected in such siMyoGEF-treated cells. Therefore, it is likely that these cells have a normal central spindle, which is critical for furrow formation and ingression. Second, the central spindle appears to be normal in siMyoGEF-treated cells that show mislocalization of p-MRLC (Figure 6), RhoA (Figures 7), and ECT2 (Figure 8). Third, decreased expression of CSPP or MyoGEF does not affect spindle formation (Figures 1E, 4D, and 5) and the localization of aurora B at the central spindle (Figure 4E and data not shown). These findings suggest that cytokinesis defects resulting from CSPP or MyoGEF depletion are unlikely to be correlated with abnormalities in central spindle formation.
Low frequency of cytokinesis defects (see Table 1) also raises a question as to whether furrow regression resulting from CSPP or MyoGEF depletion is a consequence of mitotic abnormalities, such as chromosome bridging or lagging, leading to DNA trapping at the cleavage furrow. However, one of our major findings was that active RhoA did not restrict to a narrow region at the cleavage furrow in cells depleted of CSPP or MyoGEF (see Figure 7). Consistent with these findings, we also found that p-MRLC mislocalized in cells treated with CSPP or MyoGEF siRNA (see Figure 6). In addition, we did not observe obvious DAPI-staining signals at the central spindle in cells treated with CSPP or MyoGEF siRNA (see Figures 1E, 4D, 5, 6, 7D, and 8E). Further, immunofluorescence with an antibody specific for lamin A+C show no lamin-staining signals at the central spindle and cleavage furrow in cells depleted of CSPP or MyoGEF (data not shown). Although we do not know at present how CSPP or MyoGEF depletion results in mitotic defects, our results indicate that chromosome bridging and lagging are unlikely to be the major causes of furrow regression after RNAi-mediated depletion of CSPP or MyoGEF.
Low Penetrance Cytokinesis Defects after MyoGEF or CSPP Depletion
Our results show that MyoGEF interacts with ECT2, and depletion of MyoGEF by RNAi causes mislocalization of p-MRLC, RhoA, and ECT2 during cytokinesis (see Figures 6–8). However, only a small subset of HeLa cells depleted of CSPP or MyoGEF show cytokinesis defects (see Table 1). One possibility is that the efficiency of CSPP and MyoGEF depletion by RNAi is relatively low, i.e., we observed an average of 60–70% knockdown for MyoGEF and 70–75% for CSPP (see Figures 1G, 2D, and 7C). In contrast, we consistently observed a much higher efficiency of ECT2 depletion by RNAi (>90% knockdown; see Figure 7C). Another possibility is that cells with higher efficiency of CSPP or MyoGEF depletion may well be arrested at mitosis, even though at present we do not have direct evidence to support this speculation. Further, our results indicate that depletion of MyoGEF results in mislocalization of ECT2 and this may be the cause of cytokinesis defects. Therefore, it is also possible that recruitment of ECT2 to the central spindle by a redundant mechanism can partially compensate the absence of MyoGEF. Although it is not clear how CSPP or MyoGEF depletion causes mitotic arrest, our results demonstrate that depletion of MyoGEF or CSPP leads to mislocalization of RhoA and p-MRLC. We believe that such abnormal molecular events may explain some, if not all, of cytokinesis phenotype resulting from CSPP or MyoGEF depletion.
It should be also noted that mislocalization of RhoA and ECT2 during cytokinesis was found in 35–50% of CSPP- or MyoGEF-depleted cells. However, only 15–25% of CSPP- or MyoGEF-depleted cells showed furrow regression and/or ectopic furrowing (see Table 1). Although depletion of MyoGEF or CSPP results in mislocalization of RhoA and/or ECT2 during cytokinesis, we still observed a significant amount of active RhoA at the cleavage furrow (Figures 7A, d, f, and h) as well as a broader ECT2 band at the central spindle (Figure 8Ei) after MyoGEF or CSPP depletion. It is likely that these cells may still be able to complete cytokinesis, leading to the lower percentage of cytokinesis defects observed in live cells that were treated with MyoGEF or CSPP siRNA.
MyoGEF Interacts with CSPP and ECT2
ECT2 has been implicated as a key regulator of RhoA localization and activation at the cleavage furrow (Yoshizaki et al., 2004; Yuce et al., 2005; Niiya et al., 2006; Nishimura and Yonemura, 2006; Petronczki et al., 2007). Our results show that MyoGEF can interact with ECT2 during cytokinesis and that depletion of MyoGEF interferes with ECT2 localization at the central spindle (see Figure 8). It has been reported that CYK-4/MgcRacGAP can interact with ECT2 and that this interaction is critical for the localization of ECT2 at the central spindle as well as equatorial RhoA activation (Yuce et al., 2005; Zhao and Fang, 2005). It is not clear at present how MyoGEF and CYK-4/MgcRacGAP coordinate the regulation of ECT2 localization at the central spindle. It would be interesting to know whether MyoGEF, CYK-4, and ECT2 can form a complex at the central spindle and whether MyoGEF has an impact on ECT2-CYK-4 interaction. Nonetheless, our results indicate that interplay between MyoGEF and ECT2 may play an important role in regulating cytokinesis.
There are at least two CSPP isoforms in humans, i.e., hCSPP-S and hCSPP-L. This study has been focused on hCSPP-L. The siRNA against hCSPP-L was designed based on its 3′-UTR, which is different from that of hCSPP-S. Therefore, the siRNA against hCSPP-L used in this study is likely specific for hCSPP-L. Exogenous expression of hCSPP-L causes chromosome lagging or monopolar spindle formation, whereas exogenous expression of hCSPP-S leads to the formation of multipolar spindles (Patzke et al., 2005, 2006), suggesting that both isoforms may not be completely redundant. Exogenous expression of MyoGEF or ECT2 also causes failure in cytokinesis (Niiya et al., 2006; Asiedu et al., 2008). Therefore, it appears that optimal levels of CSPP, MyoGEF, and ECT2 are needed for the regulation of mitosis and/or cytokinesis. Our results show that CSPP interacts with MyoGEF and that depletion of CSPP interferes with MyoGEF localization at the central spindle (see Figures 3 and 5). In addition, depletion of CSPP or MyoGEF causes mislocalization of p-MRLC and RhoA (see Figures 6 and 7). Further, simultaneous depletion of both CSPP and MyoGEF increases the percentage of cells with mislocalization of RhoA during cytokinesis (siMyoGEF: 47%; sihCSPP-L: 35%; siMyoGEF+sihCSPP-L: 53%). Therefore, our findings suggest that CSPP interacts with MyoGEF and promotes its localization to the central spindle, where MyoGEF may modulate the localization of ECT2, thereby contributing to equatorial RhoA and myosin II activation during cytokinesis. However, it is not clear at present whether the GEF activity of MyoGEF directly contributes to equatorial RhoA activation during cytokinesis.
It has been shown that CSPP is phosphorylated at mitotic phase, and potential phosphorylation sites by Plk1 have been indicated (Patzke et al., 2005). Consistent with these observations, our results show that phosphorylation can enhance CSPP-MyoGEF interaction (see Figure 3D), suggesting that CSPP-MyoGEF interaction may be regulated by mitotic kinases such as Plk1. This speculation is reminiscent of the PLK1 and ECT2 interaction that was recently reported (Niiya et al., 2006; Burkard et al., 2007; Petronczki et al., 2007). Those reports clearly demonstrate that Plk1 plays an essential role in the regulation of furrow formation and ingression by recruiting ECT2 to the central spindle. Our recent studies also show that Plk1 can phosphorylate MyoGEF at Thr-574, and this phosphorylation is important for the recruitment of MyoGEF to the central spindle (Asiedu et al., 2008). Further study to illustrate the role of Plk1 and/or aurora kinases in the regulation of CSPP-MyoGEF interaction should provide new insights into the molecular mechanism of cytokinesis, with respect to the spatial and temporal regulation of myosin contractile ring assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert S. Adelstein and Dr. Mary Anne Conti for critical reading and comments on the manuscript. We thank Dr. Hans-Christian Aasheim for providing GFP-hCSPP plasmid and Dr. Michael Glotzer for providing YFP-ceRhoA plasmid. This publication was made possible by National Institutes of Health Grants P20 RR-015563 and P20 RR-017708 from the National Center for Research Resources. This work was also supported by National Institutes of Health Grant k22 HL071542 (Q.W.) as well as a fund from Terry C. Johnson Center for Basic Cancer Research (Q.W.). This is contribution 08-12-J from the Kansas Agricultural Experiment Station, Manhattan, Kansas.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0001) on January 7, 2009.
REFERENCES
- Asiedu M., Wu D., Matsumura F., Wei Q. Phosphorylation of myogef on thr-574 by polo-like kinase 1 (PLK1) promotes myogef localization to the central spindle. J. Biol. Chem. 2008;283:28392–28400. doi: 10.1074/jbc.M801801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W. M., Benink H. A., von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenfeld J., Nalbant P., Bohl B. P., Pertz O., Hahn K. M., Bokoch G. M. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev. Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D. R., Chang F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 2005;15:156–162. doi: 10.1016/j.tcb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Burkard M. E., Randall C. L., Larochelle S., Zhang C., Shokat K. M., Fisher R. P., Jallepalli P. V. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Eggert U. S., Mitchison T. J., Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 2003;161:535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Miller F. J., Cham M., Khodjakov A., Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. RHO GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kaiser B. K., Zimmerman Z. A., Charbonneau H., Jackson P. K. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell. 2002;13:2289–2300. doi: 10.1091/mbc.01-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo K., Ohara N., Abe M., Uchimura T., Hosoya H., Lee J. S., Miki T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol. Biol. Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T., Sullivan K. F., Wahl G. M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kosako H., Yoshida T., Matsumura F., Ishizaki T., Narumiya S., Inagaki M. Rho-kinase/ROCK is involved in cytokinesis through the phosphorylation of myosin light chain and not ezrin/radixin/moesin proteins at the cleavage furrow. Oncogene. 2000;19:6059–6064. doi: 10.1038/sj.onc.1203987. [DOI] [PubMed] [Google Scholar]
- Mackay D. J., Hall A. Rho GTPases. J. Biol. Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- Mailand N., Lukas C., Kaiser B. K., Jackson P. K., Bartek J., Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 2002;4:317–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- Matsui T., Amano M., Yamamoto T., Chihara K., Nakafuku M., Ito M., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Matsumura F., Totsukawa G., Yamakita Y., Yamashiro S. Role of myosin light chain phosphorylation in the regulation of cytokinesis. Cell Struct. Funct. 2001;26:639–644. doi: 10.1247/csf.26.639. [DOI] [PubMed] [Google Scholar]
- Niiya F., Tatsumoto T., Lee K. S., Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 2006;25:827–837. doi: 10.1038/sj.onc.1209124. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 2006;119:104–114. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- Patzke S., Hauge H., Sioud M., Finne E. F., Sivertsen E. A., Delabie J., Stokke T., Aasheim H. C. Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene. 2005;24:1159–1173. doi: 10.1038/sj.onc.1208267. [DOI] [PubMed] [Google Scholar]
- Patzke S., Stokke T., Aasheim H. C. CSPP and CSPP-L associate with centrosomes and microtubules and differently affect microtubule organization. J. Cell Physiol. 2006;209:199–210. doi: 10.1002/jcp.20725. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J. M. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Prokopenko S. N., Brumby A., O'Keefe L., Prior L., He Y., Saint R., Bellen H. J. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W. G., Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q. Pitx2a binds to human papillomavirus type 18 E6 protein and inhibits E6-mediated P53 degradation in HeLa cells. J. Biol. Chem. 2005;280:37790–37797. doi: 10.1074/jbc.M502974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Asiedu M., Adelstein R. S., Wei Q. A novel guanine nucleotide exchange factor MyoGEF is required for cytokinesis. Cell Cycle. 2006;5:1234–1239. doi: 10.4161/cc.5.11.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y., Yamashiro S., Matsumura F. In vivo phosphorylation of regulatory light chain of myosin II during mitosis of cultured cells. J. Cell Biol. 1994;124:129–137. doi: 10.1083/jcb.124.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Parrini M. C., Dulyaninova N. G., Bresnick A. R., Mochizuki N., Matsuda M. Cell type-specific regulation of RhoA activity during cytokinesis. J. Biol. Chem. 2004;279:44756–44762. doi: 10.1074/jbc.M402292200. [DOI] [PubMed] [Google Scholar]
- Yuce O., Piekny A., Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. M., Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA. 2005;102:13158–13163. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.