Abstract
Objectives
Biomedical named entity recognition (BNER) is a critical component in automated systems that mine biomedical knowledge in free text. Among different types of entities in the domain, gene/protein would be the most studied one for BNER. Our goal is to develop a gene/protein name recognition system BioTagger-GM that exploits rich information in terminology sources using powerful machine learning frameworks and system combination.
Design
BioTagger-GM consists of four main components: (1) dictionary lookup—gene/protein names in BioThesaurus and biomedical terms in UMLS Metathesaurus are tagged in text, (2) machine learning—machine learning systems are trained using dictionary lookup results as one type of feature, (3) post-processing—heuristic rules are used to correct recognition errors, and (4) system combination—a voting scheme is used to combine recognition results from multiple systems.
Measurements
The BioCreAtIvE II Gene Mention (GM) corpus was used to evaluate the proposed method. To test its general applicability, the method was also evaluated on the JNLPBA corpus modified for gene/protein name recognition. The performance of the systems was evaluated through cross-validation tests and measured using precision, recall, and F-Measure.
Results
BioTagger-GM achieved an F-Measure of 0.8887 on the BioCreAtIvE II GM corpus, which is higher than that of the first-place system in the BioCreAtIvE II challenge. The applicability of the method was also confirmed on the modified JNLPBA corpus.
Conclusion
The results suggest that terminology sources, powerful machine learning frameworks, and system combination can be integrated to build an effective BNER system.
Introduction
Biomedical named entity recognition (BNER) is a critical component in systems that mine biomedical knowledge embedded in free text 1–10 (e.g., protein–protein interaction or gene–disease association). Approaches to tackling BNER can be categorized into three main types: (1) rule/pattern-based recognition methods characterized by handcrafted name/context patterns and associated rules 11–15 , (2) dictionary lookup methods requiring a list of entity names 16–21 , and (3) machine learning methods utilizing named entity tagged corpora. 22–24 Among them, machine learning methods have achieved promising performance given a large entity tagged corpus. The availability of machine learning software packages (e.g., YamCha 25 or MALLET 26 ) has boosted the baseline performance of BNER systems. Many recent research studies on machine learning-based BNER focus on incorporating unique properties of biomedical terminology sources into the powerful machine learning frameworks, where machine learning algorithms that can accommodate rich domain-oriented features are preferred. 22–24 Another trend of machine learning–based BNER is to combine results from multiple systems. It has been observed that combining accurate and diverse classifiers (a classifier is said to be accurate if it performs better than a random classifier, and two classifiers are considered diverse if they do not make the same classification mistakes) can outperform a single classifier. 27,28 Most of the top BNER systems in the BioCreAtIvE II challenge combine results from multiple classifiers using simple heuristic rules. 24,29–31
In this article, we report our investigation of developing a BNER system, BioTagger-GM, using rich terminology sources, machine learning packages, and the combination of publicly available language processing and BNER tools. We have previously developed a CRF-based tagger during our participation in the BioCreAtIvE II Gene Mention (GM) shared-task challenge. 32 In the current study, we investigated the utility of individual resources considered for this tagger, which include features based on large terminology sources, BioThesaurus and UMLS Metathesaurus. The best configuration exploiting these features was sought to build high-performance systems. We then used system combination to aim further enhancement of such customized high-performance systems. Experiments were conducted on the BioCreAtIvE GM corpus, a collection of MEDLINE excerpts annotated (labeled) with gene/protein names. Our experiments showed that BioThesaurus, 33 an extensive terminology source of genes/proteins, was effective in boosting the performance of machine learning systems. We also demonstrated that publicly available BNER resources can be used to enhance the performance of such systems. In order to confirm that our system was not overly tuned to the BioCreAtIvE II annotation guidelines, we also conducted an experiment using the JNLPBA corpus.
Background
Terminology Sources
We used two terminology sources in our study. The first is BioThesaurus, which contains gene/protein names for all records in the UniProt Knowledgebase (UniProtKB), 34 a knowledgebase of protein sequences and functional annotation maintained by the UniProt Consortium. BioThesaurus version 4.0 consists of 5.8 million unique gene/protein names extracted from 35 underlying resources based on database cross-references provided in the iProClass database, 35 an integrated database providing links to more than 90 biological databases. As described in Liu et al., 33 the construction of BioThesaurus involves extraction of individual gene/protein names from underlying resources and also filtration of nonsensical gene/protein names (e.g., novel protein, fragment, and hypothetical protein).
Our other terminology source was the Unified Medical Language System (UMLS), 36 developed and maintained by the U.S. National Library of Medicine (NLM). It consists of three resources: the Metathesaurus, the SPECIALIST Lexicon, and the Semantic Network. The Metathesaurus is an integrated multi-lingual knowledge source of biomedical vocabularies. The Metathesaurus (2006AD) contains 3.3 million English phrases from 90 underlying sources. In the Metathesaurus, phrases referring to the same concept are designated by a common concept unique identifier (CUI), and each concept is associated with one or more of 135 UMLS semantic types. In this study, we used English entries of the Metathesaurus and associated them with the corresponding semantic types. Another component of the UMLS is the SPECIALIST Lexicon, an English lexicon containing biomedical terms. We used the file LRAGR (the agreement and inflection records) to normalize words. The last component of the UMLS is the Semantic Network, which defines relationships among semantic types. The semantic relation information is not used in the current study.
Machine Learning
We used two machine learning frameworks, Maximum Entropy Markov Models (MEMMs) 37 and Conditional random fields (CRFs). 38 They are probabilistic frameworks applicable to sequential labeling tasks, with the advantage of accommodating many dependent features in predicting a label sequence.
The MEMMs use exponential models to calculate probability of a label for each token, given the label(s) of the previous token(s) and an observation(s). In gene name recognition, observations may be words, word affixes, parts of speech, etc., and labels are often B, I, and O to demarcate gene name occurrences (▶). Based on the probability of labels calculated at each token, the MEMM framework derives the most likely sequence of labels for a sequence of tokens. Similarly to MEMMs, CRFs also use an exponential model, but instead of using a model to calculate probability for each token, they use a model to calculate probability of a label sequence. Both MEMMs and CRFs have achieved excellent performance in sequential labeling tasks, but CRFs have been reported to yield better performance than MEMMs. 38–40
Figure 1.
Demarcation of entity names in text using IOB (IOB2 46 ) labeling. Tokens in text constituting or not constituting entity names (e.g., small GTP-binding proteins Rac1) are marked with the labels B (beginning of a name), I (inside a name), and O (outside of a name).
Early BNER applications of CRFs include a system by McDonald and Pereira 41 and also the ABNER system. 42 McDonald and Pereira 41 used second-order CRFs, in which prediction of a label depends on labels assigned to its neighboring two tokens. Their system achieved one of the highest F-Measures in the gene/protein name recognition shared task (Task 1A) in the BioCreAtIvE I challenge. 22 The ABNER software package facilitates development of BNER systems using the CRF implementation of MALLET. 26 A system derived from ABNER marked a high F-Measure in the BNER shared-task at the JNLPBA workshop 23 (recognition of five types of biological named entities). Among 21 groups that participated in the gene mention (GM) task of the BioCreAtIvE II challenge, more than half of the teams used the CRF framework with three systems achieving the first quartile performance. 24
Labeled Corpora
We used two labeled corpora in this study, the BioCreAtIvE II GM corpus 43 and the JNLPBA corpus, 23 both created from MEDLINE abstracts for shared-task challenge workshops. We designed recognition systems on the training corpus of the BioCreAtIvE II GM corpus, without referencing its test corpus or the JNLPBA corpus. The JNLPBA corpus was reserved to test whether our system was not overly tuned to the problem settings of the BioCreAtIvE II GM corpus, e.g., particular annotation guidelines used in the BioCreAtIvE II GM shared task.
The BioCreAtIvE II GM corpus consists of 20,000 excerpts (sentences) from diverse MEDLINE citations (15,000 excerpts for training and 5,000 for testing). Gene/protein name occurrences in those excerpts were manually annotated. Note that boundaries of gene/protein names in text can be fuzzy even for human annotators. In such cases, alternative annotations were provided with the corpus. The entire GM corpus as well as the tagging results submitted by the participants of the GM task has become publicly available online after the workshop (http://biocreative.sourceforge.net/).
The JNLPBA corpus consists of 22,402 sentences from MEDLINE abstracts (18,546 sentences for training and 3,856 for testing), where five categories of entities (protein, DNA, RNA, cell line, and cell type) are labeled. These abstracts were retrieved from MEDLINE using the MeSH terms human, blood cells, and transcription factors. To test gene/gene-product name recognition systems, we merged three categories, protein, DNA, and RNA, and regarded them as a gene/gene-product type analogous to BioCreAtIvE II GM corpus. Another notable difference of the JNLPBA corpus from the BioCreAtIvE II GM corpus is that sentences are pre-tokenized in the JNLPBA corpus, e.g.,
A decreased number of calcitriol (1 , 25 (OH) 2D3) receptorsGENE has been observed in parathyroid glands of uremic animals .
Unlike BioCreAtIvE II GM corpus, alternative annotations for different name boundaries are not provided in the JNLPBA corpus.
Combining Outputs of Multiple Recognition Systems
Many of the systems with high F-Measures in the BioCreAtIvE II GM task combined outputs from two to three machine learning models. The first place system of the GM task 30 used a set-union of names detected by two regularized linear regression models trained for forward (left-to-right) and backward (right-to-left) parsing of tokens. When names recognized by the two models are partly overlapped, ones with longer spans were kept in the final output. Kuo et al. 29 built two CRF models for forward and backward parsing, and outputs of the models were combined based on scores. Huang et al. 44 combined one CRF model and two models that were trained with one-vs.-one and one-vs.-all multi-class extensions of Support Vector Machine (SVM), a machine learning algorithm that can also accommodate a rich set of features. The final output was a set-union of names detected by the CRF model and names detected by the two SVM models (i.e., set-intersection). Klinger et al. 31 built two CRF models trained with different boundaries of gene names using alternative annotations provided with the corpus, and took the union of the sets. During our participation in BioCreAtIvE II, we also combined multiple recognition systems: (1) a CRF model, (2) a dictionary lookup system based on BioThesaurus, and (3) a model derived with the LingPipe suite of Alias-i, Inc. 45 We took a set-intersection of the latter two, and then took a set-union with the first system. With this combination method, we were successful in improving the recall measure, whereas the improvement of the F-Measure was modest. 32
Methods
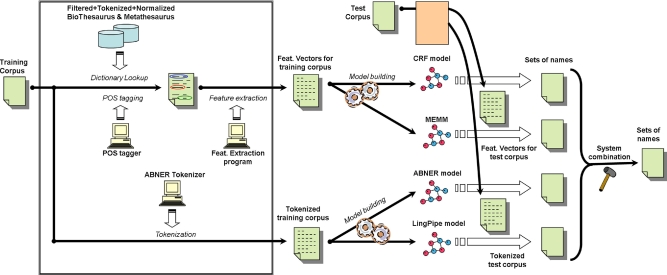
Our previous studies showed that dictionary lookup using BioThesaurus could achieve a very high recall but with a very low precision, and the resulting F-Measure was not comparable with those obtained by machine learning-based BNER systems. 32 In BioTagger-GM, we incorporate dictionary lookup into a machine learning framework where the lookup results are considered as one type of features used by machine learning algorithms. ▶ shows the overall architecture of BioTagger-GM, consisting of four main components: (1) dictionary lookup, (2) machine learning, (3) post-processing, and (4) system combination. Details of each component are shown in the following.
Figure 2.
Training/application of BioTagger-GM.
Dictionary Lookup
BioTagger-GM uses a normalized dictionary lookup method, where both input text and name entries in the dictionaries, BioThesaurus and UMLS Metathesaurus, are normalized. Nonsensical terms in BioThesaurus are ignored during dictionary lookup. Lengthy terms in both dictionaries are also ignored because they are less likely to be found in text. The following summarizes the normalized dictionary lookup method:
1 Tokenize phrases intensively (e.g., GluRdelta2 → Glu R delta 2)
2 Filter out terms that have more than 10 tokens or more than 80 characters (e.g., novel protein similar to vertebrate SH3 and multiple ankyrin repeat domains family protein)
- 3 Normalize text and terms by
- a) Converting tokens to base forms according to the SPECIALIST lexicon (e.g., localisation → localization, thioredoxins → thioredoxin, or flagella → flagellum),
- b) Changing letters to lower case (e.g., Glu R delta 2 → glu r delta 2),
- c) Ignoring punctuation marks, and
- d) Converting digit sequences and Greek alphabets to 9 and G, respectively (e.g., glu r delta 2 → glu r G 9)
4 Identify occurrences of normalized dictionary phrases in input text and associate them with their corresponding BioThesaurus or UMLS Metathesaurus entries.
All phrase occurrences, including overlapping phrases, are recorded during dictionary lookup (see the next subsection).
Machine Learning
We used the CRF and MEMM frameworks implemented in MALLET. To use these implementations, sentences need to be tokenized and specified with features characterizing tokens and token occurrences in their contexts. BioTagger-GM uses a simple tokenization strategy in which sentences are tokenized according to the following symbols: “:”, “/”, “-” as well as parentheses, brackets, and white spaces. Resulting tokens are further split if suffixes are Greek alphabets (e.g., alpha and beta), digit sequences, or one of the following three punctuation marks: “.”, “;”, and “,”. A capitalized letter at the beginning of a sentence (excerpt) is lowercased, and tokens recognized as nouns by the GENIA tagger are converted into their base forms according to the SPECIALIST lexicon. Given tokenized texts, named entity recognition (NER) tasks can be modeled as sequential labeling by associating each token (e.g., word) with an appropriate label to demarcate biomedical entity names (see ▶). 46 Each token is then transformed into a vector of features (▶). Besides features widely considered for NER/BNER, BioTagger-GM includes dictionary lookup results as features. If a phrase in text (a sequence of tokens) can be mapped to a phrase in BioThesaurus (denoted as BioT) or UMLS Metathesaurus (denoted as UMLS), the leftmost token of that phrase in text is assigned with label BioT_B or UMLS_B_SemT, respectively, where B stands for beginning of a mapped phrase and SemT is the corresponding UMLS semantic type. The remaining tokens in the mapped phrase if applicable are assigned with BioT_I or UMLS_I_SemT, where I stands for inside of a name phrase. 46 Note that it is possible that multiple labels are assigned to one token (▶).
Table 1.
Table 1 Features Considered for CRF Models and MEMMs in BioTagger-GM
| Feature Name | Description/Example |
|---|---|
| tokeni | Normalized token at the current position |
| tokeni-1 | Normalized token at the position i-1, if available |
| tokeni-2 | Normalized token at the position i-2, if available |
| tokeni+1 | Normalized token at the position i+1, if available |
| tokenj,j+1 for j=i-2 to i+1 | Normalized token bigrams |
| is tokeni a sub-word | If a token is originated from a consecutive letter sequence such as a hyphenated word, then true, or false otherwise |
| shape of normalized tokeni | Given a token at the position i (tokeni), convert an uppercase letter as ‘X', a lowercase letter as ‘x', a digit sequence as ‘9', and a Greek letter as ‘G'. A sequence of every two to five consecutive ‘X' (‘x') was converted to ‘XXX' (‘xxx') |
| suffix of normalized tokeni (length 4) | If tokeni consists only of alphabets, and its length is greater than 5, extract the last four lowercase alphabets |
| POSi | Part-of-speech for tokeni assigned by the GENIA tagger 52 |
| BioThesaurus labeli | B/I labels (“BioT_{B, I}” or none) indicating mapping of tokeni to a BioThesaurus entry |
| UMLS labeli | B/I labels with semantic type information (UMLS_{B,I}_SemT or none) indicating mapping of tokeni to a token in a UMLS entry |
CRF = conditional random fields; GM = gene mention; MEMM = maximum entropy Markov models.
Figure 3.
Mapping of gene/protein names. Gene phrases mapped to BioThesaurus entries can overlap due to the nested nature of gene/protein names and to the variation of gene/protein name phrases. This figure shows phrase mapping results, and how they are used as machine learning features at each token position in BioTagger-GM (CRF models and MEMMs).
Post-processing
In BioTagger-GM, a post-processing module developed during our participation in the BioCreAtIvE II GM shared task is used without modification. This module includes three parts: (1) correcting boundary errors, (2) exploiting the acquired long and short form information within each sentence, and (3) applying the one-sense-per-discourse heuristics. 47
For the first part of the module, BioTagger-GM uses regular expression patterns to correct name boundary errors. A number of different patterns are used, but representative patterns are summarized below (square brackets indicate name boundaries):
• Separate compound names, coordinated names, and parenthetical expressions, e.g., [DER/Egfr] → [DER] / [Egfr], [PKC alpha and PKC delta] → [PKC alpha] and [PKC delta], and [recombinant human erythropoietin (rhEPO)] → [recombinant human erythropoietin] ( [rhEPO]).
• Extend recognized names to the left when they are immediately preceded by a letter with a hyphen, e.g., m-[Stat5b] → [m-Stat5b].
• Correct unmatched parentheses, e.g., the previously reported [HMGR1 mRNA (HMGR1S mRNA] ) → the previously reported [HMGR1 mRNA] ( [HMGR1S mRNA] ) or [NADPH:cytochrome P450 oxidoreductase] ([P450R) gene] → [NADPH:cytochrome P450 oxidoreductase (P450R) gene].
The second part of the module uses a long-form detection algorithm similar to the one introduced by Schwartz and Hearst. 48 For example, given an excerpt fragment interact with pyruvate kinase (Pk), pyruvate kinase can be detected as the corresponding long form for Pk. If a long form was recognized as a gene/protein name, then the corresponding short form was also marked as a gene/protein name within the same expert, because we assume that they refer to the same entity.
The third part of the module is motivated by the one-sense-per-discourse assumption: different occurrences of one phrase string refer to the same entity within one discourse. 47 In our case, if one occurrence of a phrase is recognized as a gene/protein name, then all of the occurrences of the same phrase should be recognized as gene/protein names within the same excerpt. The module also uses hand-coded regular expression patterns to group phrases differing only by numbers, Greek letters, or single letters (e.g., H2A1, H2A2, and H2As or YAP1 uORF and YAP2 uORF1). For each group, if one member refers to a gene/protein, then the module annotates all other members in the group as genes/proteins.
BNER System Combination
Besides the CRF and MEMM models derived with MALLET, we incorporate two other recognition models derived with existing software packages in BioTagger-GM. These models are a first-order CRF model derived with the ABNER package and a CharLmRescoringChunker model built with the LingPipe suite (a character-based language model in a Hidden Markov Model [HMM] framework with a rescoring mechanism). We use the tokenizer provided in the ABNER package to generate input token sequences. The CharLmRescoringChunker model requires two sets of data to build an HMM model and to implement the rescoring mechanism. We split the training portion of the corpus into 85% and 15% for these purposes, respectively. The n-gram parameter of the model is set to 36. Outputs from these models are passed to the post-processing modules presented in the previous subsection and then combined by voting: A phrase is elected as an entity name through voting if at least two models recognize the phrase as an entity name. 49,50
Experiments and Results
We evaluated BioTagger-GM and its component models over the BioCreAtIvE II GM training corpus (15,000 excerpts) using 5×2-fold cross-validation tests, which had been used by Leaman and Gonzalez 51 in testing their gene recognition system BANNER on the same corpus. Specifically, the training corpus was split into two partitions, one for training and the other for evaluation, and then the roles of the two partitions were switched for another test (i.e., a two-fold cross-validation test). The procedure was repeated five times, and the average precision, recall, and F-Measure of the ten runs were calculated. After the cross-validation tests, we built BioTagger-GM using the entire training corpus (15,000 excerpts), and tested it on the BioCreAtIvE II GM test corpus (5,000 excerpts), which was not referenced during the development of any component system. Results of these experiments are shown in ▶ (cross-validation tests) and ▶ (the final evaluation).
Table 2.
Table 2 Recognition Performance Over the Training Corpus of the BioCreAtIvE II GM Corpus, N = 18,265
| Software | Notes | Precision | Recall | F-Measure |
|---|---|---|---|---|
| BioThesaurus | With all mapping | 0.2253 | 0.8654 | 0.3576 |
| With all mapping + false-positive list | 0.5000 | 0.8541 | 0.6308 | |
| Above w/longest first mapping | 0.6100 | 0.8378 | 0.7059 | |
| ABNER | First-order CRF model | 0.8324 | 0.7246 | 0.7753 |
| With post-processing module | 0.8361 | 0.7493 | 0.7901 | |
| LingPipe | CharLmRescoring with 36-gram | 0.7637 | 0.8204 | 0.7910 |
| With post-processing module | 0.7661 | 0.8364 | 0.7997 | |
| MEMM (MALLET) | Second-order MEMM | 0.8432 | 0.8044 | 0.8233 |
| With post-processing module | 0.8412 | 0.8175 | 0.8291 | |
| CRF (MALLET) | Without BioThesaurus | 0.8621 | 0.7765 | 0.8170 |
| Without post-processing | 0.8718 | 0.8133 | 0.8415 | |
| Without POS | 0.8717 | 0.8138 | 0.8417 | |
| Without UMLS | 0.8660 | 0.8187 | 0.8417 | |
| Without false-positive list | 0.8772 | 0.8109 | 0.8428 | |
| With longest first mapping | 0.8673 | 0.8212 | 0.8436 | |
| The best configuration | 0.8714 | 0.8261 | 0.8481 | |
| BioTagger-GM | Combination of four systems | 0.8658 | 0.8717 | 0.8687 |
GM = gene mention; MEMM = maximum entropy Markov model; CRF = conditional random field.
Reported numbers are averages of performance measures in 5×2-fold cross-validation tests.
Table 3.
Table 3 Recognition Performance over the Test Corpus of the BioCreAtIvE II GM Corpus, N = 6,331
| System | Notes | Precision | Recall | F-Measure |
|---|---|---|---|---|
| Ando | With unlabeled data | 0.8848 | 0.8597 | 0.8721 |
| BANNER | model version 2 | 0.8741 | 0.8277 | 0.8502 |
| ABNER | With post-processing | 0.8590 | 0.7999 | 0.8284 |
| LingPipe | 36-gram with post-processing | 0.7898 | 0.8735 | 0.8295 |
| MEMM | 2nd-order with post-processing | 0.8526 | 0.8379 | 0.8452 |
| CRF | The best configuration | 0.8938 | 0.8496 | 0.8712 |
| BioTagger-GM | Combination of four systems | 0.8821 | 0.8952 | 0.8887 |
GM = gene mention; CRF = conditional random field.
A concern in developing and evaluating recognition systems on one corpus is that the derived systems may be specialized in that particular corpus (e.g., overly tuned to the corpus annotation guidelines of BioCreAtIvE II). To address this issue, we evaluated the systems on the JNLPBA corpus. Namely, we re-trained the systems on the JNLPBA training corpus without adjusting their configurations, and evaluated on its test corpus. However, we had to turn off the tokenization module and also the post-processing module in the BioTagger-GM, which is not applicable to pre-tokenized sentences with extra/artificial blank spaces in text. The results of the experiment are reported in ▶.
Table 4.
Table 4 Recognition Performance over the Test Corpus of the Modified JNLPBA Corpus, N = 6,241
| System | Notes | Precision | Recall | F-Measure |
|---|---|---|---|---|
| ABNER | Without post-processing | 0.6491 | 0.7505 | 0.6961 |
| LingPipe | 36-gram without post-processing | 0.6079 | 0.7166 | 0.6577 |
| MEMM | 2nd-order without post-processing | 0.6668 | 0.7375 | 0.7004 |
| CRF | Configured for BioCreAtIvE | 0.7083 | 0.7702 | 0.7379 |
| BioTagger-GM | Combination of four systems | 0.7058 | 0.8247 | 0.7607 |
GM = gene mention; CRF = conditional random field.
The JNLPBA corpus is different from the BioCreAtIvE II GM corpus in several ways. The corpus and modules were adjusted for these differences, but the taggers were not tuned to the corpus.
In the following sections, we discuss three aspects of the obtained results, BioThesaurus lookup, machine learning, and the generalizability of BioTagger-GM on the JNLPBA corpus.
BioThesaurus Lookup
If we assume all phrases mapped to BioThesaurus are gene/protein names, the recognition performance of such a lookup system can achieve a recall of 0.8654 with a precision of 0.2253 as shown in ▶ (BioThesaurus with all mapping). A large number of false positives were attributed to the following:
• Spurious entries remained in BioThesaurus (e.g., protein, gene, DNA, and amino acid)
• Overlapping mapping (e.g., ▶), which degrades precisions in the given evaluation framework
• Gene/protein names that are also common English words (e.g., Time)
To provide high-quality features to machine learning systems, those likely-false-positive phrases should be filtered from BioThesaurus. In the training portion of the corpus, we identified phrases mapped to BioThesaurus entries that occurred three or more times with 95% of the occurrences being false positives. For example, the five most frequent false positives in the BioCreAtIvE GM training corpus were protein (1,231 times), gene (1,083), dna (679), acid (540), and human (521), whose occurrences were 100% false positives. A phrase gas was 93.3% (42 of 45) false positives (e.g., gas as in gas chromatography vs. gamma-activated sequence abbreviated as gas). Using a false-positive collection compiled in this manner (990 phrases on average during 5×2 cross-validation tests on the BioCreAtIvE II GM training corpus, and 1,528 phrases from the entire training corpus), the F-Measure of the lookup approach was significantly improved (an F-Measure of 0.6308 as in ▶). If we also consider the longest-first mapping without allowing overlaps, the F-Measure was improved to 0.7059 (see also ▶).
Machine Learning
We used MALLET version 0.4 to build CRF models exploiting the features in ▶. The best-identified settings for the BioTagger-GM CRF model were:
• Using a second-order CRF model
• Using overlapping mapping during dictionary lookup (e.g., not longest-first mapping)
• Filtering phrases in a likely-false-positive list (see the preceding section for its derivation) after BioThesaurus lookup
• Applying a post-processing module
▶ shows the performance comparison of the BioTagger-GM CRF model and CRF models that lack one of the above properties. The largest contribution to the performance came from BioThesaurus lookup features. The BioTagger-GM CRF model outperformed the MEMM model, which uses the same features as the CRF model and other models considered in the study. After combining results from the four machine learning models (ABNER, CRF, LingPipe, MEMM), BioTagger-GM outperformed the best F-Measure reported for the BioCreAtIvE II GM task by 1.66%.
Generalizability of BioTagger-GM
On the modified JNLPBA test corpus, BioTagger-GM MEMM and CRF achieved F-Measures of 0.7004 and 0.7379, respectively (▶). BioTagger-GM, combining these and the two other systems, achieved superior performance, an F-Measure of 0.7607, to its constituent systems. These results suggest that CRF and MEMM incorporating BioThesaurus information are generally effective, and also that publicly available recognition systems (ABNER and LingPipe) can be used to boost the recognition performance further. We are aware that ABNER, used in BioTagger-GM, is an advanced version of the third-place system in theJNLPBA workshop. In fact, ABNER achieved good performance (F-Measure of 0.6961) on this corpus, which is comparable to BioTagger-GM MEMM (F-Measure of 0.7004). Thus, the JNLPBA corpus is not totally new to BioTagger-GM for evaluation purposes, but the obtained results would still support the general applicability of the BioTagger-GM.
Discussion
The experiments showed that machine learning features based on BioThesaurus could significantly boost BNER performance. These features were effective when the false-positive list and the overlapping mapping method were used. The use of the false-positive list was shown to improve recall (▶). The improved performance with the overlapping mapping method might be explained by the difficulty in determining entity name boundaries and also by the nested nature of gene/protein names. By allowing overlapping mapping, subtle name boundaries would be determined by machine learning systems.
Although there are not readily generalizable errors that need individual analysis, recognition errors of BioTagger-GM include two major types. The first error type involves short forms, i.e., acronyms, abbreviations, and symbols, which lack distinctive phrase–inner features such as the headwords receptor or the suffix- -dase/nase/lase. For example, it is difficult to recognize Gs (stimulatory G protein alpha subunit) as a gene/protein name in “the third intracellular loop of the V2 receptor is required and sufficient for coupling to Gs.” The second type of errors involves name boundary detection (or the lack of comprehensive annotation of alternative boundaries). For example, when the hand-labeled name was ICS, BioTagger-GM recognized chicken ICS, although there are cases in which organism names may or may not be appropriate as a part of gene/protein names. There are also some cases in which machine learning systems identify a noun phrase spanning a gene/protein name, e.g., IgG subclass profile was recognized by BioTagger-GM, where IgG is the hand-labeled gene/protein name. Note that errors in a particular machine learning system may be overcome by the proposed ensemble method (▶ A and ▶B), but there are name phrases that may be inherently difficult to recognize (or not to falsely recognize) for machine learning systems (▶C). Further investigation is needed in these cases.
Figure 4.
Labeling of different systems. Underlined phrases were identified as genes/proteins by the listed machine learning systems in one run of the 5×2-fold cross-validation tests on the BioCreAtIvE II GM corpus. White text boxes indicate the recognized gene names are true positives, and the dark text boxes (with white fonts) indicate that the recognized gene names are false positives. Note that in the proposed ensemble approach, names recognized by two or more methods are regarded as gene names by the combined system, BioTagger-GM.
During our experiment on the JNLPBA corpus, we observed that the performance of ABNER on this corpus could be significantly improved (an F-Measure of 0.6961 to that of 0.7306) when the pre-tokenized input sentences in this corpus were further tokenized. Performance of the other recognition systems, however, was degraded on such overly tokenized inputs. We need further investigation on these unexpected phenomena, but this implies the significance and subtlety of proper tokenization in boosting BNER performance.
Conclusion
We have presented a gene/protein BNER system, BioTagger-GM, which utilizes extensive terminology sources in the domain and powerful machine learning frameworks implemented in publicly available software packages. We described our approaches to incorporating features based on BioThesaurus in detail, which consists of normalization of name phrases and filtration of false-positive words. The experiments showed that these features based on BioThesaurus contributed much to this improved performance of CRF and MEMM models. BioTagger-GM, which incorporates individual systems through a voting scheme, achieved an F-Measure of 0.8887 on the BioCreAtIvE II GM test corpus, which is higher than that of the top-ranked BNER systems in the BioCreAtIvE II GM challenge. To confirm that the proposed system is not overly tuned to the BioCreAtIvE II GM corpus, we tested BioTagger-GM on the JNLPBA corpus, and the results suggest that this system is still effective, not just on the BioCreAtIvE II GM corpus.
Although BioTagger-GM achieved good performance in these experiments, there were still a number of cases in which this ensemble system failed (e.g., ▶C). Further investigation is necessary to overcome these challenges. One future direction is to utilize unlabeled text during the training of the recognition systems, 30 which had been successfully used in the first-place system in the BioCreAtIvE II GM challenge. We also plan to improve the post-processing modules to handle the types of errors that cannot be currently handled by machine learning systems. Lastly, but importantly, we plan to evaluate BioTagger-GM in real-life problems, such as literature-based database curation projects to see if the BioTagger-GM can improve the efficiency of literature-based curation of genes or proteins.
Acknowledgments
The authors thank those who developed and made available the machine learning packages, natural language processing tools, terminological resources, and labeled corpora mentioned in this study. The authors thank the editor and the anonymous reviewers for their insightful suggestions.
Footnotes
Supported by IIS-0639062 from the National Science Foundation.
References
- 1.Ng SK, Wong M. Toward routine automatic pathway discovery from on-line scientific text abstracts Genome Inform Ser Workshop Genome Inform 1999;10:104-112. [PubMed] [Google Scholar]
- 2.Sekimizu T, Park HS, Tsujii J. Identifying the interaction between genes and gene products based on frequently seen verbs in Medline abstracts Genome Inform Ser Workshop Genome Inform 1998;9:62-71. [PubMed] [Google Scholar]
- 3.Rzhetsky A, Iossifov I, Koike T, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data J Biomed Inform 2004;37:43-53. [DOI] [PubMed] [Google Scholar]
- 4.Craven M. The Genomics of a Signaling Pathway: A KDD Cup Challenge Task SIGKDD Explorations 2003;4:97-98. [Google Scholar]
- 5.Thomas J, Milward D, Ouzounis C, Pulman S, Carroll M. Automatic extraction of protein interactions from scientific abstracts Pac Symp Biocomput 2000:541-552. [DOI] [PubMed]
- 6.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature Pac Symp Biocomput 2000:517-528. [DOI] [PMC free article] [PubMed]
- 7.Proux D, Rechenmann F, Julliard L. A pragmatic information extraction strategy for gathering data on genetic interactions Int Conf Intell Syst Mol Biol 2000;8:279-285. [PubMed] [Google Scholar]
- 8.Humphreys K, Demetriou G, Gaizauskas R. Two applications of information extraction to biological science journal articles: enzyme interactions and protein structures Pac Symp Biocomput 2000:505-516. [DOI] [PubMed]
- 9.Yakushiji A, Tateisi Y, Miyao Y, Tsujii J. Event extraction from biomedical papers using a full parser Pac Symp Biocomput 2001:408-419. [DOI] [PubMed]
- 10.Wong L. PIES, a protein interaction extraction system Pac Symp Biocomput 2001:520-531. [PubMed]
- 11.Fukuda K, Tamura A, Tsunoda T, Takagi T. Toward information extraction: identifying protein names from biological papers Pac Symp Biocomput 1998:707-718. [PubMed]
- 12.Franzen K, Eriksson G, Olsson F, Asker L, Liden P, Coster J. Protein names and how to find them Int J Med Inform 2002;67:49-61. [DOI] [PubMed] [Google Scholar]
- 13.Narayanaswamy M, Ravikumar KE, Vijay-Shanker K. A biological named entity recognizer Pac Symp Biocomput 2003:427-438. [DOI] [PubMed]
- 14.Hanisch D, Fluck J, Mevissen HT, Zimmer R. Playing biology's name game: identifying protein names in scientific text Pac Symp Biocomput 2003:403-414. [PubMed]
- 15.Chang JT, Schutze H, Altman RB. GAPSCORE: finding gene and protein names one word at a time Bioinformatics 2004;20:216-225. [DOI] [PubMed] [Google Scholar]
- 16.Tsuruoka Y, Tsujii J. Improving the performance of dictionary-based approaches in protein name recognition J Biomed Inform 2004;37:461-470. [DOI] [PubMed] [Google Scholar]
- 17.Tsuruoka Y, McNaught J, Tsujii J, Ananiadou S. Learning string similarity measures for gene/protein name dictionary look-up using logistic regression Bioinformatics 2007;23:2768-2774. [DOI] [PubMed] [Google Scholar]
- 18.Koike A, Niwa Y, Takagi T. Automatic extraction of gene/protein biological functions from biomedical text Bioinformatics 2005;21:1227-1236. [DOI] [PubMed] [Google Scholar]
- 19.Kou Z, Cohen WW, Murphy RF. High-recall protein entity recognition using a dictionary Bioinformatics 2005;21(Suppl 1):i266-i273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egorov S, Yuryev A, Daraselia N. A simple and practical dictionary-based approach for identification of proteins in Medline abstracts J Am Med Inform Assoc 2004;11:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mika S, Rost B. NLProt: extracting protein names and sequences from papers Nucleic Acids Res 2004;32:W634-W637(Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh A, Morgan A, Colosimo M, Hirschman L. BioCreAtIvE task 1A: gene mention finding evaluation BMC Bioinformatics 2005;6(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J-D, Ohta T, Tsuruoka Y, Tateisi Y, Collier N. Introduction to the bioentity recognition task at JNLPBA Proceedings of the International Workshop on Natural Language Processing in Biomedicine and its Applications (JNLPBA). 2004. pp. 70-75.
- 24.Smith L, Tanabe LK, Ando RJ, et al. Overview of BioCreative II gene mention recognition Genome Biol 2008;9(Suppl 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo T, Matsumoto Y. Chunking with Support Vector Machines Proceedings of the Second Meeting of the North American Chapter of Association for Computational Linguistics. San Francisco, CA: Morgan Kaufmann; 2001. pp. 192-199.
- 26.McCallum AK. 2002. MALLET: A Machine Learning for Language Toolkithttp://mallet.cs.umass.edu 2001. Accessed: September 10, 2007.
- 27.Dietterich TG. Ensemble methods in machine learning Proceedings of the First International Workshop on Multiple Classifier Systems, Lecture Notes in Computer Science. 1857. New York: Springer-Verlag; 2000. pp. 1-15.
- 28.Chung YS, Hsu DF, Tang CY. On the Diversity-Performance Relationship for Majority Voting in Classifier Ensembles Proceedings of the Seventh International Workshop on Multiple Classifier Systems, Lecture Notes in Computer Science. 4472. New York: Springer-Verlag; 2007. pp. 407-4207th International Workshop on Multiple Classifier Systems (MCS2007). Springer-Verlag, 2007.
- 29.Kuo C-J, Chang Y-M, Huang H-S, et al. Rich Feature Set, Unification of Bidirectional Parsing and Dictionary Filtering for High-F-Score Gene Mention Tagging Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 30.Ando RK. BioCreative II Gene Mention Tagging System at IBM Watson Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 31.Klinger R, Friedrich CM, Fluck J, Hofmann-Apitius M. Named Entity Recognition with Conditional Random Fields Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 32.Liu H, Torii M, Hu ZZ, Wu CH. Gene Mention and Gene Normalization Based on Machine Learning and Online Resources Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 33.Liu H, Hu ZZ, Zhang J, Wu C. BioThesaurus: a web-based thesaurus of protein and gene names Bioinformatics 2006;22:103-105. [DOI] [PubMed] [Google Scholar]
- 34.Bairoch A, Apweiler R, Wu CH, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res 2005;33:D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu CH, Huang H, Nikolskaya A, Hu Z, Barker WC. The iProClass integrated database for protein functional analysis Comput Biol Chem 2004;28:87-96. [DOI] [PubMed] [Google Scholar]
- 36.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology Nucleic Acids Res 2004;32:D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCallum A, Freitag D, Pereira F. Maximum Entropy Markov Models for Information Extraction and Segmentation Proceedings of the Seventeenth International Conference on Machine Learning. San Francisco, CA: Morgan Kaufmann; 2000. pp. 591-598.
- 38.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data Proceedings of the Eighteenth International Conference on Machine Learning. San Francisco, CA: Morgan Kaufmann; 2001. pp. 282-289.
- 39.Grover C, Haddow B, Klein E, et al. Adapting a relation extraction pipeline for the BioCreAtIvE II task. The BioCreAtIvE II Workshop 2007, Madrid, Spain. 2007.
- 40.Sha F, Pereira F. Shallow Parsing with Conditional Random FieldsConference of Human Language Technology and North American Chapter of Association of Computational Linguistics. 2003.
- 41.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields BMC Bioinformatics 2005;6(Suppl 1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text Bioinformatics 2005;21:3191-3192. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe L, Xie N, Thom LH, Matten W, Wilbur WJ. GENETAG: a tagged corpus for gene/protein named entity recognition BMC Bioinformatics 2005;6(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H-S, Lin Y-S, Lin K-T, et al. Hign-Recall Gene Mention Recognition by Unification of Multiple Backward Parsing Models Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 45.Alias-i. 2007. LingPipe 3.1.2. http://alias-i.com/lingpipe/ [computer program]. Accessed: November 1, 2007.
- 46.Sang EFTK, Veenstra J. Representing text chunksNinth Conference of the European Chapter of the Association for Computational Linguistics. 1999. pp. 173-179.
- 47.Gale W, Church K, Yarowsky D. A Method for Disambiguating Word Senses in a Large Corpus Computers and the Humanities 1992;26:415-439. [Google Scholar]
- 48.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text Pac Symp Biocomput 2003:451-462. [PubMed]
- 49.Baumgartner WA, Lu Z, Johnson HL, et al. An integrated approach to concept recognition in biomedical text Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: Centro Nacional de Investigaciones Oncologicas (CNIO); 2007. pp. 257-271.
- 50.Torii M, Liu H. At-least-N voting over biomedical named entity recognition systems. Paper presented at: The Annual Meeting of the ISMB BioLINK Special Interest Group on Text Mining; July 18, 2008; Toronto, Canada.
- 51.Leaman R, Gonzalez G. BANNER: An executable survey of advances in biomedical named entity recognition Pac Symp Biocomput 2008:652-663. [PubMed]
- 52.Tsuruoka Y, Tateishi Y, Kim J-D, et al. Developing a Robust Part-of-Speech Tagger for Biomedical Text Proceedings of the Tenth Panhellenic Conference on Informatics, Lecture Notes in Computer Science. Berlin, Germany: Springer-Verlag; 2005. pp. 382-392.