Abstract
AtTCP20 is a transcription factor belonging to the Arabidopsis (Arabidopsis thaliana) TCP-P subfamily, characterized by its capacity to bind to site II motifs (TGGGCY). Our aim was to understand the role of AtTCP20 in plant development. The expression pattern of a translational fusion of PromTCP20:CDS20∷GUS∷GFP suggested a function for AtTCP20 in several plant organs and stages of development. The role of AtTCP20 was challenged in planta by inducing expression of AtTCP20 proteins fused with either a transcriptional activator domain (VP16) or a repressor domain (EAR). Expression of both modified proteins led to severe developmental phenotypes. In-depth analysis suggested that AtTCP20 may participate in the regulation of cell expansion, cell division, and cell differentiation. Gene expression profiling in roots and hypocotyls revealed that 252 genes were down-regulated in both organs after induction of the AtTCP20∷EAR repressor gene. Site II motifs (TGGGCY) were underrepresented in their promoters. Conversely, GG(A/T)CCC sequences related to binding sites identified for TCP proteins in rice (Oryza sativa) were overrepresented, and a TCP20 fusion protein was shown to bind to these sequences in vitro. Gene ontology indicated that many targeted genes were involved in cell wall biogenesis and modification during expansion and also encoded numerous transcription factors controlling plant development. Our results are consistent with the previous proposal that AtTCP20 is involved in cell division and growth coordination. Moreover, they further suggest that AtTCP20 also contributes to cell expansion control and indicate a different involvement of this protein in plant morphogenesis depending on the organ and the developmental stage.
TCP transcription factors constitute an ancient plant-specific family (Navaud et al., 2007). The best-characterized members of this family have been shown to act on several aspects of plant development. It has been demonstrated that the maize (Zea mays) teosinte branched1 gene (ZmTB1) and its orthologous rice (Oryza sativa) gene OsTB1 repress lateral branching in maize and rice, respectively (Doebley et al., 1995, 1997; Takeda et al., 2003). The CYCLOIDEA gene together with the homologous gene DICHOTOMA have been shown to be involved in the dorsoventral asymmetry of flowers by arrest of the uppermost stamen in the Antirrhinum flower (Luo et al., 1996, 1999). CINCINNATA and related proteins have been shown to be associated with cell division arrest and differentiation during leaf development (Nath et al., 2003).
The TCP family can be subdivided into two subfamilies (TCP-C and TCP-P) based on the structure of the DNA-binding domain of the proteins (Cubas et al., 1999; Navaud et al., 2007). All genes described above are members of the TCP-C subfamily. The role of TCP-P proteins in plant development is much less documented. The first TCP-P proteins, namely PCF1 and PCF2 factors, were identified in rice by Kosugi and Ohashi (1997). They were described as putative regulatory proteins that bind to cis-acting elements named site II motifs in the promoter of the Proliferating Cell Nuclear Antigen (PCNA) gene. These motifs had been shown to be essential for the proliferating cell-specific transcriptional activity of the gene (Kosugi et al., 1995). We identified, in the promoter of the Arabidopsis (Arabidopsis thaliana) PCNA-2 gene, homologous site II motifs. In vitro, these site II motifs are targets for several DNA-binding activities of cellular proteins prepared from exponentially growing Arabidopsis cell suspension cultures and bind an Arabidopsis TCP-P protein produced in Escherichia coli, AtTCP20 (At3g27010; Tremousaygue et al., 2003). Li et al. (2005) showed that AtTCP20 bound in vivo to sequences corresponding to site II motifs in the promoter of genes coding for PCNA (PCNA-2), several ribosomal proteins, and cyclin CYCB1;1. In addition to these binding properties, the only functional data available for the TCP-P subfamily, obtained recently by RNA interference, demonstrated a role for AtTCP16 in pollen development (Takeda et al., 2006).
Using a random binding site selection approach, Kosugi and Ohashi (2002) defined, in rice, two different consensus sequences for site II motifs: GGNCCCAC for the class 1 motif recognized by TCP-P proteins, and GTGGNCCC for the class 2 motif recognized by TCP-C proteins. In Arabidopsis, bioinformatic analyses revealed that site II motifs were present in the promoter of genes regulated during the G1-S transition (like PCNA-2) and in 70% of the promoters of ribosomal protein genes (RP genes). By compiling all of the motif sequences identified in these promoters, we proposed TGGGCY (Y = C or T) as a consensus sequence for site II motifs in Arabidopsis (Tremousaygue et al., 2003). In these promoters, the site II motifs were seen to be associated with another cis-acting element, a telomeric internal motif named telo box (Tremousaygue et al., 2003). Related site II motifs (consensus GGCCCAWW [W = A or T]) and telo boxes were associated in the promoter of genes up-regulated during the initiation of axillary shoot outgrowth in Arabidopsis (Tatematsu et al., 2005). This is in good agreement with the dividing cell restricted expression pattern of a GUS reporter gene driven by these motifs in Arabidopsis (Tremousaygue et al., 2003). Site II motifs were identified also in the upstream regions of several nuclear genes in Arabidopsis encoding components of the mitochondrial oxidative phosphorylation machinery (Welchen and Gonzalez, 2006). The authors proposed that these motifs could participate in the expression coordination of nuclear genes encoding components of cytochrome c-dependent respiration. Further analysis revealed that approximately 5% to 10% of the entire set of Arabidopsis genes show a high frequency of site II motifs in their promoters (Welchen and Gonzalez, 2006). By contrast, few motifs proposed as TCP-binding sites in rice have been detected in the Arabidopsis genome (Kosugi and Ohashi, 2002).
In conclusion, several GC-rich consensus sequences, which partially match each other, have been proposed as target sequences for TCP proteins, but specificity of binding is not well documented and repercussions on gene regulation and consequently plant development are far from clear. In rice, because the class 1 and 2 consensus sequences overlap, Kosugi and Ohashi (2002) suggested that the TCP-P and TCP-C proteins could coordinately or competitively regulate transcription through the binding of consensus site II motifs. A similar model was proposed by Li et al. (2005) based on AtTCP20 binding properties; the authors suggested that TCP proteins support organ growth rate and shape via binding to GCCCR (R = A or G) sequences. However, because this motif was found in a high number of gene promoters, and because in some promoters these motifs failed to bind AtTCP20 in vivo (Li et al., 2005), it is not clear which genes will be regulated in planta in response of AtTCP20 binding.
The purpose of this study was to gain insight into the function of AtTCP20 and to identify its target genes in planta. To this end, we induced expression of AtTCP20-modified proteins in transgenic Arabidopsis. Plant phenotypes were analyzed. The transcriptomes of plants before and after induction of the modified protein were compared. Our results demonstrate the involvement of AtTCP20 in the control of cell shape and plant growth and led to the identification of a new set of target genes that have a rice class 1-related consensus sequence in their promoters that is bound by AtTCP20 in vitro.
RESULTS
Tissue Localization of the AtTCP20∷GUS∷GFP Fusion
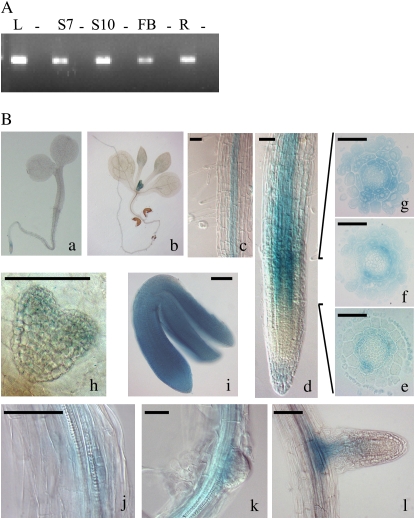
Reverse transcription (RT)-PCR analysis of RNA extracted from wild-type (ecotype Columbia [Col-0]) plants showed that the AtTCP20 gene is expressed in seedling, leaf, root, and flower bud (Fig. 1A). To study the localization of AtTCP20 in the tissues, wild-type (Col-0) Arabidopsis plants were transformed with a construct encoding a translational fusion between the promoter and the coding sequence of AtTCP20 and a GUS∷GFP reporter gene. Identical patterns of GUS staining were observed between several independent F2 lines. Two of these lines were propagated, and GUS staining analyses were performed on homozygous F4 lines grown in long-day light. In the embryo, GUS staining was detected from the heart stage of development, and mature embryos were totally stained (Fig. 1, Bh and Bi). In seedlings, GUS staining was present in the emerging young leaves, in the apex, and in the vascular tissue of roots (Fig. 1, Ba–Bc). In mature plants, GUS staining was mainly detected in vascular tissue. A detailed GUS analysis of the root tip of 6-d-old seedlings revealed GUS staining in the root cap and in the elongation zone (EZ; Fig. 1Bd). To specify the cellular layers stained in the EZ, roots were embedded in resin and transverse sections were made. At the top of the zone of cell division, the staining was mainly in the endodermis (Fig. 1Be). In the zone of cell elongation, progressively all of the cellular layers were stained (Fig. 1, Bf and Bg). The first cell layers providing lateral root initiation sites were stained (Fig. 1Bj). This staining disappeared in the emerging meristem, and later the lateral roots presented a similar GUS expression pattern to that of the primary root (Fig. 1, Bk and Bl). These results show that AtTCP20 expression is probably developmentally regulated from seed formation to young plantlet.
Figure 1.
Gene expression and tissue localization of AtTCP20. A, RT-PCR was carried out on leaf (L), seedling of 7-d-old (S7) or 10-d-old (S10) plants, flower buds (FB), and root (R) of ecotype Col-0 of Arabidopsis. In each case, a control using RNA without RT was made (−). B, GUS expression was observed in 5-d-old seedling (a), 10-d-old seedling (b), root of 6-d-old seedling (c and d), transversal sections of the EZ (e–g), and cell layers of the initiation site of lateral root (j) but not meristem (k). The expression profile of AtTCP20 in lateral root was similar to that observed in the primary root (l). Embryos from heart stage (h) showed light GUS staining, whereas mature embryos were strongly stained (i). Bars = 50 μm.
AtTCP20 Plays a Role in Plant Development
We identified and characterized T-DNA insertion mutants (salk_000843 and salk_016203) of the AtTCP20 gene without any phenotype, but mRNA expression of the gene was not abolished in these plants even though the T-DNA was inserted in the promoter and at the end of the coding sequence, respectively. Li et al. (2005) have postulated that it was not possible to characterize knockout mutant lines from AtTCP20 showing a phenotype because of probable gene redundancy. Our results did not permit us to confirm or refute this hypothesis. However, the closely related protein in Arabidopsis, AtTCP6, showed only 49.1% similarity with AtTCP20, which is not very high to provide functional redundancy. In addition, expression profiles of AtTCP-P genes are not similar in all organs and at all steps of development (according to Genevestigator data; Zimmermann et al., 2004). Therefore, to access the function of AtTCP20, the protein was overexpressed. Thirty independent plants transformed with P35S:TCP20 were selected on kanamycin medium and were grown to be self-fertilized. Nine developed normally, and the others showed reduced fertility with variable severity. The F2 seeds of all of these lines were sown on selective medium. Numerous lines had a low germination rate, and no clear phenotype was observed on young resistant plantlets after 15 d of cultivation in vitro (data not shown). An overexpression of the AtTCP20 protein was detected in 50% of the lines tested by western blotting without any obvious correlation with the germination rate (Fig. 2A). Plants overexpressing the AtTCP20 protein were grown in a culture room, and six revealed an abnormal development. The phenotypes observed were pleiotropic; plants were small with small, wrinkled, or curled leaves (Fig. 2B). The initiation of inflorescence was delayed; many siliques aborted, and consequently few seeds were obtained (Fig. 2B). However, it was difficult to get a clear and reproducible impact of AtTCP20 overexpression on adult plant development because the phenotype was leaky and was not maintained during whole vegetative development. Ultimately, the fertility was severely affected; therefore, the ability to characterize progeny was difficult. For all of these reasons, another approach to clarify the role of the AtTCP20 protein was tried. Under the control of an estrogen-inducible promoter (Zuo and Chua, 2000), the coding sequence of AtTCP20 was fused to a dominant negative repressor domain named EAR (Hiratsu et al., 2003) or to an activator domain named VP16 (Parcy et al., 1998). This approach should prevent lethality due to AtTCP20 gene deregulation and should induce a strong dominant phenotype.
Figure 2.
Analysis of overexpression of AtTCP20 protein in planta. A, Quantification of AtTCP20 by western-blot analysis of protein extract from rosette leaves of 35S:TCP20 lines (lanes 1–8) and the wild type (WT). B, Phenotypes of plants overexpressing the AtTCP20 proteins. WT, The wild type; 20-1 and 20-4, two independent lines showing dwarf development and delay in the initiation of inflorescence corresponding to lines 1 and 4 analyzed by western blotting in A. On the left, plants are at the same magnification; for 20-1, magnification is 9×; for 20-4, magnification is 2.5×. [See online article for color version of this figure.]
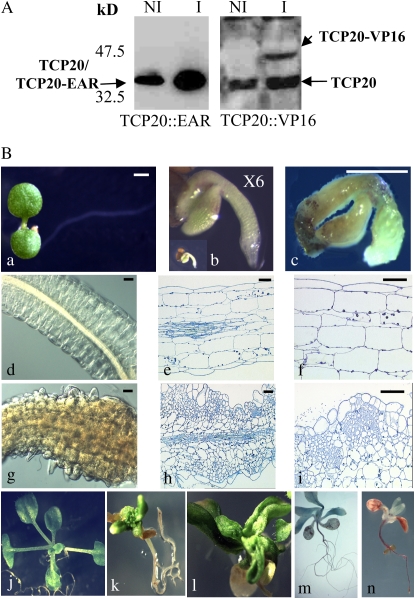
Seedlings of transgenic plants, transformed by a construct containing estradiol-inducible promoters driving the expression of AtTCP20-modified proteins, were selected on hygromycin medium. To investigate the effect of the modified protein expression on plant growth and development, F2 and F3 seeds of several lines were germinated on GM medium with dimethyl sulfoxide (DMSO) as a control (see “Materials and Methods”) or with 17β-estradiol. No phenotype was observed in plants germinated and grown with DMSO alone (data not shown). On inductive medium, five independent lines with AtTCP20∷EAR constructs had a similar strong developmental phenotype (100% of the tested lines). On eight independent lines tested containing the AtTCP20∷VP16 construct, four had their development greatly altered. These lines were analyzed by western blotting (Fig. 3A), and we observed that the phenotype was correlated with the expression of the two modified AtTCP20 proteins (Figs. 3B, 4, and 5).
Figure 3.
The modified protein AtTCP20∷EAR has dramatic pleiotropic effects on plant development. A, Detection of AtTCP20∷EAR and AtTCP20∷VP16 proteins by western-blot analysis of protein extracts from seedlings treated with (I) or without (NI) β-estradiol (5 μm) for 24 h. B, The seeds were directly sown on noninducible medium (a, d, e, and f) or on inducible medium (b, c, and g–i). a, Six-day-old noninduced seedling; b, 6-d-old induced seedling at 6× magnification (the inset shows a 6-d-old induced seedling at the same magnification as in a); c, 11-d-old induced seedling; d, 12-d-old noninduced hypocotyl section; e and f, 12-d-old noninduced longitudinal sections; g, 12-d-old induced hypocotyl section; h and i, 12-d-old induced longitudinal sections. Bars = 50 μm. j, Twenty-four-day-old noninduced plant; k and l, 7-d-old plants transferred on inductive medium and cultivated on this medium for 26 d; m and n, phloroglucinol staining of plantlets growing on inductive medium (n) or on noninductive medium (m).
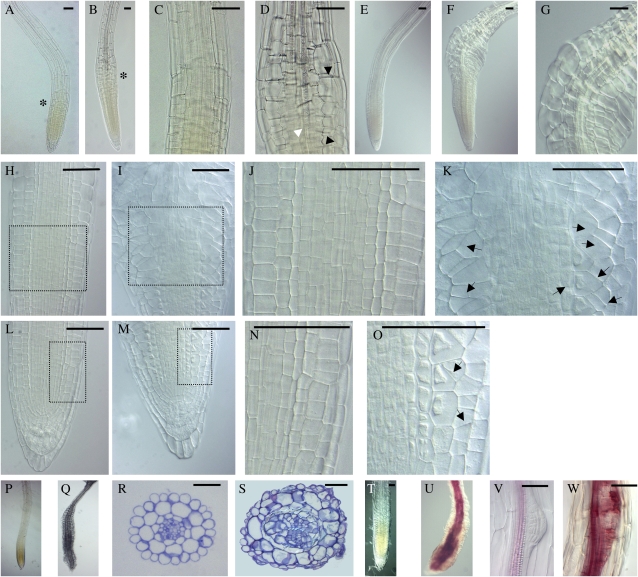
Figure 4.
AtTCP20∷EAR protein disrupts root development. For induction, 3-d-old seedlings were transferred onto inductive medium; otherwise they were transferred at the same age on new noninducible medium. A, Four-day-old noninduced primary root tip. B, Twenty-four-hour induced primary root tip. C and D, Magnification of the region marked with asterisks in A and B, respectively. Black arrowheads indicate abnormal expansion of epidermal and cortical cells in the rapid EZ, and the white arrowhead indicates discontinuous vascular elements that also appeared in this region. E, H, J, L, and N, Five-day-old noninduced primary root tip. F, G, I, K, M, and O, Forty-eight-hour induced primary root tip. G, Magnification of the EZ of F. J, K, N, and O, Magnification of the square zone defined in H, I, L, and M, respectively. Arrows plot the aberrant cell division planes observed in epidermal, cortex, and endodermis layers. H to K, EZ. L to O, Cell division zone. P and Q, Primary roots of 20-d-old noninduced and induced plants, respectively. R and S, Transverse sections in the EZ of 20-d-old noninduced and induced plants. T to W, Phloroglucinol staining of plantlets growing on noninductive (T and V) and inductive (U and W) medium. Seedlings in the induced condition reveal an excessive and ectopic lignification of vascular bundles clearly visible in primary root (U) and lateral root (W) meristems. Bars = 50 μm. [See online article for color version of this figure.]
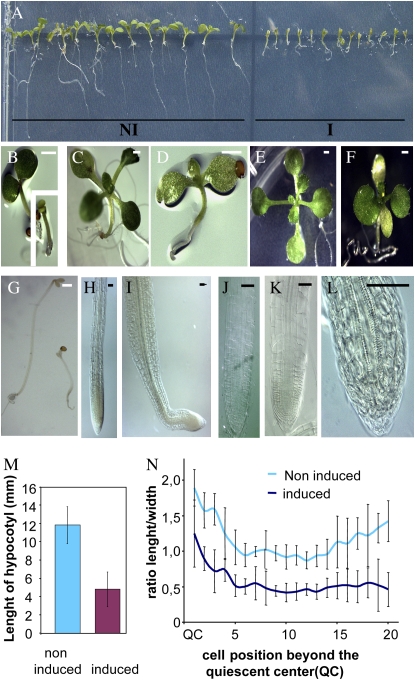
Figure 5.
The modified protein AtTCP20∷VP16 disrupts growth by reducing cell elongation. A, Nine-day-old induced plants (I) and 9-d-old noninduced plants (NI). B, Five-day-old noninduced plant; the inset shows a 5-d-old induced plant. C, Eleven-day-old noninduced plant. D, Eleven-day-old induced plant. E, Seventeen-day-old noninduced plant. F, Seventeen-day-old induced plant. G, Five-day-old seedlings grown in the dark: on the left is a noninduced seedling, and on the right is an induced seedling. B to G: Bar = 1 mm. H and I, Three-day-old primary root tip in noninduced (H) and induced (I) conditions. J and K, Ten-day-old primary root tip in noninduced (J) and induced (K) conditions. L, Twenty-day-old primary root tip in induced conditions. H to L: Bar = 50 μm. M, Hypocotyl lengths of 5-d-old dark-grown seedlings under induced or noninduced conditions. Each bar represents the average ± sd of 20 noninduced hypocotyls and 28 induced hypocotyls. N, The ratio of length to width of the first 20 cells located upstream of the quiescent center of roots of 10-d-old noninduced and induced plants. To calculate this ratio, median lengths and median widths of cells of five noninduced and seven induced roots were used. The vertical bars represent sd.
AtTCP20∷EAR Protein Induces Strong Developmental Phenotypes
Induced AtTCP20∷EAR seeds germinated with a delay of 3 to 4 d compared with noninduced seeds (data not shown). Seedlings emerging from the seed coats looked like mature embryos (Fig. 3, Ba and Bb) but showed rapidly a drastic growth arrest in shoot apical and root apical meristems (Fig. 3Bc). Cotyledons became yellowish, and the hypocotyl region was swollen (Supplemental Fig. S1). Within a few days, the hypocotyls developed green calli and occasionally embryo-like structures (Supplemental Fig. S1). Visual microscopic comparison of longitudinal sections of hypocotyls from induced or noninduced plants revealed a loss in cell polarity of epidermal, cortical, and endodermal cells and abnormal islands of cell proliferation and growth in cortical tissue (Fig. 3, Bd–Bi). The typical cellular organization of the hypocotyl was completely lost, and in addition cells often lost their adhesion capacity, creating big lacunas in the tissues. Induction was also carried out on 7-d-old plants, in which both root and shoot are differentiated, by transferring the plants onto the inductive medium. The aerial part of the plants was severely affected. In the shoot apical meristem, abnormal cell proliferation appeared, leading to a disorganized meristem (Fig. 3Bk; Supplemental Fig. S1). The shapes of differentiated leaves and cotyledons were abnormal (Fig. 3Bl; Supplemental Fig. S1). Adventitious roots were initiated from hypocotyls, and contrary to what was observed on immature hypocotyls, no callus appeared (Supplemental Fig. S1). This deregulation was obviously restricted to developmental stages prior to the differentiation of the hypocotyl, suggesting that embryo-specific cofactor(s) are required for this process. A root phenotype appeared 24 h after induction, and in the rapid EZ of the primary root the cells swelled (Fig. 4, A–D). We first observed a deregulated isotropic expansion of epidermal and cortical cells in the rapid EZ (Fig. 4, B and D). Discontinuous vascular elements appeared also in this region (Fig. 4D). Forty-eight hours after induction, the root tip presented an abnormal bending, and epidermal and cortical layers expanded asymmetrically in the EZ of induced roots (Fig. 4F). On one side of the root, the cells had an isotropic expansion, and on the other side, the cells expanded preferentially in width (Fig. 4G). In the zone of cell division, abnormal planes of division appeared in epidermal and cortical cell layers. In roots induced for 5 d, more and more aberrant planes of cell division were detected in epidermal, cortical, and also endodermal layers (Fig. 4, H–O).
After several days on inductive medium, roots were shorter and had a noticeably larger diameter than wild-type roots. In roots induced for 20 d, the degree of cell expansion varied along the length of the root (Fig. 4Q). Transverse sections through the abnormal expanded zone revealed irregular growth affecting the number and form of cells in all cellular layers (Fig. 4S). Young secondary roots began to expand after emerging from the primary root and had a similar phenotype as the primary root (Supplemental Fig. S1). Phloroglucinol staining of seedlings revealed an excessive and ectopic lignification of vascular bundles (Figs. 3Bn and 4U) and lateral root meristems (Fig. 4W).
AtTCP20∷VP16 Protein Reduces Cell Elongation
On inductive medium, the AtTCP20∷VP16 protein showed an inhibitory effect on plant growth and development, although less drastic than the AtTCP20∷EAR protein. Because phenotypes were easy to analyze directly after sowing on inductive medium, transfer experiments were not carried out. On inductive medium, the plants were dwarf (Fig. 5A). The cotyledons were unable to expand and were light green (Fig. 5, B and F). Leaf development was delayed and leaves stayed small (Fig. 5, D and F). In the light, hypocotyls were slightly shorter than in the noninduced condition (data not shown). In the hypocotyl of plants cultivated in the dark for 5 d, reduced cell elongation was clearly observed (Fig. 5G). On inductive medium, the hypocotyl measured about one-third the length of the hypocotyl of noninduced plants (Fig. 5M). At the beginning, the primary root had a typical root tip organization but the root was very short, and after 3 d on the inductive medium, the cell division zone of the root appeared shorter than in noninduced plants (Fig. 5I) and then progressively disappeared (Fig. 5L). In plants cultivated for 10 d, in the first 20 cortical cells beyond the quiescent cells, the cell length was significantly reduced and the width was slightly increased (Fig. 5K). The ratio of cell length to width indicates the polarity of cell expansion. Cell elongation was clearly arrested in roots, and for this reason the polarity of cells rapidly appeared to be reversed (Fig. 5N). The root meristem lost its ability to maintain growth, the root became determinate, and vascular bundles reached the tip of the primary root (Fig. 5L).
AtPCNA-2 Gene Expression Is Not Modified When the Expression of AtTCP20∷EAR Is Induced in Planta
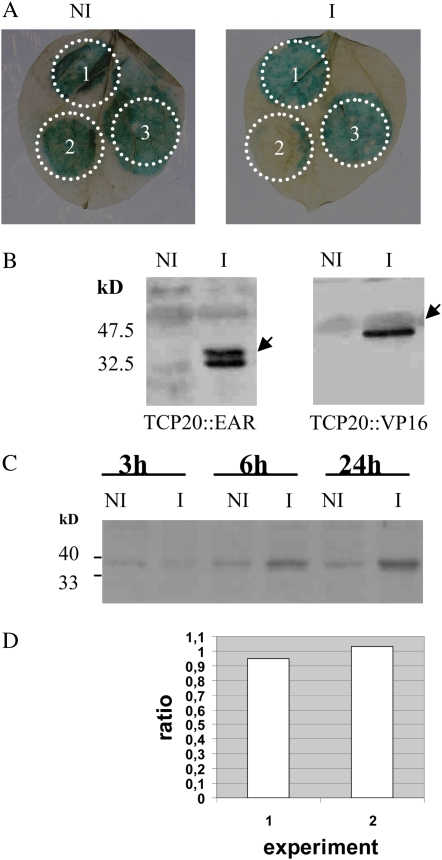
Our previous studies demonstrated that a purified recombinant protein, MBP∷TCP20, specifically interacts in electrophoretic mobility shift assay (EMSA) experiments with site II motifs from the AtPCNA-2 gene promoter. In addition, Li et al. (2005) demonstrated the binding of AtTCP20 to site II motifs from the AtPCNA-2 gene promoter in vivo by chromatin immunoprecipitation. While these motifs have been shown to associate with gene expression in dividing cells (Tremousaygue et al., 2003), there is still no evidence that expression regulation driven by site II motifs is due to AtTCP20 binding. Indeed, site II motifs obviously are recognized by several protein complexes, and only one of them contains AtTCP20. The transgenic lines expressing AtTCP20∷EAR gave us the opportunity to examine the effect of AtTCP20 on gene expression regulation in planta. We first verified, by transient expression in Nicotiana benthamiana leaves, that the AtTCP20∷EAR protein retained its binding capacity: it was possible to abolish GUS reporter gene expression, controlled by site II motifs, by cotransfecting the reporter gene and a plasmid expressing the AtTCP20∷EAR protein (Fig. 6, A and B). Then, AtTCP20∷EAR was induced in Arabidopsis transgenic lines for 24 h and the transcription level of AtPCNA-2 was analyzed in roots by quantitative real-time (Q-RT)-PCR. Whereas the AtTCP20∷EAR protein was detected by western blot (Fig. 6C) and a clear phenotype was observed on roots (Fig. 4), the transcription level of AtPCNA-2 was not modified after induction (ratios of 0.95 and 1.03 in two independent experiments; Fig. 6D).
Figure 6.
Recognition of site II motifs by AtTCP20-modified proteins. A, Effect of transient expression of AtTCP20∷EAR or AtTCP20∷VP16 on the level of GUS activity in N. benthamiana leaves. Leaves were infiltrated with a culture of Agrobacterium strain containing the GUS gene under the control of the site II motif described by Tremousaygue et al. (2003; 1); a mix in a 1:1 ratio of the culture used for 1 with a culture from a recombinant Agrobacterium strain expressing the AtTCP20∷EAR protein (2); or a mix in a 1:1 ratio of the culture used for 1 with a culture from a recombinant Agrobacterium strain expressing the AtTCP20∷VP16 protein (3). B, Detection of AtTCP20-modified proteins by western-blot analysis of protein extracts from transfected N. benthamiana treated with (I) or without (NI) β-estradiol. Protein expression was induced at 48 h by painting leaves, 2 d after infiltration, with 5 μm β-estradiol in the presence of 0.002% Silwett. The arrows highlight the induced AtTCP20∷EAR and AtTCP20∷VP16 proteins. C, Detection of induced AtTCP20∷EAR protein by western-blot analysis of proteins extracted from seedlings cultivated for 3, 6, and 24 h on β-estradiol (5 μm). D, Quantification of PCNA-2 expression level in roots by Q-RT-PCR in response to induction of AtTCP20∷EAR protein. Plants were induced for 24 h with β-estradiol as described in “Materials and Methods.” In each independent experiment (1 and 2), the ratio of induced over noninduced values of expression was calculated as described in “Materials and Methods” using the average threshold cycle and efficiency values from two replicates obtained for PCNA-2, taking into account the housekeeping gene (At5g08290) expression level in each case. [See online article for color version of this figure.]
Repression of Gene Transcription in Plants Expressing the AtTCP20∷EAR Protein
Transcriptome analyses were performed in order to identify genes repressed in response to AtTCP20∷EAR induction. RNA was prepared from roots or from hypocotyls of induced and noninduced plants. cDNA was hybridized to Affymetrix ATH1 genome arrays. Raw data were processed as described in “Materials and Methods,” and results are shown in an Excel file as Supplemental Table S2. All genes repressed in the induced tissues above a threshold of 2 with P < 0.05 in both duplicates were first selected. As a check of the microarray data, the expression level of a set of 30 genes was analyzed by Q-RT-PCR (Table I). Repression was confirmed for all genes in roots and for 25 genes in hypocotyls; however, five genes that had a repression threshold value below 4 were found activated in hypocotyls. Therefore, the threshold value was raised to 4 to perform the quantitative and qualitative analysis of Affymetrix data on hypocotyls. This resulted in the selection of 1,214 genes repressed in hypocotyls, 594 genes repressed in roots, and 252 genes repressed in both organs. Subsequent work was done on the three sets of genes (repressed in hypocotyls, in roots, or in both organs), corresponding to probes on the microarray associated with unique sequences.
Table I.
Q-RT-PCR validation of the microarray data for 30 genes
AGI, Arabidopsis Genome Initiative numbers of the selected genes. For hypocotyls, Q-RT-PCR data were obtained from averages of three independent values, and microarray data were normalized with Bioconductor (BIOC). For roots, Q-RT-PCR data were obtained from averages of three independent values, and microarray data were normalized with Bioconductor. Validation is scored as yes (y) or no (bold n; corresponding Q-RT-PCR values are in italic type). All data are expressed as log2 of the ratio of induced over noninduced values.
| AGI | Hypocotyls
|
Validation | Roots
|
Validation | ||
|---|---|---|---|---|---|---|
| Q-RT-PCR | BIOC | Q-RT-PCR | BIOC | |||
| At1g03870 | −4.9 | −5.6 | y | −3.4 | −4.0 | y |
| At1g05850 | −4.2 | −4.3 | y | −2.4 | −2.1 | y |
| At1g12110 | −2.9 | −2.4 | y | −2.0 | −2.5 | y |
| At1g20010 | −7.5 | −6.3 | y | −2.0 | −1.7 | y |
| At1g53580 | −4.5 | −4.0 | y | −1.0 | −1.0 | y |
| At1g71030 | −2.3 | −3.4 | y | −1.6 | −2.0 | y |
| At2g15570 | −4.6 | −3.9 | y | −1.2 | −1.6 | y |
| At2g35190 | 0.2 | −2.0 | n | −0.9 | −0.8 | y |
| At2g46270 | −2.4 | −1.3 | y | −0.8 | −0.8 | y |
| At2g46830 | −3.7 | −1.7 | y | −1.7 | −1.6 | y |
| At2g47070 | −4.1 | −3.7 | y | −1.5 | −1.5 | y |
| At3g01560 | 0.9 | −0.6 | n | −0.2 | −0.9 | y |
| At3g12110 | −2.4 | −1.9 | y | −1.3 | −2.0 | y |
| At3g13870 | −0.9 | −2.9 | y | −1.4 | −1.5 | y |
| At3g26520 | −5.6 | −3.6 | y | −0.9 | −0.7 | y |
| At3g58730 | −0.7 | −1.9 | y | −0.7 | −1.3 | y |
| At3g63120 | −5.1 | −4.2 | y | −1.6 | −1.7 | y |
| At4g02590 | −0.2 | −1.0 | y | −0.5 | −0.7 | y |
| At4g13770 | −8.0 | −5.0 | y | −2.4 | −2.7 | y |
| At4g21280 | −0.9 | −1.5 | y | −0.2 | −0.7 | y |
| At4G24780 | −3.3 | −4.2 | y | −1.1 | −2.1 | y |
| At4g25570 | −4.8 | −4.0 | y | −1.8 | −1.6 | y |
| At4g29080 | −3.7 | −3.7 | y | −3.2 | −2.8 | y |
| At5g15490 | 1.6 | −0.8 | n | −0.5 | −0.9 | y |
| At5g40450 | −10.0 | −7.8 | y | −2.5 | −2.4 | y |
| At5g44340 | 1.0 | −0.6 | n | −0.3 | −0.3 | y |
| At5g49720 | −0.5 | −1.7 | y | −1.1 | −1.0 | y |
| At5g51110 | −6.4 | −4.8 | y | −2.2 | −1.6 | y |
| At5g62540 | 0.7 | −0.7 | n | −1.0 | −1.0 | y |
| At5g65670 | −0.3 | −1.2 | y | −1.0 | −1.0 | y |
Four Motifs Are Overrepresented in the Promoters of Genes Repressed in Plants Expressing AtTCP20∷EAR
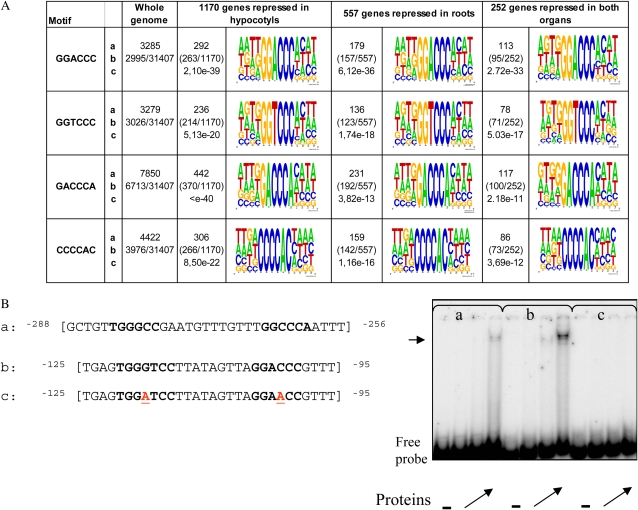
The 1-kb promoter DNA sequences of all genes that were significantly down-regulated in roots, in hypocotyls, or in both organs were retrieved, and a statistical motif analysis of these sequences was performed at The Arabidopsis Information Resource (TAIR). The program compares the frequencies of 6-mer “words” in the set of sequences (on both strands) with the frequencies of the words in the current promoter sequence set of 31,407 Arabidopsis genes (TAIR 6 release). Several randomly generated promoter sets were also used as controls. It appeared from this analysis that four motifs (GACCCA, GGTCCC, GGACC, and CCCCAC) were overrepresented in the promoters of repressed genes, in all sets, and the highest overrepresentation was obtained in promoters of genes repressed both in roots and in hypocotyls (Fig. 7A). These motifs were not overrepresented in control sets (data not shown). All of them match to the class 1 (GGNCCCAC) or the class 2 (GTGGNCCC) consensus motif defined in rice by Kosugi and Ohashi (2002). To extend nucleotide homologies around the 6-mer words defined by the motif analysis program, four bases were recovered upstream and downstream of each motif occurrence as described in “Materials and Methods.” The graphical representation of a complete set of extended motifs found in each data batch was displayed using WebLogo (Fig. 7A). The logo displays the frequencies of bases at each region as the relative heights of letters, along with the degree of sequence conservation as the total height of a stack of letters, measured in bits of information. When surrounding bases were added, the homology to the class 1 motif was confirmed for GACCCA, GGTCCC, and GGACCC, and the most probable core consensus sequence deduced from these sequences for the Arabidopsis class 1 motif was GG(A/T)CCC, with a prefix before the core that is rather T(T/G) and the suffix rather AC. However, the fourth motif, CCCCAC, no longer matched the rice binding sites, as nucleotides in positions −1 and −2 were adenines rather than guanines. Therefore, in our work, this latter motif will be treated separately. It is worth noting that a search for site II motifs (TGGGCY according to Tremousaygue et al. [2003] or GCCCR according to Li et al. [2005]) in promoters from the three sets of genes led to the detection of very few occurrences, even less than theoretically expected (data not shown). The binding of AtTCP20 to class I motifs from one of the putative target gene promoters (At2g36870) was tested by a gel retardation experiment. The results demonstrated a high affinity of MBP-TCP20 protein for double-stranded oligonucleotides containing these motifs (Fig. 7B). Moreover, mutations in the motifs completely abolished the binding (Fig. 7B). Thus, the genes repressed in our experiments were considered as putative targets of AtTCP20.
Figure 7.
Definition of putative motifs in the query set of promoter sequences of genes repressed in hypocotyls, in roots, or in both organs according to the Motif Analysis program at TAIR and test of binding activity to MBP-TCP20. A, Sequence logos of the overrepresented motifs found in the AtTCP20∷EAR-repressed genes: a, absolute number of motifs in the query set; b, number of promoter sequences in the query set containing a motif; c, P value for binomial distribution. The logo was created based on motif instances in the query set and surrounding bases using WebLogo (Crooks et al., 2004). The height of the symbols within the stack reflects the relative frequency of the corresponding nucleic acid at that position. B, EMSA for binding of MBP-TCP20 to class I motifs: a, sequence of the wild-type-CC probe from the AtPCNA-2 promoter used as a control; b and c, sequences of wild-type and mutated probes from the At2g36870 promoter used for EMSA as indicated. Class II and I motifs are shown in boldface, and the mutated nucleotides are underlined (in red) in the mutant probe. The − lanes at right correspond to migration of the probe without protein. Other lanes correspond to migration obtained with increasing quantities of the recombinant protein (angled arrows). The horizontal arrow indicates the complex obtained with the probe corresponding to oligonucleotides a and b. [See online article for color version of this figure.]
Functional Classification of Putative Target Genes
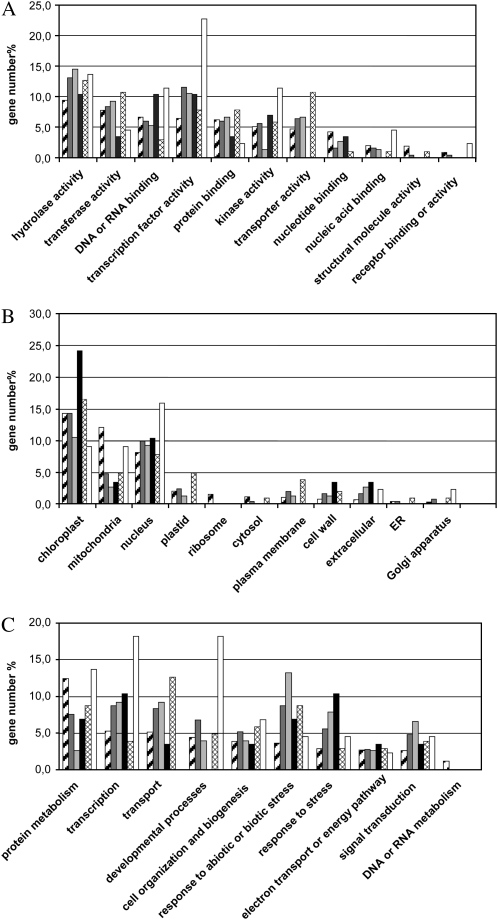
To tentatively identify primary targets and determinate biological pathways disturbed in our experiment, the 252 genes repressed in both organs were further analyzed. Using the promomer program (http://bar.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi), subsets of genes were defined that had in their 1-kb sequence promoters (1) none of the motifs (76 genes), (2) only CCCCAC motifs (29 genes), (3) only class 1-like motifs (103 genes), or (4) at least one class 1 motif and one CCCCAC motif (44 genes). A functional categorization of the 252 genes and of each gene subset was performed according to Gene Ontology categories proposed at TAIR (Supplemental Table S3). A graphical representation highlights modifications of distributions in comparison with the whole genome categorization (Fig. 8). Genes with no informative annotation were not included in the representation (e.g. 22.8% of the 252 genes have unknown molecular functions). The results indicated clearly a different distribution for each subset, reinforcing the idea that class 1-like and CCCCAC motifs do not have the same function. We focused the analysis on 147 genes (members of subset 3 or subset 4) having within their promoters at least one motif potentially recognized by AtTCP20, which were thus good candidates to be targets of AtTCP20∷EAR. All annotations were manually checked, and genes were associated with a biological keyword. This classification and the promoter structure of each gene are reported in Supplemental Table S4. It appeared from annotation of the 147 genes that the largest category comprises 23 genes associated with cell wall structure, among which 19 are from subset 3. In addition, 18 transcription factors, 11 proteins regulating transcription (interacting with transcription factors or nucleic acids), and 14 genes involved in protein modifications were identified among the putative direct targets. Several other genes, mostly from subset 3, were associated with transport (14 genes), metal handling, and membrane functioning.
Figure 8.
Bar graph representation of gene functional classifications based on Gene Ontology at TAIR. Classification was performed according to keyword category (A, molecular function; B, cellular component; C, biological process), and the gene number percentage of each category is shown as a hatched bar for the whole genome, as a dark gray bar for 252 repressed genes, as a light gray bar for subset 1, as a black bar for subset 2, as a cross-hatched bar for subset 3, and as a white bar for subset 4.
DISCUSSION
In Vivo Expression of AtTCP20-Modified Proteins Produces Pleiotropic and Drastic Phenotypes
A strategy capable of overcoming the infertility probably caused by constitutive expression was developed using an estradiol-inducible promoter. This approach was coupled to the expression of modified AtTCP20 proteins containing either a dominant negative repressor domain, EAR, or a strong activator domain, VP16. In this way, the modified proteins could interfere in vivo with the normal functions of AtTCP20 and of functionally redundant proteins. Previous work showed that the expression of transcription factor∷VP16 fusions activates specific target genes in a gain-of-function approach (Parcy et al., 1998; Bensmihen et al., 2004). It was also demonstrated for several transcription factors that chimeric repressors with an EAR domain motif suppressed the expression of original target genes and that suppression was dominant, superseding the endogenous activities of the native protein and functionally redundant transcription factor relatives (Hiratsu et al., 2003; Fujita et al., 2004, 2005; Matsui et al., 2005; Koyama et al., 2007). Recently, Koyama et al. (2007) used a chimeric repressor from AtTCP3 to study the functions of this gene. Their results demonstrated that AtTCP3 plays a role in the control of morphogenesis of shoot lateral organs via the negative control of expression of boundary-specific genes. These results were obtained with both a 35S promoter and the TCP3 native promoter, even if phenotypes observed with the native promoter were milder. They also demonstrated redundant functions of eight members of the AtTCP-C family, which are clustered in the same subgroup in a phylogenetic tree (Navaud et al., 2007). In addition, five of them are regulated by the same miR319. Using individual lines containing chimeric repressors derived from each of seven other TCPs, they found phenotypes similar to that obtained with the AtTCP3 repressor and modification of the expression of the same targets. In this case, TCP3SRDX protein also affects the target genes of other TCPs, but only those that are involved in similar functions. In our case, we cannot exclude that functions of AtTCP6 and AtTCP11, which are clustered in the same group with AtTCP20, overlap functions of AtTCP20. Even if this is the case, at the least AtTCP20 will be involved in physiological processes (cell division, cell elongation, and differentiation) affected by the chimeric repressor, and inversely, the repressor will not affect processes other than the ones involving AtTCP20 and any functional homologs. Therefore, we evaluated functions involving the AtTCP20 protein by investigating the phenotypes induced by expression of the chimeric AtTCP20 repressor or activator. Strong developmental phenotypes were obtained. These phenotypes were not related to those obtained by interfering in TCP-C protein function (Koyama et al., 2007). This observation supports the idea that TCP-P proteins have specific functions and that functions of TCP-C proteins are not affected in our approach. Following the induction of either modified protein, the development of roots, hypocotyls, cotyledons, and leaves of young seedlings was disrupted and plant development was hindered. Observed phenotypes were not opposite, even if a repressor domain was added in one case and an activator domain in the other. Moreover, responses to induction of the modified proteins differed according to organ and stage of development when estradiol was applied. To date, whether AtTCP20 acts as an activator or a repressor has not been demonstrated. Our results suggest that AtTCP20 probably has different functions in different contexts, according to promoter sequences or depending on partners in regulatory complexes. In this case, any modification of its activity will alter the expression profile of some of its target genes, and induction of any of the two constructs will disturb plant development, although the same genes will not be deregulated. This indicates that a fine adjustment of AtTCP20 activity is necessary for normal development.
In the aerial parts of AtTCP20∷EAR transgenic lines, after induction, the balance between proliferation and differentiation was deregulated and the plant failed to develop properly. In our study, we focused in detail on the root phenotype, which was the first visible phenotype after induction of AtTCP20∷EAR protein. In the EZ of roots, where the AtTCP20 protein accumulated, the phenotypes obtained are reminiscent of those observed in conditional root expansion mutants (Hauser et al., 1995) or other mutants affected in cell expansion (Benfey et al., 1993). As in Cobra or Lion's tail mutants, the cell expansion of the induced AtTCP20∷EAR lines is related to the rate of root growth of the primary root (Hauser et al., 1995). Under conditions that provide maximal wild-type root growth (high concentrations of Suc in the culture medium), AtTCP20∷EAR-induced plants exhibited a maximal cell-expansion phenotype (data not shown). Abnormal cell plane division represented an additional phenotype observed in roots of AtTCP20∷EAR plants, which are similar to the strong allele of the Arabidopsis KORRIGAN gene, an endo-1,4-β-d-glucanase involved in cell elongation and cytokinesis (Zuo et al., 2000; Lane et al., 2001). Ectopic lignification was also observed in AtTCP20∷EAR-induced plants. Previous works have reported that ectopic lignification could be correlated with the degree of altered cell expansion in several mutants, suggesting a linkage between cell expansion and the initiation of secondary cell wall formation and subsequent lignification (Cano-Delgado et al., 2000; Newman et al., 2004). Cell expansion in plants requires the coordinated assembly of cytoskeleton and cell wall and the control of the turgor pressure to provide the mechanical force for expansion (Cosgrove, 2005; Smith and Oppenheimer, 2005). Homozygous AtTCP20∷EAR lines expressing a constitutive Microtubules-Associated Protein4 fused to the GFP reporter protein were used to visualize the cytoskeleton in the root epidermal cell of the EZ. In the noninduced condition, cortical microtubule arrays were transversely oriented to the major axis of cell expansion. In induced plants, the typical orientation of microtubules disappeared (data not shown). However, at this stage, it is difficult to say if it is a cause or a consequence of the abnormal cell expansion.
Previous molecular studies proposed that AtTCP20 was involved in the coordination of cell proliferation and cell growth genes (Tremousaygue et al., 2003; Li et al., 2005; Tatematsu et al., 2005). This study suggests that AtTCP20 might play a role in the regulation of genes controlling cell expansion. Altogether, these data suggest a role of AtTCP20 in the coordination of cell expansion, cell division, and cell growth genes. However, from our data, we do not know if other TCP proteins are involved.
The Nature of Target Genes Is Consistent with Observed Phenotypes
The transcriptomes of roots or hypocotyls from plantlets expressing or not expressing the AtTCP20∷EAR protein were compared. We used a set of stringent criteria to analyze the transcriptome results in order to narrow down the data set to the most probable direct targets and concentrated on genes repressed in both roots and hypocotyls, even though this would eliminate the identification of specific targets. By so doing, 252 genes were identified matching these criteria. AtTCP20 has been shown to bind directly to a DNA fragment containing the site II motif (TGGGCC) of AtPCNA-2 in 12-d-old seedlings (Li et al., 2005). Surprisingly, the consensus sequence for this site is in fact underrepresented in the 252 promoters in comparison with the distribution observed for the promoter sequences in the whole genome (data not shown). In our sets, we found an enrichment of a consensus sequence, GG(A/T)CCC, matching the class 1 motif characterized by Kosugi and Ohashi (2002) as a binding site to rice TCP proteins, and AtTCP20 was shown to bind this motif in vitro. Our previous data indicated that the site II motif drives transcription in root meristems and in young emerging leaves (Tremousaygue et al., 2003). By analyzing the expression pattern of AtTCP20 using GUS staining of translational fusion, we showed that the protein was detected in young leaves but not in root meristems of young plantlets. Therefore, AtTCP20 colocalizes only partially with the activation pattern driven by the site II motif in dividing cells, and its localization appears inconsistent with a direct activation via the cis-acting element II in root meristems. Coherently, we showed that AtPCNA-2 was not repressed in roots of plants expressing AtTCP20∷EAR. We propose that another TCP protein could play this role in Arabidopsis root meristems. According to expression profiling analysis, AtTCP19 is preferentially expressed in the root tip and could be a good candidate (Zimmermann et al., 2004). Interestingly, the major root phenotype observed in our experiment was localized in the EZ, where the AtTCP20 protein was detected by the translational GUS fusion expression analysis. By comparing promoter sequences of the 252 repressed genes, four motifs were found to be overrepresented (58%), which thus could be considered as signatures for putative target genes at least in our experiment. The sequences of three of these motifs and their flanking sequences were related to the class 1 TCP-binding site previously identified in rice (Kosugi and Ohashi, 2002). This suggests that, at least in roots and hypocotyls, developmental defects observed in the plants expressing AtTCP20∷EAR are related in part to the deregulation of the genes possessing class 1 motifs in their promoters. The involvement of AtTCP20∷EAR in this repression is highly probable, especially since direct binding of the MBP-TCP20 protein to the class 1 site has been demonstrated in vitro.
Two major classes can be distinguished among the 147 repressed genes having at least one class 1 TCP binding site within their promoters. The first one contains genes coding for transcription factors (18 genes) and for proteins interfering with transcription (11 genes). The second major class of AtTCP20∷EAR-repressed genes contains 23 genes involved in primary and secondary cell wall biogenesis and modification. Genes from the first class are mostly members of transcription factor families, known to be important for development. Among them we found Rav1, a negative growth regulator, another TCP gene, AtTCP21, shown to be a repressor of the Arabidopsis clock transcriptional network (Pruneda-Paz et al., 2007), and a NAM-like gene involved in the control of multicellular organism development. There are three homeodomain genes; two are paralogous class I genes (Henriksson et al., 2005), HB6 and HB16, which were previously proposed to play a role in cell expansion (Wang et al., 2003), and one from class III, HB8, has been suggested to be involved in vascular development (Ohashi-Ito and Fukuda, 2003). Many other genes from this class are well-known regulators involved in hormone signaling. For example, AUX/IAA13, -16, and -27 have been shown to repress Auxin Response Factor transcription factors in the auxin signaling pathway (Tiwari et al., 2004; Weijers et al., 2005). ARR4, ARR6, and ARR7 are A-type Arabidopsis Response Regulator genes of cytokinin two-component signaling described by several biochemical and genetic studies as negative regulators (To et al., 2004; Ferreira and Kieber, 2005; Lee et al., 2007). Recently, data obtained by Sabatini's team suggested that the size of the Arabidopsis root meristem may be established by a balance between the antagonistic effects of cytokinins, which mediate cell differentiation at the transition zone, and auxin, which mediates cell division (Ioio et al., 2007). Because of the AtTCP20 expression pattern, it will be interesting to know if this protein could play a role in these processes. Ethylene response is also probably affected, as attested by the repression of three ERF/AP2 transcription factors, including TINY (Wilson et al., 1996).
Deregulation of such a large number of transcription factors probably has a broad impact on gene expression and is consistent with the pleiotropic developmental abnormal phenotypes observed in plants expressing the AtTCP20∷EAR protein.
Among the 23 genes forming the second most important category of repressed genes, four of the 10 cellulose synthase genes (CESA) of Arabidopsis that synthesize the cellulosic component of walls are found. CESA2, CESA5, and CESA6 are involved in primary wall synthesis but have nonessential roles (Somerville, 2006). The CESA6 mutant procuste1 shows reduced elongation and radial swelling of hypocotyl and root tissues (MacKinnon et al., 2006), and insertion mutations in CESA2 and CESA5 result in similar phenotypes (Somerville, 2006). The fourth gene, CESA4, is required for secondary cell wall synthesis in Arabidopsis. In all of these cellulose-deficient mutants, the cells swell in expansion zones, probably reflecting loss of the ability to restrain turgor, and sometimes an ectopic lignification is associated. These phenotypes are in good agreement with our observations of AtTCP20∷EAR-induced plants (see above). In addition, nine genes encoding glycosyl hydrolases, of which six belong to the xyloglucan endotransglucosylase/hydrolase family (XTH), are also repressed. XTHs are enzymes that mediate the construction and restructuring of cross-links among cellulose microfibrils, a process that is essential in the regulation of the orientation and extent of cell expansion in plants. Functional studies of the loss of function of AtXTH18 (repressed in our experiment) propose a principal role in root elongation. Two other down-regulated AtXTH genes, AtXTH17 and AtXTH20, are phylogenetically closely related to AtXTH18 (Osato et al., 2006). Four other repressed genes encode proteins containing a fasciclin domain, which is an ancient, putative adhesion domain in plants (Johnson et al., 2003). In plant cells, adhesion events are likely important within the primary cell wall, as attested by the sos5 mutant phenotype, which shows abnormal cell expansion and cell wall structure (Shi et al., 2003). The Root Hair Defective3 gene, previously reported to be involved in cell expansion by regulating the traffic of wall or plasma membrane determinants (Wang et al., 1997; Yuen et al., 2005), is also one of the 23 repressed genes. Clearly, all of these genes down-regulated by the induction of repressor AtTCP20∷EAR contribute to the normal continuous synthesis, breaking, and remaking of bonds necessary to maintain the integrity of the cell wall during cell expansion. Deregulation of these genes would be enough to account for the major phenotypes observed in our induced plants.
In many cases (17.5%), the class 1 motif appears to be associated with another sequence motif (CCCCAC) for which no hit was retrieved in AGRIS (http://Arabidopsis.med.ohio-state.edu/AtcisDB). Site II motifs and related sequences were also identified in regulatory modules, for example in association with another cis-acting element named the telo box (Tremousaygue et al., 2003; Tatematsu et al., 2005; Vandepoele et al., 2006). This module was associated with ribosome biogenesis. It has been shown in yeast that the design of transcription factor binding sites is affected by combinatorial regulation (Bilu and Barkai, 2005). Whether the obvious diversity of TCP target sites is linked to combinatorial regulation driven by TCP proteins and their partners is an interesting question.
In conclusion, our results are consistent with AtTCP20 binding different targets depending on the organ, tissue, or cellular context. In plants, there is no morphogenetic cell movement; morphogenesis is thus entirely dependent on how and when cells divide and expand. The coordination of these developmental processes is critical for coherent development. From our results, we propose that AtTCP20 is indeed a common regulator of many effectors involved in cell division, cell elongation, and differentiation. AtTCP6 and AtTCP11 may have related functions, which will be interesting to decipher. It will be of great importance to learn more about the expression and function of TCP transcription factors, which constitute an ancient plant-specific transcription factor family that expanded in parallel to the increasing complexity of plant morphogenesis.
MATERIALS AND METHODS
Plasmid Construction
The construct ProTCP20:CDS20∷GUS∷GFP contains a sequence of 2.9 kb of the AtTCP20 promoter amplified from the genomic sequence (Col-0) using the specific primers PromTCP20-NcoI and PromTCP20-EcoRI. The primer sets used in the different experiments are listed in Supplemental Table S1. This fragment was introduced between the corresponding sites of the plasmid pCambia 1303 (CAMBIA). The coding sequence of AtTCP20 without the termination codon was amplified with the specific primers TCP20-5′-NcoI and TCP20-3′-NcoI and cloned at the NcoI site to achieve a translational fusion between the TCP20∷GUS∷GFP coding sequence driven by the AtTCP20 promoter.
The coding sequence of AtTCP20 was amplified from pBD-TCP20 with the specific primers TCP20-5′-KpnI and TCP20-3′-StuI and introduced between the corresponding sites of pBLTI121 after the 35S promoter (Kiefer-Meyer et al., 1996). The coding sequence of AtTCP20 was amplified from pBD-TCP20 (Tremousaygue et al., 2003) by T7 primer and the specific primer TCP20-3′-BglII designed to suppress the termination codon and introduced a BglII site in frame. This fragment was cloned into pGEM-T vector (Promega). The EAR motif repression domain described by Hiratsu et al. (2003; LDLDLELRLGFA) was produced using two complementary oligonucleotides containing in addition a TGA codon at the 3′ end and the digested BglII and SpeI sites at the 5′ and 3′ ends, respectively. The transactivation domain (amino acids 413–490) of herpes viral protein VP16 was obtained by amplification with the specific primers VP16-1 and VP16-2 from pER8 vector (Zuo et al., 2000). These primers contain the BglII site and termination codon and the SpeI site, respectively. The two regulatory domains were introduced in frame at the 3′ end of the AtTCP20 coding sequence between the BglII and SpeI sites located in the pGEM-T vector. The modified AtTCP20 sequences were cloned between the ApaI and SpeI sites of the pER8 vector, and expression was driven by an estradiol-inducible promoter.
Plant Transformation
Some of these constructs were used to transform Agrobacterium tumefaciens strain C58 pMP90 and were introduced by floral dipping (Clough and Bent, 1998) into Arabidopsis (Arabidopsis thaliana) plants. Wild-type plants (Col-0) were transformed by the construct ProTCP20:CDS20∷GUS∷GFP or P35S:CDS20, and transgenic plants (ecotype Wassilewskija) containing GUS reporter gene, described by Tremousaygue et al. (2003), were transformed with the inducible constructs. For each construct, several independent transformants were selected by resistance to hygromycin (50 mg L−1) or kanamycin (50 mg L−1). These lines were selfed one or several times, and T2 homozygous or heterozygous plant and T3 and T4 homozygous plants were used in the experiments.
Plant Materials and Growth Conditions
Arabidopsis seeds were sown aseptically in petri dishes containing GM medium (1× MS salts [Murashige and Skoog, 1962], 10 g L−1 Suc, 100 mg L−1 inositol, 0.5 g L−1 MES, and 0.8% agar, pH 5.7) and cultivated under a 16/8-h photoperiod at 75 μE m−2 s−1, 15 W m−2, and 19°C. The inductions were carried out by directly germinating seeds on a medium supplemented with 1 or 5 μm 17β-estradiol (Sigma) or transferring seedlings from noninductive medium onto inductive medium. GM medium supplemented by DMSO was used as a control because 17β-estradiol is prepared in DMSO. In long-term experiments, plantlets were transferred every 2 d on fresh medium containing estradiol.
Histology and Histochemistry
The plantlets were observed using a Leica MZFLIII stereomicroscope with a Leica DC200 camera and Leica DC Viewer software. Detailed observations were realized using differential interference contrast optics on a Zeiss Axiophot I microscope with a Leica DC200 camera and Leica DC Viewer software.
The tips of roots were observed after brief clearing in sodium hypochloride solution (3% NaOCl) for 3 min, followed by five washings with water and mounting in Hoyer's medium (chloral hydrate:water:glycerol, 8:3:1). About 20 cortical cells located upstream of the quiescent center of 10-d-old primary root (induced or not) were photographed using an Axiophot I microscope coupled to a Leica DC200 camera. The length and width of each cortical cell were measured using ImageJ software. To visualize the lignin, seedlings were stained with a phloroglucinol-HCl solution (VWR). For GUS staining, seedlings and seeds were analyzed as described by Tremousaygue et al. (1999). Embryos were observed after seed fixation in formaldehyde/acetic acid buffer for 4 h and dehydrated in a gradual ethanol series, and ethanol was gradually replaced by benzyl benzoate. They were mounted in benzyl benzoate and observed by microscopy using differential interference contrast optics. Roots and GUS-stained roots were embedded in Technovit 7100 resin (Haereus-Kulzer) according to the manufacturer's instructions. Unstained root sections were dried onto glass slides and stained with toluidine blue. Hypocotyl samples were prepared as described by Pineau et al. (2005).
Transient Expression in Nicotiana benthamiana
Overnight-grown bacteria were pelleted and resuspended in a medium containing 10 mm MgCl2, 10 mm MES/KOH, pH 5.6, and 150 μm acetosyringone to a final optical density at 600 nm of about 0.7, and agrobacteria were left standing for at least 2 h. Two leaves per plant were infiltrated with appropriate agrobacteria strains. In all cases, a strain expressing the p19 protein of TBSV, a viral silencing suppressor plasmid (Voinnet et al., 1999), was coinfiltrated. The infiltrated area was infiltrated again, 2 d later, with a solution containing 5 μm 17β-estradiol. Histological GUS stainings were performed after 2 d of induction by infiltrating 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in 50 mm phosphate buffer, pH 7, and 0.001% Triton X-100 into the leaves.
Western Blotting
For the isolation of total protein fractions from Arabidopsis seedlings, leaves or one leaf disc from transfected N. benthamiana were ground in liquid nitrogen and homogenized in 100 μL of 2× Laemmli buffer. Twenty microliters of total protein extracts was separated by SDS-PAGE according to Laemmli (1970) and subsequently transferred to nitrocellulose membranes (Protran; Schleicher & Schuell). Western blotting was conducted with a rabbit polyclonal antibody against synthetic peptides (Eurogentec) corresponding to amino acids 58 to 72 and 166 to 180 of AtTCP20 (1:3,000 dilution) followed by secondary probing with anti-rabbit horseradish peroxidase (1:3,000). Secondary antibody-horseradish peroxidase conjugates and detection kits (ECL Plus) were from Amersham Biosciences.
EMSA
MBP-TCP20 binding assays were done according to Tremousaygue et al. (2003), with double-stranded oligonucleotides corresponding to a 30-bp fragment from the At2g36870 gene promoter or to a mutated sequence that did not contain any more class 1-like binding sites.
RT-PCR and Q-RT-PCR Analysis
For classical RT-PCR, total RNA was extracted from young seedling, leaf, root, and flower buds in Extract-All (Eurobio) following the manufacturer's instructions. For Q-RT-PCR, total RNA was extracted with the Nucleospin RNAII kit following the supplier's recommendations (Macherey-Nagel). RNA was extracted from roots of 6-d-old plants induced (I) for 6 or 24 h or not induced (NI; for control) or from hypocotyls of plantlets grown on inductive medium (I) or noninductive medium (NI) for 1 week. RNAs were treated with RNAse-free DNAse I (Ambion), and cDNAs were synthesized using SuperScriptII reverse transcriptase (BRL) according to the instructions of the supplier. RT-PCR was performed on 5 μL of cDNA product using 1 unit of Taq polymerase (Invitrogen), 2.5 mm MgCl2, 0.25 mm deoxynucleoside triphosphate, and 0.25 μm of primers TCP20-F and TCP20-R (see Supplemental Table S1 for sequences of primers) with the polymerase manufacturer's buffer in a 50-μL final volume. Amplification was performed as follows: 3 min of initial denaturation at 94°C; followed by 32 cycles of 1 min at 95°C, 1 min at 50°C, and 1 min at 72°C; and a final elongation of 2 min at 72°C. Q-RT-PCR was performed in a 10-μL reaction using the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche) in the LightCycler software version 3.5 sequence detection system (Roche) under the following conditions: 95°C for 9 min, 95°C for 5 s, 65°C for 10 s, and 72°C for 20 s. The reaction was performed for up to 40 cycles, and threshold cycle (CT) values were obtained. DIM1 (At5g08290) and UBQ (At4g05050) genes, with a constitutive expression, were used for data normalization. The efficiency of amplification of the target gene (TG) and the housekeeping gene (HKG) was calculated using the LinReg program (Ramakers et al., 2003), and relative quantification was calculated according to Pfaffl (2001) as follows: ratio = efficiencyTGΔCT(NI-I)/efficiencyHKGΔCT(NI-I)).
Functional Classification of Genes
For functional classification of the repressed genes, Gene Ontology annotations at TAIR (http://www.arabidopsis.org/; Berardini et al., 2004) were used. For a query gene set, classification was obtained and a percentage of genes in each category was calculated based on the number of genes in the category and the set size. The percentage of genes in each category for the whole genome was calculated from the total number of annotated genes (25,108 genes based on process ontology, 26,044 genes based on molecular function ontology, and 26,055 genes based on component ontology).
Motif analysis was used at TAIR to identify overrepresented motifs within the −1,000 promoter sequences. The surrounding four bases upstream and downstream of a DNA motif of interest were recovered using the emacs-lisp program given in Supplemental File S1. The sequence logo was made at http://weblogo.berkeley.edu/logo.cgi .
Affymetrix Hybridization and Data Analysis
Total RNA of roots or hypocotyls was prepared as described for Q-RT-PCR analysis. All experiments were duplicated. Hybridization was carried out at the Microarray Platform of the IGBMC and Génopole Alsace-Lorraine (http://www-microarrays.u-strasbg.fr/). Processing of RNA, ATH1 GeneChip (Affymetrix) hybridization, and raw data collection were performed as described by Redman et al. (2004). The raw data from the .cel files were Robust Multiship Averaging normalized (Irizarry et al., 2003) and analyzed with the LIMMA package of Bioconductor (Gentleman et al., 2004) using an empirical Bayes linear modeling approach (Smyth, 2005), and P values were corrected for multiple testing according to Benjamini and Hochberg (1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes observed in plants expressing the AtTCP20∷EAR protein.
Supplemental Table S1. Primer list.
Supplemental Table S2. Results from microarray experiments after Bioconductor processing of raw data (.cel files).
Supplemental Table S3. Functional classification of 252 repressed genes and associated gene subsets.
Supplemental Table S4. Data on 147 genes having at least one class I-like motif in their promoters and repressed in both organs.
Supplemental File S1. The emacs-lisp program.
Supplementary Material
Acknowledgments
C.H. and D.T. are grateful to Olivier Navaud for his bibliography survey and technical support. We thank Ton Timmers for helpful discussion, Prof. Nam Chua for providing the pER8 plasmid, Daniel Sidobre for writing the emacs-lisp program, and Thomas Kroj for his help in using the Bioconductor package. We thank Deborah Goffner for her helpful comments on the analysis of regulated genes involved in the cell wall and Julie Cullimore for critical reading of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dominique Tremousaygue (dominique.tremousaygue@toulouse.inra.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119 57–70 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57 289–300 [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561 127–131 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilu Y, Barkai N (2005) The design of transcription-factor binding sites is affected by combinatorial regulation. Genome Biol 6 R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Delgado AI, Metzlaff K, Bevan MW (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127 3395–3405 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6 850–861 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18 215–222 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ (2005) Cytokinin signaling. Curr Opin Plant Biol 8 518–525 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39 863–876 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN (1995) Conditional root expansion mutants of Arabidopsis. Development 121 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engstrom P, Soderman E (2005) Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol 139 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34 733–739 [DOI] [PubMed] [Google Scholar]
- Ioio RD, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17 678–682 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiol 133 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer-Meyer MC, Gomord V, O'Connell A, Halpin C, Faye L (1996) Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene 178 205–207 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30 337–348 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y (1995) Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J 7 877–886 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Hofte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J (2007) Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) overexpression in cytokinin response. Mol Genet Genomics 277 115–137 [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383 794–799 [DOI] [PubMed] [Google Scholar]
- MacKinnon IM, Sturcova A, Sugimoto-Shirasu K, His I, McCann MC, Jarvis MC (2006) Cell-wall structure and anisotropy in procuste, a cellulose synthase mutant of Arabidopsis thaliana. Planta 224 438–448 [DOI] [PubMed] [Google Scholar]
- Matsui K, Hiratsu K, Koyama T, Tanaka H, Ohme-Takagi M (2005) A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol 46 147–155 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioessays with tobacco cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O, Dabos P, Carnus E, Tremousaygue D, Herve C (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65 23–33 [DOI] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM (2004) Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 37 239–250 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44 1350–1358 [DOI] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119 153–162 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395 561–566 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau C, Freydier A, Ranocha P, Jauneau A, Turner S, Lemonnier G, Renou JP, Tarkowski P, Sandberg G, Jouanin L, et al (2005) hca: an Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant J 44 271–289 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J, Ghislain Breton G, Kay S (2007) A functional genomics approach reveals TCP21 as a new component of the Arabidopsis clock transcriptional network. In 18th International Conference on Arabidopsis Research, Beijing, China. The Arabidopsis Information Resource, Stanford, CA, p 172
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38 545–561 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21 271–295 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2005) Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420
- Somerville C (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22 53–78 [DOI] [PubMed] [Google Scholar]
- Takeda T, Amano K, Ohto MA, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C (2006) RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol Biol 61 165–177 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33 513–520 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremousaygue D, Garnier L, Bardet C, Dabos P, Herve C, Lescure B (2003) Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33 957–966 [DOI] [PubMed] [Google Scholar]
- Tremousaygue D, Manevski A, Bardet C, Lescure N, Lescure B (1999) Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J 20 553–561 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Casneuf T, Van de Peer Y (2006) Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome Biol 7 R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11 799–811 [DOI] [PubMed] [Google Scholar]
- Wang Y, Henriksson E, Soderman E, Henriksson KN, Sundberg E, Engstrom P (2003) The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Dev Biol 264 228–239 [DOI] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2006) Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol 141 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G (1996) A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CY, Sedbrook JC, Perrin RM, Carroll KL, Masson PH (2005) Loss-of-function mutations of ROOT HAIR DEFECTIVE3 suppress root waving, skewing, and epidermal cell file rotation in Arabidopsis. Plant Physiol 138 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Chua NH (2000) Chemical-inducible systems for regulated expression of plant genes. Curr Opin Biotechnol 11 146–151 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH (2000) KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.