T-cell acute lymphoblastic leukemia (T-ALL) is a malignancy of T-cell precursors that mainly occurs in children and adolescents. A variety of oncogenic events that are involved in the pathogenesis of T-ALL have been identified, including NOTCH1 and PTEN mutations, overexpression of TAL1, LYL1 and TLX1, and deletion of CDKN2A (p16).1 Apart from mutations in FLT3 and NRAS, and chromosomal aberrations generating the NUP214-ABL1 fusion, mutations that drive proliferation and survival of T-ALL cells are still unknown in the majority of patients. Recently, activating point mutations in the JAK1 gene were identified in patients with ALL, and rarely also in acute myeloid leukemia (AML) patients.2–4 In T-ALL, JAK1 mutations were identified in approximately 20% of adult T-ALL cases, with a much lower frequency in childhood T-ALL.2 These mutations are very heterogeneous in the sense that they are dispersed over several JAK1 domains, and differ in their ability to transform hematopoietic cells and to activate downstream signaling pathways such as the STAT, PI3K and MAPK cascades.2–4
Leukemia cell lines with mutations in FLT3, JAK2 and NOTCH1 have been described as useful models for pre-clinical testing of small molecule inhibitors.5–8 Given the recent identification of JAK1 mutations in T-ALL, we investigated if JAK1 mutations could be detected in a panel of 18 common T-ALL cell lines. By sequencing of the JAK1 open reading frame at cDNA level in these cell lines, we identified 2 transcript variants, one non-synonymous substitution, as well as several synonymous substitutions (Table 1).
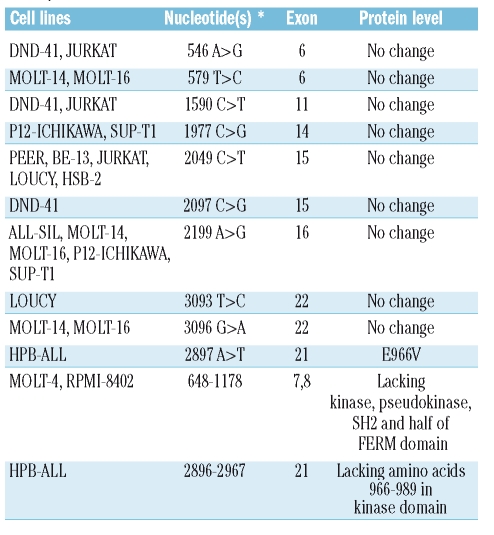
Table 1.
Overview of synonymous and non-synonymous variations detected in the T-cell acute lymphoblastic leukemia cell lines analyzed in this study (nucleotides are numbered relative to the start codon).
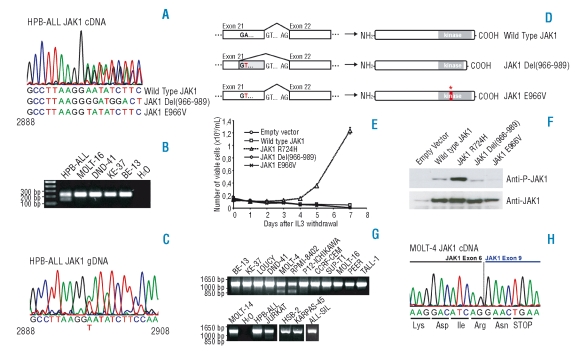
A first transcript variant was identified in the HPB-ALL cell line (Figure 1A, 1B). This transcript lacks nucleotides 2896–2967, encoding amino acids 966–989 that are located between the P-loop and the activation loop in the kinase domain. When sequencing HPB-ALL genomic DNA, we could not detect the presence of a deletion, but we detected a single nucleotide change (2897 A>T) generating a novel GT splice donor site in exon 21 (Figure 1C). As a consequence, the second half of exon 21 is spliced out, resulting in the absence of amino acids 966–989 in the protein. Due to the fact that this splice site is not 100% effective, a normally spliced JAK1 transcript is also present harboring a 2897 A>T nucleotide change with corresponding E966V amino acid change (Figure 1A, 1D).
Figure 1.
Analysis of JAK1 sequence variants detected in T-ALL cell lines. (A) Chromatogram corresponding to the JAK1 transcript in the HPB-ALL cell line indicating the presence of wild type JAK1 and two variant transcripts. (B) RT-PCR performed with primers in exon 21 and 22 confirms the presence of an alternative transcript in HPB-ALL. (C) Genomic DNA sequence of JAK1 in the HPB-ALL cell line reveals a 2897A>T substitution. (D) Scheme illustrating the mechanism for generating the observed transcript variant in the HPB-ALL cell line. (E) Proliferation of Ba/F3 cells expressing the indicated constructs in absence of IL3 (IL3 was removed at day 0). JAK1 Wild Type, R724H, Del(966–989) and E966V cDNA sequences were amplified by PCR from T-ALL cell lines and cloned into the pMSCV-GFP vector. All constructs were verified by sequencing. Data are presented as mean ± st dev. (F) Western blots illustrating that only the JAK1(R724H) mutant displays increased JAK1 phosphorylation as compared to wild type JAK1 when overexpressing the indicated constructs in HEK293T cells. (G) RT-PCR in T-ALL cell lines with primers amplifying the 5’ region of JAK1 reveals the presence of shorter JAK1 transcripts in the MOLT-4 and RPMI-8402 cell lines. (H) Chromatogram representing the sequence of the 850 bp band amplified from cDNA of the MOLT-4 cell line that is shown in part G. of this figure. The chromatogram shows fusion of exon 6 and 9, resulting in frame-shift and premature stop-codon formation.
To examine the functional consequences of the identified variants, the JAK1(Del966–989) and JAK1(E966V) variants, as well as wild type JAK1 and the known activating mutant JAK1(R724H), were expressed in 293T and in interleukin 3 (IL3) dependent Ba/F3 cells. While we could confirm transformation of Ba/F3 cells to IL3 independent proliferation by JAK1(R724H), as was reported by Flex et al., the E966V and Del(966–989) variants were unable to confer IL3 independence to Ba/F3 cells (Figure 1E). Western blot analysis of JAK1 in 293T cells showed that the E966V and Del(966–989) variants displayed weak JAK1 auto-phosphorylation, comparable to wild type JAK1, and in contrast to the strongly phosphorylated R724H mutant (Figure 1F). Furthermore, Western blot analysis of STAT5 phosphorylation in Ba/F3 cells showed increased phospho-STAT5 signal only in the R724H mutant (data not shown). In agreement with these findings, JAK1 phosphorylation in HPB-ALL, the cell line expressing JAK1 E966V and Del(966–989), was not increased as compared to other T-ALL cell lines. JAK1 was found to be expressed in all examined T-ALL cell lines (data not shown). These data show that it remains critically important to test potential mutant forms of kinases to discriminate between driver and passenger mutations, as was also shown recently in the context of FLT3.9
In the MOLT-4 and RPMI-8402 cell lines, we identified another transcript variant, which lacks exons 7 and 8 (nucleotides 648–1178). This results in a shift in the open reading frame with generation of a premature stop codon in exon 9. Consequently, a truncated form of JAK1 is expressed in these cell lines, lacking the entire kinase, pseudokinase and SH2 domain as well as part of the FERM domain (Figure 1G, 1H). Due to the absence of a kinase domain, no further functional studies were performed on this variant.
To investigate if the 18 T-ALL cell lines were dependent on JAK1 signaling for their proliferation and survival, we examined the effect of treatment with a small molecule JAK inhibitor (JAK inhibitor I, Calbiochem, San Diego, CA, USA). This compound inhibited JAK1 autophosphorylation in control cells with an IC50 value of 100 nM (data not shown). The majority of T-ALL cell lines were completely insensitive to treatment with this inhibitor (IC50 values ≥10 μM), while other T-ALL cell lines (ALL-SIL, SUP-T1, DND-41, TALL-1) displayed an increased sensitivity with IC50 values below or around 1 μM. In none of the T-ALL cell lines was proliferation completely inhibited at a concentration of 10 μM of the JAK inhibitor (data not shown). Despite the presence of JAK1 variants in the cell lines HPB-ALL, MOLT-4 and RPMI-8402, these cell lines were not more sensitive to JAK1 inhibition, confirming that the observed variants were unlikely to contribute to JAK1 activation. As a final experiment, we knocked down JAK1 expression in the HPB-ALL and RPMI-8402 cell lines using a JAK1 siRNA (Validated Stealth siRNA, Invitrogen, Carlsbad, CA, USA), which again confirmed that these T-ALL cell lines were not dependent on JAK1 expression for their survival and proliferation (data not shown).
Flex et al. reported association of hyperactive JAK1 mutants with advanced age at diagnosis in ALL patients. Unfortunately, we were not able to identify a T-ALL cell line with an activating mutation in JAK1 that could be used as a T-cell model to study JAK1 signaling and study the effect of JAK1 inhibition. As most of the cell lines we tested correspond to samples from childhood/adolescent T-ALL, the results obtained in this study are in line with the data presented by Flex et al. Furthermore, our observation that concentrations in the range of 1 μM or more of a JAK kinase inhibitor are required to inhibit T-ALL cell lines indicate that T-ALL cell lines in general are not critically dependent on JAK1 activity for their proliferation and survival.
Footnotes
Funding: KDK is an ‘Aspirant’ of the ‘FWO-Vlaanderen’. This work was supported by the Foundation against Cancer, foundation of public interest (SCIE2006-34) and the Interuniversity Attraction Poles (IAP) granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium.
References
- 1.De Keersmaecker K, Marynen P, Cools J. Genetic insights in the pathogenesis of T-cell acute lymphoblastic leukemia. Haematologica. 2005;90:1116–27. [PubMed] [Google Scholar]
- 2.Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–8. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Z, Zhao Y, Mitaksov V, Fremont DH, Kasai Y, Molitoris A, et al. Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood. 2008;111:4809–12. doi: 10.1182/blood-2007-05-090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ, et al. Somatic Mutations of JAK1 and JAK3 in Acute Leukemias and Solid Cancers. Clin Cancer Res. 2008;14:3716–21. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 5.Lierman E, Lahortiga I, Van Miegroet H, Mentens N, Marynen P, Cools J. The ability of sorafenib to inhibit oncogenic PDGFRbeta and FLT3 mutants and overcome resistance to other small molecule inhibitors. Haematologica. 2007;92:27–34. doi: 10.3324/haematol.10692. [DOI] [PubMed] [Google Scholar]
- 6.Pardanani A, Hood J, Lasho T, Levine RL, Martin MB, Noronha G, et al. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21:1658–68. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- 7.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, Bekaert S, et al. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:533–42. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- 9.Fröhling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–13. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]