Little is known about cytogenetic abnormalities in patients with light chain (AL) amyloidosis. The findings of this study suggest that interphase FISH coupled to cytoplasmic staining of specific Ig on bone marrow cells could be useful in light chain (AL) amyloidosis, and that t(11;14) is an adverse prognostic factor in these patients.
Keywords: amyloidosis, cIg-FISH, t(11;14)
Abstract
Background
Light chain amyloidosis is a rare plasma cell dyscrasia. Interphase fluorescence in situ hybridization (FISH) coupled to cytoplasmic staining of specific Ig (cIg-FISH) on bone marrow plasma cells has become well established in the initial evaluation of multiple myeloma, a related disorder. Little, however, is known about cytogenetic abnormalities in patients with light chain amyloidosis.
Design and Methods
We reviewed 56 patients with light chain amyloidosis who had cIg-FISH performed as part of their routine clinical testing using the standard screening panel employed in multiple myeloma at our institution.
Results
Seventy percent of patients had abnormal cIg-FISH, with the most common abnormalities being IgH translocations [48%] – including t(11;14) [39%], and t(14;16) [2%] – and del13/del13q [30%]. No t(4;14) or deletions of 17p (p53) were observed. Patients with t(11;14) had the lowest levels of clonal plasma cells, and those with del13 had the highest. The risk of death for patients harboring the t(11;14) translocation was 2.1 (CI 1.04–6.4), which on multivariate analysis was independent of therapy.
Conclusions
Although preliminary, our data would suggest that cIg-FISH testing is important in patients with light chain amyloidosis and that t(11;14) is an adverse prognostic factor in these patients.
Introduction
Light chain amyloidosis (AL) is a clonal plasma cell disorder whose clinical manifestations are the result of extracellular amyloid fibril deposition in vital organs. Despite its devastating clinical phenotype, based on serum M-protein size and extent of bone marrow plasma cell infiltration, AL is more akin to monoclonal gammopathy of undetermined significance than to multiple myeloma. Given the low clonal plasma cell burden and low proliferative index in AL, little is known about cytogenetic abnormalities in this disorder.
The first report of metaphase analysis in AL was that made by Liang et al. in 1979;1 the one patient studied had a normal karyotype. In 1985, Dewald et al.2 reported metaphase cytogenetic abnormalities in five of 11 patients tested; however, four of the five abnormalities seen were non-specific (absent Y chromosome) or treatment-related (abnormalities of chromosome 7). Only one had a specific abnormality (14q+). The first report of the application of fluorescence in situ hybridization (FISH) coupled to cytoplasmic staining of specific IgH (cIg-FISH) to the evaluation of amyloidosis was reported by Fonseca et al. in 1998.3 Bone marrow samples from 21 patients were studied for numerical abnormalities of chromosomes 7, 9, 11, 15, 18 and X using centromere-specific probes. Trisomies for chromosomes 7, 9, 11, and 15 were observed in one-third to one-half of the patients studied. Monosomy 18 was seen in 72% of cases. Hayman et al. demonstrated that 21 of 29 patients harbored an abnormality of 14q32, with 16 having t(11;14).4 At the same time, Perfetti et al. reported the presence of IgH/MMSET(FGFR3) hybrid transcripts – t(4;14) – in six of 42 patients (14%) with amyloidosis using a reverse transcriptase polymerase chain reaction assay.5 Harrison et al. were the first to report the detection of del13/del13q in amyloidosis by FISH. Ten of 32 patients had del13/del3q, and 11 had 14q32 translocation, of whom nine had t(11;14).6 No patient had t(4;14). Five of the patients with del13/del3q had a concomitant IgH translocation, of whom three had t(11;14).
Over the past 15 years, information about genetic abnormalities in myeloma has been growing exponentially, especially since the advent of FISH.7 Recurrent abnormalities of particular significance in multiple myeloma are t(4;14), t(11;14), t(14;16), del13/del13q, del17p, and hyperdiploidy. The t(11;14) carries a neutral to improved prognosis, while t(4;14), t(14;16), and del17p all portend shortened survival.8,9
The finding of del13/del13q is unfavorable when detected by metaphase cytogenetics but has a less certain significance when detected by interphase FISH.7,10 Hyperdiploidy confers an improved prognosis,11 which may merely be secondary to the association of del13/del13q and IgH translocations with non-hyper-diploidy.12,13
Because of the pathogenetic and prognostic significance of chromosomal abnormalities in multiple myeloma, we sought to define the frequency and significance of abnormalities in AL patients tested in the routine clinical laboratory. Herein, we report our analysis of 56 cases of AL by metaphase cytogenetics and a standard myeloma cIg-FISH panel.
Design and Methods
Patients
Using our Dysproteinemia database, we reviewed all cases of AL amyloidosis seen at our institution who had had cIg-FISH or cytogenetics performed as part of their routine clinical testing. Clinical data were culled from the database and through abstraction by two of the authors (AB and AD). Between March 1, 1998 and October 31, 2006, 515 AL patients were seen, 226 of whom had had their diagnosis made within 30 days of presentation to the Mayo Clinic. During this same interval, metaphase cytogenetic studies had been performed in 339 patients with AL, 68 of whom also had the myeloma cIg-FISH panel ordered in the clinical laboratory. Testing was guided by physician preference rather than clinical picture. Patients who had clear multiple myeloma at any point in their clinical course, as established by lytic bone lesions or infiltrative anemia (n=10), were excluded as were the three patients in whom cIg-FISH testing could not be done due to insufficient plasma cells, yielding the 56 patients who are analyzed herein. Patients whose cIg-FISH was done remote from diagnosis (in 4 patients more than 1 year after diagnosis) or after initiating chemotherapy were not excluded (n=14). Of the seven patients who had more than one cIg-FISH test panel performed, none had additional abnormalities on repeat testing.
Interphase cytogenetic analysis
The cIg-FISH test uses commercially available and in-house chromosome-specific fluorescent-labeled DNA probes for FISH. Bone marrow samples were processed to keep the cytoplasm of the leukocytes intact. Slides were prepared using a cytospin centrifuge. Each probe set was hybridized to a separate hybridization site. Plasma cells were specifically detected by using immunoglobulin staining techniques with commercially available antibodies (cIg) for κ and λ.
Deletions or monosomies of chromosomes 13 and 17 were detected using FISH enumeration strategies employing 13q14 (RB1) and 13q34 (LAMP1) and 17p13.1 (p53). Centromere probes were used to detect chromosomal aneusomies for chromosomes 3, 7, 9, 15 and 17 using probes D3Z1, D7Z1, D9Z1, D15Z4, and D17Z1, respectively. Translocations involving the immunoglobulin heavy chain locus (IGH) were investigated using probes for 14q32 (IGH-XT) and 14q32 (5'IGH,3'IGH). Partner translocations for chromosomes 4p16.3 (FGFR3), 11 (CCND1) or 16q23 (c-MAF) were detected by double fusion FISH strategies.
For each probe set, 100 plasma cells (if possible) were scored. For translocations, at least three abnormal cells had to be present for the sample to be considered positive. For trisomies, at least five cells needed to contain the trisomy to be scored positive. Finally, for monosomies, the number of affected cells required for a positive score depended on the number of probes used for the given chromosome. If two probes were used, as was the case for chromosomes 13 and 14, then only three cells with the monosomy were required for a positive score. In contrast, five cells containing the abnormality were required for all other chromosomes tested to be scored as containing a monosomy. For the purposes of analysis, less than 1% plasma cells in the bone marrow were considered insufficient numbers for testing. This probe set and scoring system constitute the standard screening panel used in multiple myeloma at our institution.
Statistical analyses
Qualitative differences were analyzed by the χ2 and Fisher’s exact tests. p<0.05 was used to indicate statistical significance. Survival was estimated from the time of diagnosis to last follow-up or death. Survival curves were generated with the Kaplan-Meier method and differences analyzed with the log-rank test. A Cox proportional hazards model was used to analyze the interaction of all potential patients’ characteristics with survival. All analyses were performed on JMP statistical software (SAS Institute Inc. Cary, NC, USA).
Results
Patients
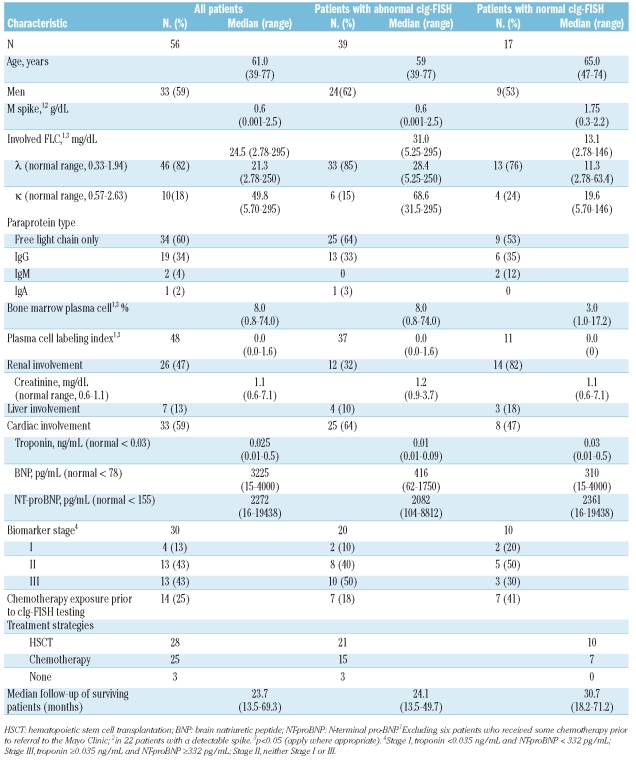
The baseline characteristics of the patients are shown in Table 1. The median age of the cohort was 61 years old, with approximately two thirds being men. As defined by standard criteria,14 at diagnosis 33 patients had cardiac involvement. 26 had renal involvement, and 7 had liver involvement. There was no measurable serum M spike by serum electrophoresis in the majority of patients; rather, the presence of a monoclonal protein was detected by immunofixation and/or serum immunoglobulin free light chains. Forty-six patients had a monoclonal λ clone. The median bone marrow plasma cell percentage of patients was 8.0 (2.0–65.0). The median plasma cell labeling index (PCLI) was low (0%) in the 48 patients in whom it was measured.
Table 1.
Baseline characteristics of study patients upon first presentation at the Mayo Clinic.
Chromosome analysis and interphase cytogenetics
Of the 56 patients who underwent cIg-FISH analysis, 50 had also undergone productive conventional metaphase chromosome karyotyping, only two of whom had abnormal karyotypes. One patient had t(11;14) in one of 30 metaphases, and one patient had a single (non-clonal) complex abnormal karyotype.
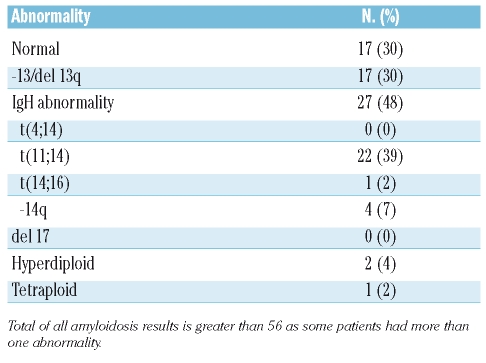
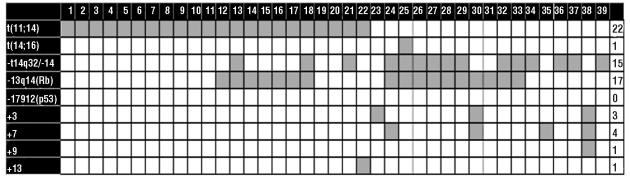
In contrast, 39 patients had abnormal cIg-FISH results (Tables 1 and 2). The abnormalities demonstrated by cIg-FISH included IgH translocations in 27 patients, including 22 with t(11;14) and one with t(14;16), and del13/del13q in 17 patients. No patients had a del17/del17p or t(4;14). More than one cIg-FISH abnormality was detected in 18 patients (Figure 1). Eight patients had del13/del13q with an IgH translocation, with seven of these having t(11;14). The one patient with t(14;16) also had del13/del13q. Twenty of 22 patients with t(11;14) had a third cIg-FISH signal for 11q23 (CCND1).
Table 2.
Presence of chromosomal abnormalities in amyloidosis as detected by cIg-FISH.
Figure 1.
Overlap of cIg-FISH abnormalities. Each column represents an individual patient. Each row indicates a specific abnormality. The gray color represents the presence of a cIg-FISH abnormality.
Survival
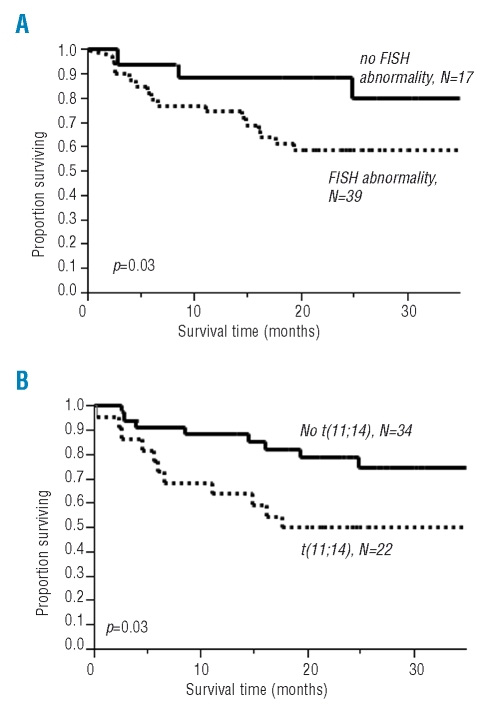
As of July 8, 2008, the median follow-up of surviving patients was 24 months (range, 0.2 to 69 months). Eighteen of 39 patients with abnormal cIg-FISH results had died, as opposed to three of 17 in whom cIg-FISH did not reveal abnormalities (p=0.03) (Figure 2A). Similarly, there was a significant survival disadvantage (HR 2.1, 95% CI 1.04–6.39, p=0.04) for patients harboring the t(11;14) translocation (Figure 2B), but not for those patients with del13/del13q (data not shown). On univariate analysis, survival was also related to cardiac involvement (HR 2.1, 95% CI 0.83–6.1, p=0.11), and there was a trend toward significance for treatment administered, that is, standard chemotherapy versus high dose chemotherapy with peripheral blood stem cell transplant (HR 2.4, 95% CI 0.92–7.07, p=0.07). On Cox multivariate modeling the t(11;14) translocation retained its significance despite the addition of treatment administered.
Figure 2.
Kaplan-Meier survival estimates based on cIg-FISH abnormalities. (A) Kaplan-Meier estimate based on the presence or absence of any cIg-FISH abnormality. (B) Kaplan-Meier estimate based on the presence or absence t(11;14)
Treatment
Despite the fact that the prognostic significance of the presence of t(11;14) was independent of treatment on multivariate analysis, we carefully scrutinized the interventions patients received. Three patients were deemed to ill to tolerate any therapy, 22 received chemotherapy alone, and 28 underwent autologous stem cell transplantation. The patients who underwent chemotherapy alone received a combination of melphalan and steroids with or without thalidomide. The patients who underwent transplantation either had no treatment or a short course of corticosteroids plus or minus melphalan prior to stem cell collection. Overall, of the patients with t(11;14), eight received chemotherapy alone, 12 underwent stem cell transplantation, and two died without receiving any therapy. Among the patients without the t(11;14) translocation, 17 received chemotherapy alone, 16 underwent stem cell transplantation, and one died without receiving therapy. The difference in treatments between patients with or without t(11;14) was not significant (p=0.41)
Relationship of cIg-FISH results to plasma cell burden and other known high risk features
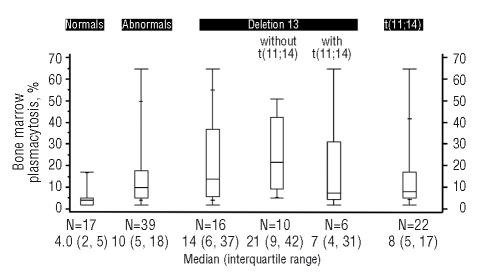
We evaluated whether there was any relationship between bone marrow plasmacytosis and FISH abnormalities (Figure 3). Patients with abnormal FISH results were more likely to have more bone marrow plasma cells (p=0.0007). Among patients with FISH abnormalities, those with t(11;14) had the lowest levels of bone marrow infiltration. Patients with del13 appeared to have the highest bone marrow plasma cell counts. Of the 33 patients seen at the Mayo clinic within 100 days of their diagnosis and in whom the PCLI (a measure of proliferative rate) was evaluated, one patient had a high PCLI, and nine had intermediate PCLI levels. None of these patients had normal FISH. Six had t(11;14), four del13 (three of whom also had an IgH translocation), and three had non-t(11;14) IgH translocations. The relative percentages of elevated PCLI were different across the groups.
Figure 3.
Bone marrow plasmacytosis relative to FISH abnormalities.
The frequency of organ involvement and abnormal cardiac biomarkers was analyzed according to cytogenetic category. There was no significant difference in organ involvement or the levels of either brain natriuretic peptide (BNP) or N-terminal probrain natriuretic peptide (NT-BNP) between patients with normal versus abnormal cytogenetics, with the exception of t(11;14). Among the 36 patients seen within 100 days of amyloid diagnosis, and in whom troponin measurements were performed, there was an association between elevated troponin (greater than 0.035 pg/mL) and t(11;14) (p=0.04). Patients with t(11;14) were more likely to have cardiac involvement with 17/22 having cardiac involvement versus 16/34 patients without the translocation (p=0.02). Renal involvement and liver involvement were not related to the presence of t(11;14).
Discussion
In a cohort of 56 patients with AL whose bone marrow plasma cells were subjected to routine cIg-FISH testing in the clinical laboratory, we demonstrated that 70% of patients harbor the same cytogenetic abnormalities found in myeloma patients. Forty-eight percent of patients had an IgH abnormality, 39% of patients had the t(11;14) translocation, and 30% of patients had del13. The most important finding of this study is the potentially negative prognostic impact that translocation t(11;14) had on survival. On multivariate analysis, the hazard ratio for death for patients with this abnormality was 2.5 times that of the other patients without this abnormality. This finding is distinct from what has been shown for patients with multiple myeloma in whom t(11;14) has been considered either a neutral or favorable prognostic factor. The importance of this finding is mitigated by the retrospective nature of the study, which may introduce unintended bias; however, this observation is novel, and should be investigated in a larger, prospectively collected cohort for confirmation.
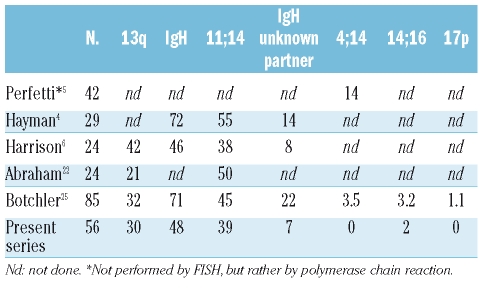
Our cytogenetic findings in AL are similar to those previously published4–6,15,16 (Table 3). Some of the minor differences in results between studies may be explained by different techniques and scoring methodologies used. Considering the distribution of cytogenetic abnormalities among AL patients relative to that among patients with other plasma cell disorders, the most striking differences as compared to multiple myeloma are as follows: hyperdiploidy, 4% versus 50%; t(11;14), 39% versus 15%; del17p, 0% versus 11%; and t(4;14), 0% versus 10–15%.7–9,17–21 The distribution of FISH abnormalities observed in AL is more akin to that seen in patients with monoclonal gammopathy of undetermined significance with the exception of t(11;14), which appears to be more common in AL than in monoclonal gammopathy of undetermined significance (incidence of 15–30%).
Table 3.
Literature comparison of FISH abnormalities in amyloidosis; percentage of patients with genetic abnormalities detected by FISH.
It is unclear whether the apparent gain of 11q13 (CCND1) in association with t(11;14) observed in the present series is a true dose effect, or merely a function of a complex three-way translocation generating the double fusion FISH pattern of 2R2G1F. In this case the total number of 11q13 signals is three, with two by themselves and one involved in a fusion. Regardless of whether it is a true gain of all or part of chromosome 11q or merely heightened expression of CCND1 due to apposition to the IgH promoter, CCND1 has been shown to be significantly upregulated in patients with amyloidosis as compared to in those with multiple myeloma both by gene expression profiling and by confirmatory polymerase chain reaction analysis.22 One could postulate that if pathogenic, this heightened expression of cyclin D1 in AL may manifest in terms of transcription factor regulation or cell cycle promotion.16 Although patients with t(11;14) had the lowest levels of bone marrow plasmacytosis among the group with abnormal FISH findings, they were no more or less likely to have elevated rates of proliferation.
Given the fact that the poor survival experienced by AL patients is the result of end organ damage from relatively low levels of light chain intermediates and fibrils23,24 rather than tumor burden, it is not immediately obvious how differences in plasma cell biology result in worse outcomes. One can speculate that the indolent nature of the t(11;14) clones – and perhaps their relative resistance to chemotherapeutic intervention –may explain our findings. However, only prospective studies will be able to clarify and substantiate our results.
Footnotes
Funding: this research was supported in part by the Hematologic Malignancies Fund of the Mayo Clinic and CA062242.
Authorship and Disclosures
AD, RKP, and AHB designed the research. AHB and RK performed research and collected data. AHB analyzed and interpreted data, performed the statistical analysis, and drafted the manuscript. AD and RKP reviewed the analysis and developed the manuscript. ML, MAG, RF, RAK, PRG, SRZ, JAL, SJR, SR, SRH, and FB reviewed the data and manuscript.
The authors reported no potential conflicts of interest.
References
- 1.Liang W, Hopper JE, Rowley JD. Karyotypic abnormalities and clinical aspects of patients with multiple myeloma and related paraproteinemic disorders. Cancer. 1979;44:630–44. doi: 10.1002/1097-0142(197908)44:2<630::aid-cncr2820440233>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 2.Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66:380–90. [PubMed] [Google Scholar]
- 3.Fonseca R, Ahmann GJ, Jalal SM, Dewald GW, Larson DR, Therneau TM, et al. Chromosomal abnormalities in systemic amyloidosis. Br J Haematol. 1998;103:704–10. doi: 10.1046/j.1365-2141.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 4.Hayman SR, Bailey RJ, Jalal SM, Ahmann GJ, Dispenzieri A, Gertz MA, et al. Translocations involving the immunoglobulin heavy-chain locus are possible early genetic events in patients with primary systemic amyloidosis. Blood. 2001;98:2266–8. doi: 10.1182/blood.v98.7.2266. [DOI] [PubMed] [Google Scholar]
- 5.Perfetti V, Coluccia AM, Intini D, Malgeri U, Vignarelli MC, Casarini S, et al. Translocation t(4;14) (p16.3; q32) is a recurrent genetic lesion in primary amyloidosis. Am J Pathol. 2001;158:1599–603. doi: 10.1016/S0002-9440(10)64115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison CJ, Mazzullo H, Ross FM, Cheung KL, Gerrard G, Harewood L, et al. Translocations of 14q32 and deletions of 13q14 are common chromosomal abnormalities in systemic amyloidosis. Br J Haematol. 2002;117:427–35. doi: 10.1046/j.1365-2141.2002.03438.x. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–58. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–75. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 9.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Franco-phone du Myelome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia. 2007;21:143–50. doi: 10.1038/sj.leu.2404413. [DOI] [PubMed] [Google Scholar]
- 11.Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229–38. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 12.Smadja NV, Leroux D, Soulier J, Dumont S, Arnould C, Taviaux S, et al. Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid cases. Genes Chromosomes Cancer. 2003;38:234–9. doi: 10.1002/gcc.10275. [DOI] [PubMed] [Google Scholar]
- 13.Fonseca R, Debes-Marun CS, Picken EB, Dewald GW, Bryant SC, Winkler JM, et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood. 2003;102:2562–7. doi: 10.1182/blood-2003-02-0493. [DOI] [PubMed] [Google Scholar]
- 14.Gertz MA, Comenzo R, Falk RH, Fermand JP, Hazenberg BP, Hawkins PN, et al. Definition of organ involvement and treatment response in immunoglobulin light chain amyloidosis (AL): a consensus opinion from the 10th International Symposium on Amyloid and Amyloidosis, Tours, France, 18–22 April 2004. Am J Hematol. 2005;79:319–28. doi: 10.1002/ajh.20381. [DOI] [PubMed] [Google Scholar]
- 15.Bochtler T, Hegenbart U, Cremer FW, Heiss C, Benner A, Hose D, et al. Evaluation of the cytogenetic aberration pattern in amyloid light chain amyloidosis as compared with monoclonal gammopathy of undetermined significance reveals common pathways of karyotypic instability. Blood. 2008;111:4700–5. doi: 10.1182/blood-2007-11-122101. [DOI] [PubMed] [Google Scholar]
- 16.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 17.Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA, et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood. 2002;100:1417–24. [PubMed] [Google Scholar]
- 18.Avet-Loiseau H, Facon T, Daviet A, Godon C, Rapp MJ, Harousseau JL, et al. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe Francophone du Myelome. Cancer Res. 1999;59:4546–50. [PubMed] [Google Scholar]
- 19.Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99:2185–91. doi: 10.1182/blood.v99.6.2185. [DOI] [PubMed] [Google Scholar]
- 20.Facon T, Avet-Loiseau H, Guillerm G, Moreau P, Genevieve F, Zandecki M, et al. Chromosome 13 abnormalities identified by FISH analysis and serum β2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood. 2001;97:1566–71. doi: 10.1182/blood.v97.6.1566. [DOI] [PubMed] [Google Scholar]
- 21.Zojer N, Konigsberg R, Ackermann J, Fritz E, Dallinger S, Kromer E, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000;95:1925–30. [PubMed] [Google Scholar]
- 22.Abraham RS, Ballman KV, Dispenzieri A, Grill DE, Manske MK, Price-Troska TL, et al. Functional gene expression analysis of clonal plasma cells identifies a unique molecular profile for light chain amyloidosis. Blood. 2005;105:794–803. doi: 10.1182/blood-2004-04-1424. [DOI] [PubMed] [Google Scholar]
- 23.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–96. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 24.Brenner DA, Jain M, Pimentel DR, Wang B, Connors LH, Skinner M, et al. Human amyloidogenic light chains directly impair cardiomyocyte function through an increase in cellular oxidant stress. Circ Res. 2004;94:1008–10. doi: 10.1161/01.RES.0000126569.75419.74. [DOI] [PubMed] [Google Scholar]
- 25.Bochtler T, Hegenbart U, Cremer FW, Heiss C, Benner A, Hose D, et al. Evaluation of the cytogenetic aberration pattern in amyloid light chain amyloidosis as compared to monoclonal gammopathy of undetermined significance reveals common pathways of karyotypic instability. Blood. 2008;111:4700–5. doi: 10.1182/blood-2007-11-122101. [DOI] [PubMed] [Google Scholar]