Abstract
The evolutionarily conserved adaptor protein-3 (AP-3) complex mediates cargo-selective transport to lysosomes and lysosome-related organelles. To identify proteins that function in AP-3–mediated transport, we performed a genome-wide screen in Saccharomyces cerevisiae for defects in the vacuolar maturation of alkaline phosphatase (ALP), a cargo of the AP-3 pathway. Forty-nine gene deletion strains were identified that accumulated precursor ALP, many with established defects in vacuolar protein transport. Maturation of a vacuolar membrane protein delivered via a separate, clathrin-dependent pathway, was affected in all strains except those with deletions of YCK3, encoding a vacuolar type I casein kinase; SVP26, encoding an endoplasmic reticulum (ER) export receptor for ALP; and AP-3 subunit genes. Subcellular fractionation and fluorescence microscopy revealed ALP transport defects in yck3Δ cells. Characterization of svp26Δ cells revealed a role for Svp26p in ER export of only a subset of type II membrane proteins. Finally, ALP maturation kinetics in vac8Δ and vac17Δ cells suggests that vacuole inheritance is important for rapid generation of proteolytically active vacuolar compartments in daughter cells. We propose that the cargo-selective nature of the AP-3 pathway in yeast is achieved by AP-3 and Yck3p functioning in concert with machinery shared by other vacuolar transport pathways.
INTRODUCTION
Subcellular compartmentalization by membrane-bounded organelles is a fundamental feature of eukaryotic cells. This organization allows for physical and functional segregation of subcellular processes. An important example of this compartmentalization is the lysosome, which is an acidic organelle that serves as a major site for protein degradation within eukaryotic cells. Degradative enzymes are sequestered within lysosomes ensuring that only material delivered to the organelle is subject to destruction.
Compartmentalization of lysosome functions necessitates transport pathways for lysosomal biogenesis, maintenance of function, and transfer of molecules targeted for turnover. Genetic analysis of proteins that are targeted to the lysosome-like vacuole of Saccharomyces cerevisiae has defined six such trafficking routes, each involving vesicle-mediated transport (Bryant and Boyd, 1993). Two of these pathways involve protein transport from the cytoplasm to the vacuole: the cytoplasm to vacuole transport pathway (CVT) and starvation-induced autophagy. A third pathway, the endocytic pathway, delivers cell surface and extracellular molecules to the vacuole. Yet another route provides for vacuole inheritance during cell division. The inheritance pathway directs vesicles derived from the maternal vacuole into the new bud where vesicle fusion seeds formation of a daughter cell vacuole. Finally, there are two vacuolar biosynthetic pathways that originate from the secretory pathway, one clathrin dependent and the other clathrin-independent. Newly synthesized vacuolar components destined for both pathways are transported from the endoplasmic reticulum (ER) to the Golgi complex. At the trans-Golgi network (TGN), the proteins enter into either the clathrin-dependent pathway that passes through endosomes to the vacuole or into a clathrin-independent pathway that proceeds directly to vacuole.
Clathrin-dependent transport between the TGN and endosomes relies on several evolutionarily conserved clathrin adaptors: monomeric GGA proteins and the heterotetrameric adaptor protein-1 (AP-1) complex (Bonifacino, 2004; Traub, 2005). These adaptors function as protein interaction platforms by binding to the coat protein clathrin and other coat-associated proteins, as well as recognizing sorting signals in the cytoplasmic domains of transmembrane cargo proteins. In this way, adaptors play central roles in vesicle formation by coupling cargo selection to assembly of a protein coat.
Most vacuolar proteins are transported through the clathrin-dependent pathway, including the soluble protease carboxypeptidase Y (CPY), the membrane protease carboxypeptidase S (CPS), and membrane components of the V-type H+-ATPase responsible for acidifying the vacuole (Bowers and Stevens, 2005). This pathway flows through multivesicular endosomes, where membrane proteins destined for the lumen of the vacuole are sorted into luminal vesicles that are subsequently delivered to the vacuole. There is limited information on sorting signals for the clathrin-dependent pathway in yeast, in part because this pathway seems to act as a default route for membrane proteins lacking cytoplasmic sorting signals (Roberts et al., 1992; Redding et al., 1996).
The clathrin-independent pathway to the vacuole is distinguished by a requirement for the AP-3 adaptor complex, a four-subunit complex with homology to AP-1. AP-3 is evolutionarily conserved and consists of two large subunits, β3 and δ; a medium subunit μ3; and a small subunit ς3 (Odorizzi et al., 1998). Only a few proteins that transit via the AP-3 pathway have been identified in yeast; among these are alkaline phosphatase (ALP) (Cowles et al., 1997a; Stepp et al., 1997), the target membrane-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (t-SNARE) Vam3p (Cowles et al., 1997a), yeast casein kinase 3 (Yck3p) (Sun et al., 2004), and the Niemann-Pick Type C homologue Ncr1p (Berger et al., 2007). Each of these proteins is localized to the limiting vacuolar membrane rather than intravacuolar vesicles, suggesting that the AP-3 pathway bypasses multivesicular endosomes and functions as a selective and direct route from the TGN to the vacuolar membrane. Cargoes of the AP-3 pathway contain either acidic dileucine-like or YXXΦ (where Φ is a bulky hydrophobic amino acid and X is any amino acid) signals that are necessary for sorting into the pathway (Darsow et al., 1998; Vowels and Payne, 1998a; Sun et al., 2004).
Formation of AP-3 vesicles in yeast, unlike vesicles with AP-1 and GGA proteins, seems to be independent of clathrin. AP-3 does not strongly associate with clathrin, nor do mutations in clathrin alter the rate of ALP transport to the vacuole (Seeger and Payne, 1992; Vowels and Payne, 1998a; Yeung et al., 1999). Only one other protein in yeast has been characterized with selective function in AP-3 vesicle formation in yeast, Vps41p. Vps41p is a member of the homotypic vacuole fusion and vacuole protein sorting (HOPS) complex (Nakamura et al., 1997; Seals et al., 2000; Wurmser et al., 2000). The HOPS complex functions in tethering vesicles to the vacuole, and, in this capacity, it is necessary for delivery of both clathrin-dependent and AP-3–dependent cargo (Bowers and Stevens, 2005; Ostrowicz et al., 2008). However, Vps41p also seems to play a selective role in AP-3 vesicle formation through an interaction with the AP-3 δ subunit (Rehling et al., 1999; Darsow et al., 2001). Vps41p has features similar to clathrin, but whether this protein is a structural component of the AP-3 coat remains unresolved (Conibear and Stevens, 1998; Ybe et al., 1999).
Mammalian AP-3 also participates in protein transport to lysosomes (Robinson and Bonifacino, 2001; Di Pietro and Dell'Angelica, 2005). Much like the yeast adaptor, mammalian AP-3 seems to act in a pathway to the lysosomal limiting membrane that bypasses multivesicular late endosomes. However, in mammalian cells, AP-3 localizes predominantly at early endosomes instead of the Golgi (Peden et al., 2004; Theos et al., 2005), suggesting that AP-3 pathways in yeast and mammalian cells originate at distinct organelles. Another difference between yeast and mammalian AP-3 is that the mammalian adaptor binds clathrin (Dell'Angelica et al., 1998), although the functional significance of this interaction has not been fully established (Newell-Litwa et al., 2007).
In addition to a ubiquitous role in lysosomal protein traffic, mammalian AP-3 acts in cell type-specific pathways to specialized lysosome-related organelles such as melanosomes and platelet dense granules (Di Pietro and Dell'Angelica, 2005; Huizing et al., 2008). Defects in transport to lysosomes and lysosome-related organelles caused by mutations in the AP-3 β subunit lead to the inherited human disease Hermansky–Pudlak syndrome (HPS) (Dell'Angelica et al., 1999; Huizing et al., 2002; Jung et al., 2006), a disease characterized by oculocutaneous albinism and prolonged bleeding times (Di Pietro and Dell'Angelica, 2005; Huizing et al., 2008). A disease similar to HPS is manifested in mice bearing mutations in AP-3 subunits (Di Pietro and Dell'Angelica, 2005; Newell-Litwa et al., 2007; Huizing et al., 2008).
Mutations in other genes also cause HPS and HPS-like diseases in both humans and mice, and the products of these genes have been associated with trafficking pathways to lysosomes and lysosome-related organelles (Di Pietro and Dell'Angelica, 2005; Huizing et al., 2008). One such protein, Vps33a, is a component of the mammalian HOPS complex that also contains Vps41 (Suzuki et al., 2003). Most of the other proteins do not seem to have homologues in yeast, although a recent report describes limited sequence homologies between HPS4 and the yeast Ccz1p protein involved in vacuolar protein transport (Hoffman-Sommer et al., 2005). Thus, although not readily apparent by sequence conservation, there is the possibility that other HPS proteins will have functional analogues in yeast.
To more fully understand the mechanisms of transport through the AP-3 pathway and identify additional candidate disease genes, having a complete catalogue of genes/proteins involved in AP-3 pathway function would be advantageous. For this reason, we have carried out a systematic, genome-wide screen in yeast to identify genes involved in AP-3–dependent ALP transport to the vacuole. We identified a complement of genes that serve a variety of functions from processing enzymes, to vesicle budding, targeting, and vesicle fusion. Many of these genes have well established roles in protein transport through both the clathrin and AP-3 pathways. Only deletions in AP-3 subunit genes and YCK3 selectively affected the AP-3 pathway. We also observed that the ER export receptor for ALP, Svp26p, is required for transport of a subset of type II membrane proteins. Finally, our results indicate that the vacuolar inheritance pathway is important for timely production of a proteolytically mature vacuole in newly budded cells.
MATERIALS AND METHODS
Yeast Media, Plasmids, and Strains
Yeast strains were grown in YPD [1% Bacto-yeast extract (Difco, Detroit, MI), 2% Bacto-peptone (Difco), 2% dextrose], YPD + 200 μg/ml G418, or SD (0.67% yeast nitrogen base [Difco], 2% dextrose) supplemented with 20 μg/ml l-histidine, uracil, and l-tryptophan, and 30 μg/ml l-leucine, adenine, and l-lysine. Solid media contained 2% agar. Yeast transformations were performed using the lithium acetate method (Ito et al., 1983). Cell densities were measured by spectrophotometry. One OD600 corresponds to 107 cells per milliliter. Sequences of oligonucleotides used in this study are available on request.
Plasmids pYCK3, pyck3-1, and pyck3-2 are described in Sun et al. (2004). pyck3-1 bears an in frame deletion of a portion of YCK3 encoding Yck3pΔ409-462. pyck3-2 bears a substitution mutation changing amino acid 444 from tyrosine to histidine. pGFP-ALP is a multicopy plasmid encoding a fusion of GFP to the N terminus of full-length ALP (Cowles et al., 1997a). pBS207, a low copy centromeric plasmid contains a fragment of chromosome XIV (coordinates 67948–78118) encoding full-length MON2 (Jochum et al., 2002).
Strains used in this study were obtained from the single gene deletion libraries constructed by the international deletion consortium (Winzeler et al., 1999) constructed in BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) and BY4742 (MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0) (Brachmann et al., 1998). Deletion of SVP26 was performed by standard polymerase chain reaction (PCR)-based methods (Longtine et al., 1998) to generate GPY4865 (MATa ura3-52 his3Δ200 leu2-3 lys2-801 trp1Δ901 suc2Δ9 mel svp26Δ::HIS3) in SEY6210 (Robinson et al., 1988). MATa cps1Δpho8Δ was generated by crossing MATα pho8Δ with MATa cps1Δ from the single gene deletion collections, sporulating, and dissecting the sporulated diploid to generate GPY4866.
Preparation of Cell Extracts from Single-Gene Deletion Libraries
The 96-well format S. cerevisiae MATa and MATα single gene deletion libraries were obtained from Open Biosystems (Huntsville, AL) and ResGen (now Invitrogen, Carlsbad, CA), respectively. Each plate of the 96-well format gene deletion libraries was replicated into a 96-well plate (Cole-Parmer Instrument, Vernon Hills, IL) containing 200 μl/well of YPD + 200 μg/ml G418. Cells were grown for 2 d at 30°C. Fifty microliters from each well was transferred to 2 ml of YPD in a 96-deep well plate (Matrix, Hudson, NH). Individual magnetic stir-bars (VP Scientific, San Diego, CA) were added and stirred continuously using a magnetic stir device (VP Scientific) for 5 h at 30°C. Cells were then sedimented by centrifugation at 1500 × g for 5 min. The supernatant was discarded, and 100 μl of cracking buffer (10% glycerol, 5% SDS, 2% β-mercaptoethanol, 50 mM Tris-Cl, pH 6.8, and 0.02% bromphenol blue) was added to each well then transferred to 96-well PCR plates (Thermo Fisher Scientific, Waltham, MA). Plates were incubated at 99°C for 3 min, and samples were frozen at −80°C until further processing.
Immunoblotting ALP for Single-Gene Deletion Library Extracts
Five microliters of each lysed strain in cracking buffer was subjected to electrophoresis through a 10% SDS-polyacrylamide gel. Gels were transferred onto polyvinylidene difluoride (PVDF) membranes at 100 V for 60 min in 24 mM Tris base, 192 mM glycine, and 20% methanol buffer. Membranes were treated with blocking buffer (5% milk, 0.1% Tween 20 in phosphate-buffered saline [PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 2 mM KH2PO4]) for 1 h and then incubated overnight at 4°C in a rabbit anti-Pho8p antibody (Seeger and Payne, 1992b) diluted in blocking buffer. Next, 25 μl/ml concentrated cps1Δpho8Δ cell extract was added to antibody dilutions, and the mixture was preincubated for 30 min before incubation with PVDF membranes to reduce nonspecific signals. Bound rabbit antibodies were detected with goat anti-rabbit horseradish peroxidase conjugated antibody (Bio-Rad, Hercules, CA). 10 μL/ml pho8Δ extract was added to the secondary antibody dilution in blocking buffer and preincubated for 30 min. Membranes were washed in PBS and 0.1% Tween 20 and developed using ECL-Plus (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. Digital images were acquired with a fluorescence imager (GE Healthcare).
To prepare concentrated cps1Δpho8Δ extract, MATa cps1Δpho8Δ cells were grown in YPD to 10 OD600/ml, 2 l were harvested by centrifugation, and frozen in liquid nitrogen. The frozen cells were mechanically lysed in a blender, and the lysate was resuspended in 75 ml of 1% SDS and heated at 100°C for 5 min. The lysate was then clarified by centrifugation at 100,000 × g for 45 min at 4°C.
Cell Extract Preparation, Immunoblotting, and Immunoprecipitation for Analysis of Individual Strains
Three stationary overnight cultures in YPD from each mating type of a gene deletion strain were diluted to 0.125 OD600 in YPD and grown to 0.5–1 OD600/ml, yielding six samples per deletion strain. Approximately 5 OD600 cells were harvested by centrifugation at 1500 × g for 3 min in 13- × 100-mm glass test tubes (Thermo Fisher Scientific). Lysates were prepared by mechanical disruption of the pellet by adding 50 μl of LSB (10% glycerol, 2% SDS, 2% β-mercaptoethanol, 50 mM Tris-Cl, pH 6.8, and 0.02% bromphenol blue) and 250 μl of 0.5-mm glass beads (Biospec, Bartlesville, OK) to the pellet and agitating with a Vortex mixer (Thermo Fisher Scientific). Lysates were resuspended in 400 μl of LSB and incubated at 100°C for 5 min. Immunoblots were performed as described above using rabbit anti-Pho8p, rabbit anti-Cps1p (generated using glutathione transferase fused to Cps1p amino acids [aa]46–576 as antigen), or rabbit anti-Gda1p (generated using 6 histidine-tagged Gda1p aa25–518). Pho8p and Cps1p antibodies and secondary antibodies were treated with concentrated pho8Δcps1Δ extract as described above. Gda1p antibodies and secondary antibodies were treated with a concentrated gda1Δ cell extract prepared as described above for pho8Δcps1Δ extract.
Radiolabeling and immunoprecipitations for ALP and CPS were performed as described in Seeger and Payne (1992b) and Costaguta et al. (2006).
Quantitation of Immunoblots and Immunoprecipitations
Digital images of immunoblots and immunoprecipitations were analyzed using ImageJ (http://rsb.info.nih.gov/ij/). Bands were quantified by measuring the total integrated density of a band after background subtraction. The precursor ALP (pALP) to total ALP (tALP) ratio represents the pALP band density divided by the sum of the pALP, mature ALP (mALP), and soluble ALP (sALP) band densities. A ratio of pALP/tALP was determined for each of the six samples per gene deletion strain. These six calculated ratios per strain were averaged together, and a standard deviation was calculated. Wild-type strains were also grown in sextuplicate and a pALP/tALP ratio, and standard deviation was determined. Two tailed t tests were performed for each deletion strain pALP/tALP ratio compared with the wild-type ratio. A strain was determined to display a processing defect if the deletion strain pALP/tALP was greater than wild-type, with p < 0.05. The pALP/tALP ratio for the deletion strain was then normalized to the wild-type ratio by dividing the deletion strain ratio by the wild-type ratio. The normalized pALP/tALP ratio is reported in Table 1.
Table 1.
ALP maturation defects in strains from gene deletion libraries
| Single gene deletion strain | Gene product function | ALP defect | CPS defect | Significance of ALP defect, p value |
|---|---|---|---|---|
| AP-3 | ||||
| apl5Δ | δ Subunit of AP-3 | 3.1 | − | <0.001 |
| apl6Δ | β Subunit of AP-3 | 3.1 | − | <0.001 |
| apm3Δ | μ Subunit of AP-3 | 2.3 | − | <0.001 |
| aps3Δ | ς Subunit of AP-3 | 3.1 | − | <0.001 |
| Vesicle formation | ||||
| vps1Δ | Dynamin homologue | 2.7 | M | <0.001 |
| arf1Δ | ARF GTPase, acts in vesicle coat assembly | 2.3 | M | <0.001 |
| drs2Δ | Lipid flippase | 3.2 | M | <0.001 |
| Vacuolar proteases | ||||
| pep4Δ | Vacuolar serine protease, activates other vacuolar proteases | 7.2 | S | <0.001 |
| prb1Δ | Vacuolar serine protease | 5.3 | M | <0.001 |
| HOPS/CORVET | ||||
| vps41Δ | HOPS subunit/AP-3 vesicle formation | 6.3 | S | <0.001 |
| vam6Δ(vps39Δ) | HOPS subunit/Ypt7 guanine nucleotide exchange factor | 6.5 | S | <0.001 |
| pep5Δ (vps11Δ) | HOPS/CORVET core subunit | 7.2 | S | <0.001 |
| pep3Δ (vps18Δ) | HOPS/CORVET core subunit | 7.4 | S | <0.001 |
| vps16Δ | HOPS/CORVET core subunit | 7.5 | S | <0.001 |
| vps8Δ | CORVET subunit, interacts with Vps21p | 2.3 | M | <0.001 |
| vps3Δ | CORVET subunit | 3.8 | S | <0.001 |
| Rabs and vesicle targeting factors | ||||
| ypt7Δ | Rab GTPase for tethering/fusion to vacuole | 5.6 | S | <0.001 |
| vps45Δ | Sec1/Munc-18 family, interacts with Pep12p | 3.8 | S | <0.01 |
| vps21Δ | Rab GTPase for tethering/fusion to endosome | 1.8 | M | <0.001 |
| vps9Δ | Vps21 guanine nucleotide exchange factor | 3.6 | S | <0.001 |
| SNAREs and fusion | ||||
| vam3Δ | Vacuole membrane t-SNARE | 6.2 | S | <0.001 |
| vam7Δ | Vacuole membrane t-SNARE | 6.2 | S | <0.001 |
| pep12Δ | Endosome t-SNARE | 4.6 | S | <0.001 |
| Vacuolar inheritance | ||||
| vac8Δ | Vacuolar membrane protein that binds Vac17p | 3.4 | M | <0.001 |
| vac17Δ | Links Vac8p to the myosin, Myo2p | 4.8 | S | <0.001 |
| Phosphatidylinositol synthesis | ||||
| fab1Δ | Phosphatidylinositol-3-phosphate 5-kinase | 4.0 | M | <0.01 |
| vac14Δ | Activator of Fab1p | 2.9 | M | <0.001 |
| vps15Δ | Protein kinase subunit of phosphatidylinositol 3 (PI3)-kinase complexes I and II | 5.0 | S | <0.001 |
| vps34Δ | PI3-kinase | 6.2 | S | <0.001 |
| vps30Δ | Subunit of PI3-kinase complexes I and II | 2.9 | M | <0.001 |
| vps38Δ | Subunit of PI3-kinase complex I | 3.2 | M | <0.001 |
| Retromer | ||||
| pep8Δ (vps26Δ) | Retromer subunit | 1.7 | M | <0.01 |
| vps5Δ | Retromer subunit | 1.6 | M | =0.001 |
| vps35Δ | Retromer subunit, cargo binding | 2.3 | M | <0.001 |
| vps29Δ | Retromer subunit | 2.4 | M | <0.01 |
| vps17Δ | Retromer subunit | 1.4 | M | =0.02 |
| v-type H+-A TPase | ||||
| tfp1Δ | Subunit A of V1 domain | 4.0 | S | =0.02 |
| vma2Δa | Subunit B of V1 domain | 1.7 | M | <0.01 |
| vma5Δa | Subunit C of V1 domain | 1.4 | M | <0.001 |
| vma8Δa | Subunit D of V1 domain | 1.6 | M | <0.001 |
| vma7Δ | Subunit F of V1 domain | 3.2 | S | <0.001 |
| vma10Δ | Subunit G of V1 domain | 1.7 | M | <0.05 |
| vph1Δ | Subunit a of V0 domain | 3.3 | M | <0.001 |
| cup5Δ | Subunit c of V0 domain | 3.7 | S | <0.001 |
| tfp3Δ | Subunit c′ of V0 domain | 2.9 | S | =0.1 |
| ppa1Δ | Subunit c″ of V0 domain | 2.0 | S | <0.05 |
| vma6Δ | Subunit c‴ of V0 domain | 5.4 | S | <0.05 |
| vph2Δ | v-ATPase assembly | 5.3 | S | <0.01 |
| vma22Δ | v-ATPase assembly | 3.8 | S | =0.01 |
| Miscellaneous | ||||
| yck3Δ | Type I caseine kinase involved in homotypic vacuole fusion | 3.2 | − | <0.001 |
| svp26Δ | Adaptor for sorting ALP into COP-II vesicles at ER | 4.2 | − | <0.001 |
| ccz1Δ | Acts in vesicle fusion to the vacuole in a complex with Mon1p | 4.4 | M | <0.001 |
| mon1Δ | Acts in vesicle fusion to the vacuole in a complex with Ccz1p | 4.4 | M | <0.001 |
Three colonies from each mating type of a single-gene deletion strain were individually analyzed for ALP and CPS maturation defects by immunoblotting. Band densities for ALP were quantified and an average ratio of pALP to total ALP was determined for the six samples (see Materials and Methods). The ALP defect for a gene deletion strain is expressed as the pALP/tALP ratio of the deletion strain divided by the pALP/tALP ratio of wild-type cells. Statistical significance of the ALP defect was calculated from the values and standard deviations. CPS maturation is reported as having no defect (−), moderate defect (M), or severe defect (S) as determined by the presence or absence of precursor and mature forms of CPS (see Materials and Methods). Neither MATa or MATα library contained valid deletion strains for vps33Δ (HOPS subunit), pep7Δ (vesicle targeting to endosomes), vma4Δ (subunit E of V1 domain of ATPase), or vma21Δ (v-ATPase assembly) as determined by sequencing the bar codes of the relevant strains.
a ALP maturation defects in these strains were not detected in the genome-wide screens.
Defects in CPS maturation were scored in a qualitative manner due to overlap between precursor and mature forms of the protein. Both pCPS and mCPS migrate as two differentially glycosylated forms. The lower molecular weight form of pCPS comigrates with the higher molecular weight form of mCPS—we term this the intermediate form. Thus, only the higher molecular weight precursor form can be used as a reliable diagnostic for maturation defects. Strains were scored as having no CPS maturation defect if a two band pattern corresponding to the intermediate and mature forms was detected. Strains were scored as having a severe defect if a two-band pattern was detected consisting of the higher-molecular-weight precursor and the intermediate-molecular-weight form, with little or no lower-molecular-weight mCPS. Strains were scored as having a moderate defect if three bands were detected—the higher-molecular-weight precursor form, the intermediate-molecular-weight form, and the lower-molecular-weight mature form.
Cell Lysis and Differential Centrifugation
Cell lysis and differential centrifugation was carried out as described in Yeung and Payne (2001), with the following modifications: 50 OD600 cells were harvested; cells were converted to spheroplasts with 0.05 U of Zymolyase (Seikagaku, Tokyo, Japan) per OD600 cells; and the lysis buffer was 200 mM sorbitol, 50 mM Tris, pH 6.8, 50 mM NaCl, 2 mM EDTA, and protease inhibitors.
N-[3-Triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] Pyridinium Dibromide (FM4-64) Staining and Fluorescence Microscopy
The MATa wild-type strain BY4741 and MATa deletion strains yck3Δ, apl6Δ (β3Δ), and vac17Δ, all harboring pGFP-ALP were grown in liquid SD medium with appropriate supplements overnight to 0.2–0.4 OD600/ml. Cells were harvested by centrifugation at 1500 × g for 3 min, the supernatant was decanted, and the cells resuspended in the residual liquid (200 μl). Cells were stained using 200 μM FM4-64 for 20 min, washed, and incubated for 60 min at room temperature as described in Vida and Emr (1995) and imaged using an Axioobserver Z1 1.0 spinning disk confocal microscope (Carl Zeiss, Oberkochen, Germany).
RESULTS
Genome-wide Screens for ALP Maturation Defects
To screen for AP-3 pathway defects, we monitored the proteolytic maturation of ALP. ALP, a type II membrane protein, is synthesized as a 74-kDa glycosylated zymogen (pALP) that is proteolytically processed upon delivery to the vacuole to yield a 72-kDa integral membrane mature form (mALP) and a 66-kDa soluble mature form (sALP) (Klionsky and Emr, 1989). In wild-type cells, virtually all ALP is present in the mature forms as a consequence of the rapid rate of pALP transport to the vacuole and proteolytic processing. In contrast, cells with defects in AP-3–mediated transport accumulate pALP, which is easily distinguished from the mature forms by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting (Lorenz et al., 2007). Thus, as a systematic approach to identify genes involved in ALP trafficking, we used immunoblotting to assess pALP accumulation in the sets of haploid MATa and MATα strains bearing single deletions of all nonessential genes.
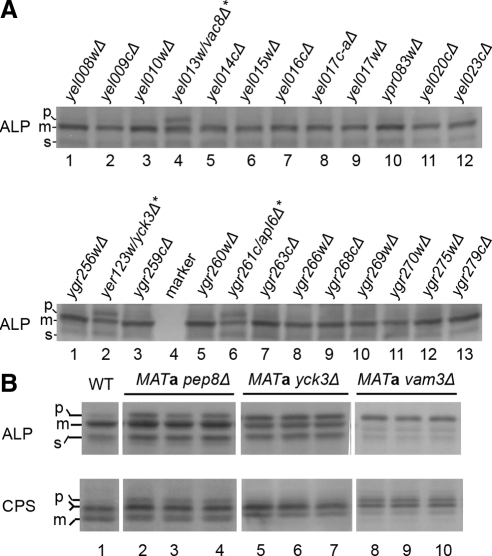
Deletion strains were grown in 96-well plates to mid-logarithmic phase, and then lysates were prepared and analyzed by SDS-PAGE and immunoblotting with an ALP antibody. A strain with a partial ALP maturation defect (yck3Δ) was included in each 96-well plate for quality control to ensure reproducibility of the assay. Strains displaying pALP to ALP ratios significantly greater than wild-type strains, using yck3Δ as a general guide, were initially scored as positive. Examples of the data are presented in Figure 1, revealing pALP accumulation in vac8Δ, yck3Δ, and apl6Δ cells (Figure 1A, top, lane 4; and bottom, lanes 2 and 6).
Figure 1.
Example of results from analysis of ALP maturation in strains from the MATa deletion library. (A) Indicated strains were grown in YPD, extracts were prepared, and ALP maturation was assessed by SDS-PAGE and immunoblotting. Strains scored as positive for pALP accumulation are designated with an asterisk. The positions of precursor (p), mature (m), and soluble (s) forms of ALP are indicated. The abundance of the sALP form can vary between experiments depending on growth state and other undetermined factors. (B) Example of results from analysis of ALP and CPS maturation in multiple samples of each strain scored as positive in the genome-wide screen. Three individual colonies from each of the indicated gene deletion strains and WT cells were independently cultured, cell extracts were prepared, and ALP and CPS were detected by immunoblotting. Forms of ALP are indicated as in A. The position of precursor (p) and mature (m) forms of CPS are indicated. The forked line indicates the intermediate band position where pCPS and mCPS forms comigrate. Examples of strains scored in Table 1 as no defect (yck3Δ), moderate defect (pep8Δ), and severe defect (vam3Δ) are shown.
Strains scored as positive in the initial screen were grown individually and retested for pALP accumulation by immunoblotting. In addition, we tested a few strains with deletions of vacuolar H+-ATPase subunits that did not display ALP maturation defects in the genome-wide screens but that were expected to score positive because deletions of other subunits resulted in maturation defects. In total, 4848 single gene deletion strains in the MATa library and 4871 strains in the MATα library were screened; 49 strains exhibited ALP maturation defects when retested (Table 1).
The specificity of the ALP maturation defect was assessed by analyzing in parallel the processing of ALP and another type II vacuolar membrane protein, CPS. CPS, like ALP, is synthesized in a precursor form that is proteolytically matured in the vacuole (Spormann et al., 1992). However, CPS is transported via the clathrin-dependent pathway (Cowles et al., 1997b). Consequently, defects specific for the AP-3 pathway should affect ALP but not CPS maturation. Three independent samples of each mating type of each positive strain were monitored for ALP and CPS maturation by immunoblotting (Figure 1B). Quantitation of the fraction of pALP present in each sample demonstrated that all the strains exhibited statistically significant accumulation of pALP compared with wild-type cells (Table 1). For CPS, the presence of partially overlapping glycosylated precursor and mature forms necessitated a more qualitative measure of maturation (Figure 1B and Table 1). Nevertheless, each strain exhibited reproducible patterns of CPS that could be reliably assessed for maturation defects (Table 1).
Most strains with ALP maturation defects could be grouped according to established functional categories or protein complexes (Table 1). In all cases except for AP-3 subunit gene deletions, svp26Δ, and yck3Δ, deletions of genes that affected ALP maturation also affected CPS maturation. Thus, the analysis yielded only six gene deletions with selective effects on ALP maturation, four of which were AP-3 subunits. Additionally, vac8Δ and vac17Δ strains displayed defects in both ALP and CPS maturation that have not been reported previously. Vac8p and Vac17p function in vacuole inheritance (Weisman, 2006).
Among the deletions affecting both ALP and CPS maturation are genes expected to influence ALP maturation through direct roles in either maturation or transport. These include the master vacuolar maturation proteases Pep4p and Prb1p that are required for ALP and CPS processing in the vacuole (Jones, 1984), proteins that act in vesicle formation such as Vps1p, and Arf1p (Odorizzi et al., 1998; Bowers and Stevens, 2005), and components of the machinery responsible for AP-3 and endosomal pathway vesicle docking and fusion with the vacuole such as the HOPS complex, the rab GTPase Ypt7p, and SNAREs (Bowers and Stevens, 2005; Ostrowicz et al., 2008).
Other gene deletions identified in the screen are likely to inhibit ALP transport/maturation indirectly. For example, proteins involved in the clathrin-dependent pathway such as Rab Vps21p, the Vps21p guanine nucleotide exchange factor Vps9p, the endosomal t-SNARE Pep12p, and the Vps34p phosphatidylinositol 3-kinase complex I, are necessary for transport of the maturation proteases Pep4p and Prb1p to endosomes (Bowers and Stevens, 2005). In these mutants, reduced levels of Pep4p and Prb1p in the vacuole can account for the ALP maturation defects. A similar explanation may apply to the weak ALP maturation defects observed in strains with deletions of genes encoding subunits of the retromer complex, which mediates endosome to Golgi retrieval of vacuolar protein sorting receptors (Seaman et al., 1998). Retromer mutations could also affect ALP maturation by blocking a retrieval pathway from the vacuole membrane that passes through endosomes to the TGN (Bryant et al., 1998). This pathway is involved in retrieval of the v-SNARE Vti1p that participates in ALP delivery to the vacuole (Vti1p acts in several transport steps and is essential for viability, thus vti1Δ is not represented in the haploid deletion mutant collection).
Deletions of FAB1 and VAC14 inhibit production of phosphatidylinositol 3,5 bisphosphate, resulting in perturbations in several vacuole pathways that could affect ALP and CPS maturation, including the vacuole to endosome retrieval pathway described above and the vacuolar inheritance pathway (Cooke, 2002; Shaw et al., 2003; Weisman, 2006). However, we did not observe defects in ALP or CPS maturation in either MATa or MATα cells lacking Vac7p, an activator of the Fab1p lipid kinase (Bonangelino et al., 1997), and we also noted variable maturation defects in different fab1Δ strains, suggesting further analysis is needed to determine whether Fab1p and its activators are necessary for ALP and CPS maturation.
Finally, deletions of many of the genes encoding subunits or assembly factors for the V-type H+ ATPase were identified. Acidification by the V-ATPase is required for optimal activity of the vacuolar maturation enzymes and for efficient protein transport to the vacuole, particularly through the endosomal pathway (Kane, 2006). Additionally the V0 membrane component has been proposed to play a more direct role in vacuole membrane fusion (Peters et al., 2001). Whether the variability observed between different subunit deletions reflects distinct functions will require additional investigation.
ALP Transport Is Specifically Affected in yck3Δ Cells
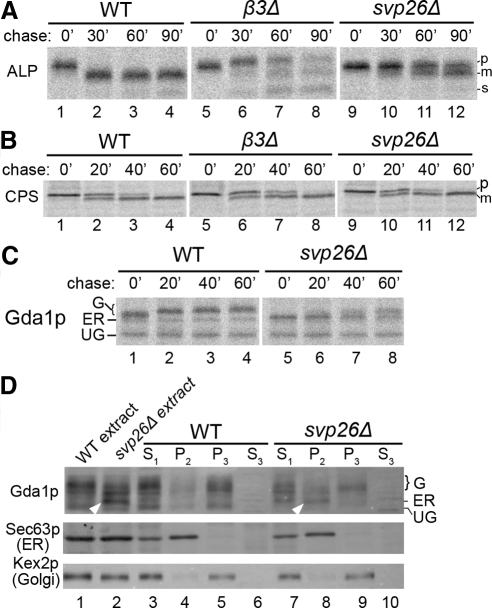
Our immunoblotting analysis of ALP and CPS in yck3Δ suggested a specific defect in ALP traffic in this strain. Yck3p is a type I casein kinase that is localized to the vacuole membrane and plays a role in regulating vacuole morphology and inheritance (Sun et al., 2004; LaGrassa and Ungermann, 2005). A role for this kinase in protein traffic to the vacuole has not been described. To more carefully assess the specificity of the trafficking defect we characterized ALP and CPS maturation by pulse-chase immunoprecipitation. Wild-type, yck3Δ, and apl6Δ cells were pulse labeled for 10 min, and then they were subjected to chase regimens before lysis and immunoprecipitation of ALP and CPS. In wild-type cells, maturation of ALP was >50% complete after the labeling period and complete by 10 min of chase (Figure 2A, lanes 1–4). In contrast, in the yck3Δ cells, >50% of ALP was in the precursor form after the 5-min chase and significant levels of precursor persisted at the 15-min chase point (Figure 2A, lanes 9–12). This corresponds to roughly a threefold delay in ALP maturation in yck3Δ cells. By comparison, cells lacking the AP-3 β subunit (β3Δ) displayed a severe ALP maturation defect, with the precursor form predominant throughout the chase period (Figure 2A, lanes 5–8). These results suggest an important but not required function of Yck3p in ALP transport to the vacuole. CPS is matured more slowly than ALP in wild-type cells, presumably because of the intermediate endosome stage in the clathrin-dependent pathway (Figure 2, A and B, lanes 1–4). Maturation of CPS was not significantly delayed in β3Δ or yck3Δ cells (Figure 2B, lanes 5–12). Together, the results from immunoblotting and pulse-chase immunoprecipitation provide evidence that maturation of ALP is selectively affected in yck3Δ cells. This phenotype is similar to, but less severe than, those of AP-3–deficient cells, suggesting that Yck3p acts in the AP-3 pathway.
Figure 2.
Specific defect in ALP transport in yck3Δ cells. (A) Pulse-chase immunoprecipitation of ALP. Cells from WT, apl6Δ-AP-3 β subunit gene deletion strain (β3Δ), and yck3Δ strains were metabolically labeled with [35S]methionine for 7 min, and labeling was quenched with nonradioactive amino acids. Samples of cells were removed at the indicated times after initiation of the chase. Cells were lysed, ALP was immunoprecipitated, and the immunoprecipitates were subjected to SDS-PAGE. Gels were exposed to a phosphor screen and digitally imaged. (B) Pulse-chase immunoprecipitation of CPS. Pulse-chase analysis of CPS was performed as described in A, except that samples were removed at 0, 10, 20, and 40 min after addition of the chase. (C) Subcellular fractionation of yck3Δ cells. Cell lysates from wild-type (WT), apl6Δ (β3Δ), and yck3Δ strains were fractionated by differential centrifugation to generate low-speed (300 × g for 5 min) supernatant (S1), medium-speed (10,000 × g for 15 min) supernatant and pellet (S2, P2), and high-speed (200,000 × g for 17 min) supernatant and pellet (S3, P3) fractions. Fractions were analyzed by SDS-PAGE and immunoblotted for ALP, the Golgi protein Kex2p, and the vacuole membrane (Vac) protein Vph1p. (D) ALP maturation in sorting defective Yck3p mutants. Cells from the MATa yck3Δ strain harboring empty vector, or plasmids containing wild-type YCK3 or two mutant yck3 genes (yck3-1, yck3-2) that lack a YDSI tyrosine-based AP-3 pathway sorting sequence (Sun et al., 2004). Cells were cultured in selective media, cell extracts were prepared and subjected to immunoblotting for ALP.
To determine whether the ALP maturation delay in yck3Δ cells is due to a defect in ALP transport to the vacuole, we analyzed the subcellular distribution of pALP by differential centrifugation. Extracts from wild-type, yck3Δ and β3Δ cells were first sedimented at 300 × g to remove unbroken cells, and then the resulting supernatant (S1) was sedimented at 13,000 × g to pellet larger organelles including the ER and vacuoles (P2). The 10,000 × g supernatant (S2) was then sedimented at 200,000 × g to pellet smaller organelles such as the Golgi and transport vesicles (P3). Fractions were then analyzed by immunoblotting for ALP, Kex2p (TGN/endosome marker), and Vph1p (vacuole membrane marker). In all three strains, mALP pelleted together with the Vph1p in P2 (Figure 2C, lanes 2, 6, and 10), whereas sALP was mostly present in S3 due to release from vacuoles during the lysis procedure (Figure 2C, lanes 4, 8, and 12). In β3Δ and yck3Δ cells, pALP, like Kex2p, sedimented predominantly in P3, indicating a defect in ALP transport to the vacuole in the two strains.
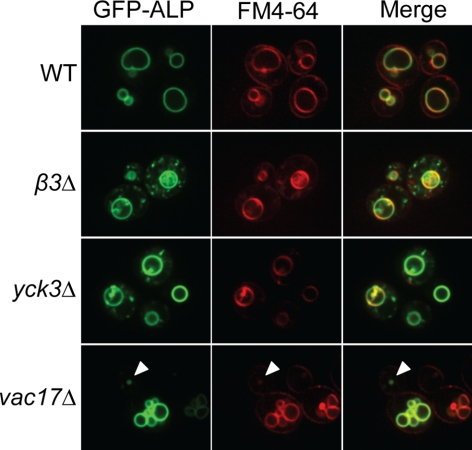
As an independent test of ALP traffic, localization of GFP-tagged ALP was assessed by fluorescence microscopy. To visualize vacuoles, cells were also stained with FM4-64, which is delivered to the vacuole via the endocytic pathway (Vida and Emr, 1995). In wild-type cells, GFP-ALP colocalized with FM4-64 at vacuole membranes (Figure 3, WT). In contrast, in β3Δ cells GFP-ALP was present both at the vacuole together with FM4-64 and also at cytoplasmic puncta that did not label with FM4-64 (Figure 3, β3Δ). A similar but less severe localization defect was apparent in yck3Δ cells (Figure 3, yck3Δ). The punctate distribution of GFP-ALP in β3Δ and yck3Δ cells is consistent with the pALP transport defect observed by differential centrifugation and likely reflects accumulation of pALP in Golgi and/or endosomes. Together, results from analysis of yck3Δ cells identify Yck3p as a factor specifically required for ALP transport between the Golgi and the vacuole.
Figure 3.
Mislocalization of GFP-ALP in yck3Δ but not vac17Δ cells. WT, β3Δ, yck3Δ, and vac17Δ cells harboring a plasmid encoding GFP-ALP were grown to early-logarithmic phase in minimal media, collected by centrifugation, and stained with FM4-64 for 20 min. The cells were then washed and allowed to internalize the dye for 60 min. Cells were imaged by confocal microscopy to visualize GFP-ALP and FM4-64 localization. Arrowhead indicates a vacuole probably generated de novo.
The normal localization of Yck3p to the vacuole limiting membrane suggests a role in regulating transport vesicle targeting/fusion to the vacuole. The established role of Yck3p in controlling vacuole morphology supports this idea because many proteins involved in vacuole morphology also participate in transport vesicle targeting and fusion to the vacuole membrane (Ostrowicz et al., 2008). Through such functions, these proteins are required for both the AP-3 and clathrin-dependent pathways (Bowers and Stevens, 2005). Indeed, our screen identified severe pALP and pCPS accumulation in strains lacking components of the HOPS complex and vacuole fusion machinery (Table 1). However, the selective effects of yck3Δ on pALP transport imply a more specific role for Yck3p in the AP-3 pathway. Yck3p is a palmitoylated, membrane-associated protein that is delivered to the vacuole through the AP-3 pathway (Sun et al., 2004), raising the possibility that sorting into the pathway confers functional specificity. An example of this mechanism was recently reported for the mammalian AP-3 pathway cargo, phosphatidylinositol-4-kinase type IIα (Craige et al., 2008). To test whether Yck3p function in ALP transport requires sorting into the AP-3 pathway, we analyzed ALP maturation in cells expressing mutant forms of Yck3p that carry alterations of the YDSI signal that directs sorting into the AP-3 pathway. Although these mutants are not efficiently sorted into the AP-3 pathway, they still localize to the vacuole membrane, at least in part because of default transport through the endosomal route (Sun et al., 2004). As assessed by immunoblotting, cells expressing these mutants as the sole source of Yck3p displayed no accumulation of pALP (Figure 2D). These data suggest that sorting into the AP-3 pathway is not required for Yck3p function in ALP transport.
ER Export of a Subset of Type II Integral Membrane Proteins Is Delayed in svp26Δ Mutants
In addition to yck3Δ and AP-3 subunit deletions, svp26Δ affected maturation of ALP but not CPS (Table 1). A specific effect of svp26Δ on ALP transport was reported previously in a study that characterized Svp26p as a transmembrane adaptor required for incorporation of ALP into COP-II vesicles at the ER (Bue et al., 2006). In this study, svp26Δ did not affect transport of CPY, a soluble luminal protein, or Gas1p, a GPI-linked membrane protein. A separate study described mislocalization of the Golgi mannosyltransferase Ktr3p to the ER in svp26Δ cells (Inadome et al., 2005), suggesting that Svp26p may serve as an adaptor for multiple proteins. Both ALP and Ktr3p are type II membrane proteins, a feature that could be important for recognition by Svp26p. To further define the specificity of Svp26p, we assessed maturation of ALP and CPS, another type II vacuole membrane protein, in svp26Δ cells by pulse-chase immunoprecipitation. ALP maturation was severely hindered in svp26Δ cells, as expected, with a delay commensurate with that displayed in AP-3–deficient cells (Figure 4A). CPS maturation was unaffected in svp26Δ cells, indicating that not all type II membrane proteins require Svp26p for transport from the ER (Figure 4B).
Figure 4.
Selective defects in type II membrane protein transport in svp26Δ cells. (A) Pulse-chase immunoprecipitation of ALP. Wild-type (WT), apl6Δ (β3Δ), and svp26Δ cells were analyzed by pulse-chase immunoprecipitation of ALP as described in the legend to Figure 2A, except that chase samples were removed at 0, 30, 60, and 90 min. (B) Pulse-chase immunoprecipitation of CPS. Pulse-chase immunoprecipitation of CPS was performed as described in the legend to Figure 2B, except that samples were removed at 0, 20, 40, and 60 min after addition of the chase. (C) Pulse-chase immunoprecipitation of Gda1p. Pulse-chase immunoprecipitation of Gda1p from the indicated strains was performed as described in the legend to Figure 2A, except that samples were removed at 0, 20, 40, and 60 min after addition of the chase. UG, unglycosylated form of Gda1p (does not diminish in size after treatment with endoglycosidase H [data not shown]); ER, endoplasmic core glycosylated form of Gda1p; G, Golgi glycosylated forms of Gda1p (Vowels and Payne, 1998). (D) Subcellular fractionation of svp26Δ cells. Subcellular fractionation was performed as described in the legend to Figure 3. White arrowheads indicate the accumulated, glycosylated form of Gda1p not present in WT extract. Endoglycosidase H treatment reduces all forms of Gda1p (G, Golgi form; ER, endoplasmic reticulum form; UG, unglycosylated form) in both WT and svp26Δ strains to the UG form (data not shown). Fractions were immunoblotted for Gda1p, the ER protein Sec63p, and Kex2p.
We also examined trafficking of another type II membrane protein, the Golgi guanosine diphosphatase Gda1p. In wild-type cells, residence of Gda1p in the Golgi complex allows continued exposure to glycosyltransferases that extend oligosaccharide chains on Gda1p, causing a progressive size increase over time (Vowels and Payne, 1998b; Figure 4C, lanes 1–4). Mutations that prevent ER-to-Golgi transport result in accumulation of a core-glycosylated form of GDPase that does not change size (Vowels and Payne, 1998b). Accordingly, the size of Gda1p over time was monitored in wild-type and svp26Δ cells by pulse-chase immunoprecipitation. Unlike Gda1p in wild-type cells, which gradually increased in size during the chase period (Figure 4C, lanes 1–4), Gda1p in svp26Δ cells did not display a size shift until 40 min after the chase and even at the 60-min chase time exhibited substantial amounts of the ER form (Figure 4C, lanes 5–8). These data are consistent with an impediment in export of Gda1p from the ER in svp26Δ cells.
To more directly assess Gda1p localization, the subcellular distribution of Gda1p was examined in wild-type and svp26Δ strains by differential centrifugation. In wild-type cell extracts, several Gda1p species were detected by immunoblotting, including a major 70-kDa form and minor 67- and 57-kDa forms (Figure 4D, Gda1p, lane 1). Treatment with endoglycosidase H reduced all forms to 57 kDa, indicating that the higher-molecular-weight forms are glycosylated (unpublished data). All Gda1p forms in wild-type cells cofractionated with the Golgi marker Kex2p in P3 (Figure 4D, Gda1p and Kex2p, lane 5). In svp26Δ cell extracts, a new species of ≈60 kDa was detected which corresponded in size to that expected for ER form (Figure 4D, Gda1p, lane 2, arrowhead). Additionally, there was a shift in the ratio of the higher-molecular-weight forms with the 67-kDa form most prominent. The 60-kDa form sedimented with the ER marker Sec63p (Figure 4D, Gda1p and Sec63p, lane 8), whereas the higher-molecular-weight species fractionated mostly in P3. Mislocalization of the 60-kDa Gda1p in svp26Δ cells is consistent with accumulation in the ER, and together with the results of the pulse-chase immunoprecipitation, provides evidence that Gda1p requires Svp26p for export from the ER. This result, combined with the earlier studies, indicates that Svp26p plays a role in transport of only a subset of type II membrane proteins from the ER to the Golgi, including Gda1p, Ktr3p, and ALP but not CPS.
ALP Maturation Is Defective in Vacuole Inheritance Mutants
Our original screen detected both ALP and CPS maturation defects in vac8Δ and vac17Δ strains. VAC8 and VAC17 encode proteins involved in vacuole inheritance during cell division (Weisman, 2006). Vac8p is associated with the vacuole membrane and also participates in cytoplasmic to vacuole transport and homotypic vacuole fusion (Wang et al., 1998, 2001). Vac17p, which acts preferentially in vacuole inheritance, links Vac8p to the type-V myosin Myo2p (Ishikawa et al., 2003). Myo2p directs movement of tubulovesicular vacuole inheritance structures along the actin cytoskeleton from the mother cell vacuole into the newly forming bud in which they seed formation of a new vacuole (Hill et al., 1996). Roles for Vac8p or Vac17p in vacuolar protein maturation and/or vesicle-mediated protein transport have not been reported.
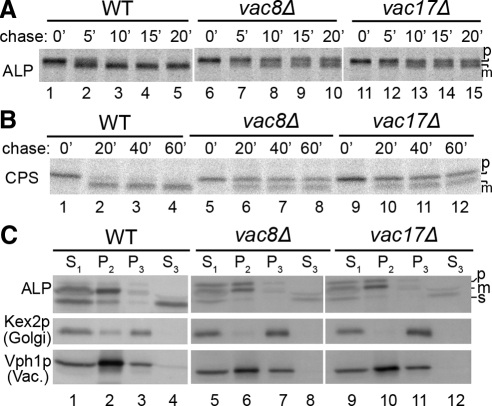
To further evaluate the ALP and CPS maturation defects, vac8Δ and vac17Δ cells were analyzed by pulse-chase immunoprecipitation. The results of this analysis revealed an unusual maturation pattern. In the case of ALP, ∼50% was mature by the 10-min chase point but the level of precursor did not decline substantially at later chase points (Figure 5A). Similar biphasic maturation kinetics were observed for CPS (Figure 5B).
Figure 5.
ALP and CPS maturation is delayed in vac8Δ and vac17Δ cells. (A) Pulse-chase immunoprecipitation of ALP. Pulse-chase immunoprecipitation of ALP from the indicated strains was performed as described in the legend to Figure 2A except that chase samples were removed at 0, 5, 10, 15, and 20 min. (B) Pulse-chase immunoprecipitation of CPS. Pulse-chase immunoprecipitation of CPS was performed as described in the legend to Figure 4B. (C) Subcellular fractionation of vac8Δ and vac17Δ cells. Subcellular fractionation vac8Δ and vac17Δ cells and immunoblotting analysis was performed as described in the legend to Figure 2C.
To determine the subcellular localization of the accumulated pALP in the inheritance mutants, we performed subcellular fractionation of vac8Δ and vac17Δ cells. Unlike pALP in trafficking mutants such as β3Δ and yck3Δ, which fractionated in P3, pALP in vac8Δ, and vac17Δ strains sedimented in P2 with mALP and the vacuolar membrane marker Vph1p (Figure 5C, ALP and Vph1p, lanes 6 and 10). Consistent with these results, GFP-ALP in vac17Δ cells was localized to the vacuolar limiting membrane with FM4-64 (Figure 3). These results provide evidence that pALP in vac8Δ and vac17Δ cells is transported to vacuole membranes but a portion is inefficiently matured.
The inheritance defect in vac8Δ and vac17Δ cells provides a possible explanation for the unusual maturation kinetics of ALP and CPS. Vacuole inheritance mutants can generate vacuoles de novo through an unknown mechanism (Weisman et al., 1987; Gomes De Mesquita et al., 1997). However, such vacuoles do not receive contents from the maternal vacuole that contains the active form of protease A responsible for maturation of ALP and CPS. Consequently, in contrast to inherited vacuoles, de novo-generated vacuoles are not expected to be immediately proteolytically active. Thus, the relatively stable population of pALP and pCPS in vac8Δ and vac17Δ cells may represent those precursors delivered to de novo-formed vacuoles. In contrast, newly synthesized pALP and pCPS that is delivered to the maternal vacuoles is expected to be matured with essentially normal kinetics.
Over time, vacuoles formed de novo can become proteolytically active through autocatalytic activation of proteases A and B that are delivered from the biosynthetic pathway (Woolford et al., 1986; Nebes and Jones, 1991). If the stable population of pALP is indeed present in de novo-generated vacuoles, then pALP should be matured once the vacuole acquires sufficient levels of active protease A. To test this prediction, we monitored maturation of ALP in vac17Δ cells by using a pulse-chase regimen that extended the chase period over 3.5 h (Figure 6). The level of pALP maturation after 30 min reached a plateau that persisted for 90 min (Figure 6, A and B). However, after 2 h ALP processing resumed, and virtually all of the pALP was mature by the 3.5-h time point. A similar result was obtained with vac8Δ cells (unpublished data). These data conform to the model that a percentage of pALP in vac8Δ and vac17Δ cells is transported to proteolytically inactive vacuoles formed de novo in the absence of the inheritance pathway. Our results suggest that the vacuole inheritance pathway is important for the timely biogenesis of functional vacuoles in newly forming daughter cells during cell division.
Figure 6.
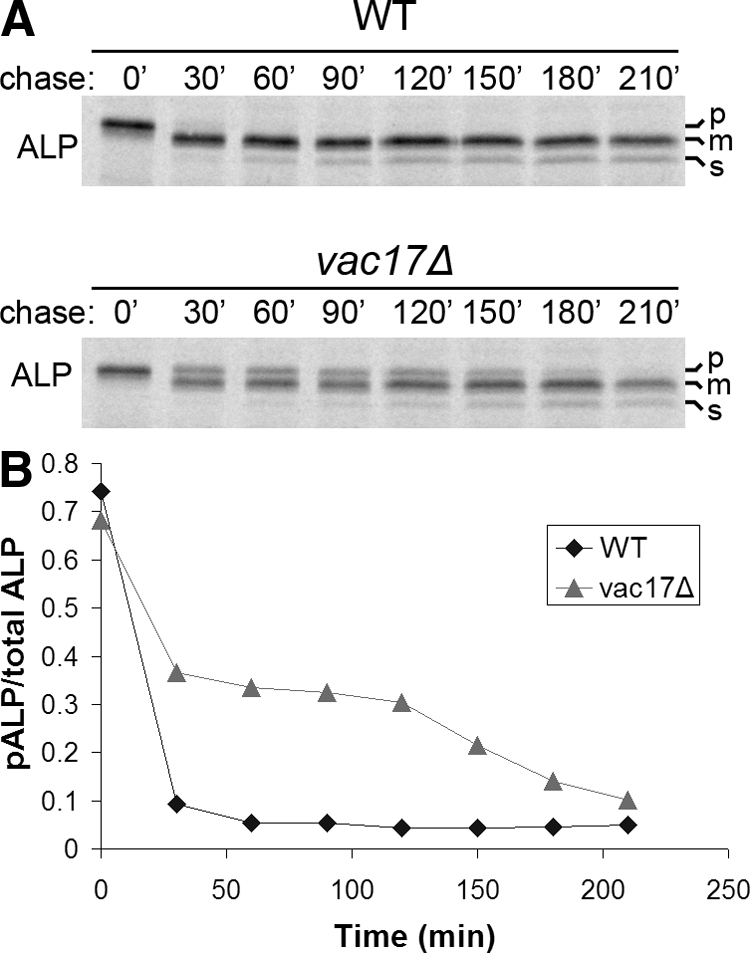
Accumulated pALP in vac17Δ cells matures after 120 min delay. (A) Pulse-chase immunoprecipitation of ALP from WT and vac17Δ cells was performed as described in the legend to Figure 2A except that chase samples were removed at 0, 30, 60, 90, 120, 150, 180, and 210 min. (B) Bands in A were quantified as described in Materials and Methods, and the ratio of pALP to total ALP was calculated.
DISCUSSION
As an approach to identify the full complement of proteins required for transport through the AP-3 pathway in yeast, we have systematically surveyed collections of viable gene knockout strains by using maturation of vacuolar ALP as an indicator of AP-3 pathway function. Among the genes identified through this strategy are those that encode proteins involved in vesicle budding, targeting, and fusion, as well as other processes such as proteolytic processing in the vacuole, organelle acidification, vacuole inheritance, and protein export from the ER. Our study uncovered a selective role for the vacuolar casein kinase Yck3p in AP-3 pathway function, the only protein other than AP-3 that was required specifically for AP-3–dependent traffic.
Yck3p is one of four type I casein kinases in yeast and is unique in localizing to the vacuole (Wang et al., 1996; Sun et al., 2004). Recently, LaGrassa and Ungermann (2005) provided initial insight into Yck3p function, reporting that yck3Δ cells display a defect in maintenance of vacuole fragmentation triggered by hyperosmotic stress. The study presented evidence that the fragmentation maintenance defect results from an up-regulation of vacuole fusion driven by increased tethering activity of the HOPS complex in the absence of Yck3p. Phosphorylation of Vps41p, a HOPS subunit, was demonstrated to depend on Yck3p. Based on these findings, it was proposed that Yck3p-mediated Vps41 phosphorylation inhibits HOPS tethering activity, thereby decreasing vacuole fusion and maintaining the fragmented state in hypertonic conditions. By analogy to the role of Vps41p in AP-3 vesicle formation, the authors also speculated that Vps41p might act in vacuole fragmentation and suggested that phosphorylation could control the distribution of Vps41p functioning in vacuole fragmentation versus fusion. Our finding that yck3Δ cells are defective in transport of ALP to the vacuole is consistent with a role for Vps41p phosphorylation in regulating vesicle formation and, importantly, indicates that Yck3p plays a role under normal growth conditions as well as in response to hypertonic stress.
We envision a model for Yck3p function in the AP-3 pathway that integrates the mechanism proposed to control stress-induced vacuole fusion (LaGrassa and Ungermann, 2005). In this model, Yck3p serves to regulate transition of the bifunctional Vps41p between two functional states—one state involved in AP-3 vesicle formation (phosphorylated) and the other state involved in vesicle targeting/fusion (dephosphorylated) at the vacuole (Figure 7). According to this model, deletion of YCK3 would shift the balance of Vps41p toward targeting/fusion at the vacuole. Consistent with this prediction, yck3Δ cells display increased levels of vacuole-associated Vps41p in vivo under normal growth conditions and more avid association of Vps41p with vacuole membrane in vitro (LaGrassa and Ungermann, 2005). Redistribution of unphosphorylated Vps41p to the vacuole would allow fusion of vesicles from both the AP-3 pathway and the clathrin-dependent pathway but would deplete the pool of Vps41p available to participate in AP-3 vesicle formation, thereby accounting for the specific defect in ALP transport in yck3Δ cells. To determine whether the yck3Δ defect can be overcome simply by increasing levels of Vps41p, ALP maturation was monitored in yck3Δ cells expressing VPS41 from a multicopy plasmid (Supplemental Figure S1). However, the ALP defect persisted in these cells, suggesting that phosphorylation by Yck3p may be required for Vps41p function in AP-3 vesicle formation. Alternatively, Vps41p may function in AP-3 dependent traffic at the Golgi as part of a complex, conceivably HOPS. In this scenario, overexpression of Vps41p without other members of the complex would not increase AP-3–specific Vps41p activity.
Figure 7.
Model for Yck3p function. Vps41p phosphorylation by Yck3p targets Vps41p for function at the Golgi in AP-3–dependent vesicle formation. Unphosphorylated Vps41p functions as a component of HOPS in vesicle docking/fusion at the vacuole membrane. See text for details. Phosphorylated Vps41p may function at the Golgi as part of the HOPS complex although it is pictured acting independently. The site of Vps41p dephosphorylation and release from AP-3 vesicles is unknown.
The apparently stable membrane association of Yck3p imposes a spatial constraint on Yck3p activity that may be important for models of AP-3 pathway-specific function. As an AP-3–dependent cargo, it is possible that the pathway-specific activity of Yck3p occurs during transport through the pathway. However, our results indicate that sorting-defective Yck3p mutants supported wild-type levels of ALP maturation, making it unlikely that Yck3p transport through the AP-3 pathway confers specificity. The Yck3p sorting mutants are reported to localize primarily at the vacuole membrane, probably due to transport through the clathrin-dependent endosomal pathway (Sun et al., 2004). Thus, function of the sorting mutants in ALP transport suggests that the vacuole membrane is the site from which Yck3p provides AP-3 pathway-specific activity. This possibility is compatible with the Vps41p phosphorylation model described in Figure 7—the pool of unphosphorylated Vps41p involved in tethering could serve as substrate for vacuole-localized Yck3p, thereby generating the phosphorylated form of Vps41p proposed to function in AP-3 vesicle formation.
As an approach to address the importance of membrane association in Yck3p function, we assessed ALP maturation in cells expressing a palmitoylation-defective mutant of Yck3p (Sun et al., 2004). This mutant supported normal ALP maturation (Supplemental Figure S1). However, differential fractionation revealed significant but not complete mislocalization to the soluble fraction. Similarly, Vancura et al. (1994) reported that expression of a palmitoylation-defective mutant of the plasma membrane casein kinase I, Yck2p, rescued defects in Yck2p-deficient cells, although the mutant kinase was substantially mislocalized. It seems that, in these cases, a small fraction of properly localized kinase may be sufficient to retain function, or these kinases can provide function from the cytoplasm.
Casein kinase I activity has also been associated with AP-3 function in mammalian cells (Faundez and Kelly, 2000). In this case, however, the β3 subunit of AP-3 was identified as a kinase target. Inhibition of the kinase reduced AP-3 recruitment to synaptic vesicles in vitro and synaptic vesicle budding in vivo. In these studies, the role of Vps41 was not examined. Conversely, the possibility that AP-3 is a target of Yck3p has not been addressed in yeast. A more complete identification of kinase targets and functional tests of phosphorylation site mutants in both yeast and animal cells will be needed to define the regulatory mechanisms and determine the extent of similarity across species.
Our screen also identified Svp26p as a factor required for ALP maturation. During the course of our work, Svp26p was reported to function as an ER receptor for ALP, necessary for sorting ALP into COP II vesicles (Bue et al., 2006). Another group observed that Svp26p interacted with the Golgi glycosyltransferase Ktr3p and svp26Δ caused mislocalization of Ktr3p to the ER, results consistent with a role for Svp26p as an ER export receptor for Ktr3p (Inadome et al., 2005). Our results provide evidence that another Golgi membrane protein, Gda1p, also accumulates in the ER in svp26Δ cells, supporting the view that Svp26p is a general ER cargo receptor, directing multiple cargoes into COP II vesicles. It is currently unknown how Svp26p recognizes cargo. ALP, Ktr3p, and Gda1p do not share significant sequence homology, yet all are type II integral membrane proteins, raising the possibility that membrane topology could constitute part of the binding determinant for Svp26p. However, our observation that CPS, another type II membrane protein, is not affected by svp26Δ indicates that membrane topology is not the sole factor for Svp26p recognition. Importantly, the common topologies of CPS and the three Svp26-dependent proteins should now allow a straightforward chimeric protein approach to map the elements that target cargo to Svp26p. Single homologues of Svp26p are present in a number of eukaryotic species, including humans (Bue et al., 2006). Considering the evolutionary conservation of both Svp26p and COP II proteins, analysis of Svp26p cargo recognition has the potential to provide insight into a fundamental mechanism of protein export from the ER in eukaryotic cells.
During the polarized growth of yeast cells, the vacuole inheritance pathway directs transfer of tubular/vesicular elements of the mother cell vacuole into the emerging daughter bud through an actin and myosin-based mechanism (Hill et al., 1996). This pathway promotes distribution of vacuoles to both mother and daughter cells during cell division. The role of vacuole inheritance is not completely clear, because daughter cells can synthesize vacuoles de novo in vacuole inheritance mutants. Our analysis of the ALP and CPS maturation defects discovered in vac8Δ and vac17Δ cells provides evidence that the vacuole inheritance pathway is important for the timely generation of a proteolytically active vacuole. In these vacuole inheritance mutants we observed that precursor ALP is delivered to vacuole membranes but only a fraction undergoes rapid maturation. Based on these findings, we suggest that precursor vacuolar enzymes are efficiently transported to two populations of vacuoles, one population that is proteolytically active and the other population that becomes active only after several hours. Slowly activating vacuoles would arise in daughter cells through de novo biogenesis of organelles that, because of the inheritance pathway defect, would initially lack active proteases normally transferred from the mother cell vacuole. Without acquisition of preactivated forms of the master processing proteases, generation of a fully active hydrolytic compartment would be delayed until newly synthesized protease zymogens are delivered by the biosynthetic transport pathways and undergo autocatalytic activation. Thus, it seems that the vacuolar proteolytic maturation system, which restricts activation of hydrolase precursors to the appropriate compartment, imposes a significant time delay in de novo formation of active vacuoles. Our study suggests that the inheritance pathway surmounts this problem by seeding daughter cell vacuoles with preactivated maturation proteases and other vacuolar hydrolases from the maternal vacuole.
Our use of a direct screen for ALP maturation in two collections of genome-wide yeast gene deletions identified a total of forty-nine strains with maturation defects. Notably, only five of these mutants exhibited selective defects in post-ER ALP transport, those with deletions of single AP-3 subunit genes and yck3Δ. Does this represent the complete set of factors required exclusively for traffic through the AP-3 pathway in yeast? In addition to our systematic screen for ALP maturation defects, several other genetic approaches have been applied to identify AP-3 pathway-specific proteins. A systematic screen of ∼15% of an independent gene deletion library by immunoblotting did not reveal new AP-3 pathway-specific mutations (Avaro et al., 2002). Emr and colleagues carried out two independent screens of randomly mutagenized yeast for AP-3 pathway mutants (Darsow et al., 2001). These approaches were sufficiently powerful to identify an AP-3 pathway-specific allele of VPS41 but did not yield mutations in genes other than those encoding AP-3. Finally, a systematic screen for mutants with defects in sorting through the endosomal pathway also identified AP-3 mutants, probably due to indirect effects of AP-3 mutations on the endosomal pathway (Bonangelino et al., 2002). In contrast to our work, this study reported mon2Δ as a mutation that caused stronger defects in ALP maturation than in sorting of carboxypeptidase Y through the clathrin-dependent pathway. We specifically tested MATa and MATα haploid mon2Δ strains but did not detect ALP maturation defects by immunoblotting and pulse-chase immunoprecipitation. However, the screen by Bonangelino et al. (2002) was carried out with the library of diploid strains homozygous for each gene deletion. We confirmed that the mon2Δ strain from the homozygous diploid collection accumulates ALP precursor. However, a plasmid expressing wild-type Mon2p that complemented the monensin sensitivity of the mon2Δ diploid strain did not restore ALP maturation, indicating that the ALP maturation defect in this strain is not due to the absence of Mon2p (unpublished data). These results suggest that Mon2p is not an AP-3 pathway-specific factor, a conclusion strongly supported by the recently reported role of Mon2p in Gga protein localization (Jochum et al., 2002).
Considering the combined results of our direct systematic screen together with previous systematic and random screens, it is reasonable to suppose that AP-3 and Yck3p are the only proteins that function preferentially in the AP-3 pathway in yeast. Full AP-3 pathway function in yeast is therefore achieved by a very small set of pathway-specific components acting in concert with factors common to multiple trafficking pathways, some of which, like Vps41p, may be multifunctional with AP-3 pathway-specific activities. The strong evolutionary conservation of both specific and shared elements of the AP-3 pathway in yeast suggests that this set of proteins serves as a core foundation for elaboration of the machinery necessary to accommodate more complicated trafficking patterns in specialized metazoan cells.
Supplementary Material
ACKNOWLEDGMENTS
We are most grateful to Randy Schekman (University of California, Berkeley), Scott Emr (Cornell University), Nicholas Davis (Wayne State University), and Birgit Singer-Krüger (Universität Stuttgart) for gifts of antibodies, strains, and plasmids. We especially thank Giancarlo Costaguta, Mara Duncan, and Esteban Fernandez for helpful insight. This work was supported by National Institutes of Health grants R01 GM-39040 and GM-67911 (to G.S.P.), National Institutes of Health Medical Scientist Training Program T32 GM-08042 (to V.C.A.), and National Institutes of Health National Research Service Award T32 GM-007104 (to L.D.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0819) on December 30, 2008.
REFERENCES
- Avaro S., Belgareh-Touze N., Sibella-Arguelles C., Volland C., Haguenauer- Tsapis R. Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast. 2002;19:351–371. doi: 10.1002/yea.838. [DOI] [PubMed] [Google Scholar]
- Berger A. C., Salazar G., Styers M. L., Newell-Litwa K. A., Werner E., Maue R. A., Corbett A. H., Faundez V. The subcellular localization of the Niemann-Pick Type C proteins depends on the adaptor complex AP-3. J. Cell Sci. 2007;120:3640–3652. doi: 10.1242/jcs.03487. [DOI] [PubMed] [Google Scholar]
- Bonangelino C. J., Catlett N. L., Weisman L. S. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C. J., Chavez E. M., Bonifacino J. S. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Boyd A. Immunoisolation of Kex2p-containing organelles from yeast demonstrates colocalisation of three processing proteinases to a single Golgi compartment. J. Cell Sci. 1993;106:815–822. doi: 10.1242/jcs.106.3.815. [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Piper R. C., Weisman L. S., Stevens T. H. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bue C. A., Bentivoglio C. M., Barlowe C. Erv26p directs pro-alkaline phosphatase into endoplasmic reticulum-derived coat protein complex II transport vesicles. Mol. Biol. Cell. 2006;17:4780–4789. doi: 10.1091/mbc.E06-05-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cooke F. T. Phosphatidylinositol 3,5-bisphosphate: metabolism and function. Arch. Biochem. Biophys. 2002;407:143–151. doi: 10.1016/s0003-9861(02)00487-3. [DOI] [PubMed] [Google Scholar]
- Costaguta G., Duncan M. C., Fernandez G. E., Huang G. H., Payne G. S. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol. Biol. Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Odorizzi G., Payne G. S., Emr S. D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997a;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997b;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Salazar G., Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol. Biol. Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Burd C. G., Emr S. D. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Katzmann D. J., Cowles C. R., Emr S. D. Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol. Biol. Cell. 2001;12:37–51. doi: 10.1091/mbc.12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Klumperman J., Stoorvogel W., Bonifacino J. S. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol. Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Di Pietro S. M., Dell'Angelica E. C. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- Faundez V. V., Kelly R. B. The AP-3 complex required for endosomal synaptic vesicle biogenesis is associated with a casein kinase Ialpha-like isoform. Mol. Biol. Cell. 2000;11:2591–2604. doi: 10.1091/mbc.11.8.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes De Mesquita D. S., Shaw J., Grimbergen J. A., Buys M. A., Dewi L., Woldringh C. L. Vacuole segregation in the Saccharomyces cerevisiae vac2–1 mutant: structural and biochemical quantification of the segregation defect and formation of new vacuoles. Yeast. 1997;13:999–1008. doi: 10.1002/(SICI)1097-0061(19970915)13:11<999::AID-YEA151>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Hill K. L., Catlett N. L., Weisman L. S. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Sommer M., Grynberg M., Kucharczyk R., Rytka J. The CHiPS Domain–ancient traces for the Hermansky-Pudlak syndrome. Traffic. 2005;6:534–538. doi: 10.1111/j.1600-0854.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W. A. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M., Scher C. D., Strovel E., Fitzpatrick D. L., Hartnell L. M., Anikster Y., Gahl W. A. Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr. Res. 2002;51:150–158. doi: 10.1203/00006450-200202000-00006. [DOI] [PubMed] [Google Scholar]
- Inadome H., Noda Y., Adachi H., Yoda K. Immunoisolaton of the yeast Golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments. Mol. Cell. Biol. 2005;25:7696–7710. doi: 10.1128/MCB.25.17.7696-7710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Catlett N. L., Novak J. L., Tang F., Nau J. J., Weisman L. S. Identification of an organelle-specific myosin V receptor. J. Cell Biol. 2003;160:887–897. doi: 10.1083/jcb.200210139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum A., Jackson D., Schwarz H., Pipkorn R., Singer-Kruger B. Yeast Ysl2p, homologous to Sec7 domain guanine nucleotide exchange factors, functions in endocytosis and maintenance of vacuole integrity and interacts with the Arf-Like small GTPase Arl1p. Mol. Cell. Biol. 2002;22:4914–4928. doi: 10.1128/MCB.22.13.4914-4928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu. Rev. Genet. 1984;18:233–270. doi: 10.1146/annurev.ge.18.120184.001313. [DOI] [PubMed] [Google Scholar]
- Jung J., et al. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood. 2006;108:362–369. doi: 10.1182/blood-2005-11-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrassa T. J., Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz T. C., Anand V. C., Payne G. S. High through-put protein extraction and immunoblotting analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2008;457:13–27. doi: 10.1007/978-1-59745-261-8_2. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Hirata A., Ohsumi Y., Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- Nebes V. L., Jones E. W. Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J. Biol. Chem. 1991;266:22851–22857. [PubMed] [Google Scholar]
- Newell-Litwa K., Seong E., Burmeister M., Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J. Cell Sci. 2007;120:531–541. doi: 10.1242/jcs.03365. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Cowles C. R., Emr S. D. The AP-3 complex: a coat of many colours. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Ostrowicz C. W., Meiringer C. T., Ungermann C. Yeast vacuole fusion: a model system for eukaryotic endomembrane dynamics. Autophagy. 2008;4:5–19. doi: 10.4161/auto.5054. [DOI] [PubMed] [Google Scholar]
- Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C., Bayer M. J., Buhler S., Andersen J. S., Mann M., Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Redding K., Seeger M., Payne G., Fuller R. S. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol. Biol. Cell. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P., Darsow T., Katzmann D. J., Emr S. D. Formation of AP-3 transport intermediates requires Vps41 function. Nat. Cell Biol. 1999;1:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- Roberts C. J., Nothwehr S. F., Stevens T. H. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J. Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., Bonifacino J. S. Adaptor-related proteins. Curr. Opin. Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., McCaffery J. M., Emr S. D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J. Cell Biol. 1992b;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. D., Hama H., Sohrabi F., DeWald D. B., Wendland B. PtdIns(3,5)P2 is required for delivery of endocytic cargo into the multivesicular body. Traffic. 2003;4:479–490. doi: 10.1034/j.1600-0854.2003.t01-1-00106.x. [DOI] [PubMed] [Google Scholar]
- Spormann D. O., Heim J., Wolf D. H. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J. Biol. Chem. 1992;267:8021–8029. [PubMed] [Google Scholar]
- Stepp J. D., Huang K., Lemmon S. K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Chen L., Cao W., Roth A. F., Davis N. G. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol. Biol. Cell. 2004;15:1397–1406. doi: 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Oiso N., Gautam R., Novak E. K., Panthier J. J., Suprabha P. G., Vida T., Swank R. T., Spritz R. A. The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc. Natl. Acad. Sci. USA. 2003;100:1146–1150. doi: 10.1073/pnas.0237292100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Vancura A., Sessler A., Leichus B., Kuret J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J. Biol. Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998a;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol. Biol. Cell. 1998b;9:1351–1365. doi: 10.1091/mbc.9.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hoekstra M. F., DeMaggio A. J., Dhillon N., Vancura A., Kuret J., Johnston G. C., Singer R. A. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 1996;16:5375–5385. doi: 10.1128/mcb.16.10.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Catlett N. L., Weisman L. S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Kauffman E. J., Duex J. E., Weisman L. S. Fusion of docked membranes requires the armadillo repeat protein Vac8p. J. Biol. Chem. 2001;276:35133–35140. doi: 10.1074/jbc.M103937200. [DOI] [PubMed] [Google Scholar]
- Weisman L. S. Organelles on the move: insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 2006;7:243–252. doi: 10.1038/nrm1892. [DOI] [PubMed] [Google Scholar]
- Weisman L. S., Bacallao R., Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J. Cell Biol. 1987;105:1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Biol. Cell. 1986;6:2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe J. A., Brodsky F. M., Hofmann K., Lin K., Liu S. H., Chen L., Earnest T. N., Fletterick R. J., Hwang P. K. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature. 1999;399:371–375. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- Yeung B. G., Payne G. S. Clathrin interactions with C-terminal regions of the yeast AP-1 beta and gamma subunits are important for AP-1 association with clathrin coats. Traffic. 2001;2:565–576. doi: 10.1034/j.1600-0854.2001.20806.x. [DOI] [PubMed] [Google Scholar]
- Yeung B. G., Phan H. L., Payne G. S. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.