Abstract
Chloroplast NADPH-thioredoxin reductase (NTRC) belongs to the thioredoxin systems that control crucial metabolic and regulatory pathways in plants. Here, by characterization of T-DNA insertion lines of NTRC gene, we uncover a novel connection between chloroplast thiol redox regulation and the control of photoperiodic growth in Arabidopsis (Arabidopsis thaliana). Transcript and metabolite profiling revealed severe developmental and metabolic defects in ntrc plants grown under a short 8-h light period. Besides reduced chlorophyll and anthocyanin contents, ntrc plants showed alterations in the levels of amino acids and auxin. Furthermore, a low carbon assimilation rate of ntrc leaves was associated with enhanced transpiration and photorespiration. All of these characteristics of ntrc were less severe when plants were grown under a long 16-h photoperiod. Transcript profiling revealed that the mutant phenotypes of ntrc were accompanied by differential expression of genes involved in stomatal development, chlorophyll biosynthesis, chloroplast biogenesis, and circadian clock-linked light perception systems in ntrc plants. We propose that NTRC regulates several key processes, including chlorophyll biosynthesis and the shikimate pathway, in chloroplasts. In the absence of NTRC, imbalanced metabolic activities presumably modulate the chloroplast retrograde signals, leading to altered expression of nuclear genes and, ultimately, to the formation of the pleiotrophic phenotypes in ntrc mutant plants.
Thiol redox regulation is a universal mechanism to control biochemical processes in living cells. Among thiol redox regulators, the thioredoxin superfamily consists of regulatory proteins that mediate dithiol-disulfide exchange of Cys residues, thereby modulating the activity of their target proteins. Members of the thioredoxin superfamily include thioredoxins, glutaredoxins, protein disulfide isomerases, as well as a group of thioredoxin-like proteins with mostly unknown function (Buchanan and Balmer, 2005; Gelhaye et al., 2005; Holmgren et al., 2005; Meyer et al., 2005, 2006). Members of the thioredoxin superfamily are characterized by the presence of a conserved redox-active site, where two Cys residues are separated from each other by two amino acids (Lemaire, 2004; Meyer et al., 2005). Thioredoxins with the redox-active site WC(G/P)PC constitute the best-characterized subclass of the thioredoxin superfamily and are found in all cellular compartments (Meyer et al., 2005). Numerous studies have revealed crucial functions for thioredoxins in the regulation of developmental and acclimation processes in plants (for review, see Buchanan and Balmer, 2005), and the vital roles for thioredoxin superfamily members in oxidative stress responses have also become evident (Hirt et al., 2002; Perez-Ruiz et al., 2006; Vieira Dos Santos and Rey, 2006).
Oxidized thioredoxins become rereduced via the action of thioredoxin reductases that, together with the target thioredoxins, constitute the cellular thioredoxin systems. Plants differ from other organisms in having a wide spectrum of possible thiol redox mediators. Recent analyses of Arabidopsis (Arabidopsis thaliana) gene sequence databases revealed more than 100 genes encoding members of the thioredoxin superfamily (Buchanan and Balmer, 2005; Houston et al., 2005; Meyer et al., 2005). A particularly divergent composition of thioredoxin systems is found in plastids, in which four types of thioredoxins (m, f, x, and y), several thioredoxin-like proteins, and two distinct types of thioredoxin reductases have been identified (Meyer et al., 2005; Perez-Ruiz et al., 2006). The classical ferredoxin-dependent thioredoxin reductase (FTR) is an iron-sulfur protein unique to chloroplasts and cyanobacteria (Dai et al., 2004). The second type, NADPH-dependent thioredoxin reductase (NTR), belongs to the flavoprotein family of pyridine nucleotide disulfide oxidoreductases present in all living cells (Hirt et al., 2002). In Arabidopsis, two genes called NTRA and NTRB encode NTR isoforms that are dually localized in cytosol and mitochondria (Reichheld et al., 2007). The Arabidopsis NTRC gene encodes a chloroplast-localized NTR that comprises a unique isoform found in oxygenic photosynthetic organisms and in Mycobacterium leprae (Hirt et al., 2002; Serrato et al., 2004; Perez-Ruiz et al., 2006). Besides the conserved NTR domain, NTRC possesses an additional thioredoxin domain at the C terminus of the protein (Serrato et al., 2004).
The light-activated ferredoxin/thioredoxin system has been linked to the regulation of primary photosynthetic processes during diurnal dark/light transitions of plants (Buchanan, 1991). In contrast, the action of the NADPH-dependent thioredoxin system in plastids has remained less well characterized. Besides photosynthesis, chloroplasts perform a number of other metabolic processes, including biosynthesis of amino acids, fatty acids, hormones, and secondary compounds. Indeed, proteomic approaches have revealed numerous chloroplastic enzymes that are potential targets for regulation by yet unidentified thioredoxins (Motohashi et al., 2001; Balmer et al., 2003, 2006; Marchand et al., 2004).
In chloroplasts, NADPH can be produced via photosynthetic electron transfer reactions in light and via NADP dehydrogenase activity in darkness. These different pathways provide alternative interactions for NADPH-dependent thioredoxin systems and metabolic pathways in chloroplasts. Thus, it is intriguing that Arabidopsis ntrc knockout mutants showed distinctly reduced growth, and a shift of plants to continuous darkness further pronounced the mutant phenotype (Perez-Ruiz et al., 2006). Furthermore, in vitro assays showed that NTRC was capable of utilizing NADPH to mediate the reduction of oxidized chloroplast 2-Cys peroxiredoxin (Moon et al., 2006; Perez-Ruiz et al., 2006; Alkhalfioui et al., 2007). Peroxiredoxins are small hydrogen peroxide-scavenging enzymes coupled to the thioredoxin or glutaredoxin system (Rouhier et al., 2001). These findings led to the conclusion that NTRC is required for protection against oxidative stress during the dark period (Spinola et al., 2008).
In this paper, we report that NTRC has a critical role in the regulation of photoperiod-dependent metabolic and developmental processes in Arabidopsis. Transcript and metabolite profiling of plants grown under short-day (8 h of light/16 h of dark) and long-day (16 h of light/8 h of dark) photoperiods revealed that ntrc plants display severe photoperiod- and age-dependent developmental disorders. The absence of NTRC induced alterations in chloroplast biogenesis, which in turn interfered with photoperiodic development in Arabidopsis. Thus, apart from being a source of energy, functional chloroplasts also contribute to the regulation of plant development in response to changing environmental cues.
RESULTS
Identification of the ntrc Knockout Mutants and Accumulation of NTRC in Different Plant Organs
Homozygous ntrc plants were identified from the SALK institute's T-DNA insertion mutant collection (SALK_096776 and SALK_114293). To confirm the absence of the NTRC enzyme in ntrc plants, a polyclonal antibody was raised against amino acids 475 to 488 of Arabidopsis NTRC. In total leaf extracts isolated from wild-type plants, the antibody specifically recognized NTRC, which was detected as a polypeptide with an apparent molecular mass of 60 kD on SDS-PAGE (Fig. 1B). NTRC was also present in the cotyledons and in the stem of the inflorescence but not in the roots of wild-type plants (Fig. 1B). Clearly, ntrc knockout plants showed no accumulation of NTRC (Fig. 1B).
Figure 1.
Identification of ntrc knockout mutants. A, T-DNA insertion site of the NTRC (At2g41680, SALK_096776) gene. The insertion site is indicated with a triangle, and black and gray bars represent exons and introns, respectively. B, Detection of NTRC in plant organs. Total plant extracts were separated with SDS-PAGE, and NTRC was immunodetected with an Arabidopsis anti-NTRC antibody. The left panel demonstrates the linear range of the antibody when 5 to15 μg of protein of the leaf extract was loaded in the wells. The right panel demonstrates the presence of NTRC in the cotyledons and in the stem of inflorescence but not in the roots of Col-0.
The ntrc Mutants Show Photoperiod-Dependent Phenotypes
Growth of plants under various light rhythms with daily 8-h, 16-h, or 24-h illumination periods revealed distinct photoperiod-dependent alterations in the growth rate, pigmentation, and flowering time of ntrc plants as compared with wild-type plants (Fig. 2; Table I; Supplemental Fig. S1; Supplemental Table S1). The most profound mutant phenotype was observed when ntrc plants were grown under a short 8-h photoperiod. Cotyledons of ntrc plants, however, were visually indistinguishable from those of the wild-type plants, and the clear mutant phenotype became evident upon the emergence of the first true leaves. During the first month of growth, ntrc plants formed small rosettes with pale green leaves (Fig. 2). In 4-week-old ntrc rosette leaves, the total concentration of chlorophyll per leaf area was 50% lower than in the wild-type plants (Table I). Upon aging, however, the pale green ntrc leaves started to green (Fig. 2; Table I) and the ntrc rosette gained the size typically observed for mature wild-type plants (Fig. 2). Intriguingly, the greening of ntrc leaves occurred gradually via a transient phase of the reticulate phenotype, with dark green cells surrounding the vascular tissue (Fig. 2). Furthermore, the life cycle of ntrc plants was significantly extended with delayed flowering time (Table II).
Figure 2.
Phenotypes of ntrc plants grown under various photoperiods. A, Five-day-old Col-0 and ntrc seedlings grown in short-day conditions. B, Ten-day-old Col-0 and ntrc seedlings grown in short-day or long-day conditions. C, Four-week-old Col-0 and ntrc grown in short-day conditions. D, Rosette and leaf phenotypes of 10-week-old Col-0 and ntrc in short-day conditions. The leaves were excised from the same rosette to demonstrate the gradual greening of ntrc leaves during aging. The oldest leaves are shown on the left side. E, Three-week-old Col-0 and ntrc grown in long-day conditions. F, Twelve-day-old Col-0 and ntrc grown in continuous light. G, Col-0 and ntrc grown in short-day conditions at 10°C. Below the photographs (C–F), confocal microscopy images of chlorophyll autofluorescence from mesophyll cells are shown. The small cell size of short-day-grown ntrc (C) is indicated with circles. Bars = 100 μm.
Table I.
Specific leaf weight and pigment content in ntrc and Col-0 grown under different photoperiods
CL, Continuous light; na, not analyzed; nd, not detected; SD and LD, short-day and long-day conditions, respectively; SD old, short-day-grown 10-week-old plants. Data are means ± se of three to eight independent measurements. *** P < 0.005, ** P < 0.05 by Student's t test. For age of the plants, see Figure 2.
| Sample and Conditions | Specific Leaf Weight | Total Chlorophyll | Chlorophyll a/b | Anthocyanins |
|---|---|---|---|---|
| g (dry weight) m−2 | μg cm−2 | [(A530 − A657) × 103] cm−2 | ||
| Col-0 SD | 14.1 ± 0.4 | 10.8 ± 0.1 | 3.37 ± 0.02 | nd |
| ntrc SD | 11.0 ± 0.4*** | 4.8 ± 0.1*** | 3.07 ± 0.04*** | nd |
| Col-0 SD old | na | 40.0 ± 1.0 | 3.38 ± 0.03 | 71.3 ± 14.4 |
| ntrc SD old, pale leaves | na | 7.0 ± 1.2*** | 3.20 ± 0.29 | na |
| ntrc SD old, green leaves | na | 14.4 ± 0.5*** | 3.35 ± 0.08 | 8.0 ± 1.15*** |
| ntrc SD old, dark green leaves | na | 19.0 ± 0.2*** | 3.28 ± 0.02 | na |
| Col-0 LD | 16.1 ± 0.7 | 18.3 ± 1.4 | 3.83 ± 0.05 | 197 ± 37.0 |
| ntrc LD | 15.6 ± 0.1 | 10.4 ± 0.3** | 3.57 ± 0.07** | 13.0 ± 1.30*** |
| Col-0 CL | 18.1 ± 0.3 | 23.5 ± 1.2 | 3.90 ± 0.04 | na |
| ntrc CL | 15.2 ± 0.3*** | 17.5 ± 0.6*** | 3.80 ± 0.02** | na |
Table II.
Number of stomata and flowering time of ntrc and Col-0 grown under different photoperiods
CL, Continuous light; LD, long day; na, not analyzed; SD, short day. Data are means ± se; n = 5 in all experiments. *** P < 0.005 by Student's t test.
| Sample and Conditions | Stomata | Stomatal Index | Flowering Time | No. of Leaves upon Budding |
|---|---|---|---|---|
| mm−2 | d | |||
| Col-0 SD | 50 ± 4 | 19.3 ± 1.4 | 64 ± 2 | 50 ± 5 |
| ntrc SD | 82 ± 8*** | 22.1 ± 0.9 | 80 ± 2 *** | 42 ± 2 |
| Col-0 LD | 120 ± 13 | 27.5 ± 0.9 | 19 ± 1 | 11 ± 1 |
| ntrc LD | 148 ± 13 | 27.8 ± 1.0 | 20 ± 1 | 10 ± 1 |
| Col-0 CL | na | na | 15 ± 1 | 9 ± 1 |
| ntrc CL | na | na | 17 ± 1 | 10 ± 1 |
Under a long 16-h photoperiod or under continuous light, the ntrc mutant phenotype was less distinct and the biomass production by the ntrc line was partially restored (Fig. 2; Table I). Both the ntrc and wild-type plants responded to long photoperiods by increasing the total chlorophyll concentration per leaf area and the chlorophyll a/b ratio (Table I). Moreover, the transition to flowering took place approximately at the same time in ntrc and wild-type plants (Table II).
Imaging of chlorophyll autofluorescence by confocal microscopy revealed that the retarded growth and diminished chlorophyll content of ntrc plants were accompanied by structural changes in the mesophyll tissue. In plants grown under the short photoperiod, the mesophyll cells of the diminutive ntrc leaves were significantly smaller and contained fewer chloroplasts than wild-type leaves (Fig. 2). Notably, the greening of ntrc leaves during aging was accompanied by the accumulation of chloroplasts in the mesophyll cells, while the small cell size was still retained despite the expansion of the leaves (Fig. 2). In plants grown under the long photoperiod or under continuous light, both the mesophyll cell size and the number of chloroplasts per cell were partially restored (Fig. 2).
The accumulation of anthocyanins was dramatically reduced in ntrc leaves under both short-day and long-day conditions (Table I; Supplemental Table S1). When the plants were grown in the short photoperiod under low temperature of 10°C, the difference in pigmentation was even visually observable (Fig. 2). Wild-type plants responded to low temperature by enhancing the accumulation of anthocyanins, whereas in ntrc leaves, only modest anthocyanin accumulation became evident (Fig. 2). Morphologically, the cold-acclimated 3-month-old ntrc plants were quite similar to wild-type plants (Fig. 2).
Molecular and Functional Characterization of Photosynthesis in ntrc Plants
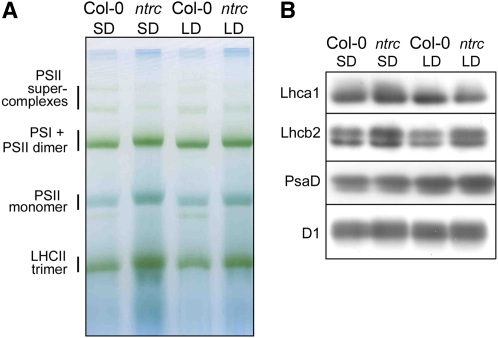
To gain insights into possible photoperiod-dependent adjustments in photosynthetic light reactions, thylakoid membrane protein complexes of ntrc and wild-type plants were studied by blue-native gel electrophoresis (Fig. 3). When equal amounts of chlorophyll were loaded in the wells, the pattern of thylakoid protein complexes in ntrc was comparable to that of the wild-type plants under both short-day and long-day conditions (Fig. 3A). In fact, the amount of trimeric PSII light-harvesting antenna (LHCII) complexes was even slightly higher in ntrc than in wild-type plants (Fig. 3A). Consistently, the ntrc plants showed a slightly increased level of the LHCII protein Lhcb2 (Fig. 3B) and a lower chlorophyll a/b ratio than the wild-type plants (Table I). No significant adjustments were observed in the levels of PsaD and D1 proteins, representatives of PSI and PSII core complexes, respectively.
Figure 3.
Photosynthetic thylakoid protein complexes in ntrc and Col-0 plants grown under short-day (SD) or long-day (LD) conditions. A, Chlorophyll-binding protein complexes in the thylakoid membranes of ntrc and Col-0 plants. Thylakoid membranes corresponding to 5 μg of chlorophyll were separated by blue-native PAGE, and the pigment protein complexes were identified according to Aro et al. (2005). B, Levels of representative PSI and PSII proteins in ntrc and Col-0 leaves grown under short-day or long-day conditions. Thylakoid membranes corresponding to 1 μg of chlorophyll were separated on SDS gels, blotted on a polyvinylidene difluoride membrane, and immunodetected with protein-specific antibodies. LHCII, Light-harvesting complexes of PSII; Lhca1 and Lhcb2, representatives of light-harvesting complexes of PSI and PSII, respectively; PsaD and D1, representatives of PSI and PSII core proteins, respectively.
In contrast to the basic structures of the light reactions that were not affected, ntrc plants showed a decrease in the rate of net CO2 assimilation under low and ambient CO2 concentrations. CO2 assimilation varied in individual ntrc rosettes (Supplemental Fig. S2). However, the CO2 compensation point was distinctly higher in all analyzed short-day ntrc plants than in the wild-type plants. The ntrc plants also suffered from enhanced photoinhibition of PSII, measured as a decrease in Fv/Fm (see “Materials and Methods”), when grown under the short photoperiod (Table III). Under the long photoperiod, the differences in the net CO2 assimilation, in the CO2 compensation point, and in PSII photoinhibition between ntrc and wild-type plants were less distinct (Table III; Supplemental Fig. S2B). Modeling of the response of net CO2 assimilation to increasing atmospheric CO2 concentrations revealed only slight differences in photosynthetic parameters between ntrc and wild-type plants, regardless of the length of the photoperiod (Table III). Indeed, the calculations indicated that maximal electron transport rate (Jmax) and maximal carboxylation rate of Rubisco (Vcmax) were not significantly impaired in ntrc leaves.
Table III.
Photosynthetic parameters of ntrc and Col-0 grown under different photoperiods
Maximal carboxylation rate of Rubisco (Vcmax), maximal electron transport rate (Jmax), and the rate of mitochondrial respiration in light (Rd) were obtained by modeling the CO2 assimilation responses presented in Supplemental Figure S2 according to Farquhar et al. (1980). Fv/Fm represents the photochemical efficiency of PSII. Activation state of MDH was calculated as a proportion of initial activity to maximal activity. LD, Long day; SD, short day. Data are means ± se; n = 3 to 4 in all experiments. *** P < 0.005, ** P < 0.05 by Student's t test.
| Sample and Conditions | Fv/Fm | Vcmax | Jmax | Rd | Activation State of MDH |
|---|---|---|---|---|---|
| μmol CO2 m−2 s−1 | μmol m−2 s−1 | μmol m−2 s−1 | % | ||
| Col-0 SD | 0.80 ± 0.004 | 21.1 ± 1.8 | 44.1 ± 4.0 | 2.0 ± 0.4 | 51 ± 4 |
| ntrc SD | 0.66 ± 0.016*** | 27.0 ± 3.2 | 44.5 ± 5.1 | 4.1 ± 0.6** | 41 ± 6 |
| Col-0 LD | 0.82 ± 0.002 | 19.6 ± 0.8 | 44.0 ± 0.4 | 0.9 ± 0.2 | 31 ± 7 |
| ntrc LD | 0.80 ± 0.008** | 22.5 ± 1.4 | 37.9 ± 1.9 | 1.8 ± 0.2** | 24 ± 7 |
Next, we studied whether the absence of NTRC had influenced the activities of two thioredoxin-regulated enzymes of chloroplasts, the chloroplast fructose-1,6-bisphosphatase (FBPase) and the malate dehydrogenase (MDH; Buchanan and Balmer, 2005). Chloroplast FBPase is a Calvin cycle enzyme, while MDH controls the export of reducing equivalents through the chloroplast envelope via the malate shuttle. The ntrc and wild-type plants showed no differences in the estimated activation state of FBPase (data not shown), and also the activation state of MDH was only slightly lower in ntrc as compared with wild-type plants (Table III). Thus, both the unchanged FBPase activity and the unaltered values of Jmax and Vcmax (Table III) indicated that the low net CO2 assimilation rate of ntrc leaves under the short photoperiod was not due to an imbalance in Calvin cycle reactions. Instead, the low net assimilation of CO2 in short-day ntrc could be explained by the low number of chloroplasts in leaves (Fig. 2) and, importantly, by enhanced respiration (Rd), which was distinctly higher in ntrc than in wild-type plants (Table III).
Finally, we explored whether the retarded growth of ntrc plants under short-day conditions was associated with imbalances in diurnal starch metabolism. The accumulation or degradation of starch in ntrc leaves did not differ to a large extent from the cycling of starch in wild-type leaves (Supplemental Fig. S3). Indeed, iodine staining of intact leaves indicated only slightly lower accumulation of starch in ntrc plants grown under the short photoperiod, probably due to the lower number of chloroplasts in mesophyll cells (Fig. 2; Supplemental Fig. S3). Moreover, both the ntrc and wild-type plants were capable of degrading starch during the following dark period.
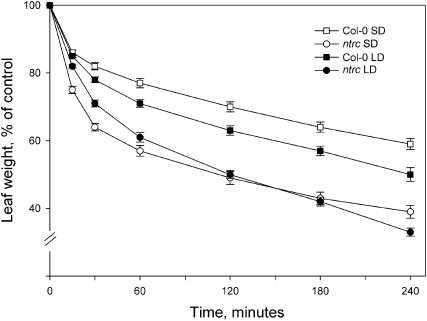
Gas exchange and transpiration through stomata are also crucial determinants of photosynthetic performance. Notably, wild-type plants showed distinct photoperiod-dependent adjustments in stomatal density, with a clear increase in stomatal index upon growth under the long photoperiod (Table II). Moreover, the stomatal density was higher in ntrc than in wild-type plants, especially in short-day conditions (Table II). Accordingly, ntrc plants lost significantly more water from excised rosettes than the wild-type plants during the light period (Fig. 4). At the end of the diurnal dark period, before the onset of illumination, water loss from excised rosettes of both ntrc and wild-type plants was significantly reduced, indicating that both wild-type and ntrc plants were capable of closing the stomata during the dark period (data not shown).
Figure 4.
Water loss from Col-0 and ntrc rosettes grown under short-day (SD) or long-day (LD) conditions. The loss of water was followed in light by weighing excised rosettes at regular intervals and is expressed as a percentage of the value at time point 0. Data are means ± se of three independent experiments.
Transcript Profiling of ntrc Plants under Different Physiological and Developmental Stages
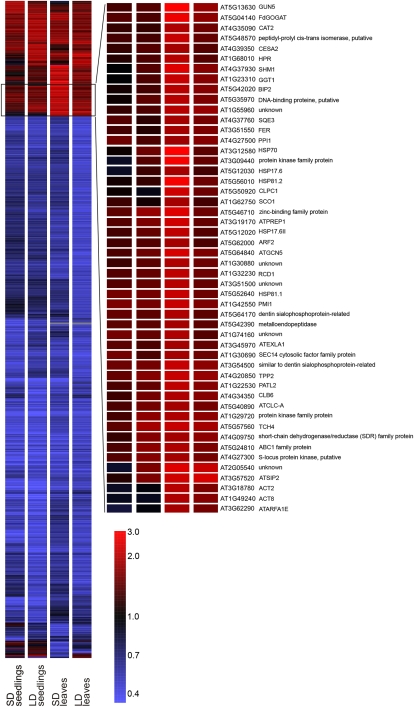
To identify the metabolic processes that contribute to the age- and photoperiod-dependent ntrc phenotype, we carried out comparative transcript profiling of ntrc and wild-type plants. Ten-day-old seedlings and expanded rosettes of plants grown under short-day or long-day conditions were used for microarray analysis. A total of 621 genes with higher (>1.8-fold and P < 0.05) or lower (<0.55-fold and P < 0.05) transcript levels in ntrc mutants relative to wild-type plants were chosen for further analysis. This revealed ntrc-specific, age-specific, and photoperiod-dependent alterations in gene expression of ntrc plants (Fig. 5; Table IV; Supplemental Fig. S4; Supplemental Table S2). Under long-day conditions, the transcriptomic adjustments of ntrc plants were much more pronounced than under short-day conditions. However, in short-day ntrc rosette leaves, 68% of the differentially expressed genes were up-regulated, whereas in long-day ntrc rosette leaves, the proportion of up-regulated genes was only 4%. Furthermore, the transcript profile of short-day ntrc rosette leaves comprised a unique combination of induced genes, which differed from the transcript profiles of both the young short-day seedlings and all of the long-day-grown ntrc plants (Fig. 5; Supplemental Fig. S4).
Figure 5.
Differentially expressed genes in ntrc relative to Col-0 in 10-d-old seedlings and rosette leaves grown under short-day (SD) or long-day (LD) conditions. Genes up-regulated more than 1.8-fold or down-regulated more than 0.55-fold with P < 0.05 in at least one of the conditions were clustered using an average linkage clustering algorithm with standard correlation as a similarity measure. The 50 up-regulated genes indicated constitute a unique transcript profile of ntrc rosette leaves in short-day conditions.
Table IV.
Mutant phenotype-associated genes differentially expressed in ntrc relative to Col-0
Fold change values for genes of particular interest are shown in boldface. Values are means of three independent biological replicates. For se values, P values, and a complete list of differentially expressed genes, see Supplemental Table S2. AGI, Arabidopsis Genome Initiative; GO, gene ontology; LD, long day; SD, short day.
| Gene | GO Cellular Component | AGI Code | SD, Seedlings | SD, Leaves | LD, Seedlings | LD, Leaves |
|---|---|---|---|---|---|---|
| fold change | ||||||
| Response to light | ||||||
| CRY2; Cryptochrome 2 | AT1G04400 | 0.36 | 0.46 | 0.39 | 0.47 | |
| FRS3; Far1-related sequence 3 | AT2G27110 | 0.40 | 0.44 | 0.36 | 0.42 | |
| Chloroplast biogenesis + chlorophyll biosynthesis + photosynthesis | ||||||
| FZL; Thylakoid membrane organization-like | Chloroplast | AT1G03160 | 0.30 | 0.38 | 0.33 | 0.46 |
| CLB6; Chloroplast biogenesis 6 | Chloroplast | AT4G34350 | 1.11 | 1.92 | 1.41 | 1.55 |
| GUN5; Genomes uncoupled 5 | Chloroplast | AT5G13630 | 1.15 | 3.32 | 1.24 | 1.72 |
| HEMA1; Glutamyl-tRNA reductase | Chloroplast | AT1G58290 | 0.99 | 2.07 | 1.41 | 1.43 |
| LHCB1B1; PSII light-harvesting complex gene 1.4 | Chloroplast | AT2G34430 | 0.44 | 0.47 | 0.46 | 0.35 |
| LHCB1B2; PSII light-harvesting complex gene 1.5 | Chloroplast | AT2G34420 | 0.44 | 0.47 | 0.51 | 0.35 |
| LHCB2.2; PSII light-harvesting complex gene 2.2 | Chloroplast | AT2G05070 | 0.73 | 0.61 | 0.60 | 0.52 |
| LHCB3; Light-harvesting chlorophyll binding 3 | Chloroplast | AT5G54270 | 0.78 | 0.53 | 1.06 | 0.62 |
| Rubisco small subunit 1A | Chloroplast | AT1G67090 | 0.68 | 0.50 | 0.48 | 0.40 |
| Photorespiration | ||||||
| CAT2; Catalase 2 | Peroxisome | AT4G35090 | 1.09 | 1.90 | 1.09 | 1.35 |
| GGT1; Ala-2-oxoglutarate aminotransferase 1 | Peroxisome | AT1G23310 | 0.98 | 2.20 | 1.18 | 1.57 |
| HPR; Hydroxypyruvate reductase | Peroxisome | AT1G68010 | 1.05 | 2.04 | 1.17 | 1.24 |
| SHM1; Ser hydroxymethyltransferase 1 | Mitochondrion | AT4G37930 | 0.99 | 2.57 | 1.22 | 1.58 |
| CDCP1; Gly decarboxylase P-protein 1 | Mitochondrion | AT4G33010 | 1.00 | 2.21 | 1.08 | 1.31 |
| FdGOGAT; Fd-dependent Glu synthase 1 | Chloroplast | AT5G04140 | 1.21 | 2.81 | 1.16 | 1.63 |
| Proteases | ||||||
| ATPREP1/ATZNMP; Zinc metalloprotease pitrilysin subfamily A | Chloroplast/mitochondrion | AT3G19170 | 1.22 | 2.36 | 1.46 | 1.31 |
| CLPC1; Clp protease, heat shock protein 93-V | Chloroplast | AT5G50920 | 1.00 | 2.31 | 0.95 | 1.38 |
| TPP2; Tripeptidyl peptidase II | Chloroplast | AT4G20850 | 1.32 | 2.17 | 1.52 | 1.70 |
| Metalloendopeptidase | Chloroplast | AT5G42390 | 1.27 | 1.82 | 1.26 | 1.27 |
| Stomatal development | ||||||
| SDD1; Stomatal density and distribution | Secretion | AT1G04110 | 0.47 | 0.62 | 0.40 | 0.53 |
| EPF1; Epidermal patterning factor 1 | Secretion | AT2G20875 | 0.50 | 0.49 | 0.37 | 0.47 |
| Heat shock proteins + stress responses | ||||||
| HSP70; Heat shock protein 70 | AT3G12580 | 1.01 | 3.24 | 1.37 | 1.26 | |
| HSP81-3; Heat shock protein 81-3 | Secretion | AT5G56010 | 1.01 | 2.85 | 1.08 | 1.37 |
| HSP70-3, Heat shock cognate 70-kD protein 3 | AT3G09440 | 0.90 | 2.83 | 1.21 | 1.30 | |
| HSP81-1; Heat shock protein 81-1 | AT5G52640 | 1.41 | 2.20 | 1.44 | 1.39 | |
| HSP17.6-CII; 17.6-kD class II heat shock protein | AT5G12020 | 1.14 | 2.09 | 1.30 | 1.20 | |
| AT-HSP17.6A; Heat shock protein 17.6A | AT5G12030 | 0.91 | 1.86 | 1.12 | 1.14 | |
| BIP; Luminal binding protein | Secretion | AT5G42020 | 1.01 | 2.02 | 1.16 | 1.52 |
| GPX7; Glutathione peroxidase | Chloroplast | AT4G31870 | 1.39 | 1.92 | 1.76 | 1.85 |
| RCD1; Radical-induced cell death 1 | AT1G32230 | 1.23 | 2.11 | 1.31 | 1.38 | |
Gene Expression Changes Associated with the Morphogenic Phenotypes of ntrc Plants
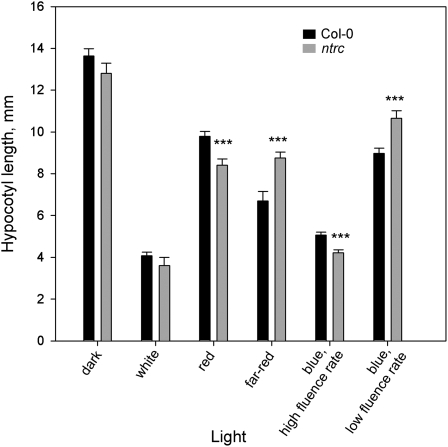
Two genes belonging to families that regulate photoperiodic growth in plants, CRYPTOCHROME2 (CRY2) and the FAR-RED-IMPAIRED RESPONSE REGULATOR (FRS3), were down-regulated in ntrc plants (Table IV). CRY2 is a blue-light receptor that mediates the inhibition of hypocotyl growth and the timing of flowering in Arabidopsis (for review, see Li and Yang, 2007). FRS3 belongs to the FHY3/FAR1 (far-red elongated/far-red impaired response) protein family that mediates phytochrome A-controlled far-red light responses (Lin and Wang, 2004). Loss-of-function mutants for members of this family displayed defects in the inhibition of hypocotyl elongation, accumulation of anthocyanins, and photoperiod-dependent timing of flowering (Hsieh et al., 2000; Lin and Wang, 2004), the symptoms observed also in ntrc plants. Thus, to reveal potential dysfunction of photoreceptors in ntrc plants, we carried out classical deetiolation experiments. Wild-type and ntrc seedlings were germinated in darkness and under white, blue, red, and far-red light. Under high fluence rates of blue or red light, ntrc hypocotyls were significantly shorter than in wild-type plants, whereas no differences were detected in seedlings germinated in white light or in darkness (Fig. 6). However, the hypocotyls of ntrc seedlings germinated under low fluence rates of blue light and under far-red light were significantly longer than in wild-type plants (Fig. 6). As CRY2 and PHYA act as photoreceptors for low fluence rates of blue light and far-red light in deetiolation, respectively (Fankhauser and Casal, 2004), these results verify the mircoarray data on defects in light signaling in ntrc plants.
Figure 6.
Hypocotyl length of 7-d-old ntrc and Col-0 grown under different light qualities in short-day conditions. n = 20 in all experiments. *** P < 0.005 by Student's t test.
The ntrc plants also showed lowered accumulation of transcripts for genes assigned to the development of structural components in leaves (Table IV). Expression of DYNAMIN-RELATED MEMBRANE REMODELING-LIKE (FZL), STOMATAL DENSITY AND DISTRIBUTION1-1 (SDD1-1), and EPIDERMAL PATTERING FACTOR1 (EPF1) was repressed in ntrc plants under all conditions tested. Of these genes, FZL encodes a protein that controls the organization of grana and stroma thylakoids in chloroplasts (Gao et al., 2006). SDD1-1 and EPF1 encode putative plasma membrane proteins that control the early signaling steps in stomatal development (Casson and Gray, 2008). Accordingly, sdd1-1 mutants showed increased stomatal density, which was also the case in ntrc plants (Table II; Von Groll et al., 2002).
Notably, despite the stunted phenotype of ntrc plants, the mutant phenotype was not accompanied by any significant induction of marker genes related to hydrogen peroxide-dependent (Vanderauwera et al., 2005) or singlet oxygen-dependent (Gadjev et al., 2006) stress responses. Neither did the expression of genes for thioredoxin superfamily members (thioredoxins, thioredoxin-like proteins, glutaredoxins, and protein disulfide isomerases) show any significant changes in ntrc plants (Supplemental Table S2).
Short-Day ntrc Rosette Leaves Constitute a Unique Transcript Profile
Short-day ntrc rosette leaves with the most severe mutant phenotype also showed a distinct transcript profile, reflecting the unique metabolic state of the ntrc leaves under these conditions (Fig. 2; Supplemental Fig. S4). Related to the pale green leaves and the low net CO2 assimilation of short-day-grown ntrc leaves, two distinct metabolic processes came up from the transcript profiling: chlorophyll biosynthesis and photorespiration (Table IV). Two genes encoding enzymes of the chlorophyll biosynthesis pathway, glutamyl-tRNA reductase (HEMA1) and the H subunit of the Mg-chelatase (GUN5), were among the most up-regulated genes in short-day-grown ntrc leaves (Table IV). Slightly increased accumulation of the transcript for GUN5 was also observed in long-day-grown ntrc rosette leaves. The increased accumulation of GUN5 transcripts in ntrc leaves was verified by northern blotting (Supplemental Fig. S5). Furthermore, transcripts for CHLOROPLAST BIOGENESIS6 (CLB6) accumulated in short-day-grown ntrc rosette leaves. CLB6 is an enzyme of plastid isoprene biosynthesis, which provides phytol chains for chlorophyll molecules.
Consistent with the high CO2 compensation point of ntrc leaves (Supplemental Fig. S2), transcript levels of six photorespiratory genes were induced in short-day-grown ntrc rosette leaves: peroxisomal CATALASE2, ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE, and NAD+-HYDROXYPYRUVATE REDUCTASE, mitochondrial GLYCINE DECARBOXYLASE P SUBUNIT and SERINE HYDROXYMETHYL TRANSFERASE, and plastidial FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE (Table IV). Thus, enhanced photorespiration may account for draining of excess light energy in the slowly growing ntrc leaves, thereby hindering more severe damage to photosynthetic structures (Kozaki and Takeba, 1996).
It is worth noting that CATALASE2 and GLUTATHIONE PEROXIDASE7 were the only hydrogen peroxide metabolism-linked genes that were differentially expressed in short-day-grown ntrc plants (Table IV). However, eight genes related to various stress responses were markedly induced in short-day ntrc leaves. Four of them belong to heat shock proteins (HSP70, HSP81-1, HSP17.6-CII, and HSP17.6A) that were also induced by high-light treatment of catalase-deficient plants (Vanderauwera et al., 2005). Up-regulation of RADICAL-INDUCED CELL DEATH1, which is associated with hormonal signaling and stress responses (Ahlfors et al., 2004), and three ATP-dependent chaperones (HSP70-3, HSP81-3, and BIP) also referred to specific stress responses in short-day ntrc leaves (Table IV). Finally, transcript levels for four chloroplast-located proteases, including a Clp protease (CLPC1), were increased in short-day ntrc rosette leaves, indicating stress responses and/or modified activity of proteolysis in ntrc chloroplasts.
Auxin and Abscisic Acid Contents in ntrc Plants
Chloroplast-derived metabolites serve as precursors for biosynthesis of the growth hormone auxin (indole-3-acetic acid [IAA]) and the growth inhibitor abscisic acid (ABA). The auxin content is highest in the first true leaves of Arabidopsis seedlings after 8 to 10 d of germination (Ljung et al., 2001). Therefore, we measured the amounts of auxin and ABA in 5- and 10-d-old ntrc seedlings, which began to display the visible mutant phenotype. Consistent with the earlier measurements (Ljung et al., 2001), auxin content in young seedlings of wild-type Arabidopsis was independent of photoperiod (Fig. 7). Notably, ntrc plants grown under the short photoperiod contained significantly less auxin than wild-type plants, whereas under the long photoperiod, differences in auxin levels were less distinct (Fig. 7). Thus, the small size of ntrc leaves may be related to the restriction of auxin-dependent cell elongation under short-day conditions. No differences were observed in the levels of ABA between the wild-type and ntrc plants (data not shown).
Figure 7.
IAA content in Col-0 and ntrc seedlings. IAA content was determined from 5- and 10-d-old seedlings grown in short-day (SD) or long-day (LD) conditions. Data are means ± se of three independent experiments. ** P < 0.05 by Student's t test. FW, Fresh weight.
Photoperiod-Dependent Accumulation of Amino Acids in Young ntrc and Wild-Type Plants
The clear deficiency in the accumulation of auxin and anthocyanins in short-day ntrc plants (Table I; Fig. 7) prompted us to address possible photoperiod-dependent alterations in the accumulation of amino acids in the wild-type and ntrc plants. Aromatic amino acids, which serve as precursors for the biosynthesis of auxin and flavonoids, are synthesized in chloroplasts via the shikimic acid pathway (Herrmann, 1995; Woodward and Bartel, 2005). The length of the daily photoperiod distinctly modulated the amino acid composition of the wild-type Arabidopsis plants (Fig. 8; Supplemental Table S3). The levels of Gly, Ala, and Thr were significantly lower in plants grown in the long-day photoperiod, and this trend was similar in wild-type and ntrc plants. Interestingly, however, ntrc plants accumulated more aromatic amino acids (Trp, Phe, and Tyr) than the wild-type plants, and the differences were more pronounced in plants grown under the short-day photoperiod. Furthermore, short-day-grown ntrc plants had higher amounts of Arg, Asn, and His than wild-type plants, whereas growth under the long photoperiod diminished the differences. Finally, ntrc plants accumulated less Leu and Ile than wild-type plants under short-day conditions. The opposite was observed under long-day conditions, where the levels of Leu and Ile were higher in ntrc as compared with wild-type plants.
Figure 8.
Free amino acid concentrations (nmol g−1 fresh weight) of Col-0 and ntrc seedlings grown under short-day (SD) and long-day (LD) conditions.
Seedling Development on a Medium Supplemented with External Auxin and Amino Acids
As demonstrated above, the absence of NTRC resulted in photoperiod-dependent imbalances in both amino acid and auxin metabolism in the developing ntrc seedlings. Thus, we tested whether the small size and the pale green phenotype of ntrc observed under short-day conditions could be rescued by the addition of external auxin or amino acids on the growth medium.
When grown on Murashige and Skoog agar plates, the ntrc phenotype was somewhat less distinct than in ntrc grown on soil (Supplemental Fig. S6). Addition of external auxin did not significantly enhance the growth of ntrc seedlings (Supplemental Fig. S6A). An enlargement of mesophyll cell size and an increase in the number of chloroplasts per cell, however, were observed when the growth medium was supplemented with aromatic amino acids (Supplemental Fig. S6B). The most distinct recovery of cell size and a particularly enhanced accumulation of chloroplasts were observed when ntrc seedlings were supplemented with Ile (Supplemental Fig. S6B). Notably, the level of this chloroplast-derived amino acid was reduced in ntrc seedlings (Fig. 8). A similar but slightly less pronounced effect was observed upon the addition of Phe (Supplemental Fig. S6B). The enlargement of ntrc cell size was also obtained by the addition of Trp in the growth medium (Supplemental Fig. S6B), but its impact on the recovery of cell size was not systematic on every plate. Moreover, it is worth emphasizing that none of the externally added amino acids fully restored the greening process of ntrc leaves. The addition of Ala had no effect on the growth and development of ntrc plants (Supplemental Fig. S6B). The external supply of auxin or amino acids did not significantly modulate the growth and development of wild-type seedlings.
DISCUSSION
Plants possess two main types of thioredoxin reductases, the universal NTRs and the plastidial FTR, which is unique for photosynthetic organisms (Hirt et al., 2002; Dai et al., 2004). The family of NTRs includes NTRA and NTRB, dually localized in cytosol and mitochondria, and chloroplastic NTRC (Hirt et al., 2002). Functional analysis of the Arabidopsis single mutants ntra and ntrb revealed no phenotypic deficiencies under standard growth conditions (Reichheld et al., 2007). Surprisingly, the ntra ntrb double knockout mutant also was viable, even though the absence of the two important regulatory enzymes, NTRA and NTRB, led to a notable reduction in growth rate (Reichheld et al., 2007). In this paper, we demonstrate that single mutants deficient in NTRC display severe photoperiod- and age-dependent developmental disorders, indicating that the two types of plastidial thioredoxin reductases, FTR and NTRC, are functionally nonredundant. Moreover, the pleiotropic phenotype of ntrc plants suggests that NTRC contributes to multiple important metabolic processes in chloroplasts, such as the biosynthesis of chlorophyll and amino acids. NTRC is present solely in green tissues (Fig. 1; Perez-Ruiz et al., 2006), emphasizing its impact on light-dependent functions of plastids. Loss of NTRC, however, does not interfere with the primary photosynthetic reactions (Fig. 3; Table III), implying that the FTR-dependent ferredoxin/thioredoxin system acts as the primary regulator of photosynthetic electron transfer reactions and the Calvin cycle in chloroplasts (Buchanan and Balmer, 2005).
NTRC Is Required for Chlorophyll Biosynthesis and for the Proper Biogenesis of Chloroplasts
The ntrc lines showed substantial reduction of chlorophyll content and of the number of chloroplasts per cell, particularly under short-day conditions. In line with the pale green phenotype, microarray analysis revealed a specific up-regulation of genes related to chlorophyll biosynthesis in ntrc leaves: the genes encoding glutamyl-tRNA reductase (HEMA1), the H subunit of Mg-chelatase (GUN5), an enzyme of plastid isoprene biosynthesis (CBG6), and chloroplast clip protease (CLPC1; Table IV). Glutamyl-tRNA reductase and Mg-chelatase catalyze the important regulatory steps of tetrapyrrole biosynthesis (Tanaka and Tanaka, 2007), whereas CBG6 and ClpC1 act farther downstream in chlorophyll biosynthesis. ClpC1 regulates the biosynthesis of chlorophyll b through the destabilization of chlorophyll a oxidase (CAO), the enzyme that catalyzes the synthesis of chlorophyll b from chlorophyll a (Nakagawara et al., 2007). Finally, a mutation in the CBG6 gene resulted in an albino phenotype (de la Luz Gutierrez-Nava et al., 2004), emphasizing the key function of the enzyme in chloroplast biogenesis.
The activity of Mg-chelatase is regulated via thioredoxin-mediated disulfide/dithiol exchange in the CHL-I subunit (Ikegami et al., 2007); therefore, the enzyme may represent a potential target for thiol-redox regulation by NTRC. Furthermore, in vitro experiments showed that in the presence of chloroplast 2-Cys peroxiredoxin, NTRC can stimulate the activity of Mg-protoporphyrin IX-monomethyl ester cyclase (MgCy), the enzyme catalyzing the reaction of chlorophyll biosynthesis downstream from Mg-chelatase (Stenbaek et al., 2008). Feeding of 5-aminolevulinic acid in darkness also resulted in increased accumulation of protoporphyrin IX (PPIX), Mg-PPIX, and Mg-PPIX-monomethylester in ntrc plants compared with wild-type plants (Stenbaek et al., 2008), demonstrating that the lack of NTRC imbalances the chlorophyll biosynthesis reactions. In the absence of NTRC, decreased enzyme activities in the chlorophyll branch of tetrapyrrole biosynthesis may induce imbalanced accumulation of chlorophyll precursors and finally reduced production of chlorophyll in developing ntrc chloroplasts. Recent expression analyses of the chlorophyll biosynthesis genes showed that HEMA1, GUN5, MgCy, and CAO constitute the most important regulatory gene cluster in tetrapyrrole biosynthesis (Matsumoto et al., 2004). It is conceivable, therefore, that the expanding ntrc leaves attempt to compensate for the low enzyme activities by up-regulating the regulatory genes in the chlorophyll biosynthesis (Table IV).
Reduced activity of chlorophyll biosynthesis enzymes may promote signals that contribute to the attenuation of chloroplast biogenesis in ntrc leaves (Nott et al., 2006). Accordingly, reduced accumulation of LHCB and RBCS transcripts in ntrc plants (Table IV) may be a consequence of the imbalanced biosynthesis of chlorophyll in ntrc leaves. In line with this hypothesis, the ntrc plants also show 3-fold repression of a gene encoding the chloroplast-targeted membrane-remodeling protein FLZ, which is involved in the regulation of thylakoid membrane networks (Table IV; Gao et al., 2006). Knockout flz plants resemble short-day-grown ntrc in having pale green leaves, delayed flowering, and a reduced number of chloroplasts, albeit with heterogeneous size (Gao et al., 2006; Fig. 2). These similarities suggest that the lowered expression level of FLZ in ntrc plants may contribute to the formation of the ntrc phenotype under short-day conditions.
Interestingly, a null mutant for chloroplast ATP/ADP transporters (NTTs) also resembles ntrc with respect to chlorophyll biosynthesis and photoperiod-dependent reduction in growth (Reinhold et al., 2007). Short-day-grown ntt null mutants show a dwarf phenotype with reduced Mg-chelatase activity and generate reactive oxygen species (ROS) after the shift from darkness to light. This raises the question of whether the accumulation of chlorophyll precursors in darkness (Stenbaek et al., 2008) also induces the formation of ROS during the subsequent reillumination of ntrc plants. NTRC was recently demonstrated to act in the reduction of chloroplast 2-Cys peroxiredoxins (Moon et al., 2006; Perez-Ruiz et al., 2006; Alkhalfioui et al., 2007; Stenbaek et al., 2008) and may thereby be crucial for antioxidant activities in chloroplasts. Accumulation of photodamaged PSII complexes further speaks for photooxidative stress in illuminated short-day-grown ntrc leaves (Table III). However, we did not detect any differential accumulation of superoxide or hydrogen peroxide in ntrc leaves as compared with wild-type leaves, regardless of the length of the dark period or the duration of the subsequent daytime illumination period (A. Lepistö, unpublished data). In fact, substantial increase in the accumulation of hydrogen peroxide in ntrc leaves was reported to occur solely upon reillumination of plants after a prolonged 3-d dark treatment (Perez-Ruiz et al., 2006). It is also worth noting that the mutants with lower contents of chloroplast 2-Cys peroxiredoxins showed stronger phenotypes under long-day than under short-day growth conditions and that the stress symptoms became more severe in older plants (Heiber et al., 2007). Yet, the ntrc phenotype showed the opposite trend. Evidently, the exact role of NTRC in chloroplast ROS metabolism still requires experimental clarification.
Chloroplast Biogenesis Is Connected with the Control of Photoperiodic Growth in Plants
Regulation of developmental processes by light intensity is well documented in plants. We found that short-day-grown wild-type Arabidopsis plants possess thin leaves, low chlorophyll a/b ratio, and low stomatal density typical of shade-grown plants, whereas the opposite was observed for long-day-grown plants with adaptations typical of high-light-acclimated leaves (Tables I and II). Thus, the daily light period and the light intensity seem to be comparable in terms of developmental regulation.
Both the photoperiodic and photomorphogenic development are regulated by phytochromes and cryptochromes in Arabidopsis (Li and Yang, 2007; Bae and Choi, 2008). In photoperiodic development, the photoreceptors provide light input signals to the circadian clock, which controls developmental signaling pathways in plants (Hotta et al., 2007). It is noticeable that a distinct repression of genes coding for CRY2 and the far-red light-impaired response regulator FRS3 occurred in ntrc plants, and ntrc also showed long-hypocotyl phenotypes both under low fluence rates of blue light and under far-red light (Fig. 6). Since CRY2 and PHYA act as photoreceptors for deetiolation under these light conditions (Cashmore et al., 1999; Fankhauser and Casal, 2004; Li and Yang, 2007), we conclude that light perception by CRY2- and PHYA-mediated signaling is affected by the knockout of NTRC.
Together, the defects in light receptor-mediated signaling, altered expression of chlorophyll biosynthesis genes, and lower number of chloroplasts in short-day ntrc plants suggest that disturbances in chloroplast metabolism interfere with the circadian clock-mediated regulation of plant growth. This interpretation is consistent with a recent report by Hassidim et al. (2007). They showed that malfunction of chloroplasts in Arabidopsis mutant lines disturbed the expression pattern of genes controlled by the circadian clock, indicating that chloroplast signals may regulate the circadian system in plants.
Photoperiodic Regulation of Stomatal Development in ntrc and Wild-Type Plants
Light intensity and CO2 partial pressure are well-known environmental factors that control the number of stomata in plants. Shading and elevated CO2 partial pressure both decrease the stomatal density in leaves (Lake et al., 2001, 2002). Here, we demonstrate that the diurnal photoperiod also contributes to the development of stomata in wild-type Arabidopsis leaves. The short photoperiod during growth equals low light intensity by decreasing both the stomatal density and the stomatal index of leaves (Table II).
The stomatal density is significantly higher in short-day ntrc leaves than in wild-type plants (Table II). This is accompanied by the repression of two genes encoding important negative regulators of stomatal development, SDD1 and EPF1, in ntrc plants (Table IV), which speaks for disturbed stomatal development in the absence of NTRC. Stomatal development starts with an asymmetric division of the epidermal cell that creates a meristemoid cell (reviewed by Bergmann and Sack, 2007). Besides SDD1 and EPF1, several other genes also control the density and spatial pattern of stomata in Arabidopsis leaves (Geisler et al., 2000; Masle et al., 2005; Shpak et al., 2005). These gene products mostly act as negative regulators of stomatal development, either by reducing the entry of epidermal cells to meristemoids or by inhibiting the formation of neighboring stomata (Bergmann and Sack, 2007). Consequently, the loss-of-function mutations of these genes result in increased stomatal density and/or changes in stomatal index (Geisler et al., 2000; Von Groll et al., 2002; Masle et al., 2005).
Interestingly, in the regulatory model of stomatal development, the membrane proteins SDD1 and EPF1 control the first steps of the signaling cascade immediately after an unknown environment-dependent initiatory factor (Casson and Gray, 2008), indicating that the repression of SDD1 and EPF1 may seriously influence the developmental pattern of stomata. Induced expression of SDD1 was also observed in experiments in which shading of mature Arabidopsis leaves was found to induce systemic signals that decreased the stomatal density in young, untreated leaves of the same plant (Coupe et al., 2006). This is consistent with the role of SDD1 as a negative regulator of stomatal development. Based on the increased stomatal density and the low levels of SDD1 and EPF1 transcripts in ntrc leaves, we propose that the ntrc mutants are not capable of correctly controlling the development of stomata. This defect in stomatal development becomes pronounced under short-day conditions, in which negative developmental control is most crucial to lower the number of stomata (Table II; Lake et al., 2001).
Reduced Accumulation of Shikimate Pathway Derivatives in ntrc Plants
The plastid-localized shikimic acid pathway serves as a biosynthetic route for aromatic amino acids (Herrmann, 1995; Ishihara et al., 2007). Besides providing building blocks for protein synthesis, aromatic amino acids also serve as precursors for important secondary metabolites, including Trp-derived indole hormones and Phe- and Tyr-derived flavonoids and lignins (Herrmann, 1995; Woodward and Bartel, 2005). Growth under short-day conditions increases the pool sizes of aromatic amino acids and decreases the content of their derivatives auxin and anthocyanins in ntrc plants (Figs. 7 and 8; Table I), suggesting that the lack of NTRC imbalances the metabolic pathways that derive from the shikimate pathway. It is likely, therefore, that the distinct structural and functional deficiencies of ntrc plants are partially due to the altered metabolism of aromatic amino acids and their derivatives. Also, the restoration of growth that was observed for short-day ntrc seedlings when grown on externally added aromatic amino acids supports this conclusion (Supplemental Fig. S6). Such restoration of ntrc growth may first sound inconsistent, considering that the levels of aromatic amino acids are increased rather than decreased in ntrc seedlings. However, shikimate-derived pathways are regulated by complex networks, including feedback control by aromatic amino acids and environmental factors (Ishihara et al., 2007). Thus, exogenous amino acids may restore the growth of ntrc plants by altering the homeostasis among biochemical pathways in chloroplasts.
Attempts to identify thiol-redox-regulated enzymes have revealed putative targets for the thioredoxin systems both in the shikimate pathway and in the biosynthesis of aromatic amino acids, including 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase and Trp synthase β (Entus et al., 2002; Balmer et al., 2006; Kolbe et al., 2006). Currently, we are exploring whether these enzymes are subjects for thiol-redox regulation by NTRC.
MATERIALS AND METHODS
Materials and Growth Conditions
Homozygous T-DNA insertion mutants deficient in NTRC (At2g41680; SALK_096776 and SALK_114293) were identified from the SALK Institute's collection by PCR analysis of genomic DNA according to the institute's protocols (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al., 2003). Homozygous SALK_096776 plants were backcrossed to wild-type Arabidopsis (Arabidopsis thaliana), selfed, and rescreened prior to usage in experiments. The homozygous ntrc mutants of SALK_114293 showed visible phenotypes identical with backcrossed SALK_096776 (for phenotypic and biochemical characterization of SALK_114293, see Supplemental Fig. S1 and Supplemental Table S1). Both SALK lines were also similar in phenotypes to ntrc mutant line SALK_012208 described previously by Serrato et al. (2004). The mutant and wild-type Arabidopsis ecotype Columbia (Col-0) plants were grown on a mixture of soil and vermiculite (1:1) under 100 μmol photons m−2 s−1 at 20°C or 10°C under short-day conditions (8/16 h of light/darkness), long-day conditions (16/8 h of light/darkness), or continuous light (no dark period).
Germination of Seeds under Different Spectral Qualities of Light
Seeds were sterilized with 50% (v/v) ethanol + 0.5% (v/v) Triton X-100 for 2 min, subsequently rinsed with 95% (v/v) ethanol, and dried. Sterilized seeds were sown on half-strength Murashige and Skoog salts in 1% (w/v) agar. After incubation at 4°C for 2 d, the plates were transferred to growth chambers under different spectral qualities of light: white, 100 μmol photons m−2 s−1; red, 30 μmol photons m−2 s−1; high-fluence-rate blue, 30 μmol photons m−2 s−1; and low-fluence-rate blue, 3 μmol photons m−2 s−1. For far-red light, germination was first induced by a 2-h white light treatment under 100 μmol photons m−2 s−1, after which the plates were kept in darkness at 20°C for 70 h and finally placed under far-red light. Hypocotyl length was measured after 7 d.
Growth on a Medium Supplemented with External Amino Acids and Auxin
Sterilized seeds were sown on half-strength Murashige and Skoog salts in 1% agar containing 40 μm Trp, 40 μm Phe, 40 μm Ile, 40 μm Ala, or 1 μm IAA. After incubation at 4°C for 2 d, the plates were transferred to the growth chamber, where the seedlings were grown under short-day conditions for 2 weeks.
Measurement of Leaf Pigment Contents, Gas Exchange, Water Loss, and Chlorophyll Fluorescence
Foliar chlorophyll content was determined by punching two leaf discs, 3 mm in diameter, into 1 mL of dimethylformamide. The leaf discs were incubated overnight at 4°C in darkness, and the chlorophyll content was measured spectrophotometrically according to Inskeep and Bloom (1985). Anthocyanin content was determined according to Neff and Chory (1998) with slight modifications. Five leaf discs, 5 mm in diameter, were extracted in 300 μL of 7% (v/v) hydrochloric acid in methanol overnight at 4°C with gentle shaking in darkness. Thereafter, 200 μL of sterile water and 500 μL of chloroform was added to each sample and mixed. Samples were centrifuged at 13,000 rpm for 2 min. The top layer was used for absorbance measurements at 530 and 657 nm. Relative anthocyanin concentrations were calculated as (A530 − A657) × 1,000 cm−2.
Gas exchange of intact ntrc and wild-type plants was measured with the CIRAS-1 combined infrared gas analysis system (PP Systems) equipped with an Arabidopsis pot chamber (PP Systems). The response of net photosynthesis to the reference CO2 was measured at 20°C under a photosynthetically active photon flux density of 500 μmol m−2 s−1, which was saturating for net photosynthesis. The parameters for maximal carboxylation rate of Rubisco (Vcmax; μmol CO2 m−2 s−1), maximal electron transport rate (Jmax; μmol m−2 s−1), and rate of mitochondrial respiration in light (Rd; μmol m−2 s−1) were obtained by modeling the response of net CO2 assimilation to increasing extracellular CO2 concentration according to Farquhar et al. (1980). Water loss from detached rosettes was measured according to Leung et al. (1997).
The photoinhibition state of PSII in intact leaves was recorded as the ratio of variable to maximal fluorescence (Fv/Fm, where Fv is the difference between maximal fluorescence [Fm] and initial fluorescence [Fo]), measured with a Hansatech PEA fluorometer after a 30-min dark incubation.
Isolation of Thylakoid Membranes and Total Root and Leaf Extracts
Rosettes of ntrc and wild-type plants were immediately frozen in liquid nitrogen. Total root and leaf extracts were collected after homogenization of the tissue in ice-cold isolation buffer (330 mm Suc, 25 mm HEPES-KOH, pH 7.4, 10 mm MgCl2, and 10 mm NaF) and filtration through Miracloth under dim light. For isolation of thylakoids, the filtrate was centrifuged at 6,000g for 5 min at 4°C. The thylakoid pellet was gently resuspended in 25 mm HEPES-KOH, pH 7.4, 10 mm MgCl2, and 10 mm NaF, centrifuged at 6,000g, for 5 min at 4°C, and finally suspended in the isolation buffer and stored at −80°C. The chlorophyll content of isolated thylakoids was determined according to Porra et al. (1989), and the protein contents of the total and soluble extracts were determined with the Bio-Rad Protein Assay Kit.
Blue-Native Electrophoresis
Blue-native PAGE was performed according to Kügler et al. (1997) with modifications according to Rokka et al. (2005). Thylakoid suspension corresponding to 5 μg of chlorophyll was centrifuged at 2,000g for 1 min at 4°C and resuspended in medium A (25 mm Bis-Tris/HCl, pH 7.0, 20% [w/v] glycerol, and 0.25 mg mL−1 Pefabloc) to a final concentration of 1 μg μL−1 chlorophyll. An equal volume of 2% (w/v) dodecyl β-d-maltoside (Sigma), freshly prepared in medium A, was added, and thylakoids were solubilized on ice for 1 min and then centrifuged at 18,000g for 12 min at 4°C. The supernatant was supplemented with one-tenth volume of loading buffer (100 mm Bis-Tris/HCl, pH 7.0, 0.5 m ɛ-amino-n-caproic acid, 30% [w/v] Suc, and 50 mg mL−1 Serva Blue G) and run on a native gel with a 5% to 12% gradient of acrylamide on the separation gel. Electrophoresis was performed with the Hoefer Mighty Small system (Amersham Biosciences) at 3°C for 3.5 h by gradually increasing the voltage from 75 to 200 V. Photosynthetic thylakoid protein complexes were identified according to Aro et al. (2005).
SDS-PAGE and Western Blotting
Soluble and total extracts corresponding to 5 to 15 μg of protein were solubilized and separated by SDS-PAGE (Laemmli, 1970), using 15% (w/v) acrylamide and 6 m urea on the separation gel, and subsequently electroblotted to a polyvinylidene difluoride membrane (Millipore; http://www.millipore.com). After blocking with 1% (w/v) bovine serum albumin (fatty acid free; Sigma-Aldrich), the polypeptides were immunodetected with protein-specific antibodies using a Phototope-Star Detection Kit (New England Biolabs). The Arabidopsis NTR-specific antibody was raised against amino acids 475 to 488 (Innovagen). Protein-specific antibodies were purchased from Research Genetics (D1; http://www.resgen.com/about/index.php3), Agrisera (Lhcb1; Lhca2; http://www.agrisera.se/), and kindly provided by Prof. H.V. Scheller (psaD).
Determination of NADP-MDH and FBPase Activities
For enzymatic assays, 500 mg of leaves was ground to a fine powder in liquid nitrogen. Subsequently, initial and maximal activities of chloroplast NADP-MDH and FBPase were measured according to Scheibe and Stitt (1988) and Kelly et al. (1982), respectively. The activation states of NADP-MDH and FBPase were calculated as a proportion of initial activity to maximal activity.
Visualization of Stomata and Leaf Mesophyll Cells by Laser Scanning Confocal Microscopy
Laser scanning confocal microscopy images were obtained with an inverted confocal laser scanning microscope (Zeiss LSM510 META; http://www.zeiss.com) with a 20×/0.50 water objective. Stomata were imaged by exciting autofluorescing compounds at 488 nm on the adaxial surface of 4-week-old leaves, followed by detection with a 420- to 480-nm passing emission filter. Stomatal density (number of stomata per square millimeter) and stomatal index (the ratio of the number of stomata to the total number of epidermal cells × 100) were calculated from the images. Chloroplasts and mesophyll cells were imaged by chlorophyll autofluorescence, which was excited at 488 nm with an argon diode laser, and detected with a 650- to 710-nm passing emission filter. Maximal projections of the sequential confocal images were created with the Zeiss LSM Image Browser version 3,5,0,376.
Hormone Analysis
Plant hormones were analyzed using a modified vapor-phase extraction method described by Schmelz et al. (2003). Briefly, 100 mg of seedlings was ground in liquid nitrogen and extracted with 300 μL of 1-propanol:water:concentrated HCl (2:1:0.002, v/v) containing isotopically labeled standards, vortexed, and extracted with 1 mL of dichloromethane. The dichloromethane fraction was transferred to 4-mL glass vials, methylated with 3 μL of (trimethylsilyl)-diazomethane in hexane (2 m; Sigma-Aldrich), evaporated after 30 min under a N2 stream at 70°C (10 min) and 200°C (150 s), and collected in traps containing 20 mg of SuperQ (Alltech Associates). Standards for each sample were 10 ng of [2H5]indole-3-acetic acid (OlChemIm) and 20 ng of [2H6]abscisic acid (Icon Isotopes). Traps were eluted with 3 × 200 μL of dichloromethane, samples were dried and silylated with 8 μL of N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich) for 30 min at 37°C and diluted with 16 μL of water-free pyridine (Sigma-Aldrich), and 1 μL was injected for gas chromatography-mass spectrometry analysis performed on a Trace-DSQ (Thermo) in the single ion monitoring mode on a ZB-5 capillary gas chromatography column (5% phenylpolysiloxane and 95% methylpolysiloxane, 30 m × 0.25 mm × 0.25 μm) with splitless injection and 230°C injector temperature. The column was held at 40°C for 1 min after injection, then heated at 15°C min−1 to 250°C, held for 4 min, and heated at 20°C min−1 to 310°C final temperature (kept for 3 min) with helium as carrier gas (flow, 1 mL min−1).
Amino Acid Analysis
Approximately 100 mg of 10-d-old plant material was frozen in liquid nitrogen and ground with the Retsch Tissue Lyser (Qiagen). The plant metabolites were extracted twice with 500 μL of 50% (v/v) methanol by shaking vigorously for 30 min with the Tissue Lyser. The extract was centrifuged for 5 min at 33,000g (Jouan MR 23i; Thermo Electron Industries), and the supernatant was dried in vacuo for 2 h (SC250 Express SpeedVac Concentrator SDP121P; Thermo Electron Industries). The dried residue was redissolved in 100 μL of 10% methanol. Amino acids in the extracts were further extracted and derivatized with the Ez:faast liquid chromatography-mass spectrometry kit (Phenomenex) using the procedure described by Husek (1998) and analyzed as propyl chloroformates with HPLC-electrospray ionization mass spectrometry (Thermo LTQ; Thermo Finnigan). Liquid chromatography was performed on the Ez:faast AAA-MS column as described in the Ez:faast kit.
Microarray Analysis
Global changes in gene expression were explored with spotted Arabidopsis 24k oligonucleotide arrays (MWG Biotech; http://www.mwg-biotech.com; ArrayExpress database accession no. A-ATMX-2; http://www.ebi.ac.uk/arrayexpress). A total of 500 mg of 10-d-old seedlings or rosette leaves (28 d old, grown in short days, and 21 d old, grown in long days) of wild-type and ntrc mutant plants was collected 4 h after the onset of the light period, and total RNA was isolated with Trizol as described previously (Piippo et al., 2006). Subsequently, DNA was removed with the Qiagen RNeasy Mini Kit, and cDNA synthesis was performed in the presence of 0.2 mm aminoallyl-dUTP using anchored poly(dT) primer (Oligomer) and the reverse transcriptase SuperScript III (Invitrogen). The aminoallyl-labeled cDNA was purified with the QIAquick PCR purification kit (Qiagen) and stained with the Cy Postlabeling Reactive Dye Pack (Amersham). cDNA corresponding to 15 pg of each dye was hybridized to the arrays in 50% (v/v) formamide, 5× SSC, 0.1% (w/v) SDS, 0.1 mg mL−1 herring sperm, and 5× Denhardt's solution at 42°C overnight.
The arrays were scanned with an Agilent scanner (G2565BA; http://www.agilent.com), and spot intensities were quantified with the ScanArray Express Microarray Analysis System 2.0 (Perkin-Elmer Life Sciences; http://las.perkinelmer.com) using the adaptive circle method. Low-quality spots were flagged and not included in the analysis. The raw data were normalized using the Lowess method in GeneSpring GX 7.3 (Agilent; http://www.agilent.com). Normalized data from three biological replicates were used, and the genes with Student's t test P values below 0.05 were chosen for further analysis. The gene annotation used was derived from The Arabidopsis Information Resource (TAIR 7; http://www.arabidopsis.org).
Northern Blotting
The probe for GUN5 was amplified from cDNA by PCR with primers 5′-CTCACGGACTCCCATTTTGT-3′ and 5′-AGGGACTGCAGCTTACCTCA-3′. Northern blotting and hybridization of the membranes with GUN5 and probe for 18S rRNA were performed according to Mulo et al. (2003).
Array design and data from this article have been deposited at ArrayExpress under accession number E-MEXP-1697.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of the SALK_114293 ntrc knockout mutant grown under short and long photoperiods.
Supplemental Figure S2. Response of net CO2 assimilation to reference CO2 concentration in Col-0 and ntrc plants.
Supplemental Figure S3. Diurnal accumulation and degradation of starch in ntrc and Col-0.
Supplemental Figure S4. Numbers of differentially expressed genes in ntrc relative to Col-0.
Supplemental Figure S5. Transcript levels of GUN5 in Col-0 and ntrc grown under short- and long-day conditions.
Supplemental Figure S6. Growth of Col-0 and ntrc seedlings on external amino acids and auxin.
Supplemental Table S1. Specific leaf weight (SLW) and pigment content in SALK_114293 plants and Col-0 grown under different photoperiods.
Supplemental Table S2. Transcript profiling of ntrc plants under different physiological and developmental stages.
Supplemental Table S3. Free amino acid concentrations of ntrc and Col-0 seedlings grown under short and long photoperiods.
Supplementary Material
Acknowledgments
We thank Jouko Sandholm, Colin Ruprecht, Briitta Ruokamo, and Kati Thiel for excellent technical assistance and Eva-Mari Aro and Paula Mulo for critical reading of the manuscript. The Cell Imaging Core of the Turku Center for Biotechnology of the University of Turku and Åbo Akademi University is acknowledged for providing laser scanning confocal microscopy. We are grateful to CSC–Scientific Computing Ltd. for providing the national license for GeneSpring. The Salk Institute Genomic Analysis Laboratory, funded by the National Science Foundation, is acknowledged for providing the sequence-indexed Arabidopsis T-DNA insertion mutants.
This work was supported by the Academy of Finland (project nos. 107039 and 204521) and the Finnish Graduate School in Plant Biology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eevi Rintamäki (evirin@utu.fi).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al (2004) Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Montrichard F (2007) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58 969–978 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamaki E (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56 347–356 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59 281–311 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD (2007) Stomatal development. Annu Rev Plant Biol 58 163–181 [DOI] [PubMed] [Google Scholar]
- Buchanan BB (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys 288 1–9 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284 760–765 [DOI] [PubMed] [Google Scholar]
- Casson S, Gray JE (2008) Influence of environmental factors on stomatal development. New Phytol 178 9–23 [DOI] [PubMed] [Google Scholar]
- Coupe SA, Palmer BG, Lake JA, Overy SA, Oxborough K, Woodward FI, Gray JE, Quick WP (2006) Systemic signalling of environmental cues in Arabidopsis leaves. J Exp Bot 57 329–341 [DOI] [PubMed] [Google Scholar]
- Dai S, Johansson K, Miginiac-Maslow M, Schurmann P, Eklund H (2004) Structural basis of redox signaling in photosynthesis: structure and function of ferredoxin:thioredoxin reductase and target enzymes. Photosynth Res 79 233–248 [DOI] [PubMed] [Google Scholar]
- de la Luz Gutierrez-Nava M, Gillmor CS, Jimenez LF, Guevara-Garcia A, Leon P (2004) CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 135 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entus R, Poling M, Herrmann KM (2002) Redox regulation of Arabidopsis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Physiol 129 1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Casal JJ (2004) Phenotypic characterization of a photomorphogenic mutant. Plant J 39 747–760 [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149 78–90 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sage TL, Osteryoung KW (2006) FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc Natl Acad Sci USA 103 6759–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Navrot N, Jacquot JP (2005) The plant thioredoxin system. Cell Mol Life Sci 62 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M, Yakir E, Fradkin D, Hilman D, Kron I, Keren N, Harir Y, Yerushalmi S, Green RM (2007) Mutations in CHLOROPLAST RNA BINDING provide evidence for the involvement of the chloroplast in the regulation of the circadian clock in Arabidopsis. Plant J 51 551–562 [DOI] [PubMed] [Google Scholar]
- Heiber I, Stroher E, Raatz B, Busse I, Kahmann U, Bevan MW, Dietz KJ, Baier M (2007) The redox imbalanced mutants of Arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiol 143 1774–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM (1995) The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt RP, Muller S, Embley TM, Coombs GH (2002) The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol 18 302–308 [DOI] [PubMed] [Google Scholar]
- Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH (2005) Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33 1375–1377 [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30 333–349 [DOI] [PubMed] [Google Scholar]
- Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS (2005) Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol 137 762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Husek P (1998) Chloroformates in gas chromatography as general purpose derivatizing agents. J Chromatogr B Biomed Sci Appl 717 57–91 [PubMed] [Google Scholar]
- Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PG, Hisabori T, Takamiya K, Masuda T (2007) The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem 282 19282–19291 [DOI] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR (1985) Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiol 77 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara A, Matsuda F, Miyagawa H, Wakasa K (2007) Metabolomics for metabolically manipulated plants: effects of tryptophan overproduction. Metabolomics 3 319–334 [Google Scholar]
- Kelly GJ, Zimmermann G, Latzko E (1982) Fructose-bisphosphatase from spinach leaf chloroplast and cytoplasm. Methods Enzymol 90 371–378 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Oliver SN, Fernie AR, Stitt M, van Dongen JT, Geigenberger P (2006) Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol 141 412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Takeba G (1996) Photorespiration protects C3 plants from photooxidation. Nature 384 557–560 [Google Scholar]
- Kügler M, Jänsch L, Kruft V, Schmitz UK, Braun HP (1997) Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth Res 53 35–44 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lake JA, Quick WP, Beerling DJ, Woodward FI (2001) Plant development: signals from mature to new leaves. Nature 411 154. [DOI] [PubMed] [Google Scholar]
- Lake JA, Woodward FI, Quick WP (2002) Long-distance CO2 signalling in plants. J Exp Bot 53 183–193 [DOI] [PubMed] [Google Scholar]
- Lemaire SD (2004) The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth Res 79 305–318 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QH, Yang HQ (2007) Cryptochrome signaling in plants. Photochem Photobiol 83 94–101 [DOI] [PubMed] [Google Scholar]
- Lin R, Wang H (2004) Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol 136 4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28 465–474 [DOI] [PubMed] [Google Scholar]
- Marchand C, Le Marechal P, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, Decottignies P (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4 2696–2706 [DOI] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870 [DOI] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T (2004) Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol 135 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Riondet C, Constans L, Abdelgawwad MR, Reichheld JP, Vignols F (2006) Evolution of redoxin genes in the green lineage. Photosynth Res 89 179–192 [DOI] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, Lee JR, Lee SY, Jung YJ, Shin MR, Lim HS, Chung WS, Yun DJ, et al (2006) The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem Biophys Res Commun 348 478–484 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P, Pursiheimo S, Hou C, Tyystjärvi T, Aro E (2003) Multiple effects of antibiotics on chloroplast and nuclear gene expression. Funct Plant Biol 30 1097–1103 [DOI] [PubMed] [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A (2007) Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J 49 800–809 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57 739–759 [DOI] [PubMed] [Google Scholar]
- Perez-Ruiz JM, Spinola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta U, Battchikova N, Aro E (2006) Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol Genomics 25 142–152 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold T, Alawady A, Grimm B, Beran KC, Jahns P, Conrath U, Bauer J, Reiser J, Melzer M, Jeblick W, et al (2007) Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J 50 293–304 [DOI] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM (2005) Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem J 388 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- Scheibe R, Stitt M (1988) Comparison of NADP-malate dehydrogenase activation, QA reduction and O2 evolution in spinach leaves. Plant Physiol Biochem 26 473–481 [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, O'Donnell P, Sammons M, Toshima H, Tumlinson JH III (2003) Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc Natl Acad Sci USA 100 10552–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Perez-Ruiz JM, Spinola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279 43821–43827 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293 [DOI] [PubMed] [Google Scholar]
- Spinola MC, Perez-Ruiz JM, Pulido P, Kirchsteiger K, Guinea M, Gonzalez M, Cejudo FJ (2008) NTRC new ways of using NADPH in the chloroplast. Physiol Plant 133 516–524 [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, Jensen PE (2008) NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett 582 2773–2778 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58 321–346 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11 329–334 [DOI] [PubMed] [Google Scholar]
- Von Groll U, Berger D, Altmann T (2002) The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.