Abstract
The reduction and the formation of regulatory disulfide bonds serve as a key signaling element in chloroplasts. Members of the thioredoxin (Trx) superfamily of oxidoreductases play a major role in these processes. We have characterized a small family of plant-specific Trxs in Arabidopsis (Arabidopsis thaliana) that are rich in cysteine and histidine residues and are typified by a variable noncanonical redox active site. We found that the redox midpoint potential of three selected family members is significantly less reducing than that of the classic Trxs. Assays of subcellular localization demonstrated that all proteins are localized to the chloroplast. Selected members showed high activity, contingent on a dithiol electron donor, toward the chloroplast 2-cysteine peroxiredoxin A and poor activity toward the chloroplast NADP-malate dehydrogenase. The expression profile of the family members suggests that they have distinct roles. The intermediate redox midpoint potential value of the atypical Trxs might imply adaptability to function in modulating the redox state of chloroplast proteins with regulatory disulfides.
Recent studies suggest that redox regulation is a major mechanism in cellular signal transduction and in control of gene expression in species as diverse as humans, plants, and bacteria (Danon, 2002; Toledano et al., 2004; Buchanan and Balmer, 2005; Holmgren et al., 2005). Members of the thioredoxin (Trx) superfamily of thiol-disulfide oxidoreductases play key roles in cellular redox regulation in almost all life forms (Debarbieux and Beckwith, 2000; Holmgren et al., 2005; Carvalho et al., 2006). Trxs share a similar fold (Trx-fold) and a common active site motif comprised of two vicinal Cys separated by two variable amino acids, the CXXC motif (Martin, 1995). Despite these common features, members of the Trx superfamily have diverse functions ranging from protein disulfide reductases to disulfide isomerases or to disulfide transferases. The versatility in function is reflected in part by the broad range of redox midpoint potentials measured for the Trxs. Redox midpoint potentials range from −300 mV for the reductive-type Trx-m (Hirasawa et al., 1999) to −120 mV for the oxidative-type Escherichia coli DsbA (Zapun et al., 1993; Aslund et al., 1997). Protein disulfide isomerases (PDIs), the bacterial DsbD, and glutaredoxins have intermediate redox midpoint potentials from −175 to −241 mV (Lundstrom and Holmgren, 1993; Aslund et al., 1997; Collet et al., 2002).
The observed differences in the redox midpoint potential between different Trxs reflect to some extent divergence in the stabilization of the negative charge of the N-terminal catalytic thiolate, and thus in the pKa, of the redox active site. One of the factors suggested to have an important role in determining the pKa of the catalytic Cys and the redox midpoint potential of the protein is the amino acid identity of the central dipeptide, located between the two Cys of the active site. The classic reductive-type Trxs share a canonical active site motif, the C(G/P)PC motif, while PDIs and the oxidative DsbA have the conserved CPHC and a CGHC motif, respectively. Mutating the central dipeptide was shown to change the redox midpoint potential of the protein, as well as its activity and its interactions with other proteins (Krause and Holmgren, 1991; Lundstrom et al., 1992; Chivers et al., 1996; Huber-Wunderlich and Glockshuber, 1998; Mossner et al., 1998; Quan et al., 2007). Additional amino acids outside of the active site were also suggested to contribute to the redox midpoint potential or the activity of different Trxs. Among them are several charged residues in the vicinity of the active site and a neighboring conserved Trp that precedes the active site. The exact contribution of each of these factors to the overall activity of Trxs is not yet clear and might differ between different members of the superfamily (Debarbieux and Beckwith, 2000).
In plants, the redox reactions of the photosynthetic machinery of the chloroplast need to adjust to both rapid and gradual environmental changes, such as photon flux, CO2 availability or water potential, and the associated free radical production. Four families of chloroplast-localized classic Trxs containing a canonical active site, Trx-f, Trx-m, Trx-x, and Trx-y, were characterized. The f-type and m-type Trxs were shown to be reduced by the ferredoxin-Trx system, which receives electrons from the light-capturing reactions of photosynthesis. The classic chloroplast Trxs are implicated in the light-dependent activation by reduction of different enzymes, including several metabolic enzymes such as NADP-malate dehydrogenase (MDH) and Fru-1,6-bisP, as well as in protection against oxidative stress (Schurmann and Buchanan, 2008). Additional roles, such as in oxidative-type reactions, are inferred indirectly by studies demonstrating that protein disulfide bonds are formed correctly in the chloroplast (Staub et al., 2000; Bally et al., 2008; Wittenberg and Danon, 2008). Further roles of atypical Trxs, containing a noncanonical redox active site or additional non-Trx domains, are just starting to emerge. For example, the recently characterized thylakoid membrane-anchored HCF164 protein has an atypical CEVC active site and a redox midpoint potential value of −224 mV (Motohashi and Hisabori, 2006). It was found that HCF164 is essential for the assembly of the chloroplast cytochrome b6f (Lennartz et al., 2001).
As studying atypical Trxs is expected to shed light on additional functions of Trxs in plants, we initiated a search in the Arabidopsis (Arabidopsis thaliana) genome for Trxs with a noncanonical active site. We characterized a small family of proteins, which we denoted AtACHTs (for atypical Cys His-rich Trxs). Notably, we found that the redox midpoint potential of three selected proteins, AtACHT1, AtACHT2a, and AtACHT4a, is significantly higher than that of the classic Arabidopsis Trxf1 (AtTrx-f1). Assays of the subcellular localization of the AtACHTs suggest that they are localized to the chloroplast. Examination of AtACHT1 and AtACHT4a showed that they are partitioned between the soluble stroma and the thylakoid membranes. AtACHT1, AtACHT2a, and AtACHT4a showed high preference in their activity toward the chloroplast 2-Cys peroxiredoxin A (2-Cys PrxA) relative to their activity toward the chloroplast NADP-MDH. The unique properties of the AtACHTs and their pattern of expression suggest distinct roles in the chloroplast.
RESULTS
Characteristics of the AtACHT Family of Proteins
To begin to look at Trxs with possible distinct activities from these of the well-characterized reductive-type chloroplast Trxs, we searched the Arabidopsis genome for small Trx-like genes with a noncanonical CXXC active site. The first protein with a noncanonical active site that emerged from a National Center for Biotechnology Information (NCBI) BLASTP search using a query of AtTrx-f1 sequence was AtACHT1. A follow-up analysis with AtACHT1 sequence identified a family of five genes denoted AtACHT1 to AtACHT5 (Fig. 1). The proteins are homologs of the previously reported Lilium longiflorum Trx-like sequence (Meyer et al., 1999). AtACHT2a and AtACHT2b are predicted splice variants that differ only in their C terminus, whereas the predicted splice variants AtACHT4a and AtACHT4b differ only in a small portion of their N terminus. Three family members, AtACHT3, AtACHT4a-4b, and AtACHT5, do not contain the highly conserved Trp that precedes the redox active site. All family members include an N-terminal leader predicted to function as an organelle-targeting signal by the computer program TargetP (Emanuelsson et al., 2000). The proteins are typified by several conserved Cys and His in addition to the active-site Cys. They also contain a C-terminal tail with varying lengths with no expected common fold or function (Fig. 1). Reverse transcription (RT)-PCR authenticated that all AtACHTs are actively transcribed (data not shown). Secondary structure calculation, using the computer program JNet (Cuff and Barton, 1999), suggests that the core of all five proteins contains the structure elements that confer the Trx fold (Martin, 1995; Capitani et al., 2000; Fig. 1).
Figure 1.
Amino acid sequence alignment of the AtACHT proteins. Secondary structure elements conferring the Trx fold are illustrated by cylinders for α-helices and arrows for β-sheet strands. The residues were color labeled: light blue (A, I, L, M, F, W, V, C), red (R, K), green (N, Q, S, T), magenta (D, E), orange (G), cyan (H, Y), and yellow (P). Conserved His are marked in red and conserved Cys are in blue. Arrows mark active site Cys. AGI accession numbers of the family members are: At4g26160 (AtACHT1), At4g29670.1 (AtACHT2a), At4g29670.2 (AtACHT2b), At2g33270 (AtACHT3), At1g08570.1 (AtACHT4a), At1g08570.2 (AtACHT4b), and At5g61440 (AtACHT5). [See online article for color version of this figure.]
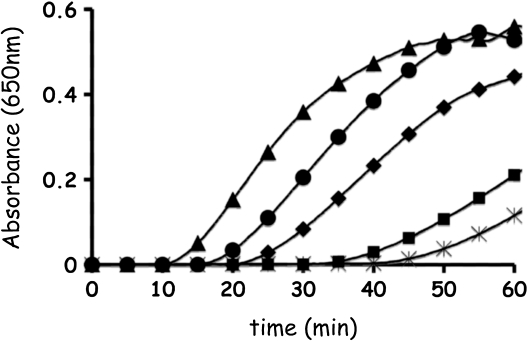
To examine whether the proteins are active thiol-oxidoreductases, leaderless forms of three selected proteins, AtACHT1, AtACHT2a, and AtACHT4a, were expressed in E. coli, purified to homogeneity, and their catalytic activity was compared to that of the classic AtTrx-f1 in the standard insulin turbidity assay (Holmgren, 1979). We found that the catalytic activity of AtACHT1 was slightly higher, and that of AtACHT2a was slightly lower, than that of AtTrx-f1 (Fig. 2). These results suggest that the AtACHT proteins preserve the catalytic activity that is common to members of the Trx oxidoreductase superfamily. Notably, the activity of the Trp-less AtACHT4a in parallel reactions was poor (Fig. 2).
Figure 2.
The activity of AtACHT proteins in the standard insulin assay. AtACHT1 (triangles), AtACHT2a (diamonds), and AtACHT4a (rectangles) were tested for their ability to catalyze the reduction of insulin disulfides. The precipitation rate of the reduced insulin by the different AtACHTs was compared to reactions catalyzed by AtTrx-f1 as positive control (circles) or DTT alone as negative control (asterisk).
The AtACHT Proteins Have a Higher Redox Midpoint Potential Than AtTrx-f1
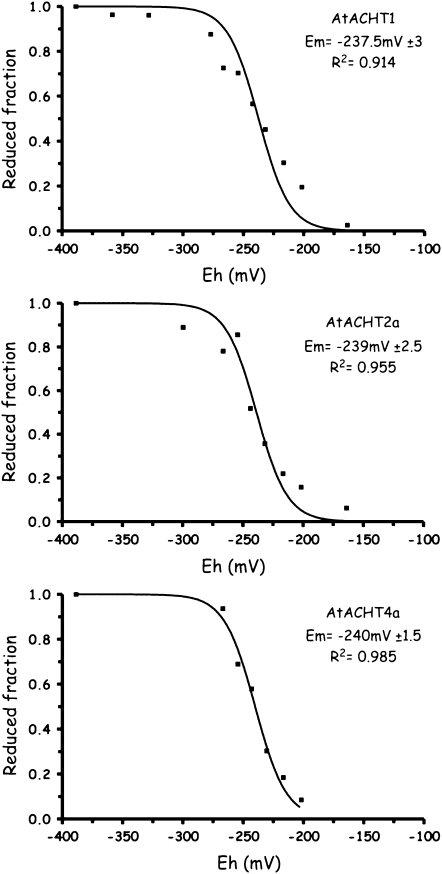
It was shown that the central dipeptide sequence of the active site could influence the redox midpoint potential as well as the protein function (Krause and Holmgren, 1991; Lundstrom et al., 1992; Huber-Wunderlich and Glockshuber, 1998; Quan et al., 2007). Thus, the noncanonical active site motifs of the AtACHTs might suggest that they have redox properties that differ from these of the classic reductive-type Trxs. We therefore estimated their redox midpoint potential by the monobromobimane (mBBr) method (Hirasawa et al., 1999). We found the redox midpoint potential of a control Trx, AtTrx-f1, to be −290 mV at pH 7.0 (data not shown) in agreement with its published value (Hirasawa et al., 1999; Collin et al., 2003). In comparison, the selected AtACHT proteins, AtACHT1, AtACHT2a, and AtACHT4a, displayed significantly higher estimated redox midpoint potential values at pH 7.0, −237 mV ± 3, −239 mV ± 2.5, and −240 mV ± 1.5, respectively (Fig. 3). This finding infers that the AtACHT proteins might have roles differing from those of the reductive-type classic Trxs.
Figure 3.
The AtACHT proteins have higher redox midpoint potentials in comparison to canonical Trxs. AtACHT1, AtACHT2a, and AtACHT4a proteins were incubated at defined redox potentials (Eh) values at pH 7.0, established by mixing various ratios of reduced and oxidized glutathione. The fluorescent thiol-labeling reagent mBBr was added and its fluorescence was measured. Estimated redox midpoint potential, se, and R2 values are presented.
The AtACHTs Are Targeted to Chloroplasts
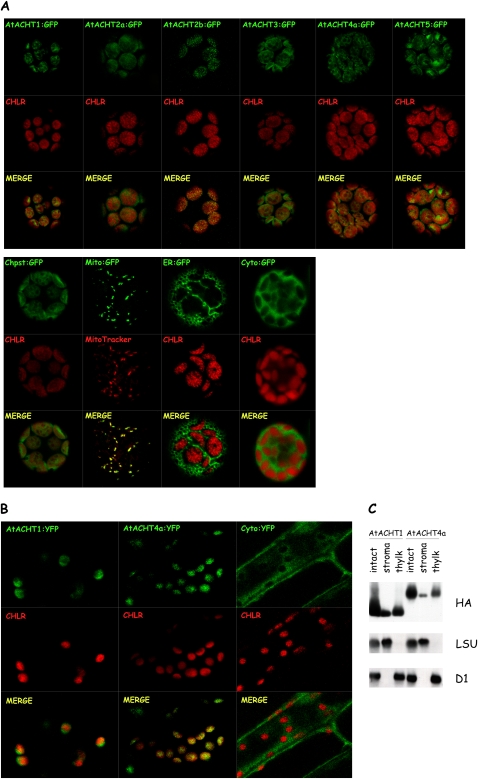
The presence of a leader peptide in the N-terminal sequence of all AtACHTs indicated that they might be localized to subcellular organelles. Thus, to study the localization of the AtACHTs, we compared the accumulation of each of the transiently expressed AtACHTs, fused with GFP at its C terminus (AtACHT1–5:GFP), to that of chloroplast-localized GFP and to the autofluorescence of chlorophyll, to mitochondrion-localized GFP, to an endoplasmic reticulum (ER)-localized GFP, and to cytoplasm-localized GFP in Arabidopsis protoplasts using confocal laser microscopy (Fig. 4A). To avoid mislocalization due to overaccumulation of expressed proteins, we imaged only protoplasts displaying the earliest signal of GFP fluorescence, as previously done in our lab (Levitan et al., 2005). Interestingly, the fluorescence images of all the AtACHTs resembled that of the chloroplast-localized GFP and overlapped the chlorophyll autofluorescence. In contrast, none of the AtACHTs fluorescence paralleled that of GFP localized to the other plant organelles, suggesting that all assayed AtACHTs are targeted to the chloroplast.
Figure 4.
All family members are localized to chloroplasts in vivo. A, AtACHT proteins fused with GFP at their C terminus were transiently expressed in Arabidopsis protoplasts. Images of the GFP-fused AtACHT (AtACHT:GFP) fluorescence and the chlorophyll autofluorescence (CHLR) were combined (MERGE) to illustrate the relative localization of the GFP fluorescence and the chloroplasts. Markers for chloroplast (Chpst), mitochondria (Mito), ER, and cytoplasm (Cyto) were visualized as well. B, Transgenic plants, each expressing AtACHT1:YFP or AtACHT4a:YFP, were compared to plants expressing cytoplasmic YFP under the control of 35S promoter. The fluorescence images corroborated that both AtACHT1 and AtACHT4a are targeted to chloroplasts in planta. C, Subfractionation of intact chloroplasts from transgenic plants expressing AtACHT fused with HA3 affinity tag into stroma and thylakoid membranes demonstrates the suborganellar localization of AtACHT1 and AtACHT4a by immunoblot using anti-HA antibody. The stromal large subunit of Rubisco (LSU) and the membranal D1 protein were blotted for quality assessment of the fractionation procedure.
We chose two proteins, AtACHT1 and AtACHT4a, for further studies. First, to authenticate the protein localization, transgenic plants expressing AtACHT1 or AtACHT4a, each fused with yellow fluorescent protein (YFP) at their C terminus, were generated and imaged using confocal laser microscopy. The fluorescence images corroborated that both AtACHT1 and AtACHT4a are targeted to chloroplasts in planta and that they do not accumulate to detectible levels in other subcellular organelles (Fig. 4B). Second, to further study the subchloroplast localization of the proteins, we purified intact chloroplasts from transgenic plants, expressing the AtACHTs with HA3 affinity tag at their C terminus, and analyzed their soluble and membranal fractions by immunoblot analysis. The AtACHT1 and AtACHT4a proteins were found partitioned in both the stromal fraction, containing the large subunit of Rubisco, and the thylakoid membranes, containing the D1 protein (Fig. 4C), suggesting that they have a role in both compartments.
The AtACHTs Show Preference toward 2-Cys Prx A
The plastidial localization of the AtACHT proteins prompted us to examine their activity toward known targets of chloroplast Trxs, such as 2-Cys Prx A and MDH. The 2-Cys Prxs detoxify peroxides by their reduction. Several Trx-like proteins were shown to reduce 2-Cys Prx (Dietz et al., 2006), of which AtTrx-x and NTRC have thus far been shown to be the most efficient (Collin et al., 2003; Perez-Ruiz et al., 2006). The ability of the AtACHTs to serve as electron donors to the recombinant mature Arabidopsis 2-Cys PrxA (At-2-Cys PrxA) was compared to that of AtTrx-f1 by measuring the rates of disappearance of hydrogen peroxide in the presence of the different Trxs. AtACHT1, AtACHT2a, and AtACHT4a all displayed high activity, with AtACHT4a showing the highest activity, toward At-2-Cys PrxA in the presence of dithiothreitol (DTT; Fig. 5A). In contrast, at a similar protein concentration, the activity of AtTrx-f1 was close to background activity, and low activity was observed at 4-fold higher protein concentration (data not shown). The higher redox potential of the ACHTs (around −240 mV; Fig. 3) suggests that these proteins might be alternatively reduced by reduced glutathione (GSH). However, when the Prx activity assay was repeated in the presence of GSH rather than DTT, the proteins were not able to efficiently increase the peroxidase activity of At-2-Cys PrxA (Fig. 5B), suggesting that a dithiol rather than a monothiol is the preferred electron donor of the AtACHTs. The Trxs did not display any peroxidase activity in the absence of At-2-Cys PrxA (data not shown).
Figure 5.
AtACHTs activation of chloroplast target enzymes. A, The AtACHTs reacted with high efficiency with At-2-Cys PrxA in the presence of DTT. At-2-Cys PrxA (5 μm) was incubated with 2.5 μm of AtTrx-f1 (circles), AtACHT1 (triangles), AtACHT2a (diamonds), or AtACHT4a (rectangles) in the presence of 0.4 mm DTT. At-2-Cys PrxA alone (asterisk) was used as a control. The reduction of H2O2 was measured at different time points. B, The ACHTs did not react efficiently with At-2-Cys PrxA when GSH was used as a reductant. The assay was performed as in A, but DTT was replaced with 0.8 mm GSH. Symbols are as in A. C, The AtACHTs reacted poorly with AtMDH. DTT-dependent activation of AtMDH (1.5 μm) was carried out with 1 μm AtTrx-f1 or the different AtACHTs. AtMDH enzyme activity after reduction was measured by monitoring the initial rate of consumption of NADPH during reaction with the substrate oxaloacetate. Symbols are as in A.
The activation of MDH, which catalyzes the reduction of oxaloacetate into malate using NADPH as a cofactor, is dependent on the light-regulated reduction of two disulfide bonds by Trx (Schurmann and Buchanan, 2008). Though originally thought to be specifically light regulated through Trx-m, Trx-f was later shown to be the most efficient activator of sorghum (Sorghum bicolor) MDH (Collin et al., 2003, 2004). Activation assays were performed by incubating recombinant mature Arabidopsis NADP-MDH (AtMDH) with either AtTrx-f1 or selected AtACHTs in the presence of DTT and measuring the AtMDH enzymatic activity as oxidation of NADPH as described before (Jacquot and Buchanan, 1981). While 1 μm of AtTrx-f1 was sufficient to efficiently activate AtMDH under our experimental conditions, the same concentration of the AtACHTs did not appreciably activate AtMDH (Fig. 5C). By increasing the AtACHT concentration to 20 μm, a poor activation of AtMDH was observed (data not shown). Thus, the AtACHTs showed higher preference in their activity toward At-2-Cys PrxA than their activity toward AtMDH.
The AtACHTs Differ in Their Expression Pattern
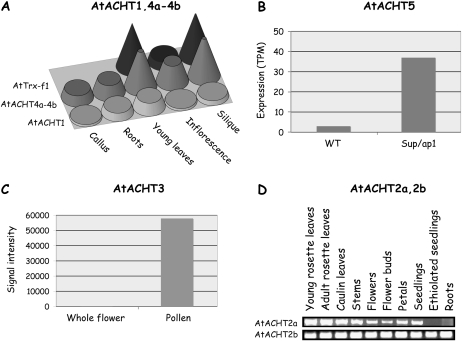
We investigated, using the MPSS (Meyers et al., 2004) and the Genevestigator (Zimmermann et al., 2004) database servers and by semiquantitative RT-PCR (sqRT-PCR) analysis, whether the expression pattern of the different AtACHTs is unique or common. The results indicated a unique pattern of expression for each gene. AtACHT1 and AtACHT4a-4b transcripts express in all examined tissues, but AtACHT4a-4b is particularly abundant in young leaves and siliques (Fig. 6A). Notably, AtACHT3 and AtACHT5 transcripts accumulate under very specific conditions. AtACHT5 is highly expressed in superman and apetala (sup/ap1) double mutant plants (Fig. 6B), which are defective in floral meristem development (Meyers et al., 2004), whereas AtACHT3 message accumulates about 400-fold more in pollen than in the whole flower (Fig. 6C), suggesting that its expression is specific to pollen tissue (Pina et al., 2005). A more detailed study of these two genes is required to determine whether they have specialized roles in reproductive tissues. We further studied the expression of the two splice variants, AtACHT2a and AtACHT2b, by sqRT-PCR analysis and found differences in their tissue specificity (Fig. 6D). AtACHT2a transcripts accumulate to high levels in green tissues, whereas AtACHT2b transcript amounts are even in all analyzed plant tissues, suggesting that AtACHT2a role is unique to chloroplasts, whereas AtACHT2b might be required for all types of plastids.
Figure 6.
Differential expression pattern of AtACHT family members. A, AtACHT1 transcript levels are similar in all tissues, while AtACHT4a-4b transcripts accumulate mainly in green tissues in units of transcripts per million (TPM; obtained from MPSS; Meyers et al., 2004). B, AtACHT5 is highly expressed in inflorescence of double mutant sup/ap1 as compared to wild-type (WT) plants (obtained from MPSS). C, AtACHT3-specific expression in pollen versus whole flower expression (obtained from Genevestigator; Zimmermann et al., 2004; Pina et al., 2005). D, sqRT-PCR analysis of the splice variants shows that AtACHT2a expression is high in green tissues, whereas AtACHT2b transcript accumulates equally in green and non-green tissues.
The ACHT Family Is Unique to Plants
The plastid localization of AtACHTs suggested plant-specific function. Thus, to determine whether this family is unique to plants, a BLAST search was made against all available sequences in NCBI and in specific databases of selected organisms. We found homologs from the green algae Chlamydomonas reinhardtii and Ostreococcus tauri, the moss Physcomitrella patens, and the higher plants Oryza sativa and Zea mays. Notably, the genomes of nonplant species as well as cyanobacteria or other prokaryotes do not seem to contain ACHT homologs, suggesting that similarly to the chloroplast Trx-f (Schurmann and Buchanan, 2008) they are plant specific and of eukaryotic origin. A phylogenetic analysis that included one-Trx-domain proteins with an atypical or classic vicinal dithiol active site showed that the AtACHTs and their homologs are clustered together on a unique node, whereas the other atypical Trxs are more closely related to the classic Trxs (Fig. 7). The finding of an AtACHT homolog in Ostreococcus (Fig. 7), one of the most ancient branches of the green lineage (Derelle et al., 2006), suggests that this family originated early before the split of higher plants from unicellular plants.
Figure 7.
A phylogenetic tree of Arabidopsis Trxs and various plant ACHT proteins, an unrooted bootstrapped tree comprised of Arabidopsis one Trx domain proteins with an atypical or classic vicinal dithiol active site, and ACHT homologs was generated in PHYLIP (Felsenstein, 2005). The ACHT proteins (AtACHT1–5, OtI, CrI, CrII, PpI, LlI, OsI, OsII, OsIII, OsIV, ZmI, ZmII, ZmIII) grouped with the Trxs with eukaryotic origin (f, AtL5, h, CxxC2, WCGVC, o) and not with the prokaryotic-origin Trxs (m, x, y, HCF164). Bootstrapped values of above 70% are indicated. Although Clot, WCRKC1, and WCRKC2 appear to associate with the eukaryotic Trxs, the bootstrapped values were not significant. The sequence of the noncanonical active-site Trxs is shown in gray letters. The organism codes are as follows: At, Arabidopsis; Ot, O. tauri; Cr, C. reinhardtii; Pp, P. patens; Ll, L. longiflorum; Os, O. sativa; Zm, Z. mays. Accession numbers are: OtI (Ot10g03450), CrI (XP_001697443), CrII (XP_001696231), PpI (EDQ64087), LlI (L18909) OsI (Os05g11090), OsII (Os03g07234), OsIII (Os03g21000), OsIV (Os07g48510), ZmI (AY104786), ZmII (BT018452), ZmIII (AY105197), and AtL5 (At1g07700, Lilium5). Abbreviations of the classic and the atypical Trxs are as in Meyer et al. (2008).
The phylogenetic analysis implies that the AtACHTs are subdivided into two classes (Fig. 7). Class 1 contains AtACHTs 1, 2a, and 2b, whereas Class 2 includes AtACHTs 3, 4a, 4b, and 5. All Class 2 AtACHTs and their homologs lack the conserved Trp preceding the redox active site and, except for one Oryza protein, have an identical active site motif, CGGC. All of them are typified by a large C-terminal extension. Class 1 AtACHTs have a C(G/A)SC active site motif and a shorter C terminus relative to Class 2 proteins. Both Class 1 and 2 proteins contain additional Cys, which are conserved in all proteins, and His, which are conserved within each class (Fig. 1). The relevance of the conserved Cys and His to possibly newly evolved activities of these proteins will have to be addressed in future studies.
The high degree of conservation of this protein family along the plant evolution course, their absence from nonplant organisms, and their chloroplast localization imply that they have a preserved role in plants. Their unique sequence elements differentiate them from the classic Trxs, and their higher redox potential raises the possibility of specialized redox function in plant chloroplasts.
DISCUSSION
The AtACHTs constitute a small family of chloroplast Trx-like proteins that display a redox midpoint potential that is significantly less reducing compared to the classic Trxs (Fig. 3), a redox active site with a different central dipeptide than the canonical sequence, and several additional conserved Cys and His residues outside of the active site (Fig. 1). Previous experiments in which the central dipeptide sequence of one Trx family member was swapped with that of a second Trx resulted in a profound effect on the activity of the Trx. Often, a shift in the redox properties of the mutated Trx toward those of the second Trx was observed (Krause and Holmgren, 1991; Lundstrom et al., 1992; Chivers et al., 1996; Huber-Wunderlich and Glockshuber, 1998; Mossner et al., 1998; Quan et al., 2007). Notably, the two classes of AtACHTs differ in their central dipeptide; the Class 1 dipeptide is C(G/A)SC and the Class 2 is CGGC (Fig. 7). Yet, in spite of the difference in the central dipeptide, the redox midpoint potentials of selected members from both Class 1 and 2 AtACHTs were found to be almost identical (Fig. 3), suggesting that other residues in addition to the redox active site dipeptide might affect the redox midpoint potential as well. This assumption is further supported by the finding that the redox midpoint potential of different Trxs, containing the same canonical central dipeptide, may vary between −300 mV and −275 mV (Collin et al., 2003, 2004). Hence, the AtACHTs have evolved a redox active site with variable central dipeptide while maintaining a similar redox midpoint potential value that has been conceivably preserved by compensatory changes in other residues than the central dipeptide.
Both prokaryotic- and eukaryotic-type Trxs contain a highly conserved Trp residue preceding the active site. In Arabidopsis, 57 out of the 64 Trx-related gene products contain this Trp (Meyer et al., 2008). Three out of the seven Arabidopsis gene products missing the conserved Trp residue are the Class 2 AtACHTs (Fig. 1). The other four Trp-less Trx-related proteins of Arabidopsis are CDSP32 and NTRC, two proteins implicated in the reduction of chloroplastic peroxiredoxin (Dietz et al., 2006), APRl7, a PDI-like protein (Houston et al., 2005) and lilium5 (Meyer et al., 2008). A structure analysis of a Trp mutant of C. reinhardtii Trx-h suggested that the Trp residue affects the active-site conformation and the substrate recognition (Menchise et al., 2001). Other studies using E. coli Trx and C. reinhardtii Trx-h suggested that replacing the Trp affects the activity of the Trx toward in vitro substrates but not its redox midpoint potential (Krause and Holmgren, 1991; Krimm et al., 1998; Menchise et al., 2001). Here, the Trp-less AtACHT4a was less active than the Trp-containing AtACHTs 1 and 2a in the standard insulin reduction assay (Fig. 2). But, similar to the Trp-less CDSP32 and NTRC Trx-like proteins, as well as the Trp-containing AtACHT1 and 2a, AtACHT4a showed high activity in assays containing the chloroplast enzyme At-2-Cys PrxA (Fig. 5). These findings suggest that in spite of the importance of the Trp to the activity of a Trx, its presence or absence from a naturally occurring Trx is not a good predictor of its activity against specific substrates.
A plastidial function for the AtACHTs is implicated by their localization (Fig. 4) and by their phylogeny (Fig. 7), which is unique to the plastid-containing viridiplantae. What might be, then, the unique function of the AtACHTs among the 20 or so Trx-like proteins shown to reside in plastids? The first clue might be found in the expression pattern of the AtACHTs. AtACHT2a and AtACHT4a-4b express mainly in photosynthetic tissue (Fig. 6), suggesting that they might function in parallel to the classic Trxs, such as Trx-f and Trx-m. In contrast, AtACHT1 and AtACHT2b transcripts accumulate quite evenly in green and non-green tissues, suggesting a more generalized role that is not limited to the chloroplast (Fig. 6). The expression of the AtACHT5 and AtACHT3 transcripts appears to be unique. AtACHT5 transcript is enriched in floral tissues and accumulates to very high levels in the double mutant of regulators of floral meristem development (Sup/ap1; Fig. 6), whereas the AtACHT3 transcript is highly specific to pollen (Fig. 6), suggesting a more specialized function for these two proteins.
A second clue might be found in the activity in vitro of the AtACHTs, which, relative to AtTrx-f1, reacted with high efficiency with At-2-Cys PrxA and poorly with AtMDH (Fig. 5). The canonical site-containing AtTrx-x and AtTrx-y were also found to be inefficient activators of sorghum MDH and to react efficiently with peroxiredoxins in vitro (Collin et al., 2003, 2004). Based on their in vitro activities, it was suggested that Trx-x and Trx-y might function specifically in resisting oxidative stress rather than in enzyme regulation (Collin et al., 2003). The similar in vitro activity profile of the AtACHTs might hint at similar roles. Furthermore, the similar partitioning between the stroma and the thylakoids of the AtACHTs (Fig. 4) and 2-Cys PrxA (Dietz et al., 2006) suggests that the two activities might be linked.
Another clue to the role of AtACHTs might be found in their redox midpoint potential, which is of an intermediate value (Fig. 3) between the two extremes of the oxidative DsbA (Zapun et al., 1993; Aslund et al., 1997) and the reductive Trx-m (Hirasawa et al., 1999) and is almost identical to the −241 mV value of the soluble Trx-like γ-domain of the membrane-bound bacterial DsbD (Collet et al., 2002). Notably, DsbD's role is to provide electrons to redox pathways supporting transient reduction for the isomerization of misformed disulfides (Nakamoto and Bardwell, 2004; Porat et al., 2004). Hence, the intermediate value of the redox midpoint potential of the AtACHTs might suggest adaptability to function in reversible reactions, such as those modulating the redox state of regulatory proteins (Buchanan, 1980; Trebitsh et al., 2000). Yet, it is important to note that while the midpoint redox potential value of a protein likely reflects its adaptation to oxidative- or reductive-type reactions, its in vivo role is not necessarily correlated with its redox potential (Ortenberg and Beckwith, 2003) and might depend on additional factors, such as dimerization, localization, or interaction with accessory proteins. Hence, further genetic and molecular studies are required to elucidate the exact redox function of the individual AtACHTs.
MATERIALS AND METHODS
Identification and Cloning of the AtACHT Proteins from Arabidopsis
The protein sequence of AtTrx-f1 was used to search the Arabidopsis (Arabidopsis thaliana) database for Trx-like proteins containing a noncanonical active site using the BLASTP program at NCBI (http://www.ncbi.nlm.nih.gov). Total RNA was extracted using Tri-reagent (MRC). The RNA was subjected to RT using Superscript II (Invitrogen, Rhenium) and oligo(dT). PCR reactions were done with gene-specific primers.
Generation of Protein Multiple Alignment and Phylogenetic Trees
Protein multiple alignments were generated using the ClustalX program (Thompson et al., 1997) and viewed through Jalview (Clamp et al., 2004). Secondary structure elements were predicted using JNet (Cuff and Barton, 1999). Homologues were identified by BLASTP or TBLASTN searches at NCBI and at specific databases of selected organisms (Chlamydomonas reinhardtii: http://genome.jgi-psf.org/cgi-bin/runAlignment?db=chlre2&advanced=1; Physcomitrella patens: http://moss.nibb.ac.jp/blast/blast.html; Oryza sativa: http://blast.jcvi.org/euk-blast/index.cgi?project=osa1). The phylogenetic bootstrapped tree was generated in PHYLIP (Felsenstein, 2005), based on protein multiple alignment of the Trx domain, and viewed in TreeView (Page, 1996).
To identify alternative splicing variants, the genome sequence of Arabidopsis was analyzed for gene structure positions (exons and introns borders). The borders between expressed sequences and introns were compared to the cDNA sequences of AtACHT2a and AtACHT2b, which were isolated by RT-PCR. The existence of the splice variants was predicted also by The Arabidopsis Information Resource (www.arabidopsis.org).
Expression and Purification of Recombinant Proteins
The cDNAs encoding AtACHTs, AtMDH (At5g58330), and At-2-Cys PrxA (At3g11630) without their putative transit peptide (primer sequences are listed in Supplemental Table S1), as predicted by TargetP (Emanuelsson et al., 2000), were cloned into the expression vector pQE30 (Qiagen, Eldan), resulting in N-terminal His-tagged proteins. M15 cells were transformed with the plasmids and protein expression induction was performed according to the manufacturer's instructions. The proteins were purified using HiTrap nickel affinity column (HiTrap Chelating HP, Amersham Pharmacia) according to the manufacturer's instructions and dialyzed overnight against 2,000 volumes of dialysis buffer, 1× phosphate saline buffer, 20% glycerol, 2 mm EDTA, and 5 mm β-mercaptoethanol for the AtACHTs, or the dialysis buffer without EDTA and β-mercaptoethanol for AtMDH and At-2-Cys PrxA. The proteins were purified to homogeneity of above 95% as judged by Coomassie Brilliant Blue gel staining.
Insulin Turbidity Assay
Recombinant proteins (5 μm) were used in the insulin reduction assay, as described previously (Holmgren, 1979). The turbidity of the reduced insulin chains was recorded using Synergy HT microplate reader at 650 nm every 30 s for 2 h.
Determination of Redox Midpoint Potential
The redox potential was evaluated using the thiol-labeling reagent mBBr, as described previously (Hirasawa et al., 1999). Different ratios of reduced to oxidized DTT or glutathione in Tricine buffer, pH 7.0, were used for AtTrx-f1 and AtACHT proteins, respectively. The redox potentials were calculated by curve fitting to the Nernst equation using GraphPad Prism version 4.0 and redox midpoint potential values of −330 mV for DTT (Hirasawa et al., 1999) and −240 mV for glutathione (Aslund et al., 1997). Experiments were repeated three times, and the obtained redox midpoint potential values were independent of equilibrium time.
Determination of Subcellular Localization
Each AtACHT open reading frame (ORF) was ligated through SmaI sites into a puc18-GFP5-containing vector, yielding a fusion protein upstream and in frame to GFP5 ORF under the control of the cauliflower mosaic virus 35S promoter. Control fusion proteins were prepared as well and included the small subunit of Rubisco (kindly provided by Yoram Eyal, Agricultural Research Organization, Volcani Center), the chitinase ER marker (kindly provided by Jean-Marc Neuhaus, University of Neuchatel, Switzerland), the mitochondrial AtTrx-o1 (At2g35010), and the mature form of the GFP protein. The transient expression assays were performed as described before (Sheen, 2002). Protoplasts were viewed 16 h after transformation.
For transgenic plant generation, AtACHT1 and AtACHT4a were ligated upstream and in frame to HA3 or YFP tags. The fragments were cloned into pART7 vector under the control of the 35S promoter and 35S terminator. As a control, the YFP ORF was ligated as well. NotI-digested fragments were ligated into the binary vector pBART, which confer glufosinate (BASTA) resistance in plants. The binary plasmids were introduced into Agrobacterium tumefaciens through electroporation. Plant transformation was made by the floral dip method (Clough and Bent, 1998).
Fluorescence images were obtained as described before (Levitan et al., 2005). Because transient expression under the 35S promoter may overproduce the expressed protein, resulting in its mislocalization, we imaged only viable protoplasts displaying the earliest signal of GFP fluorescence and performed western blots to confirm the expression of the full-length fusion proteins (data not shown).
Chloroplast Purification and Subfractionation
Intact chloroplasts were purified from transgenic plants expressing AtACHT-HA3 fusion proteins as described before (Aronsson and Jarvis, 2002) with slight modifications. Plants were grown under a 10-h photoperiod at 20°C and harvested at 6 weeks. Crude chloroplasts were overlayed on a two-step, 40/70% (w/v), Percoll gradient. After a 15-min centrifugation at 10,000g, intact chloroplasts were recovered from the interface and washed twice with chloroplast washing buffer, 0.3 m sorbitol, 20 mm HEPES, pH 8, 10 mm MgCl2, and 10 mm NaCl. Chloroplast subfractions were prepared by incubating the chloroplasts in 10 mm HEPES, pH 7.5, on ice for 15 min with vigorous vortexing. The fractions were separated by 10 min centrifugation at 10,000g. The pellet, containing membranes, was washed twice in 10 mm HEPES, pH 7.5. Equal amounts of proteins were run on SDS-PAGE gels, and immunoblots were performed using antibodies against the large subunit of Rubisco (kindly provided by Michal Shapira, Ben-Gurion University of the Negev), the D1 protein (Agrisera), and the HA tag (Sigma-Aldrich).
Peroxiredoxin Activity Assay
At-2-Cys PrxA at a concentration of 5 μm was incubated with 2.5 μm of the different AtACHTs or 2.5 or 10 μm AtTrx-f1 in a reaction mixture containing 50 mm phosphate buffer, pH 7.4, and either 0.4 mm DTT or 0.8 mm GSH. The reaction was started by adding 100 μm hydrogen peroxide (H2O2). The concentration of H2O2 was determined at different time points using the PeroXOquant reagent (Pierce).
MDH Activity Assay
DTT-dependant activation of AtMDH by 1 μm of AtTrx-f1 or of the different AtACHTs was carried out with 1.5 μm AtMDH and 10 mm DTT in 100 mm Tris-HCl, pH 7.9. At fixed times, 20-μL aliquots of the activation mixture were used to determine the activity of the AtMDH by following the initial rate of consumption of NADPH at room temperature in a 1-mL assay mixture containing 140 μm NADPH and 750 μm oxaloacetate in 100 mm Tris-HCl, pH 7.9. The oxidation of NADPH was followed spectroscopically at 340 nm. One unit of MDH activity is defined as the amount oxidizing 1 μmol NADPH/min and corresponds to an absorbance change of 6.22.
Gene Expression Analysis
The MPSS (http://mpss.udel.edu/at/java.html) and the Genevestigator database (www.genevestigator.ethz.ch/at) were used to evaluate differential gene expression. Validation of the main results was done using sqRT-PCR. Total RNA was extracted and equal amounts of RNA (2 μg) were subjected to RT using Superscript II (Invitrogen) and oligo(dT). PCR reactions were done with gene-specific primers (as listed in Supplemental Table S1). As a control, the actin and tubulin genes were amplified as well. Selected bands were excised, purified from the gel, and sequenced to authenticate their identity.
Sequence data from this article can be found in the GenBank data libraries under AGI accession numbers At4g26160 (AtACHT1), At4g29670.1 (AtACHT2a), At4g29670.2 (AtACHT2b), At2g33270 (AtACHT3), At1g08570.1 (AtACHT4a), At1g08570.2 (AtACHT4b), and At5g61440 (AtACHT5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Abdussalam Azem and his lab members at Tel Aviv University for helping us with the MDH activity assay. We are grateful for Vladimir Kiss for his assistance with the confocal microscopy. A.D. is incumbent of The Henry and Bertha Benson Chair, Weizmann Institute of Science.
This work was supported by the Israeli Science Foundation, by the Minerva Foundation, and by the Charles W. and Tillie K. Lubin Center for Plant Biotechnology at the Weizmann Institute of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Avihai Danon (avihai.danon@weizmann.ac.il).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529 215–220 [DOI] [PubMed] [Google Scholar]
- Aslund F, Berndt KD, Holmgren A (1997) Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem 272 30780–30786 [DOI] [PubMed] [Google Scholar]
- Bally J, Paget E, Droux M, Job C, Job D, Dubald M (2008) Both the stroma and thylakoid lumen of tobacco chloroplasts are competent for the formation of disulphide bonds in recombinant proteins. Plant Biotechnol J 6 46–61 [DOI] [PubMed] [Google Scholar]
- Buchanan BB (1980) Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31 341–374 [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Capitani G, Markovic-Housley Z, DelVal G, Morris M, Jansonius JN, Schurmann P (2000) Crystal structures of two functionally different thioredoxins in spinach chloroplasts. J Mol Biol 302 135–154 [DOI] [PubMed] [Google Scholar]
- Carvalho AP, Fernandes PA, Ramos MJ (2006) Similarities and differences in the thioredoxin superfamily. Prog Biophys Mol Biol 91 229–248 [DOI] [PubMed] [Google Scholar]
- Chivers PT, Laboissiere MC, Raines RT (1996) The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J 15 2659–2667 [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20 426–427 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Collet JF, Riemer J, Bader MW, Bardwell JC (2002) Reconstitution of a disulfide isomerization system. J Biol Chem 277 26886–26892 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol 136 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ (1999) Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins 34 508–519 [DOI] [PubMed] [Google Scholar]
- Danon A (2002) Redox reactions of regulatory proteins: do kinetics promote specificity? Trends Biochem Sci 27 197–203 [DOI] [PubMed] [Google Scholar]
- Debarbieux L, Beckwith J (2000) On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J Bacteriol 182 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, et al (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SMN, Baier M, Finkemeier I (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57 1697–1709 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. PHYLIP. http://evolution.genetics.washington.edu/phylip.html (January 1, 2007)
- Hirasawa M, Schurmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38 5200–5205 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254 9627–9632 [PubMed] [Google Scholar]
- Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH (2005) Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33 1375–1377 [DOI] [PubMed] [Google Scholar]
- Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS (2005) Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol 137 762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber-Wunderlich M, Glockshuber R (1998) A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold Des 3 161–171 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Buchanan BB (1981) Enzyme regulation in C(4) photosynthesis: purification and properties of thioredoxin-linked NADP-malate dehydrogenase from corn leaves. Plant Physiol 68 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G, Holmgren A (1991) Substitution of the conserved tryptophan 31 in Escherichia coli thioredoxin by site-directed mutagenesis and structure-function analysis. J Biol Chem 266 4056–4066 [PubMed] [Google Scholar]
- Krimm I, Lemaire S, Ruelland E, Miginiac-Maslow M, Jaquot JP, Hirasawa M, Knaff DB, Lancelin JM (1998) The single mutation Trp35-->Ala in the 35-40 redox site of Chlamydomonas reinhardtii thioredoxin h affects its biochemical activity and the pH dependence of C36-C39 1H-13C NMR. Eur J Biochem 255 185–195 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plucken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan A, Trebitsh T, Kiss V, Pereg Y, Dangoor I, Danon A (2005) Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc Natl Acad Sci USA 102 6225–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom J, Holmgren A (1993) Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry 32 6649–6655 [DOI] [PubMed] [Google Scholar]
- Lundstrom J, Krause G, Holmgren A (1992) A Pro to His mutation in active site of thioredoxin increases its disulfide-isomerase activity 10-fold. New refolding systems for reduced or randomly oxidized ribonuclease. J Biol Chem 267 9047–9052 [PubMed] [Google Scholar]
- Martin JL (1995) Thioredoxin: a fold for all reasons. Structure 3 245–250 [DOI] [PubMed] [Google Scholar]
- Menchise V, Corbier C, Didierjean C, Saviano M, Benedetti E, Jacquot JP, Aubry A (2001) Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism. Biochem J 359 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Siala W, Bashandy T, Riondet C, Vignols F, Reichheld JP (2008) Glutaredoxins and thioredoxins in plants. Biochim Biophys Acta 1783 589–600 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Verdoucq L, Vignols F (1999) Plant thioredoxins and glutaredoxins: identity and putative roles. Trends Plant Sci 4 388–394 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S (2004) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner E, Huber-Wunderlich M, Glockshuber R (1998) Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxidoreductases. Protein Sci 7 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T (2006) HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J Biol Chem 281 35039–35047 [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Bardwell JC (2004) Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta 1694 111–119 [DOI] [PubMed] [Google Scholar]
- Ortenberg R, Beckwith J (2003) Functions of thiol-disulfide oxidoreductases in E. coli: redox myths, realities, and practicalities. Antioxid Redox Signal 5 403–411 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Perez-Ruiz JM, Spinola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat A, Cho SH, Beckwith J (2004) The unusual transmembrane electron transporter DsbD and its homologues: a bacterial family of disulfide reductases. Res Microbiol 155 617–622 [DOI] [PubMed] [Google Scholar]
- Quan S, Schneider I, Pan J, Von Hacht A, Bardwell JC (2007) The CXXC motif is more than a redox rheostat. J Biol Chem 282 28823–28833 [DOI] [PubMed] [Google Scholar]
- Schurmann P, Buchanan BB (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10 1235–1274 [DOI] [PubMed] [Google Scholar]
- Sheen J (2002) A transient expression assay using Arabidopsis mesophyll protoplasts. Sheen Laboratory. http://genetics.mgh.harvard.edu/sheenweb (January 1, 2003)
- Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18 333–338 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano MB, Delaunay A, Monceau L, Tacnet F (2004) Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem Sci 29 351–357 [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Levitan A, Sofer A, Danon A (2000) Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol Cell Biol 20 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg G, Danon A (2008) Disulfide bond formation in chloroplasts: formation of disulfide bonds in signaling chloroplast proteins. Plant Sci 175 459–466 [Google Scholar]
- Zapun A, Bardwell JC, Creighton TE (1993) The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32 5083–5092 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.