Abstract
The binding pockets of Cys-loop receptors are dominated by aromatic amino acids. In the GABAA receptor α1Phe65, β2Tyr97, β2Tyr157, and β2Tyr205 are present at the β2/α1 interface and have been implicated in forming an important part of the GABA binding site. Here, we have probed interactions of these residues using subtle chemical changes: unnatural amino acid mutagenesis was used to introduce a range of Phe analogs, and mutant receptors expressed in oocytes were studied using voltage-clamp electrophysiology. Serial mutations at β297 revealed a ∼20-fold increase in EC50 with the addition of each fluorine atom to a phenylalanine, indicating a cation–π interaction between GABA and this residue. This is the first example of a cation–π interaction in loop A of a Cys-loop receptor. Along with previous studies that identified cation–π interactions in loop B and loop C, the result emphasizes that the location of this interaction is not conserved in the Cys-loop family. The data further show that α165 (in loop D) is tolerant to subtle changes. Conversely, mutating either β2Tyr157 (in loop B) or β2Tyr205 (in loop C) to Phe substantially disrupts receptor function. Substitution of 4-F-Phe, however, at either position, or 4-MeO-Phe at β2Tyr157, resulted in receptors with wild-type EC50 values, suggesting a possible hydrogen bond. The molecular scale insights provided by these data allow the construction of a model for GABA docking to the agonist binding site of the GABAA receptor.
Keywords: ligand-gated ion channel, Cys-loop receptor, cation–π interaction, GABAA receptor binding site, unnatural amino acids, homology model
Introduction
The GABAA receptor is a member of the Cys-loop ligand-gated ion channel family, which includes nicotinic acetylcholine (nACh), glycine, and 5-HT3 receptors. These proteins have a pentameric structure, with five subunits associating pseudosymmetrically around an ion-selective pore. Each subunit has an extracellular ligand-binding N-terminal domain containing the eponymous Cys-Cys loop, four transmembrane domains, and a large intracellular loop; the C terminus is extracellular.
Agonists bind at subunit interfaces, and of particular importance are a group of aromatic residues associated with six discontinuous loops, termed A to F (Akabas, 2004). These aromatics are clustered together in the crystal structure of the acetylcholine binding protein (AChBP), which is homologous to the extracellular domain of the GABAA receptor (Brejc et al., 2001). The resulting “aromatic box” is a source of hydrophobicity that occludes water from the binding pocket and provides sites for interactions with ligands, including a cation—π interaction identified in a number of Cys-loop receptors (Table 1). This cation< interaction has been identified between the positive ammonium moiety on the neurotransmitter and an aromatic residue on loop B in the nACh receptor (Zhong et al., 1998), the 5-HT3 receptor (Beene et al., 2002), and the GABAC receptor (Lummis et al., 2005), but on loop C in the MOD-1 receptor (Mu et al., 2003). Because the primary amine of GABA has been shown to form a cation N|gp interaction in the GABAC receptor, it is likely that there is a similar interaction with one of the aromatic residues in the GABAA receptor binding site. In the α1β2 GABAA receptor, candidate residues for a cation—π interaction include α1Phe65, α2Tyr97, α2Tyr157, and α2Tyr205, and, based on our knowledge of the other members of the Cys-loop family and the alignment of their aromatic residues (Table 1), both |gb2Tyr157 (loop B) and α2Tyr205 (loop C) are good candidates for such an interaction.
Table 1.
Cys-loop aromatic box residuesa
| Receptor | Loop A | Loop B | Loop C | Loop D | |
|---|---|---|---|---|---|
| nAChR | Y93 | W149 | Y190 | Y198 | W55 |
| 5-HT3 | E129b | W183 | F226 | Y234 | W90 |
| MOD-1 | C120 | Y180 | Y221 | W226 | F83 |
| GABAC | F138 | Y198 | Y241 | Y247 | Y102 |
| GABAA | Y97 | Y157 | F200 | Y205 | F65 |
aResidues previously identified as contributing to cation N|gp bonds are typed in bold.
bSee Sullivan et al. (2006).
Cation—π interactions can be identified by incorporation of fluorinated aromatics at prospective cation—π sites. The cation—π binding affinity of aromatics is strongly influenced by electrostatics, and addition of electron-withdrawing fluorines around the aromatic ring systematically diminishes the negative electrostatic potential on the face of the ring, and thus the cation—π binding ability (Mecozzi et al., 1996). Here, we use a series of Phe analogs that contain an increasing number of fluorine atoms on their phenyl rings to probe potential cation—π interactions at the aromatic residues that contribute to the α1/β2 GABAA receptor binding site. These Phe analogs were incorporated into GABAA receptors expressed in oocytes using nonsense suppression (Nowak et al., 1995), and GABA EC50 values were measured using two electrode voltage-clamp electrophysiology. EC50 values incorporate both binding and gating parameters and thus do not allow us to determine whether binding or gating (or both) are modified (Colquhoun, 1998). However, these residues have already been extensively investigated by other authors, who suggested they are involved in binding and not gating (Sigel et al., 1992; Amin and Weiss, 1993; Boileau et al., 2002). Changes in EC50 are therefore likely to represent changes in binding parameters.
Materials and Methods
Mutagenesis and preparation of cRNA and oocytes.
Mutant GABAA receptor subunits were developed using pcDNA3.1 (Invitrogen, Abingdon, UK) containing the complete coding sequence for either the human α1 or human β2 GABAA receptor subunit kindly provided by Dr. K. A. Wafford (Merck, Sharp, and Dohme, Harlow, Essex, UK). The codons at positions α165, β297, β2157, and β2205 were replaced by a TAG codon as described previously (Beene et al., 2002). Mutagenesis reactions were performed using the method of Kunkel (1985) and confirmed by DNA sequencing. Harvested stage V–VI Xenopus oocytes were washed in four changes of OR2 buffer (in mm: 82.5 NaCl, 2 KCl, 1 MgCl2, 5 HEPES, pH 7.5), defolliculated in 1 mg/ml collagenase for ∼1 h, washed again in four changes of OR2, and transferred to 70% Leibovitz media (Invitrogen) buffered with 10 mm HEPES, pH 7.5. The following day, they were injected with 5 ng of mRNA produced by in vitro transcription using the mMESSAGE mMACHINE kit (Ambion, Austin, TX) from DNA subcloned into pGEMHE (Liman et al., 1992) as described previously (Reeves et al., 2001; Beene et al. 2004). Electrophysiological measurements were performed 24–72 h after injection.
Synthesis of tRNA and dCA amino acids.
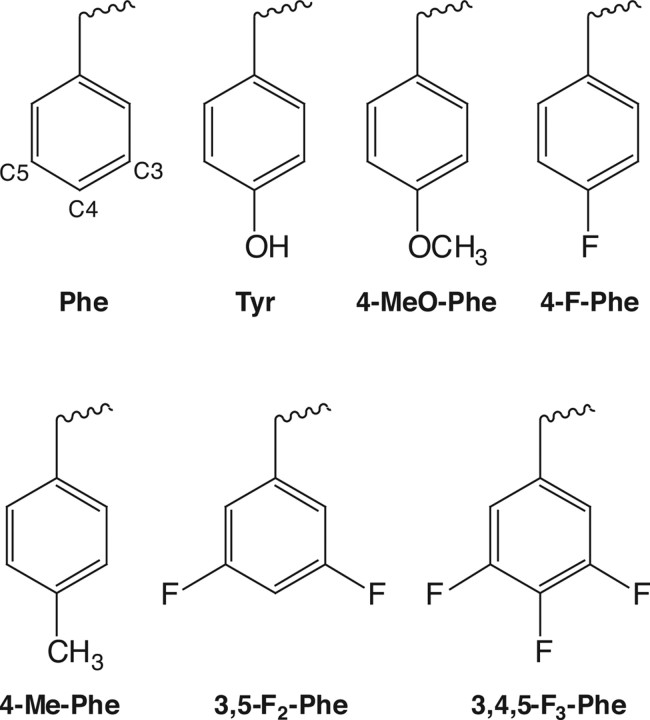
This was as described previously (Beene et al., 2004). Briefly, unnatural amino acids (Fig. 1) were chemically synthesized as NVOC (nitroveratryloxycarbonyl)-protected cyanomethyl esters and coupled to the dinucleotide dCA, which was then enzymatically ligated to 74-mer THG73 tRNACUA as detailed previously (Nowak et al., 1998). Immediately before coinjection with cRNA, aminoacyl tRNA was deprotected by photolysis (Kearney et al., 1996). Typically, 5 ng of total cRNA was injected (1 ng of wild-type α1 or β2 subunit and 4 ng of the corresponding α1 or β2 mutant subunit) with 25 ng of tRNA-aa in a total volume of 50 nl. For a control, cRNA was injected with THG 74-mer tRNA (no unnatural amino acid attached).
Figure 1.
Structures of the natural and unnatural amino acids used in this study. Carbon numbers for C3, C4, and C5 are highlighted on the phenylalanine structure.
The amino acids substituted are shown in Figure 1. Although probing tryptophan or phenylalanine residues via fluorination is straightforward, tyrosine introduces a complication. Fluorination of the aromatic ring of tyrosine substantially lowers the pKa of the ring OH, such that highly fluorinated tyrosines are expected to be ionized at physiological pH. To avoid this complication, we first studied the Tyr-to-Phe mutant, and then introduced fluorinated phenylalanines. Previous studies have validated this strategy (Lummis et al., 2005).
Characterization of mutant receptors.
Peak GABA-induced currents were recorded at 22−25°C from individual oocytes using the Opus-Xpress system (Molecular Devices, Union City, CA). GABA (Sigma, St. Louis, MO) was stored as 100 mm aliquots at −80°C, diluted in ND96 buffer (in mm: 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, 5 HEPES, pH 7.5) and delivered to cells via the automated perfusion system of the OpusXpress. Glass microelectrodes were backfilled with 3 m KCl and had a resistance of ∼1 MΩ. The holding potential was −60 mV. To determine EC50 values, GABA concentration–response data were fitted using PRISM software (GraphPad, San Diego, CA) to the four-parameter logistic equation, Y = Imin + (Imax − Imin)/1 + 10(logEC50 − [A])nH, where Imax is the maximal peak current and nH is the Hill coefficient, and with the concentration values entered as the molar concentration in scientific notation. One-way ANOVA was performed with a Dunnett's post test to determine statistical significance using the same PRISM software.
Modeling the GABA A receptor and docking of GABA.
The GABAA receptor α1 and β2 subunit sequences were aligned to the AChBP subunit sequence using FUGUE (Shi et al., 2001), a program that matches predicted secondary structures as well as amino acid characteristics for improved accuracy of alignment (Forrest et al., 2006). The loop A portion of this FUGUE alignment can be seen in Figure 2, along with an alternative version in which a 2 aa space has been imposed manually to align the β297 residue with the nACh receptor αTyr93. Manipulating the A loop improved the alignment of both 5-HT3 and GABAA receptors (Cromer et al., 2002; Sullivan et al., 2006). A three-dimensional model of the extracellular region of the GABAA α1/β2 receptor was then built using MODELLER (Sali and Blundell, 1993) based on the crystal structure of AChBP in the HEPES-bound state (Protein Data Bank ID 1i9b). GABA was docked into the GABA binding site using GOLD, version 2.2 (Jones et al., 1995). Twenty GOLDScore algorithm runs were performed to dock GABA with flexibility or rotation of the carboxyl tail permitted. The program was run with a population size of 100 and the number of generations set to 100,000. The active site cavity was defined as 10 Å radius from the α carbon of β2Tyr97, a hydrogen bond was defined with a limit of 2.5 Å and a van der Waals interaction at 4.5 Å. The docking of GABA was performed in two ways: (1) with no constraints and (2) by orientating the primary amine of GABA toward C4 carbon of β2Tyr97 by limiting the distance between the two atoms to 3.5 Å.
Figure 2.
Alignment of the A loops in the Cys-loop family. Residues underlined are Tyr89 of AChBP, αTyr93 of the nACh receptor, Glu129 of the 5-HT3 receptor, and β2Tyr97 of the GABAA receptor. The GABAA β2 (alternative) sequence has 2 aa spaces added, aligning β2Tyr97 with αTyr93 of the nACh receptor.
Results
Functional characterization of α1β2 GABAA receptors
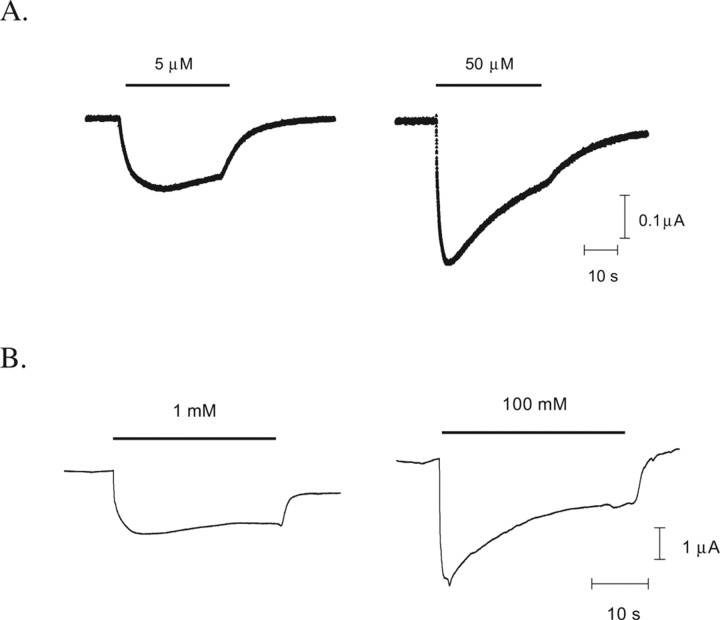
All mutant mRNAs, when coinjected into Xenopus oocytes with tRNA-Tyr (β297, β2157, and β2205) or tRNA-Phe (α165) molecules, produced functional receptors that responded to application of GABA with EC50 values and Hill coefficients similar to each other, and to previously published values (Boileau et al., 2002). This indicates that the wild-type phenotype was successfully “rescued.” Typical responses are shown in Figure 3. No currents in response to application of high concentrations of GABA (100 mm) were detected from oocytes injected with mRNA alone or with mRNA and tRNA not ligated to dCA-aa (THG73 74-mer tRNA).
Figure 3.
GABA responses. Example responses from α1β2 wild-type receptors (A) and α1β2 Phe157 mutant receptors (B) at EC50 and maximal [GABA].
Functional characterization of α1Phe65 mutations
Incorporation of phenylalanine at position α165 (loop D) through nonsense suppression reproduced the wild-type GABA EC50 (Table 2). The addition of a fluorine to the phenyl ring produced an increase in EC50 from 3.0 to 11 μm, but three fluorines (3,4,5-F3-Phe) did not further increase the EC50 (11.5 μm). There were no significant differences in Hill coefficients across the group.
Table 2.
Functional characterization of α165 mutant GABAA receptors
| Mutant | LogEC50 ± SEM | EC50 (μm) | nH | n |
|---|---|---|---|---|
| α1β2 | −5.457 ± 0.04 | 3.5 | 1.2 | 4 |
| α165-Phe β2 | −5.520 ± 0.09 | 3.0 | 1.2 | 4 |
| α165-4-F-Phe β2 | −4.962 ± 0.06a | 11 | 1.4 | 3 |
| α165-3,4,5-F3-Pheβ2 | −4.941 ± 0.14a | 11.5 | 1.5 | 3 |
aDenotes logEC50 values statistically significantly different from wild type by one-way ANOVA; p < 0.01.
Functional characterization of β2Tyr97 mutations
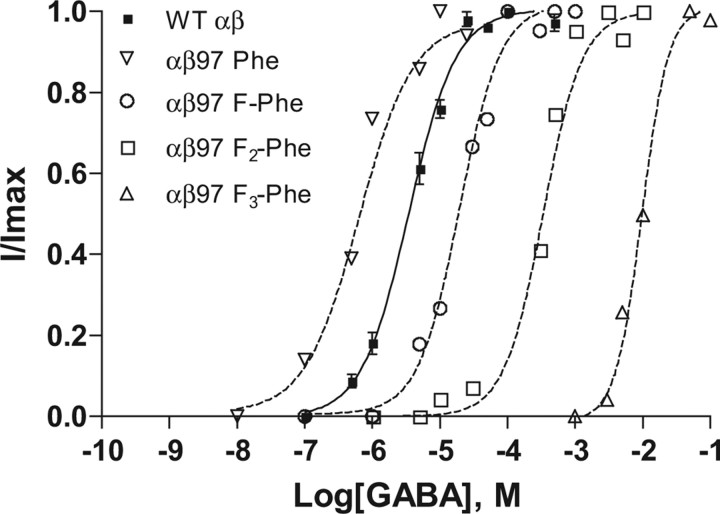
Phe functions well at position β297, giving an EC50 that is actually sixfold lower than wild type (Table 3). When referenced to the Phe variant, introduction of a single F resulted in a 33-fold increase in EC50 to 20 μm for β297-4-F-Phe. An additional 22-fold increase in EC50 to 420 μm (700-fold from Phe) was observed for β297-3,5-F2-Phe and a final 24-fold increase for β297-3,4,5-F3-Phe to EC50 9900 μm (Fig. 4). Hill coefficients were not significantly different from wild type.
Table 3.
Functional characterization of β297 mutant GABAA receptors
| Mutant | LogEC50 ± SEM | EC50 (μm) | EC50mut/EC50phe | nH | n |
|---|---|---|---|---|---|
| α1β2 | −5.457 ± 0.04 | 3.5 | 1.2 | 4 | |
| α1β297-Phe | −6.190 ± 0.16a | 0.6 | 1 | 1.4 | 4 |
| α1β297-4-F-Phe | −4.706 ± 0.07a | 20 | 33 | 1.2 | 5 |
| α1β297-3,5-F2-Phe | −3.372 ± 0.05a | 420 | 700 | 1.1 | 4 |
| α1β297-3,4,5-F3-Phe | −2.002 ± 0.04a | 9900 | 16,500 | 1.9 | 5 |
aDenotes logEC50 values statistically significantly different from wild type by one-way ANOVA; p < 0.01.
Figure 4.
Concentration–response curves for β297 mutant receptors. Normalized responses of individual oocytes expressing receptors incorporating fluorinated phenylalanine analogs at β2Tyr97. The curves were generated using the four-parameter logistic equation as described in Materials and Methods. Data are mean ± SEM; n = 3–6 oocytes. WT, Wild type.
A cation–π interaction between GABA and β2Tyr97
The systematic increase in EC50 with the addition of fluorines to the phenyl ring of receptors modified at position β297 indicates a cation–π interaction. A “fluorination” plot (Zhong et al., 1998) of the cation–π binding energy [determined as described by Mecozzi et al. (1996)], and the EC50 values compared with the parent molecule is shown in Figure 5; the direct relationship between these parameters shows there is a cation–π interaction here.
Figure 5.
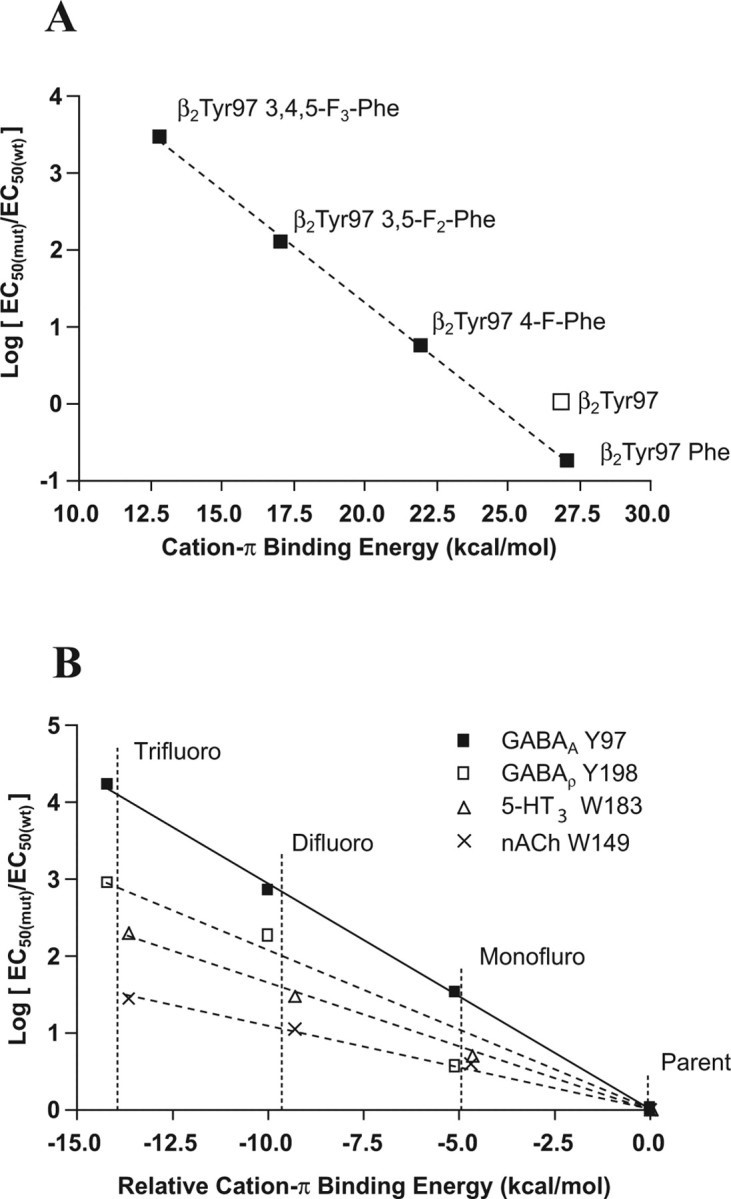
Fluorination plots. A, Plot of the β2Tyr97 mutant logEC50 values from Table 3 normalized to the wild-type logEC50 and fit by a line of best fit. B, Comparison of the cation–π interactions of the nACh, 5-HT3, GABAC, and GABAA receptors. The x-axis is offset so that the parent, nonfluorinated amino acid (Trp for nACh and 5-HT3 and Phe for GABAC and GABAA) is given a value of zero. The actual values are 32.6 kcal/mol for Trp and 26.9 kcal/mol for Phe. The values are fitted by a line of best fit to aid comparison. The gradient for the MOD-1 receptor is almost identical with that for 5-HT3 and so is removed here for clarity.
In principle, the fluorination plot of Figure 5A could signal a cation–π interaction between β2Tyr97 and some cationic moiety other than GABA. There are four positively charged amino acids within 10 Å of β2Tyr97 that could perform this role, and without detailed structural information it is difficult to exclude any of them. However, two of these, β2Lys102 and β2Lys103, face away from the binding site, suggesting they are unlikely partners for β2Tyr97, whereas interaction with α1Arg132 would change the structure of the binding site such that GABA could not access the aromatic box. The remaining positively charged amino acid is β2Arg207, which has previously been proposed to interact with the carboxylate of GABA (Wagner et al., 2004), and would therefore exclude it from contributing to a cation–π interaction at the other end of the molecule. Because all full GABA agonists have a charged amine, it is not possible to use an alternative agonist to provide definitive proof, but given the arguments described above, combined with the precedence of the ligand as the source of the cation in other Cys-loop receptors, we believe that GABA is the source of the cation.
Figure 5B presents fluorination plots for the current study and for some other members of the Cys-loop family. Small, focused ions provide the strongest electrostatic interactions, and this is illustrated by the fluorination plots; the nACh receptor has the weakest interaction as a result of the cation of ACh being a quarternary ammonium. 5-HT and GABA both have primary ammonium ions and therefore are expected to have stronger cation–π interactions. β2Tyr97 has the steepest gradient indicating the strongest cation–π interaction found to date.
Functional characterization of β2Tyr157 mutations
Removal of the hydroxyl group from β2Tyr157 (producing Phe) caused a 400-fold increase in the GABA EC50 from 3.5 to 1400 μm (Table 4). Introduction of a fluorine or methoxy group at position C4, however, resulted in near wild-type EC50 values (3 and 5 μm, respectively), whereas a methyl group at this position resulted in an intermediate EC50 (150 μm). Introducing 3,5-F2-Phe increased the EC50 120-fold to 420 μm, but β2157-3,4,5-F3-Phe increased the EC50 by only 30-fold. Hill coefficients were not significantly different from wild type.
Table 4.
Functional characterization of β2157 mutant GABAA receptors
| Mutant | LogEC50 ± SEM | EC50(μm) | EC50mut/EC50wt | nH | n |
|---|---|---|---|---|---|
| α1β2 | −5.457 ± 0.04 | 3.5 | 1.2 | 4 | |
| α1β2157-Phe | −2.846 ± 0.10a | 1400 | 400 | 0.9 | 6 |
| α1β2157-4-F-Phe | −5.514 ± 0.15 | 3.0 | 0.86 | 1.2 | 6 |
| α1β2157-3,5-F2-Phe | −3.371 ± 0.04a | 420 | 120 | 2.0 | 4 |
| α1β2157-3,4,5-F3-Phe | −3.950 ± 0.08a | 110 | 32 | 1.7 | 4 |
| α1β2157-4-MeO-Phe | −5.294 ± 0.10 | 5 | 1.4 | 1.2 | 3 |
| α1β2157-4-Me-Phe | −3.817 ± 0.15a | 150 | 43 | 0.8 | 3 |
aDenotes logEC50 values statistically significantly different from wild type by one-way ANOVA; p < 0.01.
Functional characterization of β2Tyr205 mutations
Substitutions at position β2Tyr205 (Table 5) showed some similarity to those at β2Tyr157. There was an increase of 24-fold in EC50 when this tyrosine was replaced by phenylalanine, but a wild-type EC50 when 4-F-Phe was substituted. Replacement with 3,5-F2-Phe yielded a similar EC50 to phenylalanine, as did introduction of β2205-3,4,5-F3-Phe. There were no significant differences in Hill coefficients.
Table 5.
Functional characterization of β2205 mutant GABAA receptors
| Mutant | LogEC50 ± SEM | EC50(μm) | EC50mut/EC50wt | nH | n |
|---|---|---|---|---|---|
| α1β2 | −5.457 ± 0.04 | 3.5 | - | 1.2 | 4 |
| α1β2205-Phe | −4.082 ± 0.04a | 80 | 24 | 1.4 | 5 |
| α1β2205-4-F-Phe | −5.810 ± 0.08 | 1.5 | 0.43 | 1.9 | 3 |
| α1β2205-3,5-F2-Phe | −4.174 ± 0.06a | 70 | 20 | 1.9 | 4 |
| α1β2205-3,4,5-F3-Phe | −4.039 ± 0.05a | 90 | 26 | 2.5 | 4 |
aDenotes logEC50 values statistically significantly different from wild type by one-way ANOVA; p < 0.01.
A GABAA receptor model
Images from the GABAA receptor model developed as described in Materials and Methods are shown in Figure 6. The four aromatic residues studied here have been displayed; their faces are oriented together forming the aromatic box (Fig. 6A). The model was created using the alternative loop A alignment (Fig. 2), which exposed the face of β2Tyr97 to the binding pocket. Figure 6B shows a possible hydrogen bond formed between the β2Tyr157 hydroxyl and the backbone amide of α1Thr130.
Figure 6.
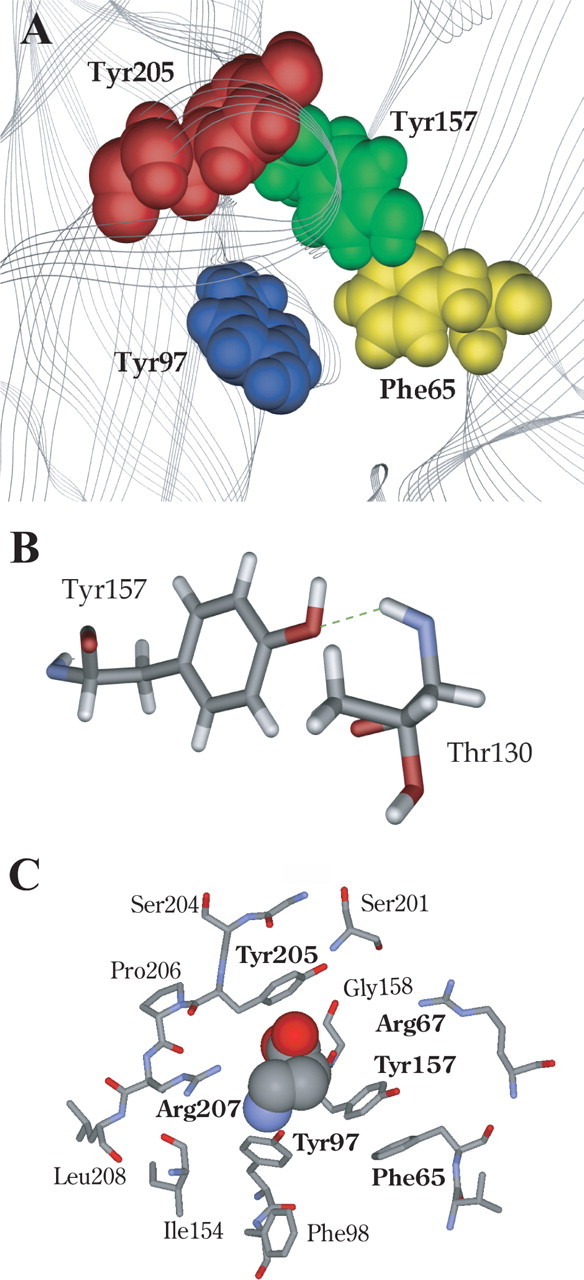
Model of the GABA binding site aromatic box. A, Model showing the positions of the aromatic box residues found at the GABA binding site between β2α1. B, Hydrogen bond formation between β2Tyr157 and α1Thr130. C, A GABA molecule docked into the aromatic pocket. Those residues discussed are shown in bold.
In docking simulations, the small size of GABA allows it to adopt many energetically favorable orientations in the binding pocket. This apparently promiscuous behavior arises primarily because current docking programs are relatively crude; they do not recognize some interactions (e.g., cation–π interactions) and most do not allow complete flexibility of side chains. It is therefore essential that some constraints, based preferably on experimental data, are used to compensate for these inadequacies. Initial studies docking GABA with no constraints revealed four distinct GABA orientations: 10% of the results located the NH4+ close to β2Tyr97, 10% located the NH4+ between β2Tyr157 and β2Tyr205, 10% located the NH4+ between β2Tyr157 and β2Tyr205, with the COO− over β2Tyr97, and 70% located the NH4+ toward the base of β2Tyr157 and the COO− toward loop C (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Thus, when unrestricted, GABA predominantly faces β2Tyr157 and/or β2Tyr205, which is not supported by the data. We therefore allowed the molecule to find an energetically favorable orientation when its ammonium was close to β2Tyr97, as implied by the fluorination data. These data (Fig. 6C) indicate that the ammonium head lies over the aromatic ring of β2Tyr97, whereas the carboxyl tail is directed upwards between β2Tyr157 and β2Tyr205 toward α1Arg67.
Discussion
The GABAA receptor binding site has been intensively investigated using a range of techniques, including mutagenesis, radioligand binding assays, and photoaffinity labeling (Sigel et al., 1992; Amin and Weiss, 1993; Smith and Olsen, 1994; Boileau et al., 1999, 2002). These techniques have implicated many amino acids that may be important in the binding site, but they cannot identify the chemical-scale interactions of each amino acid with the neurotransmitter. Here, we used unnatural amino acid mutagenesis combined with functional studies to probe the effects of subtle chemical modifications to aromatic amino acids that form a critical part of this binding site. The results indicate that the aromatic at position β297 contributes a cation–π interaction, the aromatics at positions β2157 and β2205 require electronegative groups at their C4 positions to function efficiently, and the aromatic at position α165 is relatively insensitive to subtle chemical changes. The roles of these residues are discussed further below, and also considered in relation to a model of the GABA binding pocket.
β2Tyr97 forms a cation–π interaction with GABA
Cysteine accessibility studies previously showed that β2Tyr157 lies in the binding pocket: Y97C was protected from covalent modification by the presence of GABA or the GABAA receptor antagonist gabazine (SR95531) (Boileau et al., 2002). These studies further suggested that this residue is involved specifically in GABA binding, because the Y97C mutation had the same relative effect (∼100-fold increase) on the GABA EC50, which incorporates both binding and functional parameters, as on the SR95531 IC50, which involves a purely binding phenomenon.
Our data show a strong correlation between the cation–π binding ability of tyrosine derivatives incorporated at β297 and logEC50 (Fig. 5A). The size of this effect and the systematic dependence on the number of fluorines unambiguously establish a cation–π interaction at this site. Given the precedent from other Cys-loop receptors, we propose that, when GABA binds to the GABAA receptor, the primary ammonium ionic moiety of the agonist interacts with the π-orbital of β2Tyr97. This cation–π interaction is the strongest of these relationships found in the Cys-loop family so far (Fig. 5B). The differences in the 5-HT3, GABAC, and GABAA receptor fluorination plot slopes may reflect the significance of this interaction within each of the binding sites.
Thus, the GABAA receptor becomes the first instance of a Cys-loop receptor that displays a cation–π interaction with a loop A residue. Loop A does contain an aromatic box residue in the nACh receptor, αTyr93, and in AChBP crystal structures this Tyr aligns with Tyr89, which is clearly located at the “bottom” of the aromatic box. Presumably, β2Tyr97 of the GABAA receptor plays the role of αTyr93 and Tyr89, but the alignment is problematic; a gap equivalent to 2 aa is required to bring the β2Tyr97 into alignment with the nACh receptor and AChBP Tyrs (Fig. 2). Loop A contains a highly conserved WxPDxxxxN motif, which plays a crucial role in positioning the rest of the binding site, particularly loop B (Lee and Sine, 2004; Cashin et al., 2005). It is therefore surprising that a gap insertion is necessary in a conserved region with such an important structural role, but the mutagenesis results are compelling.
The location of a cation–π interaction on loop A, contrasting to loops B or C for other members of the Cys-loop family, supports previous suggestions that the “lock-and-key” metaphor is not appropriate for such receptors (Mu et al., 2003). If they required a precise protein–ligand lock-and-key interaction, we would expect related receptors to use cation–π interactions at a conserved location in the three-dimensional structure. This is not the case in the 5-HT-gated 5-HT3 and MOD-1 receptors, in which the cation–π interactions occur on loop B and loop C, respectively (Beene et al., 2002; Mu et al., 2003). We now find similar variability in the ionotropic GABA receptor family; the cation–π interaction moves from loop B in GABAC (Lummis et al., 2005) to loop A in the GABAA receptor (the present study). These are very similar receptors, showing 39% sequence identity and 64% homology between GABAA(β2) and GABAC(ρ1) in the extracellular domain. Nevertheless, GABA binds with different orientations in the two receptors. Apparently Cys-loop receptors require only that a ligand occupies the general binding region defined approximately by the aromatic box.
Contributions of α1Phe65
There is no cation–π interaction at α165: addition of 4-F to the phenyl ring increased GABA EC50 slightly, but addition of more fluorines around the ring resulted in no additional increase. This residue has been previously reported to play a role in GABA binding: mutation of α1Phe64 (α1Phe65 by human numbering) to Leu increased the GABA EC50 from 6 to 1260 μm, with the IC50 values of bicuculline and SR95531 increasing by similar amounts (Sigel et al., 1992). This suggests that the effects of mutations at this position are attributable to disruption of binding rather than gating, and that an aromatic is preferred here, although our data indicate that α1Phe65 is tolerant to small chemical changes. Our model indicates that α1Phe65 is located on the “right-hand” face of the aromatic box (see below), and does not form significant chemical interactions with any neighboring residues or the GABA molecule. In fact, it is partially obscured by β2Tyr157and thus may solely contribute to the general hydrophobicity of the region.
β2Tyr157 and β2Tyr205
Previous data suggest that these two tyrosine residues specifically participate in GABA binding: phenylalanine mutations at both these sites significantly increased EC50 values, whereas activation of the receptors using pentobarbital, which binds at a location distinct from the GABA binding site, resulted in no change in functional response (Amin and Weiss, 1993). Similarly, we observed that removal of the OH (introducing Phe) is highly deleterious at both β2Tyr157 and β2Tyr205, resulting in ∼400- or 24-fold increases in EC50, respectively. Wild-type behavior can be rescued at β2Tyr157 by incorporation of 4-F or 4-MeO substituents, whereas 4-Me-Phe or multiply fluorinated phenylalanines increased EC50 values 35- to 120-fold. These data suggest that the OH of β2Tyr157 acts as a hydrogen bond acceptor. The strong penalty for removing the OH and the near wild-type behavior of 4-MeO-Phe support this analysis. In this light, the near wild-type behavior for 4-F-Phe is perhaps surprising. Fluorine is the most electronegative element, and as such it is reluctant to donate a lone pair of electrons to a hydrogen bond donor. As a result, organic fluorine (fluorine bonded to a carbon) hardly ever accepts a hydrogen bond, especially if an alternative, better acceptor can be accessed (Dunitz, 2004). Inspection of the region around β2Tyr157 in the receptor model we have developed (see below) suggests that there are no alternative hydrogen bond acceptors close to the hydroxyl group within the protein. In such a case, even the very poor acceptor of 4-F-Phe may be better than nothing at all (Phe). The model suggests β2Tyr157 could form a hydrogen bond with the backbone NH of α1Thr130 (Fig. 6B), and thereby support the architecture of this region of the binding pocket and help to expose β2Tyr97 for the cation–π interaction. Support for this hypothesis comes from previous modeling studies that also propose that β2Tyr157 forms a hydrogen bond with α1Thr130 (Cromer et al., 2002). The data for β2Tyr205 show no clear pattern, although there is an indication that an electronegative atom at C4 is favored.
A GABAA receptor model
The identification of a cation–π interaction between the ammonium ionic moiety of GABA and the side chain of β2Tyr97 provides an excellent starting point for locating GABA accurately in the binding site. Orientating the amine of GABA close to β2Tyr97 fits well with the probable location of the carboxyl tail of GABA close to α1Arg67 and β2Arg207. Wagner et al. (2004) showed that β2Arg207 affects GABA binding and unbinding without changing channel gating, and hypothesized that the arginine side chain directly contacts the carboxyl group of the GABA molecule in conjunction with α1Arg66 (α1Arg67 by human numbering) (Holden and Czajkowski, 2002). If the amine group of GABA is associated with β2Tyr97, the carboxyl tail is located appropriately to form such a contact with β2Arg207 and α1Arg66. This association is supported when we dock GABA into the model of the GABAA receptor: the carboxyl tail of GABA lies between α1Arg67 and β2Arg207 (Fig. 6C).
In conclusion, a cation–π interaction between GABA and a tyrosine on loop A has been identified by unnatural amino acid mutagenesis. This is the first example of a cation–π interaction with a loop A residue in a Cys-loop receptor, and along with previous studies that identified cation–π interactions with loop B and loop C residues, emphasizes the importance of this type of interaction in the binding of neurotransmitter. These results also further establish that the exact location of the cation–π interaction in the binding site is not critical, because it is not conserved among closely related members of the Cys-loop family.
Footnotes
This work was supported by the Wellcome Trust (S.C.R.L. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science), Merck Sharpe and Dohme, the Biotechnology and Biological Sciences Research Council (a CASE studentship to C.L.P.), and the National Institutes of Health (NIH) (NS11756; NS34407). A.P.H. is a recipient of NIH Predoctoral Trainee Grant ST32GMO7616.
References
- Akabas MH. GABAA receptor structure-function studies: a reexamination in light of new acetylcholine receptor structures. Int Rev Neurobiol. 2004;62:1–43. doi: 10.1016/S0074-7742(04)62001-0. [DOI] [PubMed] [Google Scholar]
- Amin J, Weiss DS. GABAA receptor needs two homologous domains of the beta-subunit for activation by GABA but not by pentobarbital. Nature. 1993;366:565–569. doi: 10.1038/366565a0. [DOI] [PubMed] [Google Scholar]
- Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- Beene DL, Price KL, Lester HA, Dougherty DA, Lummis SC. Tyrosine residues that control binding and gating in the 5-hydroxytryptamine3 receptor revealed by unnatural amino acid mutagenesis. J Neurosci. 2004;24:9097–9104. doi: 10.1523/JNEUROSCI.2429-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau AJ, Evers AR, Davis AF, Czajkowski C. Mapping the agonist binding site of the GABAA receptor: evidence for a β-strand. J Neurosci. 1999;19:4847–4854. doi: 10.1523/JNEUROSCI.19-12-04847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau AJ, Newell JG, Czajkowski C. GABAA receptor β2 Tyr97 and Leu99 line the GABA-binding site. Insights into mechanisms of agonist and antagonist actions. J Biol Chem. 2002;277:2931–2937. doi: 10.1074/jbc.M109334200. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Cashin AL, Petersson EJ, Lester HA, Dougherty DA. Using physical chemistry to differentiate nicotinic from cholinergic agonists at the nicotinic acetylcholine receptor. J Am Chem Soc. 2005;127:350–356. doi: 10.1021/ja0461771. [DOI] [PubMed] [Google Scholar]
- Colquhoun D. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol. 1998;125:924–947. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer BA, Morton CJ, Parker MW. Anxiety over GABAA receptor structure relieved by AChBP. Trends Biochem Sci. 2002;27:280–287. doi: 10.1016/s0968-0004(02)02092-3. [DOI] [PubMed] [Google Scholar]
- Dunitz JD. Organic fluorine: odd man out. Chembiochem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden JH, Czajkowski C. Different residues in the GABAA receptor α1T60-α1K70 region mediate GABA and SR-95531 actions. J Biol Chem. 2002;277:18785–18792. doi: 10.1074/jbc.M111778200. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- Kearney PC, Nowak MW, Zhong W, Silverman SK, Lester HA, Dougherty DA. Dose-response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Sine SM. Invariant aspartic acid in muscle nicotinic receptor contributes selectively to the kinetics of agonist binding. J Gen Physiol. 2004;124:555–567. doi: 10.1085/jgp.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Lummis SC, L Beene D, Harrison NJ, Lester HA, Dougherty DA. A cation-pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Mecozzi S, West AP, Jr, Dougherty DA. Cation-pi interactions in aromatics of biological and medicinal interest: electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci USA. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Lester HA, Dougherty DA. Different binding orientations for the same agonist at homologous receptors: a lock and key or a simple wedge? J Am Chem Soc. 2003;125:6850–6851. doi: 10.1021/ja0348086. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Kearney PC, Sampson JR, Saks ME, Labarca CG, Silverman SK, Zhong W, Thorson J, Abelson JN, Davidson N, Schultz PG, Dougherty DA, Lester HA. Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Gallivan JP, Silverman SK, Labarca CG, Dougherty DA, Lester HA. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- Reeves DC, Goren EN, Akabas MH, Lummis SC. Structural and electrostatic properties of the 5-HT3 receptor pore revealed by substituted cysteine accessibility mutagenesis. J Biol Chem. 2001;276:42035–42042. doi: 10.1074/jbc.M106066200. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Sigel E, Baur R, Kellenberger S, Malherbe P. Point mutations affecting antagonist affinity and agonist dependent gating of GABAA receptor channels. EMBO J. 1992;11:2017–2023. doi: 10.1002/j.1460-2075.1992.tb05258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GB, Olsen RW. Identification of a [3H]muscimol photoaffinity substrate in the bovine gamma-aminobutyric acidA receptor alpha subunit. J Biol Chem. 1994;269:20380–20387. [PubMed] [Google Scholar]
- Sullivan NL, Thompson AJ, Price KL, Lummis SCR. Defining the roles of Asn-128, Glu-129 and Phe-130 in loop A of the 5-HT3 receptor. Mol Membr Biol. 2006;23:442–451. doi: 10.1080/09687860600831539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DA, Czajkowski C, Jones MV. An arginine involved in GABA binding and unbinding but not gating of the GABAA receptor. J Neurosci. 2004;24:2733–2741. doi: 10.1523/JNEUROSCI.4316-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]