Abstract
Starch biosynthetic enzymes from maize (Zea mays) and wheat (Triticum aestivum) amyloplasts exist in cell extracts in high molecular weight complexes; however, the nature of those assemblies remains to be defined. This study tested the interdependence of the maize enzymes starch synthase IIa (SSIIa), SSIII, starch branching enzyme IIb (SBEIIb), and SBEIIa for assembly into multisubunit complexes. Mutations that eliminated any one of those proteins also prevented the others from assembling into a high molecular mass form of approximately 670 kD, so that SSIII, SSIIa, SBEIIa, and SBEIIb most likely all exist together in the same complex. SSIIa, SBEIIb, and SBEIIa, but not SSIII, were also interdependent for assembly into a complex of approximately 300 kD. SSIII, SSIIa, SBEIIa, and SBEIIb copurified through successive chromatography steps, and SBEIIa, SBEIIb, and SSIIa coimmunoprecipitated with SSIII in a phosphorylation-dependent manner. SBEIIa and SBEIIb also were retained on an affinity column bearing a specific conserved fragment of SSIII located outside of the SS catalytic domain. Additional proteins that copurified with SSIII in multiple biochemical methods included the two known isoforms of pyruvate orthophosphate dikinase (PPDK), large and small subunits of ADP-glucose pyrophosphorylase, and the sucrose synthase isoform SUS-SH1. PPDK and SUS-SH1 required SSIII, SSIIa, SBEIIa, and SBEIIb for assembly into the 670-kD complex. These complexes may function in global regulation of carbon partitioning between metabolic pathways in developing seeds.
An important question in plant physiology is the means by which glucan storage homopolymers are synthesized such that they are able to assemble into semicrystalline starch granules. The starch polymer amylopectin consists of α-(1→4)-linked Glc units in linear chains, and these are joined to each other by α-(1→6) branch linkages. A distinguishing feature of amylopectin is that the branch points are clustered relative to each other (Thompson, 2000). The functional properties of starch depend on this ordered structure, which allows crystallization of the linear glucan chains that extend from the branch clusters. Packing of insoluble Glc units provides plants with a stable and abundant energy source to maintain metabolic needs in the absence of light. Considering that crystallization draws metabolic equilibria toward carbohydrate accumulation, another important physiological question is how the flux of reduced carbon is regulated such that seeds and other storage tissues achieve the proper balance of starch compared with protein and lipids.
Biosynthesis of crystalline starch is accomplished in large part by the coordinated activities of starch synthases (SSs) and starch branching enzymes (SBEs), together with starch debranching enzymes (DBEs; Ball and Morell, 2003). SSs catalyze linear chain elongation by addition of a Glc unit donated from the nucleotide sugar ADP-Glc (ADPGlc) to the nonreducing end of an acceptor chain. Branch linkages are formed by the action of SBEs, which cleave a linear chain and transfer the released fragment to a C6 hydroxyl group of the same or a neighboring chain. DBEs hydrolyze branch linkages, and genetic evidence indicates that this function is necessary in order for plants to accumulate crystalline starch (James et al., 1995; Ball et al., 1996; Myers et al., 2000). Multiple classes of SBE, SS, and DBE are highly conserved in the plant kingdom (Ball and Morell, 2003; Li et al., 2003; Leterrier et al., 2008).
Recent evidence indicates that certain SSs and SBEs are capable of physically associating with each other (Tetlow et al., 2004a, 2004b, 2008; Hennen-Bierwagen et al., 2008). The first such evidence came from analysis of amyloplast extracts from developing wheat (Triticum aestivum) endosperm, showing that SBEI, SBEIIb, and starch phosphorylase coimmunoprecipitate and that phosphorylation of one or more of those proteins is necessary for the association (Tetlow et al., 2004b). Further studies in maize (Zea mays) and wheat utilized combinations of yeast two-hybrid assays, affinity purification with immobilized recombinant ligands, and immunoprecipitation to demonstrate a large number of pair-wise interactions involving SSI, SSIIa, SSIII, SBEIIa, and SBEIIb (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008). Gel permeation chromatography (GPC) analyses of maize amyloplast extracts demonstrated the existence of complexes in elution peaks corresponding to approximately 670 and 300 kD (referred to as C670 and C300, respectively) that contained SSIII, SSIIa, SBEIIa, and SBEIIb in varying relative concentrations (Hennen-Bierwagen et al., 2008). Essentially all of the SSIII in the amyloplast extracts is in C670, and the great majority of SSIIa is in C300, suggesting that the physiological functions of these proteins derive from these assembly states. Understanding the functions of the complexes will require detailed characterization of their constituents, including any other binding partners that may be present.
Among these enzymes, SSIII has been implicated from several observations as a regulator of starch biosynthesis, in addition to its enzymatic role. Mutations in the maize gene dull1 (du1), which codes for SSIII, eliminated SSIII enzyme activity, as expected, and in addition simultaneously caused a major reduction in the activity of SBEIIa (Boyer and Preiss, 1981). The du1− mutation of maize or the equivalent genetic defect in rice (Oryza sativa) resulted in elevated total SS activity in soluble endosperm extracts as a result of increased SSI activity (Singletary et al., 1997; Cao et al., 1999; Fujita et al., 2007). In Arabidopsis (Arabidopsis thaliana) leaves, a regulatory role was indicated by the observation that mutations eliminating SSIII caused an increased rate of starch biosynthesis (Zhang et al., 2005). The mechanism(s) by which SSIII influences the activities of other starch biosynthetic enzymes or the overall starch biosynthesis rate is unknown. Part of the explanation may be that physical association of SSIII with other enzymes provides for regulatory interactions.
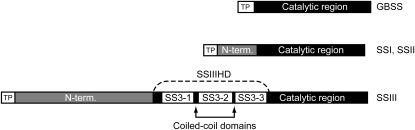
Regulatory functions of SSIII proteins may be provided by an evolutionarily conserved amino acid sequence region located adjacent to the catalytic domain responsible for SS enzyme activity. Members of the conserved SSI, SSII, and SSIII classes of starch synthase all contain an N-terminal extension relative to the conserved catalytic domain homologous to glycogen synthase. In the SSI and SSII classes, the N-terminal extension is conserved among monocots and dicots but not universally throughout the land plants or in unicellular green algae. SSIII, in contrast, contains a region of approximately 450 residues immediately upstream of the catalytic region, referred to here as the SSIII homology domain (SSIIIHD), that appears to have been fixed in evolution as far back as the emergence of land plants (Gao et al., 1998; Li et al., 2000; Dian et al., 2005). For example, SSIIIHD from the monocot maize and the unicellular green alga Chlamydomonas reinhardtii share 32% identity over 415 aligned residues, and SSIIIHD from maize and the dicot Arabidopsis are 56% identical over 458 aligned residues. In contrast, the SSI N-terminal domain from maize is 16% identical over 92 aligned residues with Arabidopsis SSI and 12% identical over 72 aligned residues with Chlamydomonas SSI. SSIIIHD in maize is involved in protein-protein interactions with SSI (Hennen-Bierwagen et al., 2008) and in addition possesses sequences that confer a glucan-binding function (Palopoli et al., 2006; Senoura et al., 2007; Valdez et al., 2008). The further N-terminal extension of maize SSIII beyond the SSIIIHD domain, which is not conserved, has also been implicated in binding to other starch biosynthetic enzymes by yeast two-hybrid data (Hennen-Bierwagen et al., 2008).
This study further characterized multisubunit complexes containing SSIII and SSIIa. Maize mutations are available that eliminate particular starch biosynthetic enzymes in vivo, and using these tools the interdependence of specific SSs and SBEs for assembly into high molecular mass complexes was assessed. The results indicated that SSIII, SSIIa, SBEIIa, and SBEIIb associate together in an enzyme complex of approximately 670 kD, as opposed to individual or pair-wise high molecular mass assemblies. Consistent with the genetic results, biochemical analyses of amyloplast extracts demonstrated stable associations of SSIII with SSIIa, SBEIIb, and SBEIIa. Proteomic analyses revealed the presence of other proteins in the starch biosynthetic enzyme complexes. Two of these were large and small subunits of ADP-Glc pyrophosphorylase (AGPase), which catalyzes formation of the substrate of SS. Other enzymes not known to be directly involved in starch synthesis also were detected, including pyruvate orthophosphate dikinase (PPDK) and Suc synthase (SUS). In the instances of PPDK and SUS, inclusion in high molecular mass assemblies required the presence of multiple starch biosynthetic enzymes, indicating that the components are likely to all be in the same complex. These data revealed that specific enzymes from apparently distinct metabolic pathways interact with starch biosynthetic enzymes, suggesting potential means of coordinating and regulating carbon metabolism during grain filling.
RESULTS
Protein-Protein Interactions Independent of Glucan Binding
SSIIIHD has been reported to bind both glucans and proteins (Palopoli et al., 2006; Senoura et al., 2007; Hennen-Bierwagen et al., 2008; Valdez et al., 2008). The proposed starch-binding domains are located within three repeat sequences distributed throughout the conserved portion of the SSIII enzyme class (Fig. 1; Li et al., 2000; Palopoli et al., 2006). Further computational analysis of the maize SSIIIHD amino acid sequence using the program COILS (Lupas et al., 1991) identified two highly predicted coiled-coil domains, which typically function to mediate protein-protein interactions (Burkhard et al., 2001; Parry et al., 2008). The two coiled-coil domains flank the center starch-binding domain (Fig. 1), which in biochemical analyses of Arabidopsis SSIII appears to be the most significant sequence for substrate affinity (Valdez et al., 2008). These computational predictions suggest the possibility that SSIII comprises structures within the conserved SSIIIHD region that could explain both protein recognition and noncatalytic glucan binding.
Figure 1.
Domain organization of SSs. The catalytic region of each enzyme is defined by a high degree of homology with bacterial glycogen synthases. Each SS class, with the exception of granule-bound starch synthase (GBSS), has an N-terminal extension beyond the catalytic region. SSIII is unique in that a portion of the N-terminal (N-term.) extension is conserved broadly in green plants (SSIIIHD). The other portion of the SSIII N terminus and the extensions of SSI and SSII are not conserved between monocots, dicots, and green algae. Within SSIIIHD are located three starch-binding domains and two predicted coiled-coil domains (see text). TP, Transit peptide.
The fact that sequences within SSIIIHD are known to affect the association with glucan substrates raises the possibility that the observed interactions between SSIII and other starch biosynthetic enzymes are mediated by common binding to the same polymer molecule. To test this hypothesis, amyloplast extracts were treated with a mixture of amyloglucosidase and α-amylase in order to completely digest any glucan polymers present. Glucan polymer concentration in the amyloplast extracts was quantified and determined to be approximately 0.4 μg mL−1. The extracts were treated with a quantity of hydrolytic enzymes that in control experiments was shown to completely digest glucan at a concentration of at least 10 μg mL−1. Complex formation was then analyzed by GPC in extracts with or without addition of the hydrolytic enzymes. Fractions were collected from the GPC column, run in SDS-PAGE, and probed by immunoblot analysis with antibodies specific for SSIII (Hennen-Bierwagen et al., 2008).
In the absence of amyloglucosidase/α-amylase treatment, SSIII migrated in GPC as expected in the fractions containing C670. The same result was observed when glucan had been eliminated from the extracts by hydrolytic enzyme treatment (data not shown). Thus, assembly of SSIII into the complex is mediated via protein-protein contacts and not by random glucan associations. Accordingly, SSIIIHD is proposed to have independent functions in glucan and protein binding.
Altered Complex Formation in the Absence of Specific SSs or SBEs
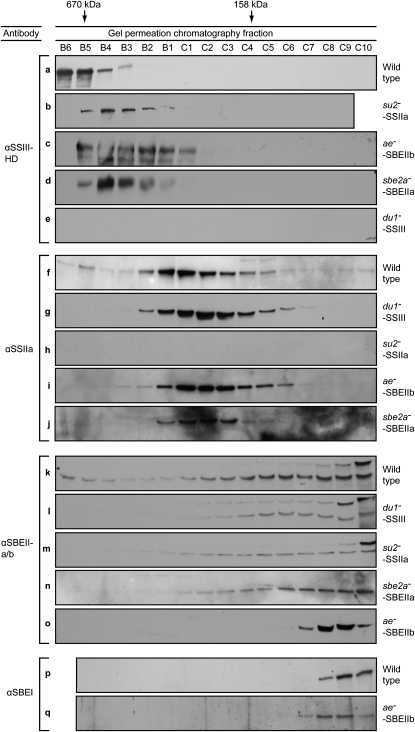
Genetic analyses were used as a means of testing whether high molecular mass forms of the starch biosynthetic enzymes are components of the same complex. If so, then loss of one component would alter the GPC mobility of other members of the complex. Maize mutants exist in which characterized null mutations cause complete loss of individual SS or SBE proteins. The mutation du1−M3 is caused by insertion of a Mutator (Mu) transposon in the first exon of the gene that codes for SSIII (M.G. James, unpublished data), and no SSIII protein is detectable in endosperm extracts from the mutant line (Hennen-Bierwagen et al., 2008; Fig. 2, row e). Mobility of SSIIa, SBEIIa, and SBEIIb in GPC/immunoblot analyses was compared between wild-type and du1−M3 extracts. The proteins were separated in a buffer containing 1 m NaCl, which favors the formation of complexes containing SSIII, SSIIa, and SBEIIb (Hennen-Bierwagen et al., 2008). In wild-type amyloplasts, a small proportion of the SSIIa migrates in the C670 peak (Fig. 2, row f); however, no signal was detected in these fractions in the mutant lacking SSIII (Fig. 2, row g). SBEIIb in wild-type extracts fractionates broadly across the gradient with a peak at approximately 670 kD and a significant concentration also in the 300-kD fractions (Fig. 2, row k). The mobility of SBEIIb is drastically changed in the du1−M3 mutant, in which the protein reaches an elution volume corresponding to approximately 200 kD (Fig. 2, row l). Little or no SBEIIb was present in the 300-kD fractions in the mutant, and the protein was undetectable in the 670-kD fractions. Only a small amount of SBEIIa is present in wild-type C670 (Hennen-Bierwagen et al., 2008), and differences in that peak between the wild type and the du1−M3 mutant could not be discerned. However, in the du1−M3 mutant, SBEIIa appeared to increase in abundance, compared with the wild type, in GPC fractions extending to approximately 300 kD (Fig. 2, row l). The effects of the du1−M3 mutation on GPC mobility of SSIIa, SBEIIa, and SBEIIb were reproducible in multiple independent analyses beginning with separate GPC fractionations (Supplemental Figs. S1–S3).
Figure 2.
GPC analyses of wild-type and mutant amyloplast extracts. Proteins present in the indicated fractions from the GPC column were separated by SDS-PAGE and then probed with specific antibodies in immunoblot analyses. The antisera used to identify SSIII, SSIIa, SBEIIa, SBEIIb, and SBEI are specified in “Materials and Methods,” as are the particular mutant alleles analyzed in each line. The peak elution volumes of molecular mass standards analyzed in the same GPC protocol are indicated at top. Letters identifying each panel are provided for ease of reference with the text. In all instances, the mobility of the indicated proteins by SDS-PAGE matched that expected based on the corresponding cDNA sequence and previous characterizations using the same antibodies.
These data indicate that assembly of both SBEIIb and SSIIa into a 670-kD complex(es) requires the presence of SSIII. SSIIa and SBEIIb differ in that the former is able to assemble into a 300-kD complex in the absence of SSIII but the latter is not. SSIII may have some effect on the assembly state of SSIIa, however, because the GPC peak fraction is shifted to a slightly smaller size in the du1− mutant (Fig. 2; Supplemental Fig. S2).
The effects of eliminating SSIIa were analyzed similarly. The allele utilized was su2-19791, which is a spontaneous mutation in the maize gene sugary2 (su2) that codes for SSIIa (Zhang et al., 2004). This mutation causes complete loss of the SSIIa protein (Hennen-Bierwagen et al., 2008; Fig. 2, row h). The apparent molecular mass of the SSIII peak, based on GPC elution volume, decreased slightly in the su2-19791 mutant compared with the wild type (Fig. 2, row b). SBEIIb mobility in the GPC column is significantly affected by loss of SSIIa, as it was by loss of SSIII. In this mutant, SBEIIb is not detected in the C670 peak, and the amount present in the C300 peak is reduced compared with the wild type (Fig. 2, row m). No obvious changes in the GPC mobility of SBEIIa were seen in the su2-19791 mutant (Fig. 2, row m). Independent repetition of these analyses yielded consistent results (Supplemental Figs. S1–S3). The fact that SSIII and SBEIIb exhibit altered GPC elution volumes when SSIIa is absent, compared with the wild type, is consistent with the proposal that all three proteins associate in the same quaternary structure.
Continuing this method of analysis, the effects of mutations eliminating SBEIIa or SBEIIb on the GPC mobility of other enzyme were observed. The alleles utilized were sbe2a∷Mu, a null mutation preventing the expression of SBEIIa, ae1, an uncharacterized mutation in the gene coding for SBEIIb, or ae−B, a deletion mutation of the SBEIIb gene (see “Materials and Methods”). Assembly of SSIIa into C300 appears to be independent of either SBEIIa or SBEIIb (Fig. 2, rows i and j). The highest molecular mass form of SSIII, in contrast, is affected in mutants lacking either SBEIIa or SBEIIb, such that its maximum apparent molecular mass is reduced slightly (Fig. 2, rows c and d). Loss of SBEIIa also affected the ability of SBEIIb to assemble into C670 (Fig. 2, row n). Again, the GPC analyses were repeated independently with consistent results (Supplemental Figs. S1–S3).
The effect of eliminating SBEIIb on the GPC migration of SBEI was determined. SBEI had previously been shown to exist entirely as a monomer, in contrast to all of the other starch biosynthetic enzymes characterized here (Hennen-Bierwagen et al., 2008). However, binding of SBEI to starch granules was significantly increased in an ae− mutant, suggesting that loss of SBEIIb might unmask a binding site for SBEI in a multisubunit complex (Grimaud et al., 2008). If so, then the mutation affecting SBEIIb could result in the presence of a high molecular mass form of SBEI. This result was not observed, however, and SBEI remained as a monomer both in the wild type and the mutant lacking SBEIIb (Fig. 2, rows p and q). The fact that SBEIIa and SBEIIb are present in the complexes, whereas SBEI is not, emphasizes the specificity of the assemblies and thus supports the assumption that they have physiological functions.
Further Purification of Complexes Containing Starch Biosynthetic Enzymes
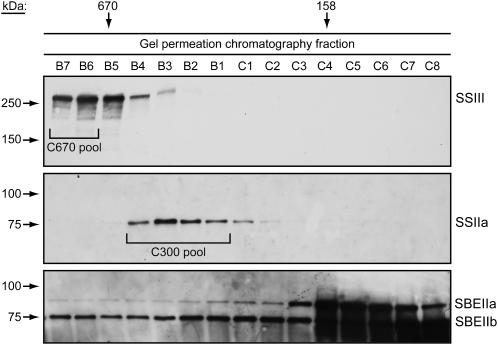
Complexes containing starch biosynthetic enzymes were partially purified in three successive steps: first amyloplast enrichment from developing endosperm tissue, then GPC, and finally anion-exchange chromatography (AEC). GPC separation of amyloplast extracts was conducted in a Tris-acetate buffer containing essentially no sodium or other positively charged ions, in order to facilitate binding to immobilized cations in the next purification step. Immunoblot analyses revealing the presence of SSIII, SBEIIa, SBEIIb, and SSIIa demonstrated that the starch biosynthetic enzyme-containing complexes present in the C670 and C300 elution peaks are stable in the low-salt condition (Fig. 3). Specific fractions from each high molecular mass peak were pooled for the following purification step (Fig. 3).
Figure 3.
Partial purification of complexes containing starch biosynthetic enzymes by GPC. GPC fractionation was performed in no-salt buffer, and fractions were probed by immunoblot analyses to identify the presence of SSIII, SSIIa, SBEIIa, and SBEIIb. SSIII served as a marker for the C670 peak complex(es), and SSIIa served to identify the C300 peak complex(es). The indicated fractions were pooled and used for further purification steps.
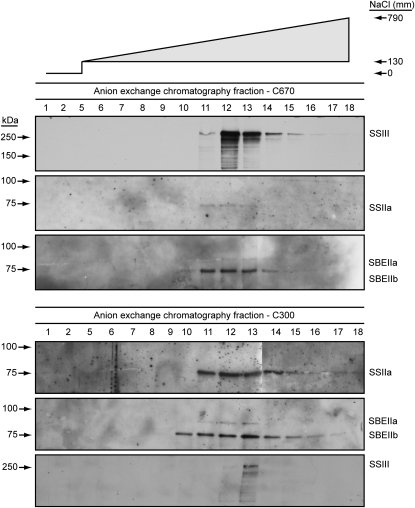
The C670 and C300 GPC pools were applied directly to a MonoQ anion-exchange column. Bound proteins were eluted in a gradient of 0 to 1 m NaCl, and fractions from the column wash and across the gradient were collected and probed by immunoblot analyses. Essentially all of the SSIII in the C670 pool bound to the AEC column and was eluted in a peak centered at approximately 0.5 m NaCl (Fig. 4). The same column fractions were probed for the presence of SSIIa, SBEIIa, and SBEIIb. All three proteins eluted in the same fractions as SSIII (Fig. 4), indicating copurification through amyloplast enrichment, GPC, and AEC.
Figure 4.
Further purification of complexes containing starch biosynthetic enzymes by AEC. The C670 and C300 GPC pools indicated in Figure 3 were separated by AEC, and the indicated fractions were probed by immunoblot analyses. The salt concentration of each fraction is indicated by the gradient at top.
The C300 GPC pool was analyzed similarly, with SSIIa serving as the primary marker for elution from the AEC column. Again, the putative complex was entirely bound to the column and eluted in a peak centered at about 0.5 m NaCl. Probing for coeluting proteins revealed that the peak fraction for SBEIIa and SBEIIb was the same as that for SSIIa (Fig. 4), again indicating copurification through three steps. The relatively low abundance of SSIII detected in the AEC separation of the C300 pool likely results from the resolution of the GPC column, such that there is slight overlap between the C670 and C300 elution peaks (Fig. 3).
Identification of Complex Components by Mass Spectrometry
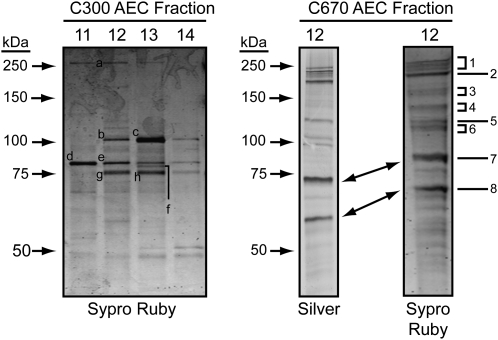
The AEC fraction containing the highest concentration of SSIII, together with coeluting starch biosynthetic enzymes, was further analyzed by silver staining after SDS-PAGE in order to visualize the proteins present (Fig. 5). The same fraction was concentrated approximately 3-fold by lyophilization, then separated by SDS-PAGE and stained with Sypro Ruby in preparation for mass spectrometry analysis. Several fractions from the AEC purification of the C300 complex(es) were analyzed similarly by Sypro Ruby staining (Fig. 5).
Figure 5.
Separation of purified proteins for mass spectrometric analyses. Proteins in the indicated fractions were separated by SDS-PAGE and visualized by the indicated staining method. Letters indicate bands from the AEC purification of complexes in the C300 GPC peak that were excised and analyzed by MS/MS, and numbers indicate the bands analyzed from the C670 GPC peak after AEC purification.
Silver staining of the AEC fraction containing proteins from the C670 peak revealed approximately 12 proteins that were highly purified from the starting material, and all of these also were clearly observed by Sypro Ruby staining. The prominent bands, labeled in Figure 5 as numbers 1 to 8, were excised from the Sypro Ruby gel and analyzed by electrospray ionization tandem mass spectrometry (MS/MS). Eight bands from three different AEC fractions containing proteins from the C300 peak were also analyzed (labeled a to h in Figure 5). The proteins identified in each band are noted in Table I and Table II.
Table I.
Identification of C670-associated proteins by mass spectrometry
| Band | Identified Protein | Protein Family | Accession No. | Unique Peptides | Predicted Molecular Massa | Observed Molecular Massb |
|---|---|---|---|---|---|---|
| kD | ||||||
| 1 | SSIII | Starch synthase | 3057120 | 8 | 188 | 200 |
| 1 | ACCIA | Acetyl-CoA carboxylase | 32264940 | 4 | 257 | 200 |
| 2 | SSIII | Starch synthase | 3057120 | 7 | 188 | 175 |
| 2 | Unknownc | Clathrin heavy chain | 115483737 | 24 | 192 | 175 |
| 3 | SSIII | Starch synthase | 3057120 | 8 | 188 | 150 |
| 3 | Unknownc | Spliceosomal-like protein | 125580741 | 2 | 135 | 150 |
| 4 | SSIII | Starch synthase | 3057120 | 5 | 188 | 140 |
| 4 | Unknownc | 26S proteosome subunit | 116310118 | 7 | 109 | 140 |
| 4 | Unknownc | RNA-binding protein RP120 | 115446411 | 2 | 108 | 140 |
| 5 | PPDK1 | Pyruvate orthophosphate dikinase | 193806357 | 9d | 95e | 105 |
| 5 | PPDK2 | Pyruvate orthophosphate dikinase | TA164186_4577 | 6f | 95e | 105 |
| 5 | Unknownc | 26S proteosome subunit | 115444199 | 4 | 98 | 105 |
| 5 | Unknownc | Cell division cycle protein CDC48 | 110289141 | 4 | 90 | 105 |
| 6 | Unknownc | 26S proteosome subunit | 115444199 | 10 | 98 | 100 |
| 6 | Unknownc | Cell division cycle protein CDC48 | 110289141 | 3 | 90 | 100 |
| 6 | Unknownc | Importin-β2 | 115489162 | 2 | 96 | 100 |
| 6 | Unknownc | Putative initiation factor eIF-3b | 115483552 | 2 | 83 | 100 |
| 7 | Unknownc | Stromal HSP70 | 115487998 | 7 | 74 | 73 |
| 7 | Unknownc | Stromal HSP70 | 115440955 | 6 | 71 | 73 |
| 7 | Unknownc | Stromal HSP70 | 125593660 | 3 | 73 | 73 |
Molecular mass values of full-length proteins are indicated (i.e. before removal of plastid or other targeting sequences), unless otherwise indicated.
Observed molecular mass values were estimated based on comparison with those of known standard proteins (Fig. 5).
Unknown proteins are those for which maize cDNA or genomic DNA sequences are not yet included in the public data set. Protein family assignment is made based on identity between the peptide sequences and known protein sequences from rice, together with BLAST analysis to identify related proteins in other species. Database accession numbers are for the predicted rice protein.
Five peptides are perfect matches with PPDK1, and four are perfect matches to both PPDK1 and PPDK2.
Predicted molecular mass is for the mature protein after cleavage of the plastid-targeting peptide.
Three peptides are perfect matches with PPDK2, and three are perfect matches to both PPDK1 and PPDK2.
Table II.
Identification of C300-associated proteins by mass spectrometry
| Band | Identified Protein | Protein Family | Accession No. | Unique Peptides | Predicted Molecular Massa | Observed Molecular Massb |
|---|---|---|---|---|---|---|
| kD | ||||||
| a | ACCIA | Acetyl-CoA carboxylase | 32264940 | 21 | 257 | 270 |
| c | Unknownc | Starch phosphorylase | 108711180 | 3 | 105 | 105 |
| d | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 8 | 92 | 85 |
| d | Unnamed | Endosperm-specific protein | 2104712 | 2 | 43 | 85 |
| e | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 2 | 92 | 85 |
| e | Unknownc | Heat shock protein 82 | 417154 | 7 | 82 | 85 |
| f | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 2 | 92 | 85 |
| f | HSP82 | Heat shock protein 82 | 477226 | 6 | 82 | 85 |
| f | Unknownc | Heat shock protein 82 | 417154 | 2 | 82 | 85 |
| g | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 7 | 92 | 75 |
| h | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 7 | 92 | 75 |
| h | HSP82 | Heat shock protein 82 | 477226 | 5 | 82 | 75 |
| h | Unknownc | Heat shock protein 82 | 417154 | 7 | 82 | 75 |
| h | Unknownc | Starch phosphorylase | 108711180 | 2 | 105 | 75 |
| h | Unknownc | Stromal HSP70 | 15233779 | 2 | 77 | 75 |
Molecular mass values of full-length proteins are indicated (i.e. before removal of plastid or other targeting sequences).
Observed molecular mass values were estimated based on comparison with those of known standard proteins (Fig. 5).
Unknown proteins are those for which maize cDNA or genomic DNA sequences are not yet included in the public data set. Protein family assignment is made based on exact matches between the peptide sequences and known protein sequences from rice or Arabidopsis, together with BLAST analysis to identify related proteins in other species.
SSIII was clearly identified by MS/MS in the AEC-purified C670 complex(es) (Table I). Four bands ranging in size from approximately 140 to 200 kD contained SSIII, indicative of fragmentation that is typical for this large protein (Hennen-Bierwagen et al., 2008). Numerous additional proteins were identified (Table I). Among these, PPDK is of particular interest because, as shown in following sections, it also associates with SSIII in affinity chromatography and coimmunoprecipitation. Two maize genes, Pdk1 and Pdk2, code for the isoforms PPDK1 and PPDK2, respectively, and Pdk1 produces alternative transcripts that encode both a cytosolic and a plastidial form (Sheen, 1991; Miyao, 2003). The PPDK1 amino acid sequence is available as GenBank accession number P11155. A partial cDNA sequence for PPDK2 is available as GenBank accession number S46967, and the full-length cDNA sequence for this protein is present in the Plant Transcript Assemblies Database as accession number TA164186_4577. Comparison of the peptide sequences identified by MS/MS with these two predicted proteins determined that both PPDK1 and PPDK2 are present in the partially purified C670 fraction (Table I; Supplemental Fig. S4). Acetyl-CoA carboxylase ACCI was also identified by MS/MS in this fraction (Table I). Neither SBEIIa, SBEIIb, nor SSIIa, all of which were observed in this fraction by immunoblot analysis (Fig. 4), was identified by MS/MS.
The proteins identified in the AEC-purified C300 complex(es) included ACCI, the SUS isoform SUS-SH1 (product of the shrunken1 gene), and starch phosphorylase (Table II). The latter protein was shown previously to coimmunoprecipitate with SBEIIb from developing wheat amyloplasts (Tetlow et al., 2004b), supporting its assignment as a component of one or more maize 300-kD complex(es) also containing SSII, SBEIIa, and/or SBEIIb. Comparison of the peptide sequences obtained by MS/MS with the predicted sequences for all three known SUS isoforms of maize specifically identified the protein that copurifies with SSII, SBEIIa, and SBEIIb in C300 as SUS-SH1 (Table II; Supplemental Fig. S5), and this same protein also associates with SSIII in affinity chromatography and coimmunoprecipitation (see following sections). Immunoblot analyses of GPC fractions from wild-type amyloplasts with isoform-specific antibodies that recognize SUS1, SUS2, or SUS-SH1 (Duncan et al., 2006) yielded a signal for only the latter protein, confirming that the starch biosynthetic enzymes associate specifically with SUS-SH1 (data not shown). Again, neither SSIIa, SBEIIa, nor SBEIIb appeared in the MS/MS analysis, despite the fact that immunoblots demonstrated the presence of these proteins in the AEC fractions.
Affinity Purification of SSIIIHD-Binding Proteins
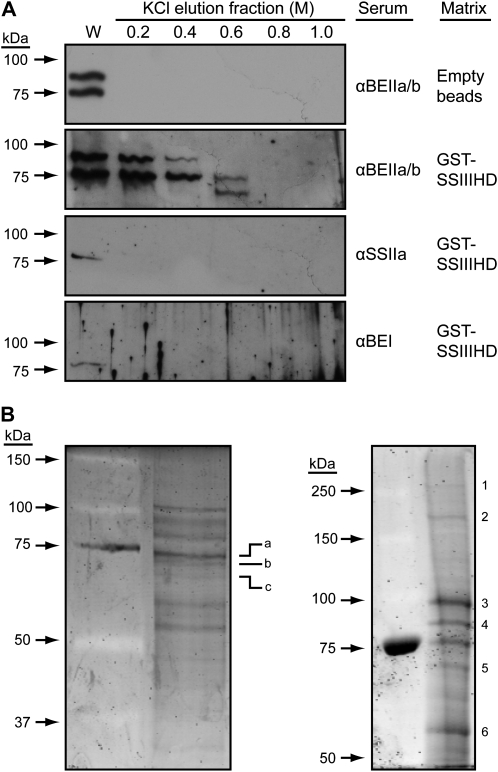
Affinity chromatography was used as a means of further investigating protein binding to SSIIIHD. A recombinant fusion protein containing glutathione-S-transferase (GST) at the N terminus and maize SSIIIHD (SSIII residues 770–1,225) at the C terminus was expressed in Escherichia coli, purified, and immobilized on glutathione-Sepharose beads. Wild-type amyloplast extracts were passed over a GST-SSIIIHD column in a low-salt buffer. After extensive washing in a buffer containing 150 mm NaCl, bound proteins were eluted in steps of increasing salt concentration from 0.2 to 1.0 m KCl. After concentration, proteins in the elution fractions were separated by SDS-PAGE and then probed for the presence of various starch biosynthetic enzymes in immunoblot analyses (Fig. 6A). Antisera recognizing both SBEIIa and SBEIIb, SBEI, or SSIIa were used for protein identification in these analyses. Immunoblot analysis of the eluates probed with αBEIIab detected signals in the 0.2 and 0.4 m elution fractions, which match the known electrophoretic mobilities of SBEIIa and SBEIIb. SBEIIb continued to elute in the 0.6 m KCl fraction, which in addition contained a smaller protein that may be an alternate form of either of these two enzymes. The same antiserum was used to probe the eluates of a control column lacking GST-SSIIIHD, and no immunoblot signals were detected (Fig. 6A), nor were any proteins seen by Coomassie Brilliant Blue staining (data not shown). SBEI and SSIIa were not detected in any of the salt fractions eluted from the GST-SSIIIHD column but were only seen in the total extract flow-through fraction of washed-out unbound proteins. Thus, neither SBEI nor SSIIa appears to bind to SSIIIHD in these conditions.
Figure 6.
Affinity purification with SSIIIHD as the solid-phase ligand. Recombinant GST-SSIIIHD was immobilized on a glutathione-Sepharose column and used as an affinity ligand to purify proteins from soluble amyloplast extracts. A, Extracts were passed through the GST-SSIIIHD column or a control column with no recombinant protein attached to the matrix. After extensive washes in 150 mm KCl, a step gradient of increasing KCl concentration was applied and fractions were collected. Proteins were separated by SDS-PAGE and probed by immunoblot analyses with the indicated antiserum. W indicates the immunoblot signal from the flow-through wash. B, The procedure of A was repeated with the differences that the column was eluted in a single step of 0.6 m KCl and the proteins were stained with Sypro Ruby after SDS-PAGE. The results from two independent biological replicates are shown. Numbers and letters indicate bands that were excised and analyzed by MS/MS.
Proteins eluted from the GST-SSIIIHD column were further characterized by mass spectrometry. For these analyses, the column was washed and bound proteins were eluted in a single step with 0.6 m KCl. The eluate was concentrated 10-fold and separated by SDS-PAGE, and gels were stained with Sypro Ruby (Fig. 6B). Two independent biological replicates were performed, and essentially the same overall banding pattern was observed. Individual bands were excised, and proteins present were defined by mass spectrometry (Table III). SBEIIb was positively identified, thus verifying the immunoblot results. Other proteins of note identified in this elution fraction were PPDK1, PPDK2, and SUS-SH1, which as mentioned in the previous section also coeluted in the AEC purifications of the C670 and C300 complexes. Full-length and proteolytic fragments of PPDK1 and PPDK2 were identified in the eluate from the SSIIIHD affinity column (Table III). The discrete SSIIIHD fragment, which is known to be present in amyloplast extracts (Hennen-Bierwagen et al., 2008), was also identified as a protein bound to the GST-SSIIIHD affinity column (Table III).
Table III.
Identification of SSIIIHD-binding proteins by mass spectrometry
| Band | Identified Protein | Protein Family | Accession No. | Unique Peptides | Predicted Molecular Massa | Observed Molecular Massb |
|---|---|---|---|---|---|---|
| kD | ||||||
| a | Unknownc | Chloroplast HSP70-1 | 15233779 | 3 | 77 | 74 |
| a | SBEIIb | Maize branching enzyme IIb | 1169911 | 2 | 91 | 73 |
| b | Unknownc | Stromal HSP70 | 115487998 | 8 | 74 | 72 |
| 3 | PPDK1 | Maize pyruvate orthophosphate dikinase | 193806357 | 16d | 95e | 95 |
| 3 | PPDK2 | Maize pyruvate orthophosphate dikinase | TA164186_4577 | 12f | 95 | 95 |
| 4 | SUS-SH1 | Sucrose synthase (sh1 gene) | 135060 | 12 | 92 | 85 |
| 4 | PPDK2 | Maize pyruvate orthophosphate dikinase | TA164186_4577 | 2g | 95e | 85 |
| 5 | PPDK2 | Maize pyruvate orthophosphate dikinase | TA164186_4577 | 2g | 95e | 70 |
| 6 | SH2 | ADPGlc pyrophosphorylase large subunit (sh2 gene) | 1707924 | 7 | 57 | 57 |
| 6 | PPDK2 | Maize pyruvate orthophosphate dikinase | 62738111 | 6h | 95e | 57 |
| 6 | SSIIIHD | Maize starch synthase III homology domain | 3057120 | 4 | 188 | 57 |
Molecular mass values of full-length proteins are indicated (i.e. before removal of plastid or other targeting sequences), unless otherwise indicated.
Observed molecular mass values were estimated based on comparison with those of known standard proteins (Fig. 6B).
Unknown proteins are those for which maize cDNA or genomic DNA sequences are not yet included in the public data set. Protein family assignment is made based on exact matches between the peptide sequences and known protein sequences from rice or Arabidopsis, together with BLAST analysis to identify related proteins in other species.
Seven peptides are perfect matches with PPDK1, and nine are perfect matches with both PPDK1 and PPDK2.
Predicted molecular mass is for the mature protein after cleavage of the plastid-targeting peptide.
Six peptides are perfect matches with PPDK2, and six are perfect matches with both PPDK1 and PPDK2.
One peptide is a perfect match with PPDK2, and one is a perfect match with both PPDK1 and PPDK2.
Three peptides are perfect matches with PPDK2, and three are perfect matches with both PPDK1 and PPDK2.
Another SSIIIHD-binding protein was the AGPase large subunit (Table III). Seven distinct peptide sequences obtained by mass spectrometry, comprising 85 total amino acids, are 100% identical with the AGPase large subunit protein encoded by the sh2 gene. Alignments of those peptides with the sh2 product and the other three putative AGPase large subunit protein sequences in the public databases specifically identified the SSIIIHD-binding protein (Supplemental Fig. S6). The sh2 gene is known to encode the large subunit of the major AGPase enzyme activity present in endosperm (Hannah, 2007).
Coimmunoprecipitation with SSIII
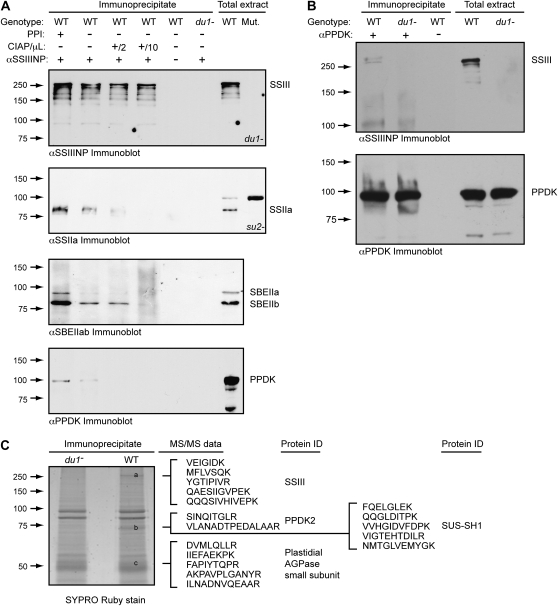
Further confirmation of the association between SSIII and SBEIIa, SBEIIb, SSIIa, PPDK, and/or AGPase was obtained by immunoprecipitation from whole cell endosperm extracts. An affinity-purified IgG fraction, termed αSSIIINP, containing antibodies that bind to the N-terminal 770 residues of SSIII, was used to immunoprecipitate that protein and any others to which it is stably bound. Proteins present in the precipitate were separated by SDS-PAGE and analyzed by immunoblotting. As expected, SSIII was detected in the immunoprecipitates from wild-type endosperm but not from a dul-M3 mutant (Fig. 7A). Using the antibody against PPDK, a signal at approximately 95 kD was detected in wild-type extracts, which matches the expected molecular mass for either mature PPDK1 or PPDK2. This signal was missing in the mutant lacking SSIII and in a control sample in which the precipitating antibody was omitted (Fig. 7A). Therefore, the presence of PPDK in the immunoprecipitate depends on SSIII. Similar results were obtained for SBEIIa, SBEIIb, and SSIIa (Fig. 7A).
Figure 7.
Coimmunoprecipitation from total endosperm extracts. A, Immunoprecipitation with αSSIIINP. Immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblot using the indicated sera. PPI indicates inclusion or absence of the protein phosphatase inhibitor NaF in the immunoprecipitation reaction, and CIAP denotes inclusion or absence of the indicated volume of calf intestinal alkaline phosphatase. The specific su2− or du1− mutant alleles lacking SSII or SSIII, respectively, used as negative controls, are indicated in “Materials and Methods.” B, Immunoprecipitation with αPPDK. Details are as in A. C, Sypro Ruby stain of αSSIIINP immunoprecipitate after separation by SDS-PAGE. The indicated bands were excised and subjected to MS/MS analysis, and peptide sequence data obtained from each band are shown. WT, Wild type; Mut., mutant.
Effects of phosphorylation state on coimmunoprecipitation were tested, considering that complexes in wheat endosperm containing specific SSs and SBEs are affected by the removal of phosphate groups (Tetlow et al., 2004b, 2008). Treatment of the maize endosperm extracts with alkaline phosphatase prior to immunoprecipitation severely reduced the amount of SSIIa, SBEIIa, SBEIIb, and PPDK that coprecipitated with SSIII (Fig. 7A). Conversely, addition of the protein phosphatase inhibitor NaF to the extracts resulted in increased amounts of all four proteins in the immunoprecipitate. Thus, protein phosphorylation plays a role in the association of these components into a high molecular mass complex.
A reciprocal immunoprecipitation was also performed using antiserum to collect PPDK from total soluble endosperm lysate along with any proteins to which it was stably bound. After SDS-PAGE, the immunoprecipitated proteins were probed with αSSIIINP to determine whether SSIII is bound to PPDK. A SSIII signal was detected from wild-type extracts but not from dul-M3 mutant endosperm tissue lacking SSIII, nor in the control assay in which the αPPDK antiserum was omitted (Fig. 7B). Thus, coimmunoprecipitation of PPDK and SSIII was detected in both orientations of the assay.
Further confirmation of PPDK coimmunoprecipitation with SSIII was provided by MS/MS data. Two Sypro Ruby-stained bands on the SDS-PAGE gel of the αSSIIINP immunoprecipitate were observed in the wild type but not in du1−M3 mutants (bands a and b in Fig. 7C). The band migrating as greater than 250 kD was identified by the amino acid sequences of multiple tryptic peptides as SSIII. The second band was identified from two peptide sequences specifically as maize PPDK2 (Supplemental Fig. S4). The molecular mass of the PPDK2 band detected by MS/MS was approximately 70 kD, indicating a proteolytic fragment of the mature protein. The PPDK2 signal in the immunoprecipitate was again dependent on the presence of SSIII in the extracts, because it was absent in the du1−M3 mutant (Fig. 7C) and in control immunoprecipitations lacking antibody (data not shown).
The association of SSIII with AGPase was also indicated from the coimmunoprecipitation data (Fig. 7C). A protein of approximately 53 kD collected from the αSSIIINP immunoprecipitate yielded five peptide sequences comprising 50 amino acids that are 100% identical to the AGPase small subunit encoded by the endosperm-expressed gene brittle2 (bt2). Four of those peptides have mismatches when compared with either the leaf or embryo form of the AGPase small subunit (Supplemental Fig. S7), so that the SSIII-binding protein can be identified specifically as the bt2 gene product.
Finally, SUS-SH1 coimmunoprecipitated with SSIII (Fig. 7C). The SUS protein was present in the same band as PPDK2, which was not detected in the mutant extracts lacking SSIII. Five peptide sequences identified the proteins specifically as SUS-SH1 from among the three known SUS sequences in maize (Supplemental Fig. S5).
Assembly Interdependence of PPDK and SUS-SH1 with SSs and SBEs
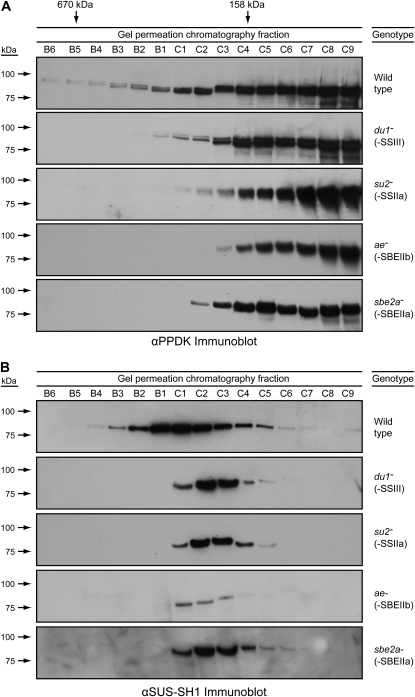
GPC analysis indicated that PPDK1 and/or PPDK2 also exist in high molecular mass forms that require multiple starch biosynthetic enzymes. In two independent replicates, the PPDK immunoblot signal fractionated broadly across the GPC gradient with a peak fraction matching the molecular mass of the monomer (Fig. 8A; Supplemental Fig. S8). PPDK also was present in presumed complexes extending all the way to the excluded volume of the column (i.e. in the C670 fractions). The intensity of the PPDK signal in the high molecular mass fractions was minor compared with the presumed monomer form, indicating that a relatively small proportion of the total PPDK associates with the starch biosynthetic enzymes in the amyloplast extracts.
Figure 8.
Dependence of PPDK and SUS-SH1 on SSIII, SBEIIa, SBEIIb, and SSIIa for assembly into high molecular mass complexes. Analyses are as in Figure 2. A, Immunoblots probed with αPPDK. B, Immunoblots probed with αSUS-SH1.
The same analysis was applied in two independent replicates to mutants lacking either SSIII, SSIIa, SBEIIa, or SBEIIb. In every instance, the highest molecular mass form of PPDK was not evident (Fig. 8A; Supplemental Fig. S8). Thus, PPDK requires all four of those starch biosynthetic enzymes in order to assemble into a high molecular mass complex. This observation is mostly likely explained by the direct association of PPDK with the starch biosynthetic machinery in the same complex (i.e. C670).
High molecular mass assemblies of SUS-SH1 were also detected by immunoblot analysis of GPC fractions (Fig. 8B). In wild-type extracts, SUS-SH1 signal exhibited a peak corresponding to approximately 300 kD, similar to C300, and also extended to a high molecular mass fraction of approximately 600 kD. Distinctly different GPC peaks were observed in the SS and SBE mutants (Fig. 8B). In amyloplast extracts from plants lacking SSIII, SSIIa, SBEIIa, or SBEIIb, SUS-SH1 migrated on the GPC column at a molecular mass of approximately 200 kD, likely corresponding to a homodimer of the 92-kD polypeptide (Duncan et al., 2006). Notably, the highest molecular mass form was not evident in any of the mutants. The GPC analyses were independently reproduced beginning with separate column runs (Supplemental Fig. S9). The data indicate that SUS-SH1 assembles into a high molecular mass form that also requires SSIII, SSIIa, and SBEIIa. Only a relatively small proportion of the total SUS-SH1 is present in these complexes in the amyloplast-enriched cell extracts, as shown by the distribution of the immunoblot signal across the GPC column elution fractions (Fig. 8; Supplemental Fig. S9).
DISCUSSION
GPC and mutational analyses of maize amyloplast extracts showed that starch biosynthetic enzymes sort into three different assembly states: as monomers, in an approximately 300-kD complex(es) (C300), and/or in a complex of approximately 670 kD (C670). SSIIa and SSIII were found predominantly in high molecular mass forms, with SSIIa almost exclusively in a C300 assembly and SSIII in C670. In contrast, SBEIIa is primarily found in monomer form, whereas SBEIIb is approximately equally distributed between monomer, C670, and C300. C670 is not a homomultimer, as relatively small quantities of SSIIa, SBEIIa, and SBEIIb are present in what assembly interdependence data show to be a single complex. C300 also contained SBEIIa and SBEIIb in minor quantities, although in this instance assembly interdependence data indicated that multiple distinct complexes could be present in that GPC elution peak. Additional proteins were identified as components of C670 and/or C300 that are known to be involved in the starch biosynthetic pathway or are proposed to be regulators of that process, namely AGPase and PPDK, respectively. The observation that so many proteins involved in the same metabolic pathway interact with each other in stable complexes suggests that these associations are physiologically significant. Nearly all of the SSIIa and SSIII in the extracts is present in these complexes, suggesting the SSs play a central role in the functions of these enzyme assemblies.
Assembly of Starch Biosynthetic Enzymes into Quaternary Structures
Previous characterization of high molecular mass forms of starch biosynthetic enzymes could not determine whether the proteins were in one complex or multiple independent complexes that happen to elute in the same GPC peaks. From the following reproducible genetic data, this study showed that C670 is a single species containing SSIII, SSIIa, SBEIIa, and SBEIIb (Fig. 2; Supplemental Figs. S1–S3). First, loss of SSIIa, SBEIIa, or SBEIIb reduced the apparent molecular mass of the SSIII-containing complex. Second, loss of SIII, SBEIIa, or SBEIIb prevented the assembly of SSIIa into C670. Third, loss of SSIII, SSIIa, or SBEIIa prevented the assembly of SBEIIb into C670. Taken together, elimination of any one of the four starch biosynthetic enzymes found in C670 resulted in altered GPC mobility for the others. These data are consistent with C670 being composed of a single high molecular mass complex containing all four proteins. The great majority of SSIII is present in C670, and this protein even in the absence of the other components remains in a high molecular mass form, although reduced in GPC mobility from the wild type. Thus, SSIII exists as a multimer whether or not it is associated with SBEIIa, SBEIIb, and/or SSIIa.
Effects of any of the mutations on SBEIIa assembly into C670 could not be discerned, owing to the low abundance of this protein in the complex. However, SBEIIa is present in the C670 GPC peak and also interacts directly with SSIII in a yeast two-hybrid test (Hennen-Bierwagen et al., 2008). This study has further shown that SBEIIa copurified with SSIII, SSIIa, and SBEIIb through AEC (Fig. 4). SBEIIa, therefore, certainly is part of a complex present in the C670 peak along with the other starch biosynthetic enzymes.
In contrast to the C670 complex, which appears to be a single species, the genetic interdependence results indicated that the C300 peak may contain multiple, distinct complexes. Copurification results showed that SBEIIa, SBEIIb, and SSIIa migrate together in the C300 GPC peak and the subsequent AEC purification step (Fig. 4). The presence of a relatively small amount of SSIII in the AEC fractions obtained from the C300 peak (Fig. 4) is likely to result from contamination in the GPC fractions. A portion of SBEIIb assembles into a complex in the C300 fractions whether or not SSIIa or SBEIIa is present, although these GPC profiles were distinct from that of the wild type (Fig. 2; Supplemental Fig. S3). The SBEII enzymes in wheat amyloplasts were shown to exist as dimers (Tetlow et al., 2004b, 2008). Presuming that this applies to SBEIIb in maize, dimerization could explain the GPC results from the SSIIa and SBEIIa mutants. Like SSIII, the great majority of SSIIa assembles into a high molecular mass complex (i.e. C300) that does not require the presence of SSIII, SBEIIa, or SBEIIb (Fig. 2; Supplemental Fig. S2). The reason that the SSIIa complex migrates at a slightly smaller molecular mass in the absence of SSIII is not known, but it could result from an indirect role of the latter protein in the assembly of C300. Taken together, these observations suggest that the material in the C300 fractions is a heterogeneous population of different assembly states involving SSIIa, SBEIIb, and SBEIIa.
The molecular mass of the proposed complex or complexes can only be very roughly estimated from the GPC data. The resolution of these columns is insufficient to define precise molecular mass, in large part because hydrodynamic volume is a critical factor in mobility through the matrix (Hernandez et al., 2008). This is especially applicable for the C670 GPC peak, which elutes close to the exclusion volume of the column, and extends broadly to a position that appears significantly larger than the 670-kD molecular mass marker. Thus, the complexes either contain multiple copies of some of these subunits and/or include other proteins in addition to SSIII, SSIIa, SBEIIa, and SBEIIb.
The enzyme complexes characterized here are relatively stable, because they remain associated through three successive purification steps (i.e. amyloplast isolation and extraction, GPC, and AEC). Another indication of the stability of the complexes is the observation that they exist in a wide range of salt conditions. In previous work, these complexes were characterized in nearly physiological ionic strength and also high-salt conditions, specifically 1 m NaCl (Hennen-Bierwagen et al., 2008). Here, the complexes were found to exist also in a buffer essentially lacking any salt ions. Based on enhancement of C670, complex abundance in 1 m NaCl hydrophobic effects was proposed as a factor in the assembly state (Hennen-Bierwagen et al., 2008). When GPC was run in the no-salt buffer, however, all of the components were maintained in both complexes through the AEC gradient (Fig. 4), thus illustrating the strength of the protein-protein interactions.
The presence of SSI was not assayed immunologically in this study, nor was it identified by MS/MS analysis. Previous evidence, however, showed interactions between SSI and SSIIIHD, SBEIIa, and SBEIIb (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008). Thus, it is likely that further analyses of the complexes partially purified here will reveal the presence of SSI. Also not detected by MS/MS in the complexes from the C670 and C300 GPC peaks were SSIIa, SBEIIa, and SBEIIb, despite their detection by immunoblot analyses. Proteomic characterizations of amyloplast proteins also failed to detect all of the starch biosynthetic enzymes (Mechin et al., 2007; Stensballe et al., 2008), even though their presence is well documented by other means. Thus, it appears that proteomics does not readily detect these starch biosynthetic enzymes, either due to resistance to proteolytic digestion or due to masking by other abundant proteins running on the same gel bands, such as the heat shock proteins.
Two aspects of these conclusions are at this point apparently discrepant from the characterization of starch biosynthetic enzyme assemblies in developing wheat amyloplasts. First, previous data demonstrated that SBEIIa and SBEIIb are unlikely to be present in the same assembly, because antibodies against either of those two proteins failed to immunoprecipitate the other (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008). This study, however, shows that C670 formation requires SBEIIa and SBEIIb. This is most simply explained by the fact that C670 contains both proteins; however, indirect effects on complex assembly cannot be ruled out. Second, in wheat, the relative abundance of SSIIa in a 300-kD complex relative to monomer varied with development and was at most approximately 50%. This is in contrast to the observation that in maize amyloplasts from 15- to 16-d-after-pollination kernels, at least 90% of the SSIIa is in C300. Further developmental studies of SSIIa in maize may reconcile these different results.
The function of SSIIa in the assembly of starch biosynthetic enzyme complexes may contribute to the genetic effects of su2− mutations. An unexplained observation in maize, Arabidopsis, and barley is that loss of SSIIa has a major effect on amylopectin structure even though the total reduction of SS enzyme activity is minimal (Gérard et al., 2001; Morell et al., 2003; Zhang et al., 2004, 2008). Thus, nonenzymatic functions of the SSIIa protein, potentially in complex formation, may contribute to the determination of amylopectin structure. Similarly, du1− mutations that affect SSIII also have drastic and specific effects on amylopectin structure, in this instance on the size and/or compactness of the molecule (Gérard et al., 2001). Such a major effect could be mediated by coordinated activities of the SSs and SBEs in C670, as opposed to the specific catalytic activity of SSIII.
Additional Proteins Associated with SSs and SBEs in Multisubunit Complexes
Partial purification of the complexes present in the C670 or C300 GPC peak by AEC, as well as SSIIIDH affinity chromatography and coimmunoprecipitation, afforded us the ability to use proteomic methods to identify other proteins also present. The identification of starch phosphorylase in C300 from MS/MS data provided validation of this technique. Starch phosphorylase is an enzyme active on glucan substrates, and it had previously been observed to coimmunoprecipitate with SBEIIa in wheat amyloplast extracts (Tetlow et al., 2004b). Proteins of interest found to be associated with the starch biosynthetic enzymes include PPDK1 and PPDK2, AGPase large and small subunits, and the SUS isoform SUS-SH1.
PPDK
The MS/MS data clearly demonstrated that both known isoforms of PPDK associate with SSIII in one or more high molecular mass complexes. Both PPDK1 and PPDK2 copurified through AEC with SSIII, SBEIIa, SBEIIb, and SSIIa from the complexes in the C670 GPC peak (Table I), and both proteins also bound to an affinity column with SSIIIHD as the immobilized ligand (Table III). PPDK2, and likely PPDK1 as well, also coimmunoprecipitated with SSIII from endosperm cell total soluble extracts using antibodies specific for either SSIII or PPDK (Fig. 7). Further evidence that PPDK associates in the same complex with starch biosynthetic enzymes is that its presence in the C670 GPC fractions required both SSIII and SSIIa (Fig. 2; Supplemental Fig. S8). These consistent results leave no doubt that PPDK is able to physically associate with SSIII, SSIIa, and most likely SBEIIa and SBEIIb in the multisubunit complex or complexes that migrate in the C670 GPC peak. The majority of PPDK migrates in GPC analyses as apparent monomer and dimer forms, with a minor fraction of the total associating with C670 and C300. Thus, PPDK is likely to have physiological roles in developing endosperm in addition to those that might be mediated by the starch biosynthetic enzyme complexes.
PPDK was initially characterized as a plastidial enzyme that generates the phosphoenolpyruvate (PEP) that is used for CO2 fixation in C4 plants (Hatch, 1987). The enzyme converts pyruvate to PEP, using ATP and inorganic phosphate, also generating AMP and inorganic pyrophosphate (PPi) as products. PPDK also is present in nonphotosynthetic tissues, including endosperm from maize, wheat, and rice (Meyer et al., 1982; Aoyagi and Bassham, 1984; Sadimantara et al., 1996; Vensel et al., 2005; Mechin et al., 2007). In maize, the Pdk1 gene codes for both cytosolic and plastidial forms of PPDK, whereas Pdk2 has been thought to code for a cytosolic protein based on the absence of a recognizable transit peptide sequence (Sheen, 1991). PPDK2, however, has been found as an internal granule-associated protein (Boren et al., 2004; F. Grimaud and V. Planchot, unpublished data), implying a plastidial location for at least a portion of the protein.
AGPase
The AGPase small subunit was found to coimmunoprecipitate with SSIII, and the large subunit of that enzyme was recovered from the SSIIIHD affinity column. Peptide sequence data revealed that the recovered AGPase small subunit is the product of the bt2 gene (Supplemental Fig. S7), and the AGPase large subunit protein was identified as the product of sh2 (Supplemental Fig. S6). Considering the plastidial location of SSIII, the AGPase isoform with which it associates is expected to also be plastidial. Approximately 5% to 15% of the total AGPase activity in cereal endosperm is plastidial, although the genetic identities of the subunits in this isoform have not been fully resolved (Denyer et al., 1996; Thorbjornsen et al., 1996; Neuhaus and Emes, 2000; Hannah, 2007). The bt2 gene coding for the AGPase small subunit produces two different transcripts, one of which contains a predicted plastid-targeting peptide and is expressed in endosperm (Rosti and Denyer, 2007), consistent with amyloplast localization. In addition, immunogold electron microscopy detected the bt2 product within amyloplasts (Miller and Chourey, 1995).
The sh2 gene encoding the AGPase large subunit is required for the major cytosolic AGPase activity in endosperm, fails to possess a predicted targeting peptide, and is translated in vitro into a protein of the same size as that found in vivo (Hannah, 2007). Thus, sh2 clearly codes for the cytosolic AGPase large subunit in endosperm. On the other hand, the protein has been observed in amyloplasts of developing endosperm by immunogold localization (Miller and Chourey, 1995), and the sh2 product was recovered in this study by affinity chromatography from amyloplast-enriched extracts. In addition, the program ChloroP yields a relatively high score compared with other AGPase large subunits thought to encode plastidial proteins in various maize tissues (Supplemental Table S1). Thus, the possibility remains that the sh2 product localizes to both the cytosol and the amyloplast. Finally in this regard, as noted in the previous section, PPDK2 is not predicted by numerous computational algorithms to contain an amyloplast-targeting peptide, yet it is found within starch granules that in higher plants are made exclusively in plastids.
Proposed Functional Associations in C670
Direct interactions in an enzyme complex between PPDK and SSs and/or SBEs could be a physiologically significant means of coordinating global aspects of metabolism as endosperm tissue develops. During maize development, a rapid rise in expression of PPDK at approximately 21 d after pollination is coincident with the maximal expression of starch biosynthetic enzymes (Mechin et al., 2007). One potential role of PPDK in endosperm, where C4 metabolism is not active, is the production of three carbon compounds used as precursors for amino acid biosynthesis (Chastain et al., 2006). Another role proposed recently is that PPDK serves as a regulator of carbon partitioning between starch and protein during grain filling in the endosperm (Mechin et al., 2007). More specifically, increased PPi concentration and reduced ATP concentration resulting from PPDK activity in the direction of pyruvate to phosphoenolpyruvate was proposed to inhibit cytosolic AGPase. In this way, PPDK could negatively regulate starch biosynthesis and divert hexoses from the carbohydrate sink toward protein and lipid production (Mechin et al., 2007). This effect had been presumed to occur in the cytosol, because the majority of AGPase in cereal endosperm is located in that compartment (Hannah, 2007) and because PPDK was presumed also to be cytosolic in endosperm (Miyao, 2003). This proposal, however, is apparently inconsistent with the fact that the steady-state cytosolic PPi concentration in plant cells is high, approximately 250 μm (Stitt, 1990; ap Rees et al., 1991). Thus, product inhibition of AGPase by changes in cytosolic PPi concentration owing to PPDK activity is unlikely. Furthermore, finding PPDK2 in maize and barley endosperm starch granules (Boren et al., 2004; F. Grimaud and V. Planchot, unpublished data) implies that at least some of this protein, previously thought to be cytosolic (Miyao, 2003), is in fact located in the plastid. This is supported by the findings that PPDK1 and PPDK2 associate with numerous starch biosynthetic enzymes in amyloplast extracts.
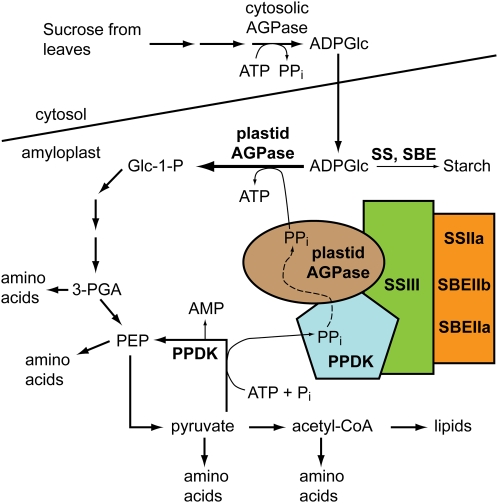
As a modification of the hypothesis described by Mechin et al. (2007), we suggest that PPDK in the amyloplast functions to regulate plastidial AGPase, either in addition to or instead of the cytosolic AGPase (Fig. 9). The fact that PPi concentration in amyloplasts is essentially zero owing to an active pyrophosphatase (Stitt, 1990; ap Rees et al., 1991) might argue against this idea. However, the data presented here suggest that PPDK and AGPase are present in a single multisubunit complex also containing at least SSIII, SSIIa, SBEIIa, and/or SBEIIb. PPDK could influence plastidial AGPase activity within C670 by supplying PPi directly through a substrate-channeling mechanism. This would shift the equilibrium of the plastidial AGPase reaction toward the direction of degrading ADPGlc to produce Glc-1-P. Such a mechanism could serve to divert Glc, originally derived from Suc and transported into the amyloplast as ADPGlc, away from the starch biosynthesis sink and into alternate metabolic fates resulting in increased amino acid production and protein accumulation that occurs late in endosperm development. A possible functional implication of the proposed association of SSs and SBEs with PPDK and AGPase could be communication between the metabolic pathways responsible for starch synthesis and protein synthesis, especially considering that ADPGlc is the substrate of both SS as it synthesizes glucan and AGPase working in the reverse direction to generate ADP and Glc-1-P (Fig. 9). Finally, existing genetic evidence for a functional relationship between SSIII and AGPase is that in Arabidopsis leaves, where AGPase is exclusively plastidial, mutations that eliminate the SS cause reduced AGPase activity as a secondary effect (Zhang et al., 2008).
Figure 9.
Model for the regulation of carbon partitioning in maize amyloplasts. Colored shapes indicate components of the C670 complex. PPi generated by PPDK activity is proposed to be channeled to AGPase, thus directing the equilibrium of the reaction toward ADPGlc degradation. Metabolites derived from the activity of PPDK and AGPase would be used as precursors for amino acid, protein, and potentially lipid biosynthesis. The model does not intend to imply specific details of stoichiometry or direct contacts within the complex. [See online article for color version of this figure.]
Support for a plastidial function of PPDK in regulating carbon flux into protein in C3 plants comes from studies of transgenic tobacco (Nicotiana tabacum; Sheriff et al., 1998). Plastidial PPDK from a Crassulacean acid metabolism plant was overexpressed either with or without its organelle-targeting peptide. When the exogenous PPDK was present in plastids of either leaves or seeds, the levels of free amino acids increased notably. In the latter case, more seeds per capsule were observed, and individual seeds increased in weight compared with wild-type seeds. In contrast, overexpression of PPDK in the cytosol caused diminished seed set and smaller seeds. Further evidence for a role of PPDK in carbon partitioning in developing endosperm comes from the characterization floury4 mutations of rice, which inactivate OsPPDKB, the rice homolog of maize Pdk1 (Kang et al., 2005). In mutant kernels, both the lipid and protein contents were increased as a percentage of dry weight. The transcript of OsPPDKB that presumably codes for the cytosolic form of the protein is preferentially expressed in rice endosperm, so this metabolic effect was presumed to result from a loss of cytosolic PPDK activity (Kang et al., 2005).
The model in Figure 9 does not imply that every metabolic function of PPDK in this tissue is mediated through physical interactions of a plastidial form in the starch biosynthetic enzyme-containing complex(es). This point is further emphasized by the fact that only a relatively small proportion of the PPDK is present in complex(es) that migrate in the C670 GPC fractions, as judged by the distribution of immunoblot signal across the column elution (Fig. 8).
SUS-SH1
The proteins discussed to this point as members of the C670 complex share the properties of being known to exist in amyloplasts, and except for PPDK they are involved directly in the same metabolic pathway of starch biosynthesis. Additionally, PPDK has been proposed to be involved in the regulation of starch metabolism. In contrast, the SUS-SH1 association with SSIII was unexpected because it has not been reported as an internal amyloplast protein (Duncan et al., 2006), nor is its function generally thought to be involved directly in committed steps of starch biosynthesis. Intriguingly, however, SUS has been proposed to function directly in the production of ADPGlc (Baroja-Fernández et al., 2003), in which case it could be part of the committed steps of starch biosynthesis.
The association of SUS-SH1 with starch biosynthetic enzymes is highly reproducible. SUS-SH1 was found to interact with SSIII by both affinity purification (Table III) and coimmunoprecipitation (Fig. 7). In addition, SUS-SH1 cofractionated with SSIIa, SBEIIa, and SBEIIb during AEC purification of the C300 complex (Table III). In each instance, the form of SUS present was identified specifically as SUS-SH1 among the three known isoforms (Supplemental Fig. S9). Assembly interdependence data showed that the highest molecular mass complex of SUS-SH1 failed to form when SSIII, SSIIa, SBEIIa, or SBEIIb was missing. Thus, as is the case for PPDK, the data indicated clearly that SUS-SH1 is associated with the starch biosynthetic enzymes in the soluble extracts of the amyloplast-enriched cell fractions. The possibility cannot be discounted that this association occurs only in the cell extracts and not in vivo, potentially owing to the fact that SUS-SH1 is a membrane-associated protein (Duncan et al., 2006). Localization of SUS-SH1 may not be fully resolved, however, because the protein has been detected in mitochondria and proposed to served a noncatalytic role in that compartment (Subbaiah et al., 2006). We suggest that the clear evidence of physical interaction in cell extracts, together with potential physiologically related roles, supports further investigation of functional interactions between SUS and starch synthase.
CONCLUSION
SUS-SH1, AGPase, and PPDK have been highlighted here because they are found associated with starch biosynthetic enzymes by multiple methods. Other proteins identified in the proteomic studies only from the copurification analyses are likely to be amyloplast proteins that happen to coelute with the starch biosynthetic enzyme complexes without being part of the same quaternary structure, such as the many heat shock proteins present. The primary argument for the physiological significance of the associations of SSIII, SSIIa, SBEIIa, SBEIIb, and AGPase is that all of these proteins function directly in starch biosynthesis, and artifactual association of such a large group of functionally related proteins is highly unlikely. By extension, finding PPDK in this complex, which is proposed to regulate AGPase, suggests that this protein as well is part of a physiologically significant metabolon.
We note the possibility that some SSIII- or SSIIa-associated proteins in C670 or C300, respectively, may regulate the starch synthase. All of the other proteins in both C670 and C300 are present in apparently substoichiometric amounts compared with the SS and also are distinct from SSIII or SSIIa in that the major form of the protein includes a monomer or native multimer state (Hatch, 1987; Duncan et al., 2006; Hannah, 2007). The possibility exists that PPDK, AGPase, SBEIIa, SBEIIb, and SUS-SH1 act as regulatory factors to influence the major activity of the complex as an SS. Such a role is not mutually exclusive with the proposed PPDK regulation of AGPase.
Finally, it is evident that phosphorylation of one or more proteins in C670 is required for their formation and/or stability. Removal of phosphate groups diminished or eliminated the coimmunoprecipitation signals, and increasing phosphorylation by inhibition of native protein phosphatase activity in the amyloplast extract enhanced the association with SSIII. This effect has been observed previously in wheat amyloplast extracts (Tetlow et al., 2004b, 2008) and so appears to be a general feature of cereal endosperm. Identification of target sites and the kinases responsible for the modifications will be required for better understanding of the role of protein phosphorylation in the regulation of starch biosynthesis or other metabolic pathways in amyloplasts.
MATERIALS AND METHODS
Plant Materials and Amyloplast Purification
Growth of maize (Zea mays) plants and preparation of amyloplast-enriched extracts from developing endosperm tissue were described previously (Hennen-Bierwagen et al., 2008). Mutations utilized were as follows: du1−M3, a null allele with a Mu transposon insertion in the first exon of the gene coding for SSIII (M.G. James, unpublished data); su2-19791, an uncharacterized, spontaneous null mutation in the gene coding for SSII (Zhang et al., 2004); sbe2a−, a Mu transposon insertion in the gene coding for SBEIIa (Blauth et al., 2001); and ae−B, an 882-bp deletion in the gene coding for SBEIIb that removes all of exon 9 (GenBank accession no. AF072725; Fisher et al., 1996; M. Yandeau-Nelson and M. Guiltinan, personal communication). All mutant alleles were backcrossed into the W64A inbred line.
Amylase Treatment of Complexes in Amyloplast Extract
Amyloplast extract from wild-type endosperm was treated with a mixture of amyloglucosidase (Megazyme catalog no. E-AMGDF) and α-amylase (Megazyme catalog no. E-ANAAM) at concentrations of 33 units mL−1 and 100 μg mL−1, respectively, for 30 min at room temperature. Glc released in the assays was quantified using a Glc oxidase assay kit (Megazyme catalog no. K-GLUC). Control experiments indicated that the glucan polymer concentration in the amyloplast extract was approximately 0.40 μg mL−1, whereas the hydrolytic enzymes in the same conditions were able to completely digest polymer at a concentration of at least 10 μg mL−1. After the amylase treatment, the amyloplast extract was analyzed by GPC and immunoblotting as described (Hennen-Bierwagen et al., 2008).
Affinity Chromatography
GST-SSIIIHD was expressed in Escherichia coli from plasmid pTB-3829 and bound to glutathione-Sepharose 4B affinity matrix, as described previously (Hennen-Bierwagen et al., 2008). The affinity matrix was equilibrated in 1× binding-wash buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, and 5 mm dithiothreitol [DTT]). Amyloplast extract from wild-type inbred line W64A, in 1× amyloplast-rupture buffer (100 mm Tricine-KOH, pH 7.8, 1 mm EDTA, 1 mm DTT, 5 mm MgCl2, and 1× protease inhibitor cocktail [Sigma catalog no. P9599]), was passed over a 1-mL bed volume column of either glutathione-Sepharose 4B bound to GST-SSIIIHD or a control column to which no protein had been bound. The columns were washed in 25 bed volumes of 1× binding-wash buffer, then eluted in 50 mm Tris-acetate, pH 7.5, with specified concentrations of KCl. Bound proteins were eluted in five1-mL sequential steps of increasing KCl concentration, from 0.2 to 1.0 m. Fractions of 1 mL were collected at each step. In other instances, the column was eluted in two steps, of 0.6 m KCl and 1.0 m KCl. Fractions were concentrated 10-fold in Millipore Ultrafree cellulose spin filters, molecular mass cutoff 5,000 D (Fisher catalog no. UFC3LCC00).
Immunoblot Analysis and Protein Staining
Protein samples from GPC fractions, AEC fractions, or affinity chromatography fractions were separated by SDS-PAGE on precast 7.5% acrylamide gels (Bio-Rad catalog no. 161-1154 or 345-0006). Some of the gels were stained with Sypro Ruby (Bio-Rad catalog no. 170-3126), according to the manufacturer's instructions, and the proteins on other gels were visualized by silver staining according to a previously published procedure (Blum et al., 1987). Immunoblot analyses were performed using the antisera αBEIIb, αBEIIab, αSSIIa, and αBEI, or the affinity-purified IgG fractions αSSIIIHDP and αSSIIINP, as described previously (Hennen-Bierwagen et al., 2008). αSSIIINP was prepared by affinity chromatography using a recombinant fragment of SSIII containing residues 1 to 770 (Cao et al., 1999) as the affinity ligand, as described previously (Hennen-Bierwagen et al., 2008). Affinity-purified antibody recognizing maize (Zea mays) PPDK, termed αPPDK, was obtained from Dr. Chris Chastain (Chastain et al., 2002). Isoform-specific antiserum recognizing SUS-SH1, termed αSUS-SH1, was obtained from Dr. Steve Huber (Duncan et al., 2006).
GPC and AEC
The GPC procedures used in this study were described previously (Hennen-Bierwagen et al., 2008). Two different column buffers were used as indicated, either high-salt buffer (50 mm sodium acetate, pH 7.0, 1 mm DTT, and 1 m NaCl) or no-salt buffer (50 mm Tris-acetate, pH 7.5, and 1 mm DTT). For successive purification steps, the GPC analysis was run in no-salt buffer, and selected fractions were pooled and applied to a 1-mL bed volume MonoQ anion-exchange column (GE Healthcare catalog no. 17-5166-01) using an AKTA-FPLC instrument. The AEC column was washed in AEC buffer (20 mm Tris-HCl, pH 8.0) and eluted in a gradient of 0 to 1 m NaCl in the same buffer. The flow rate was 1 mL min−1, the gradient volume was 20 mL, and 1-mL fractions were collected.
Immunoprecipitation
Precipitating antiserum, either αSSIIINP or αPPDK, was mixed with protein A-Sepharose CL-4B beads (Sigma catalog no. P-3391) at 4°C for 1 h, and the beads were then washed in phosphate-buffered saline. Endosperm extracts from the wild type and the du1−M3 mutant line were prepared as described previously (Hennen-Bierwagen et al., 2008). Approximately 1 mg of total protein extract was incubated with the antibody/protein A-Sepharose beads (100 μL bed volume) at 4°C for 1 h with gentle agitation. In addition, some samples had the protein phosphatase inhibitor NaF added to the cell extract prior to immunoprecipitation, and others were supplemented with calf intestinal alkaline phosphatase. After centrifugation at 500g for 5 min at 4°C to remove the unbound protein, the pelleted beads were washed extensively with phosphate-buffered saline augmented with 1% Nonidet P-40. Aliquots of the beads were boiled in SDS-PAGE loading buffer and applied to gels for immunoblot analyses and Sypro Ruby staining.
Mass Spectrometry
Sypro Ruby-stained gels were visualized by UV fluorescence on a transilluminator, and protein bands were excised with a scalpel. The acrylamide gel slices were provided to the Proteomics and Mass Spectrometry Facility, Donald Danforth Plant Science Center, and the proteins present were analyzed by electrospray MS/MS according to facility procedures (http://www.danforthcenter.org/pmsf). Data were analyzed using SCAFFOLD software (http://www.proteomesoftware.com).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Independent repetitions of GPC analyses of SSIII in the wild type and starch biosynthesis mutants.
Supplemental Figure S2. Independent repetitions of GPC analyses of SSIIa in the wild type and starch biosynthesis mutants.
Supplemental Figure S3. Independent repetitions of GPC analyses of SBEIIa and SBEIIb in the wild type and starch biosynthesis mutants.
Supplemental Figure S4. Peptide specificity between PPDK1 and PPDK2.
Supplemental Figure S5. Peptide specificity between maize SUSs.
Supplemental Figure S6. Peptide specificity between maize AGPase large subunit proteins.
Supplemental Figure S7. Peptide specificity between maize AGPase small subunit proteins.
Supplemental Figure S8. Independent repetitions of GPC analyses of PPDK in the wild type and starch biosynthesis mutants.
Supplemental Figure S9. Independent repetitions of GPC analyses of SUS-SH1 in the wild type and starch biosynthesis mutants.
Supplemental Table S1. ChloroP predictions of maize AGPase large subunit proteins.
Supplementary Material
Acknowledgments
We thank Dr. Leslie Hicks (Donald Danforth Plant Science Center) for expert assistance with proteomics analysis and Drs. Chris Chastain (Minnesota State University, Moorhead) and Steven Huber (North Carolina State University) for providing αPPDK and αSUS-SH1 antisera, respectively.
This work was supported by awards from the U.S. Department of Agriculture (grant no. 2002–35318–12646) and the U.S. Department of Energy (grant no. DE–FG02–05ER15706) to M.G.J. and A.M.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alan M. Myers (ammyers@iastate.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aoyagi K, Bassham JA (1984) Pyruvate orthophosphate dikinase of C(3) seeds and leaves as compared to the enzyme from maize. Plant Physiol 75 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees T, Entwistle TG, Dancer JE (1991) Interconversion of C-6 and C-3 sugar phosphates in non-photosynthetic cells of plants. In MJ Emes, ed, Compartmentation of Plant Metabolism in Non-Photosynthetic Tissues. Cambridge University Press, Cambridge, UK, pp 95–110
- Ball S, Guan HP, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J (1996) From glycogen to amylopectin: a model for the biosynthesis of the plant starch granule. Cell 86 349–352 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54 207–233 [DOI] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Saikusa T, Rodríguez-López M, Akazawa T, Pozueta-Romero J (2003) Sucrose synthase catalyzes the de novo production of ADPglucose linked to starch biosynthesis in heterotrophic tissues of plants. Plant Cell Physiol 44 500–509 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan MJ (2001) Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8 93–99 [Google Scholar]
- Boren M, Larsson H, Falk A, Jansson C (2004) The barley starch granule proteome: internalized granule polypeptides of the mature endosperm. Plant Sci 166 617–626 [Google Scholar]
- Boyer CD, Preiss J (1981) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol 67 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11 82–88 [DOI] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain CJ, Fries JP, Vogel JA, Randklev CL, Vossen AP, Dittmer SK, Watkins EE, Fiedler LJ, Wacker SA, Meinhover KC, et al (2002) Pyruvate,orthophosphate dikinase in leaves and chloroplasts of C(3) plants undergoes light-/dark-induced reversible phosphorylation. Plant Physiol 128 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain CJ, Heck JW, Colquhoun TA, Voge DG, Gu XY (2006) Posttranslational regulation of pyruvate, orthophosphate dikinase in developing rice (Oryza sativa) seeds. Planta 224 924–934 [DOI] [PubMed] [Google Scholar]
- Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith AM (1996) The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol 112 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56 623–632 [DOI] [PubMed] [Google Scholar]
- Duncan KA, Hardin SC, Huber SC (2006) The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol 47 959–971 [DOI] [PubMed] [Google Scholar]
- Fisher DK, Gao M, Kim KN, Boyer CD, Guiltinan MJ (1996) Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiol 110 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, et al (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard C, Barron C, Colonna P, Planchot V (2001) Amylose determination in genetically modified starches. Carbohydrate Polymers 44 19–27 [Google Scholar]
- Grimaud F, Rogniaux H, James MG, Myers AM, Planchot V (2008) Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J Exp Bot 59 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC (2007) Starch formation in the cereal endosperm. In Plant Cell Monographs, Endosperm, Vol 8. Springer, Berlin, pp 179–193
- Hatch MD (1987) C4 photosynthesis: A unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895 81–106 [Google Scholar]
- Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM (2008) Starch biosynthetic enzymes from developing Zea mays endosperm associate in multisubunit complexes. Plant Physiol 146 1892–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JM, Gaborieau M, Castignolles P, Gidley MJ, Myers AM, Gilbert RG (2008) Mechanistic investigation of a starch-branching enzyme using hydrodynamic volume SEC analysis. Biomacromolecules 9 954–965 [DOI] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42 901–911 [DOI] [PubMed] [Google Scholar]
- Leterrier MS, Holappa LD, Broglie KE, Beckles DM (2008) Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol 8 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene: motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sun F, Xu S, Chu X, Mukai Y, Yamamoto M, Ali S, Rampling L, Kosar-Hashemi B, Rahman S, et al (2003) The structural organisation of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthases in plants. Funct Integr Genomics 3 76–85 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mechin V, Thevenot C, Le Guilloux M, Prioul JL, Damerval C (2007) Developmental analysis of maize endosperm proteome suggests a pivotal role for pyruvate orthophosphate dikinase. Plant Physiol 143 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AO, Kelly GJ, Latzko E (1982) Pyruvate orthophosphate dikinase from the immature grains of cereal grasses. Plant Physiol 69 7–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Chourey PS (1995) Intracellular immunolocalization of adenosine-5′-diphosphoglucose pyrophosphorylase in developing endosperm cells of maize (Zea mays L.). Planta 197 522–527 [Google Scholar]
- Miyao M (2003) Molecular evolution and genetic engineering of C4 photosynthetic enzymes. J Exp Bot 54 179–189 [DOI] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34 173–185 [DOI] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Non-photosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51 111–140 [DOI] [PubMed] [Google Scholar]
- Palopoli N, Busi MV, Fornasari MS, Gomez-Casati D, Ugalde R, Parisi G (2006) Starch-synthase III family encodes a tandem of three starch-binding domains. Proteins 65 27–31 [DOI] [PubMed] [Google Scholar]
- Parry DAD, Fraser RDB, Squire JM (2008) Fifty years of coiled-coils and α-helical bundles: a close relationship between sequence and structure. J Struct Biol 163 258–269 [DOI] [PubMed] [Google Scholar]
- Rosti S, Denyer K (2007) Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species. J Mol Evol 65 316–327 [DOI] [PubMed] [Google Scholar]
- Sadimantara G, Abe T, Suzuki J, Hirano H, Sasahara T (1996) Characterization and partial amino acid sequence of a high molecular weight protein from rice seed endosperm: homology to pyruvate orthophosphate dikinase. J Plant Physiol 149 285–289 [Google Scholar]
- Senoura T, Asao A, Takashima Y, Isono N, Hamada S, Ito H, Matsui H (2007) Enzymatic characterization of starch synthase III from kidney bean (Phaseolus vulgaris L.). FEBS J 274 4550–4560 [DOI] [PubMed] [Google Scholar]
- Sheen J (1991) Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3 225–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff A, Meyer H, Riedel E, Schmitt JM, Lapke C (1998) The influence of plant pyruvate orthophosphate dikinase on a C3 plant with respect to the intracellular location of the enzyme. Plant Sci 136 43–57 [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL (1997) Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiol 113 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensballe A, Hald S, Bauw G, Blennow A, Welinder KG (2008) The amyloplast proteome of potato tuber. FEBS J 275 1723–1741 [DOI] [PubMed] [Google Scholar]
- Stitt M (1990) Fructose-2,6-bisphosphate as a regulatory molecule in plants. Annu Rev Plant Physiol Plant Mol Biol 41 153–185 [Google Scholar]
- Subbaiah CC, Palaniappan A, Duncan K, Rhoads DM, Huber SC, Sachs MM (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem 281 15625–15635 [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ (2004. a) Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot 55 2131–2145 [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ (2004. b) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DB (2000) On the non-random nature of amylopectin branching. Carbohydrate Polymers 43 223–239 [Google Scholar]
- Thorbjornsen T, Villand P, Denyer K, Olsen OA, Smith A (1996) Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J 10 243–250 [Google Scholar]
- Valdez HA, Busi MV, Wayllace NZ, Parisi G, Ugalde RA, Gomez-Casati DF (2008) Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47 3026–3032 [DOI] [PubMed] [Google Scholar]
- Vensel WH, Tanaka CK, Cai N, Wong JH, Buchanan BB, Hurkman WJ (2005) Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 5 1594–1611 [DOI] [PubMed] [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM (2004) Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol Biol 54 865–879 [DOI] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Szydlowski N, Delvalle D, D'Hulst C, James MG, Myers AM (2008) Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol 8 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.