Abstract
Aux/IAA proteins are proposed to be transcriptional repressors that play a crucial role in auxin signaling by interacting with auxin response factors and repressing early/primary auxin response gene expression. In assays with transfected protoplasts, this repression was previously shown to occur when auxin concentrations in a cell are low, and derepression/activation was observed when auxin concentrations are elevated. Here we show that a stabilized version of the Arabidopsis (Arabidopsis thaliana) IAA17 repressor, when expressed constitutively or in a specific cell type in Arabidopsis plants, confers phenotypes similar to plants with decreased auxin levels. In contrast, a stabilized version of IAA17 that was converted to a transcriptional activator confers phenotypes similar to plants with increased auxin levels, when expressed under the same conditions in Arabidopsis plants. Free auxin levels were unchanged compared to control (DR5:β-glucuronidase), however, in the seedlings expressing the IAA17 repressor and activator. These results together with our previous results carried out in transfected protoplasts suggest that the hormone auxin can be bypassed to regulate auxin signaling in a cell-autonomous manner in plants.
The hormone auxin plays a central role in regulating a wide variety of growth and developmental responses/processes during the life span of plants. Many of the responses to auxin are mediated by what appears to be a simple, streamlined pathway that results in degradation of Aux/IAA transcriptional repressors and derepression of auxin response genes (Guilfoyle and Hagen, 2007; Kepinski, 2007). Aux/IAA repressors are proposed to function by interacting with transcriptional activators, the auxin response factors (ARFs), which are targeted to TGTCTC auxin response elements in promoters of auxin response genes (Tiwari et al., 2001, 2003, 2004; Guilfoyle and Hagen, 2007). Entry of auxin into a cell results in its binding to the Transport Inhibitor Response1 (TIR1) receptor/F-box ubiquitin ligase or related auxin signaling F-box (AFB) receptors/F-box ubiquitin ligases in a concentration-dependent manner (Gray et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). Binding of auxin to its receptors promotes the recruitment of the Aux/IAA repressors to TIR1 or AFB auxin receptor/ubiquitin ligase complex (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Tan et al., 2007). This recruitment, in turn, triggers the destruction of the repressors via the ubiquitin-proteasome pathway (Parry and Estelle, 2006; Mockaitis and Estelle, 2008), resulting in derepression/activation of auxin response genes (Guilfoyle and Hagen, 2007).
There are 29 Aux/IAA genes in Arabidopsis (Arabidopsis thaliana; Remington et al., 2004) and 31 Aux/IAA genes in rice (Oryza sativa; Jain et al., 2006), the bulk of which encode proteins with four conserved domains, referred to as domains I, II, III, and IV (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). Domain I is an active, portable repression domain containing a LxLxL motif (Tiwari et al., 2004) that interacts with the TOPLESS corepressor, at least in the case with IAA12/BDL (Szemenyei et al., 2008), to bring about repression of auxin response genes. Domain II confers instability to Aux/IAA proteins by targeting the repressors to the TIR1/AFB auxin receptors in an auxin dose-dependent manner, where they enter the ubiquitin-proteasome pathway (Parry and Estelle, 2006; Mockaitis and Estelle, 2008). A few of the Aux/IAA proteins do not have a recognizable domain II, and those that lack a domain II display increased stability (Dreher et al., 2006) and confer defective auxin signaling phenotypes when constitutively expressed in Arabidopsis (Sato and Yamamoto, 2008). Domains III and IV are protein-protein interaction domains found in ARFs as well as Aux/IAA proteins (Kim et al., 1997; Ulmasov et al., 1997a, 1997b).
Dominant or semidominant mutations in many of the 29 Arabidopsis Aux/IAA genes have been identified (Reed, 2001; Liscum and Reed, 2002; Woodward and Bartel, 2005; Uehara et al., 2008). These mutations reside in domain II and result in enhanced stability of the repressors due to their decreased affinity for the TIR1/AFB auxin receptors (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). Domain II mutants display a range of auxin-related phenotypes with some being unique and some overlapping for each Arabidopsis iaa mutant (Reed, 2001; Liscum and Reed, 2002; Woodward and Bartel, 2005; Uehara et al., 2008). In some cases, phenotypes of different iaa mutants may be opposite to one another (e.g. root hair formation in iaa3/shy2 versus iaa17/axr3; Knox et al., 2003). The range of phenotypes may be partially explained by the different expression patterns of Aux/IAA genes, but even when expressed from the same promoter, iaa mutants have nonidentical phenotypes (Knox et al., 2003; Weijers et al., 2005; Muto et al., 2007), suggesting that intrinsic properties of different Aux/IAA repressors may also contribute to the mutant phenotypes.
In previous studies with transfected protoplasts, we showed that expression of Aux/IAA proteins from 35S promoter:effector plasmids resulted in repression of auxin-responsive reporter gene expression (Tiwari et al., 2001, 2004). Stabilized versions of Aux/IAA proteins with mutations in domain II (e.g. IAA17mII) functioned as stronger repressors than wild-type Aux/IAA proteins, while point mutations in the repression domain (e.g. IAA17mImII) compromised, but did not eliminate repression by stabilized Aux/IAA proteins (Tiwari et al., 2001). We also showed that the compromised repressor, IAA17mImII, could be converted to an activator by fusing the herpes simplex virus VP16 activation domain to its N terminus (i.e. VP16-IAA17mImII; Tiwari et al., 2003). These results suggested that a stabilized version of VP16-IAA17 can activate auxin response genes in an auxin-independent manner, and thus, bypass the hormone signal in the auxin signaling pathway. Here, we report on bypassing the auxin signal in transformed Arabidopsis plants by expressing a stabilized form of a VP16-IAA17 activator, and contrast these plants with those expressing a stabilized form of an IAA17 repressor. Our results provide support for a model in which natural Aux/IAA proteins function through an active repression domain to regulate auxin-responsive gene expression in planta and provide a strategy to control auxin signaling in an auxin-independent and cell-autonomous manner.
RESULTS
Constitutive Expression of a Stabilized IAA17 Repressor and a Stabilized IAA17 Activator Confer Low and High Auxin Phenotypes, Respectively, in Transformed Arabidopsis Plants
We used the same version of 35S:VP16-IAA17mImII gene described by Tiwari et al. (2003) that functioned as an activator of auxin-responsive reporter gene expression, to transform Arabidopsis plants (Fig. 1A). More than 10 transformed lines were selected that expressed the construct and had similar phenotypes. Analyses of two representative lines that overexpress the single-copy IAA17 transgene at levels 30- to 40-fold higher than wild-type IAA17 (Fig. 1B) are reported here.
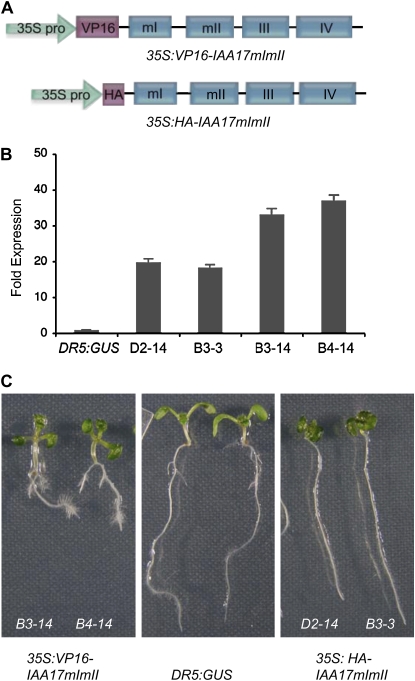
Figure 1.
Constructs and phenotypes of Arabidopsis seedlings transformed with 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII. A, Schematic diagrams of the 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII constructs used to transform Arabidopsis plants. Expression of the modified IAA17 proteins was driven by the 35S cauliflower mosaic virus promoter, and constructs are identical to those described by Tiwari et al. (2001, 2003). The 35S:HA-IAA17mImII construct has a HA epitope tag at the N terminus of IAA17 in place of the VP16 activation domain in the 35S:VP16-IAA17mImII construct. The four conserved domains, I to IV, are indicated, and m indicates a missense mutation(s) in domains I and II. In domain I, ETELCLGL was changed to ETVRCLGL, and in domain II, GWPPV was changed to GWSPV. B, Constitutive expression of HA-IAA17mImII (lines D2-14 and B3-3) and VP16-IAA17mImII (lines B3-14 and B4-14). qRT-PCR was used to determine the gene expression levels of IAA17 in control seedlings (DR5:GUS) and IAA17 plus the transgene in transformed seedlings. The expression level of IAA17 in control seedlings was set at 1.0, and expression levels of the transgenes are presented relative to the DR5:GUS control. C, Seven-day-old DR5:GUS control seedlings and seedlings transformed with 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII. A seedling from two 35S:HA-IAA17mImII (lines D2-14 and B3-3) and 35S:VP16-IAA17mImII (lines B3-14 and B4-14) independent lines is shown.
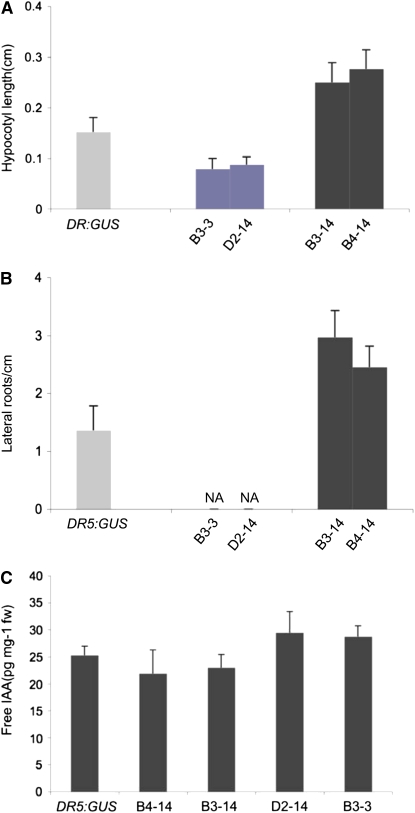
Compared to the DR5:GUS line, which had a phenotype indistinguishable from wild type and was used as a control in experiments presented here, the two 35S:VP16-IAA17mImII lines had long hypocotyls, long petioles, small epinastic cotyledons and leaves, short, highly branched roots, and dense root hairs (Fig. 1C). Hypocotyl length of 7-d-old light-grown seedlings transformed with the 35S:VP16-IAA17mImII transgene was about 60% to 70% greater than control (Fig. 2A). The number of lateral roots/cm of primary root was approximately 1.5- to 2-fold higher in 7-d-old seedlings expressing the transgene compared to control (Fig. 2B). Adult plants transformed with the 35S:VP16-IAA17mImII gene had narrow, twisted leaves, apically dominant inflorescences with a zigzag pattern of short siliques on the inflorescence stalks, and poor fertility (Fig. 3, A and C). In several ways, the seedling and adult phenotypes of the 35S:VP16-IAA17mImII lines resembled yucca1, yucca4, yucca6, CYP79B2ox, sur1, sur2, FZYox, and iaaM mutants that overproduce auxin (Klee et al., 1987; Boerjan et al., 1995; King et al., 1995; Barlier et al., 2000; Zhao et al., 2001, 2002; van der Graaff et al., 2003; Kim et al., 2007; Li et al., 2008). The phenotypes observed in the 35S:VP16-IAA17mImII seedlings did not result from increased free indole-3-acetic acid (IAA) levels, however, because free IAA levels in the 35S:VP16-IAA17mImII lines were equal to or marginally less than IAA levels in the DR5:GUS control seedlings (Fig. 2C).
Figure 2.
Quantitative characteristics of DR5:GUS and transgenic seedlings expressing 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII. A, Measurement of hypocotyl length in control (DR5:GUS), 35S:HA-IAA17mImII (lines B3-3, D2-14), and 35S:VP16-IAA17mImII 7-d-old seedlings (lines B3-14, B4-14; n ≥ 15, ANOVA test, P < 0.01). B, Lateral root numbers per centimeter in control, 35S:HA-IAA17mImII (lines B3-3, D2-14), and 35S:VP16-IAA17mImII 7-d-old seedlings (lines B3-14, B4-14). NA, 0.00 ± 0 (n ≥ 15, ANOVA test, P < 0.01). C, Free IAA levels in control, 35S:HA-IAA17mImII (lines B3-3, D2-14), and 35S:VP16-IAA17mImII (lines B3-14, B4-14) seedlings. ANOVA analyses indicate that the values are not different from each other (F = 3.693, P = 0.43) in post hoc pairwise comparisons (Tukey and SNK) or comparison to controls (Student's, Bonferroni, P = 0.285, 0.257, 0.152, 0.078, respectively). [See online article for color version of this figure.]
Figure 3.
Phenotypes of adult Arabidopsis plants expressing 35S:HA-IAAA17mImII and 35S:VP16-IAA17mImII. A, An adult DR5:GUS control plant and adult plants transformed with 35S:HA-IAA17mImII (line B3-3) and 35S:VP16-IAA17mImII (line B3-14). Inset, Zigzag pattern of the inflorescence stalk in a 35S:VP16-IAA17mImII plant. B, Small stature of a 35S:HA-IAA17mImII plant. Bar = 1 cm. C, Representative siliques of 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII compared to DR5:GUS control.
That the above phenotypes result from the addition of the VP16 activation domain to IAA17 is supported by analysis of plants transformed with a single-copy 35S:HA-IAA17mImII gene (Fig. 1A), which contains an N-terminal hemagglutinin (HA) epitope tag in place of the VP16 moiety. We have previously shown that expression of the 35S:HA-IAA17mImII construct represses auxin-responsive reporter gene expression in transfected protoplasts, and that the mutation in domain I (mI) weakened, but did not eliminate repression (Tiwari et al., 2001). Seedlings in two lines expressing the 35S:HA-IAA17mImII transgene at levels 15- to 20-fold higher than the wild-type gene (Fig. 1B) had short hypocotyls and petioles and unbranched primary roots (Fig. 1C). Seven-day-old seedlings expressing the 35S:HA-IAA17mImII transgene had hypocotyls that were about half the length of control hypocotyls (Fig. 2A) and had no lateral roots, in contrast to control seedlings (Fig. 2B). Adult plants were bushy dwarfs with short inflorescence stalks and short, wrinkled siliques (Fig. 3). The phenotypes of 35S:HA-IAA17mImII plants contrast markedly with the phenotypes observed for the 35S:VP16-IAA17mImII lines and resemble phenotypes of mutants with decreased auxin levels (Romano et al., 1991; Rampey et al., 2004). The low auxin-like phenotypes observed in the 35S:HA-IAA17mImII seedlings did not result from reduced free IAA levels, because 35S:HA-IAA17mImII lines had free IAA levels that were equal to or slightly greater than IAA levels in the DR5:GUS control seedlings (Fig. 2C).
We also examined first generation plants (T1) that were transformed with 35S:HA-IAA17mII (i.e. IAA17 with only a domain II mutation, which functions as a stronger repressor than IAA17 with mutations in both domains I and II; Tiwari et al., 2001). These plants, like plants transformed with 35S:HA-IAA17mImII had phenotypes similar to mutants with decreased auxin levels; however, the phenotypes of plants transformed with the 35S:HA-IAA17mII gene were lost in T2 generation plants (i.e. T2 lines were indistinguishable from wild type). While we have not investigated the reason for this loss of phenotype, it may be that constitutive expression of these strong repressors compromises the auxin response to such an extent that expression of the transgenes is eliminated following the T1 generation. In contrast to unstable phenotypes observed with the 35S:HA-IAA17mII, phenotypes observed with 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII lines were maintained at least through the T4 generation.
DR5:GUS Reporter Gene Expression Is Constitutively Activated in Seedlings Expressing the 35S:VP16-IAA17mImII Transgene and Constitutively Repressed in Seedlings Expressing the 35S:HA-IAA17mImII Transgene
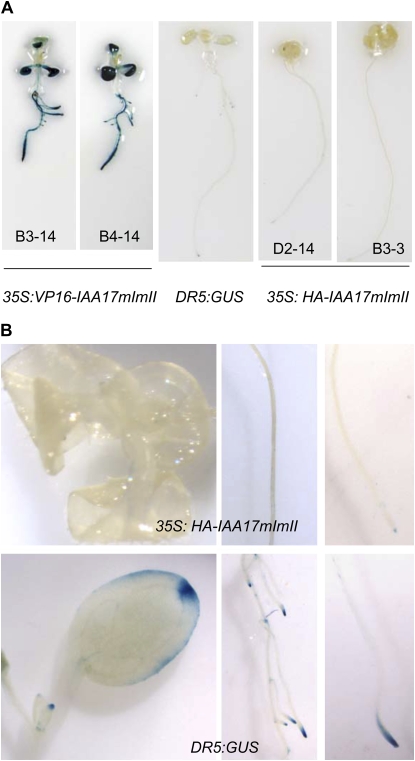
The phenotypes described above suggest that plants expressing the 35S:VP16-IAA17mImII transgene display a constitutively high auxin response in the absence of applied auxin, while plants expressing the 35S:HA-IAA17mImII transgene display a constitutively low auxin response. To examine this further, the DR5:GUS auxin-responsive reporter gene line (Ulmasov et al., 1997b) was crossed into 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII lines. Figure 4 shows DR5:GUS expression in control seedlings compared to seedlings expressing either the 35S:VP16-IAA17mImII or the 35S:HA-IAA17mImII transgene. Compared to DR5:GUS control seedlings, 35S:VP16-IAA17mImII lines showed strongly enhanced expression of DR5:GUS in seedlings that were not exposed to auxin. In contrast, DR5:GUS expression observed in leaf margins, primary root tips, and developing lateral roots (i.e. auxin maxima) in control seedlings was strongly reduced or eliminated in the 35S:HA-IAA17mImII seedlings. These results indicate that the auxin-responsive reporter gene is constitutively activated in the 35S:VP16-IAA17mImII lines and constitutively repressed in 35S:HA-IAA17mImII lines.
Figure 4.
Expression of DR5:GUS auxin-responsive reporter gene in a wild-type seedling and seedlings transformed with 35S:VP16-IAA17mImII and 35S:HA-IAA17mImII. A, Seven-day-old seedlings from two 35S:VP16-IAA17mImII (lines B3-14 and B4-14) and two 35S:HA-IAA17mImII (lines D2-14 and B3-3) independent lines are shown. Seedlings were removed from agar plates and histochemically stained for GUS. B, Loss of DR5:GUS expression (auxin maxima) in 35S:HA-IAA17mImII (B3-3) compared to expression in leaf margin, developing lateral roots, and primary root tip of DR5:GUS control seedlings.
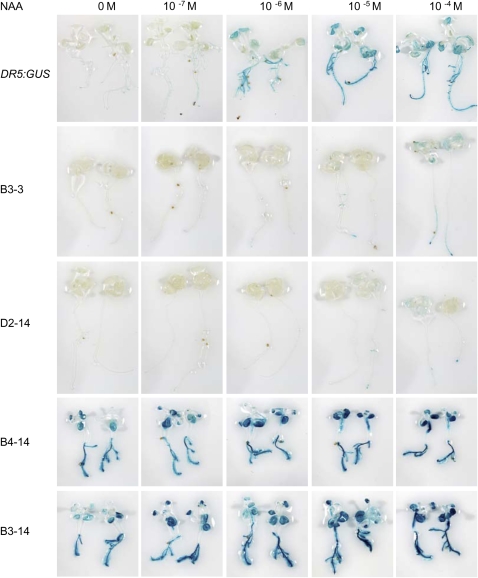
When control seedlings containing the DR5:GUS reporter gene were treated with the synthetic auxin, 1-naphthalene acetic acid (1-NAA), over a concentration range of 10−7 to 10−4 m, the reporter gene was strongly activated at 10−6 to 10−4 m 1-NAA (Fig. 5). Unlike control seedlings, the DR5:GUS reporter gene was only weakly activated by applied auxin even at 10−4 m 1-NAA in lines expressing the 35S:HA-IAA17mImII transgene. With seedlings expressing the 35S:VP16-IAA17mImII transgene, DR5:GUS gene expression was already high without applied auxin and increased only slightly in response to auxin over the concentration range tested.
Figure 5.
Comparison of auxin-responsive DR5:GUS expression in control, 35S:HA-IAA17mImII (lines B3-3, D2-14), and 35S:VP16-IAA17mImII (lines B3-14, B4-14) seedlings. Seven-day-old seedlings were removed from agar plates and incubated in a solution containing the concentration of 1-NAA indicated for 12 h and then histochemically stained for GUS.
A Variety of Natural Auxin Response Genes Are Constitutively Activated in Seedlings Expressing the 35S:VP16-IAA17mImII Transgene and Constitutively Repressed in Seedlings Expressing the 35S:HA-IAA17mImII Transgene
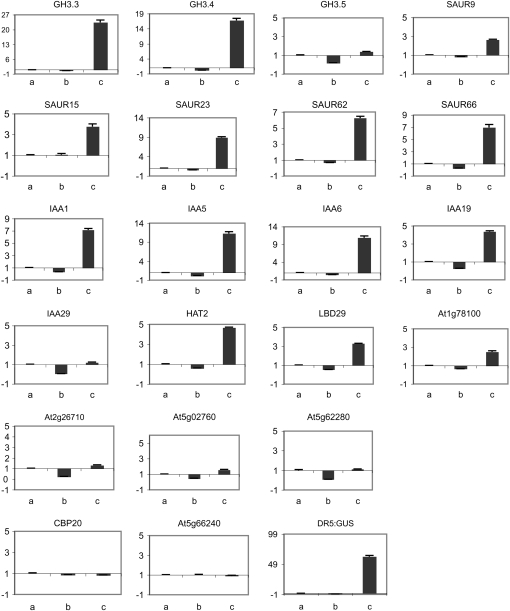
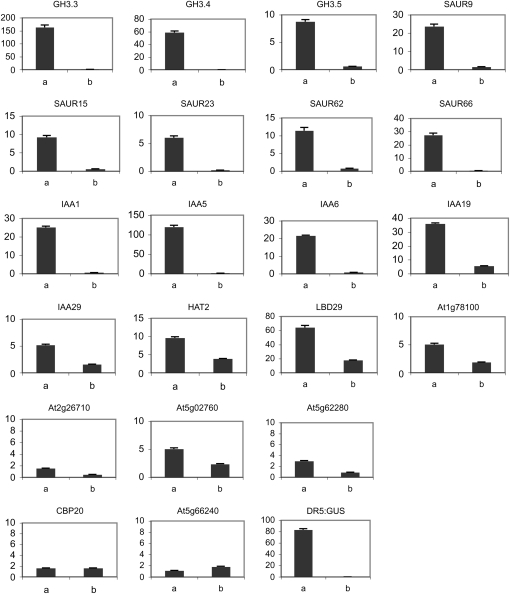
To determine if natural genes were constitutively repressed and constitutively activated like the DR5:GUS reporter gene in seedlings transformed with 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII lines, respectively, we initially carried out microarray analyses with DR5:GUS control seedlings (7-d-old) not treated or treated for 1 h with 1 μm 1-NAA (the synthetic auxin, naphthalene acidic acid), a 35S:HA-IAA17mImII line not treated or treated for 1 h with 1 μm 1-NAA, and a 35S:VP16-IAA17mImII line not treated with auxin. The microarray data were used as a guide to identify genes that were up-regulated in auxin-treated DR5:GUS seedlings, down-regulated in 35S:HA-IAA17mImII seedlings compared to DR5:GUS whether not treated or treated with auxin, and up-regulated in 35S:VP16-IAA17mImII seedlings compared to wild-type seedlings. A set of genes was then selected to carry out quantitative reverse transcription (qRT)-PCR for analysis of gene expression (Supplemental Table S1). Figure 6 shows that selected GH3, SAUR, Aux/IAA, and other auxin response genes, all of which have been identified in previous microarray analyses to be strongly induced by auxin (Nemhauser et al., 2004, 2006), were down-regulated in the 35S:HA-IAA17mImII line and up-regulated in the 35S:VP16-IAA17mImII line. The down-regulation in the 35S:HA-IAA17mImII line is not large with most genes analyzed because these genes, like the DR5:GUS reporter gene, are not highly expressed in seedlings that have not been exposed to exogenous auxin. Application of exogenous auxin to 35S:HA-IAA17mImII seedlings failed to activate the natural auxin response genes, unlike what is observed with control seedlings (Fig. 7). These results indicated that expression of the 35S:HA-IAA17mImII transgene in Arabidopsis seedlings results in constitutive repression of a spectrum of auxin response genes and that this repression is not relieved by treating seedlings with exogenous auxin, while expression of the 35S:VP16-IAA17mImII transgene results in constitutive activation of these same auxin response genes.
Figure 6.
Expression levels for selected auxin response genes in control (DR5:GUS) seedlings and seedlings transformed with 35S:HA-IAA17mImII (line D2-14) and 35S:VP16-IAA17mImII (line B3-14). Seven-day-old seedlings were used for qRT-PCR. Gene expression levels in DR5:GUS seedlings were set at 1.0, and gene expression levels in seedlings containing the modified IAA17 transgenes are presented relative to the DR5:GUS control. Expression levels of DR5:GUS and two non-auxin response genes, CBP20 (At5g44200) and At5g66240, are also shown. a, DR5:GUS control; b, 35S:HA-IAA17mImII; c, 35S:VP16-IAA17mImII.
Figure 7.
Auxin-induced expression levels for selected auxin response genes in control (DR5:GUS) seedlings and seedlings transformed with 35S:HA-IAA17mImII (line D2-14). Seven-day-old seedlings were removed from agar plates and incubated in a solution containing 1 μm 1-NAA for 1 h. qRT-PCR was used to determine the expression levels of genes indicated. See Figure 6 for additional details on the genes analyzed and presentation of the data. a, DR5:GUS control; b, 35S:HA-IAA17mImII.
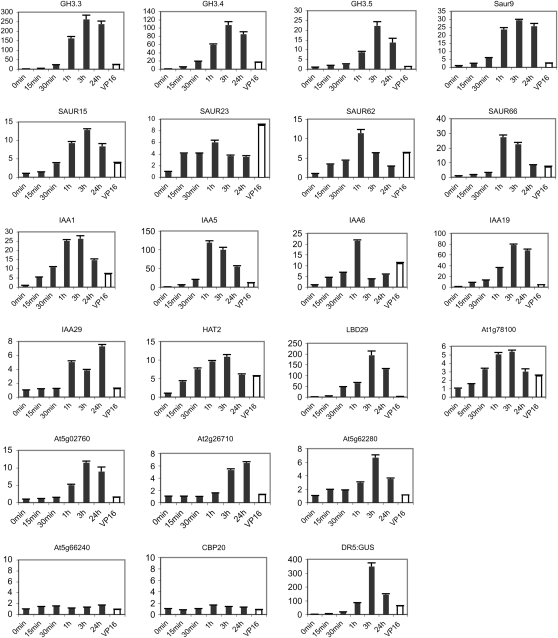
To gain insight into how robustly the 35S:VP16-IAA17mImII transgene functioned in conferring a constitutive auxin response, we compared expression of the auxin response genes in a 35S:VP16-IAA17mImII line with that in control seedlings exposed to exogenous auxin over a 24-h period. A number of early auxin response genes have been shown to display transient activation when exposed to exogenous auxin, with expression returning to basal or near-basal levels following long exposure times to auxin (Abel et al., 1995; Nakamura et al., 2003; Nemhauser et al., 2006). Figure 8 shows a time course for auxin-induced expression of genes analyzed in Figures 6 and 7. The bulk of these genes, including all of the GH3, SAUR, and IAA genes examined, were most highly expressed within 1 to 3 h of auxin treatment, and expression levels declined thereafter (i.e. 3–24 h of treatment). These results along with previous results (Abel et al., 1995; Nakamura et al., 2003; Nemhauser et al., 2006) suggest that some feedback regulation must function in desensitizing the early response genes to auxin. Figure 8 also includes data showing gene expression for seedlings containing the 35S:VP16-IAA17mImII transgene (i.e. replotted from data presented in Fig. 6). These latter results indicate that, in general (i.e. SAUR23 being an exception), seedlings containing the 35S:VP16-IAA17mImII transgene expressed the auxin response genes at levels considerably less than the maximal expression levels observed with seedlings exposed to applied auxin. With plants expressing the 35S:VP16-IAA17mImII transgene, it is possible that a full auxin response is attenuated to a level that allows the transformed plants to survive.
Figure 8.
Time course for auxin-induced expression for selected auxin response genes in control (DR5:GUS) seedlings. Seven-day-old seedlings were removed from agar plates and incubated in a solution containing 1 μm 1-NAA for the times indicated. qRT-PCR was used to determine the expression levels of genes indicated. Gene expression levels at time 0 were set at 1.0, and gene expression levels for treatments at indicated time points are presented relative to time 0 (black bars). For comparison, the expression levels of these genes are also shown for untreated (i.e. no exogenous auxin) 7-d-old seedlings expressing the 35S:VP16-IAA17mImII transgene (line B3-14; white bars, labeled as VP16). See Figure 6 for additional details on the genes analyzed.
Expression of the HA-IAA17mImII and VP16-IAA17mImII Transgenes under Control of the EXPANSIN7 Promoter Has Dramatic Effects on Root Hair Growth and Development
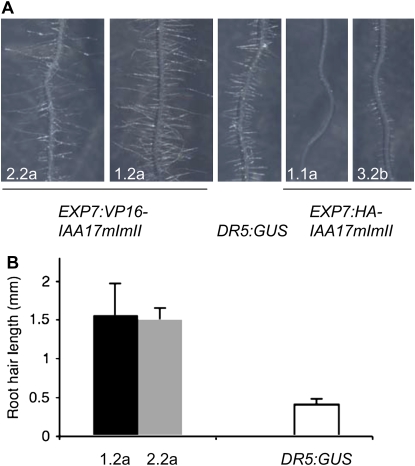
The EXPANSIN7 (EXP7) gene and promoter (i.e. EXP7:GFP and EXP7:GUS) have been reported to be expressed primarily in root hair and trichoblast cells (Cho and Cosgrove, 2002; Kim et al., 2006; Lee and Cho, 2006; Singh et al., 2008). Lee and Cho (2006) showed that when a PINOID protein kinase-GFP fusion protein (i.e. PID-GFP) or a stabilized version of IAA7 (i.e. AXR2) was expressed under control of the EXP7 promoter, root hair formation and elongation was suppressed. To determine if expression of EXP7:HA-IAA17mImII and EXP7:VP16-IAA17mImII transgenes altered root hair development and growth, homozygous seedlings expressing these transgenes were analyzed. Figure 9 shows that expression of the EXP7:HA-IAA17mImII transgene suppressed root hair formation and growth, while expression of the EXP7:VP16-IAA17mImII transgene enhanced root hair elongation compared to DR5:GUS. Our results with the EXP7:HA-IAA17mImII transgene are consistent with those of Lee and Cho (2006), which indicate that auxin signaling is required for root hair growth and development, and further suggest that there is nothing inherently different with regard to how two different Aux/IAA repressors function in Arabidopsis root hairs. In addition, our results with the EXP7:VP16-IAA17mImII transgene indicate that root hair growth can be enhanced by constitutively activating the auxin response pathway in root hairs. Our results with the EXP7:HA-IAA17mImII and EXP7:VP16-IAA17mImII transgenes taken together suggest that auxin signaling can be effectively suppressed or enhanced in a cell-autonomous manner and provide support for an important role of auxin in root hair growth and development.
Figure 9.
Root hair growth and development in DR5:GUS control seedlings and seedlings transformed with EXP7:VP16-IAA17mImII and EXP7:HA-IAA17mImII. A, Images of root hairs in control 7-d-old seedlings and seedlings transformed with EXP7:VP16-IAA17mImII (roots from two independent lines, 2.2a and 1.2a, are shown) and EXP7:HA-IAA17mImII (roots from two independent lines, 1.1a and 3.2b, are shown). B, Root hair length measurements in control 7-d-old seedlings and seedlings transformed with EXP7:VP16-IAA17mImII (two independent lines, 2.2a and 1.2a; n ≥ 20, ANOVA test, P < 0.001).
DISCUSSION
Because the auxin signal transduction pathway is streamlined, simple, and ultimately dependent on the stability of the Aux/IAA transcription factors, we reasoned that it should be possible to down-regulate the auxin response pathway in plants by constitutively expressing a stabilized version of an Aux/IAA repressor and to up-regulate the auxin response pathway by constitutively expressing a version of an Aux/IAA protein that had been converted from a repressor to a stabilized activator. Converting the repressor to an activator was made possible by mutating the repression domain of IAA17 and adding an activation domain in the form of VP16 (Tiwari et al., 2003). Previous results, using transfected protoplasts, indicated that the VP16 activation domain was ineffective when fused to an IAA17mII mutant, which contained a wild-type repression domain, suggesting that the wild-type repression domain in IAA17 was dominant over the VP16 activation domain (Tiwari et al., 2003).
Results presented here show that Arabidopsis plants constitutively expressing a stabilized version of an Aux/IAA protein that was converted into an activator have high auxin-like phenotypes, including enhanced expression of auxin response genes. This contrasts with plants that express a stabilized Aux/IAA repressor and have low auxin-like phenotypes with reduced auxin response gene expression. Free auxin levels were not significantly altered in seedlings expressing the 35S:VP16-IAA17mImII or 35S:HA-IAA17mImII transgene, however, suggesting that the high and low auxin-like phenotypes do not result from altered free auxin levels in the IAA17 transgenic lines. Therefore, the constitutively expressed, stabilized Aux/IAA activator and repressor appear to function in an auxin-independent manner to up-regulate and down-regulate auxin response genes and confer high and low auxin-like phenotypes, respectively.
In addition, our results show that by restricting expression of the modified Aux/IAA proteins to a specific cell type (e.g. in this case, trichoblast/root hair cells), auxin signaling can be suppressed or enhanced in a localized manner. By selecting appropriate promoters to drive expression of modified IAA17 proteins, it should be possible to not only shut down auxin signaling in specific cell types or under specific environmental or developmental conditions (i.e. as has been demonstrated here and previously with stabilized Aux/IAA repressors; Park et al., 2002; Fukaki et al., 2005; De Smet et al., 2007; Lucas et al., 2007; Singh et al., 2008), but to enhance auxin signaling (i.e. by using stabilized Aux/IAA activators as demonstrated here). While we have shown that root hair-specific expression of an IAA17mImII transgene can effectively suppress root hair growth, expression of IAA17mII or other Aux/IAAmII proteins (i.e. lacking a mutation in the repression domain) would likely be more effective in suppressing auxin responses when expressed in a cell/tissue-specific manner because of their greater capacity to repress auxin response genes. It is possible that by controlling the auxin signaling pathway in a cell-autonomous, developmental-specific, or environmental-specific manner, plants might be modified to enhance lateral root or root hair formation and growth, alter plant architecture, or enhance/suppress processes that require an auxin signal.
Our results indicate that a variety of auxin response genes can be targeted for repression or activation by a single, stabilized member of Aux/IAA repressor family (i.e. namely IAA17) or converted activator, respectively. Results reported here, along with our previous results using protoplast transfection assays (Tiwari et al., 2001, 2003, 2004), support a model for auxin signaling in which Aux/IAA repressors function directly on auxin response genes to actively repress their transcription. This is also supported by recent results that show that the IAA12/BDL repressor interacts with the corepressor TOPLESS in repressing auxin-responsive gene expression (Szemenyei et al., 2008). Furthermore, if Aux/IAA proteins were passive repressors and simply sequestered ARF activators away from their target sites, then expression of 35S:VP16-IAA17mImII transgenes would be expected to have the same effect as expression of 35S:HA-IAA17mImII transgenes in preventing ARFs from reaching their target sites. Because expression of 35S:VP16-IAA17mImII transgenes result in constitutive activation of auxin response genes, the modified IAA17 protein must function directly at the gene activation level. Whether the VP16 activation domain functions by recruiting a coactivator to auxin-responsive promoters or somehow prevents the IAA17 repression domain from interacting with a corepressor (e.g. TOPLESS) remains to be determined. It is possible that the VP16 activation domain simply interferes with the IAA17 repression domain, allowing the ARF activation domain to function even when an ARF is associated with the modified Aux/IAA protein.
MATERIALS AND METHODS
Vector Constructs and Transgenic Lines
The 35S:HA-IAA17mImII and 35S:VP16-IAA17mImII plasmid constructs have been previously described (Tiwari et al., 2001, 2003). For EXP7 At1g12560 promoter:IAA constructs, the EXP7 promoter was obtained by PCR amplification of Columbia genomic DNA with Fw primer 5′-TCTAGATGATTACAAAGGGGAAATTTAGG-3′, and Rv primer 5′-CCATGGTCGATAGTATCCAGCGACGA-3′, and was subcloned into pPZP221 vector harboring VP16-IAA17mImII and HA-IAA17mImII, respectively. Cloning fidelity was confirmed by sequencing.
All transgenic plants used in this report are in DR5:GUS background. EXP7 constructs were introduced into DR5:GUS background using Agrobacterium-mediated transformation by the floral-dip method (Clough and Bent, 1998). T3 homozygous lines were analyzed in this study unless indicated otherwise. 35S:HA-IAA17mImII X DR5:GUS and 35S:VP16-IAA17mImII X DR5:GUS were obtained by crossing. Plants containing the DR5:GUS transgene have been previously described (Ulmasov et al., 1997b).
Plant Growth and Treatments
Arabidopsis (Arabidopsis thaliana) plants were grown in growth chambers at 22°C under continuous, cool-white fluorescent light. For experiments with seedlings, Arabidopsis seeds were sterilized and germinated on horizontal or vertical plates as described previously (Wang et al., 2005). For auxin treatments, 7-d-old seedlings were removed from the agar plates and incubated with shaking in liquid media containing or lacking 1 μm 1-NAA for the times indicated. After incubation, seedlings were frozen in liquid nitrogen and stored at −70°C prior to RNA isolation. Root hair length was measured using ImageJ (http://rsb.info.nih.gov/ij/) from photographs taken with a Leica stereomicroscope at the University of Missouri Molecular Cytology Core. For measurements of hypocotyl length and lateral root numbers, 7-d-old seedlings were scanned and analyzed using ImageJ.
Free IAA Level Determination
Free IAA determinations were performed as described in Kim et al. (2007) using 6-d-old seedlings. Three independent determinations were made for each line using 65 seedlings for each sample. The whole assay series was repeated a second time with similar results.
Analysis of variance was performed using Student's t test followed by post hoc analyses (Tukey, Student-Newman-Keuls pairwise, Bonferroni to control).
GUS Staining
Gus staining was carried out as previously described (Ulmasov et al., 1997b). Seedlings were placed in GUS solution for 12 h at 37°C and cleared in 70% ethanol.
RNA Isolation and Real-Time RT-PCR Analysis
RNA was extracted using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocol. For the microarray analysis, hybridizations and array scanning were carried out at the University of Missouri DNA Core Facility with two biological and two technical replicates. The detection P value was generated, signal values for individual genes were obtained using statistical algorithms on Affymetrix GeneChip Microarray Suite version 5.0 software, and the resulting CEL files were further analyzed by GeneChip Operating Software 1.4 (Affymetrix). For qRT-PCR, during the RNA isolation, RNase-Free DNase (Qiagen) was used to eliminate any DNA contamination. First-strand cDNA was synthesized from 2 μg total RNA using an Omniscript RT kit (Qiagen). One microliter of the resulting cDNA was subjected to qRT-PCR using SYBR Green Supermix in a CFX96 Real-Time system (Bio-Rad).
Efficiency of each primer pair was determined by standard curves using serial DR5:GUS cDNA dilutions. PCR was performed using a three-step protocol with a melting curve. Based on primer efficiency, fold expression was generated in Bio-Rad CFX manager (version 1.0) after normalization to TUB5 (At1g20010). Triplicates for each PCR reaction and at least two biological replicates were performed for each gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primer pairs used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Shiv Tiwari for making the original IAA17 constructs and generating transgenic T1 Arabidopsis plants with the IAA17 constructs.
This work was supported by the National Science Foundation (grant no. IOB 0550417 to T.J.G. and G.H.) and the University of Missouri Food for the 21st Century Program (to T.J.G.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tom J. Guilfoyle (guilfoylet@missouri.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251 533–549 [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Montagu MV, Inzé D (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audeaert D, et al (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134 681–690 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005. a) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann J, Jürgens G, Estelle M (2005. b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9 109–119 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44 382–395 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 271–276 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10 453–460 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49 373–385 [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6 47–59 [DOI] [PubMed] [Google Scholar]
- Kepinski S (2007) The anatomy of auxin perception. Bioessays 29 953–956 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB (1995) A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee JH, Horsch RB, Hinchee MA, Hein MB, Hoffmann NL (1987) The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev 1 86–96 [Google Scholar]
- Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130 5769–5777 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho HT (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Qin GJ, Tsuge T, Hou XH, Ding MY, Aoyama T, Oka A, Chen Z, Gu H, Zhao Y, et al (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179 751–764 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49 387–400 [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L (2007) Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot 59 55–66 [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24 55–80 [DOI] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT (2007) Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2 E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim HJ, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32 669–683 [DOI] [PubMed] [Google Scholar]
- Parry G, Estelle M (2006) Auxin receptors: a new role for F-box proteins. Curr Opin Cell Biol 18 152–156 [DOI] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B (2004) A family of auxin-conjugate hydrolases that contributes to free indole-acetic acid levels during Arabidopsis germination. Plant Physiol 135 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6 420–425 [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Mein MB, Klee HJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev 5 438–446 [DOI] [PubMed] [Google Scholar]
- Sato A, Yamamoto KT (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Plant Physiol 133 397–405 [DOI] [PubMed] [Google Scholar]
- Singh SK, Fischer U, Singh M, Grebe M, Marchant A (2008) Insight into the early steps of root hair formation revealed by the procuste1 cellulose synthase mutant of Arabidopsis thaliana. BMC Plant Biol 8 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H (2008) Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49 1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997. a) ARF1, a transcription factor that binds auxin response elements. Science 276 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997. b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Boot K, Granbom R, Sandberg G, Hooykaas PJJ (2003) Increased endogenous auxin production in Arabidopsis thaliana causes both earlier described and novel auxin-related phenotypes. J Plant Growth Regul 22 240–252 [Google Scholar]
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ (2005) AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Klentz M, Wilmoth JC, Reed JW, Jürgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79Ba2 and CYP79B3. Genes Dev 16 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.