Abstract
Salicylic acid (SA) is a critical mediator of plant innate immunity. It plays an important role in limiting the growth and reproduction of the virulent powdery mildew (PM) Golovinomyces orontii on Arabidopsis (Arabidopsis thaliana). To investigate this later phase of the PM interaction and the role played by SA, we performed replicated global expression profiling for wild-type and SA biosynthetic mutant isochorismate synthase1 (ics1) Arabidopsis from 0 to 7 d after infection. We found that ICS1-impacted genes constitute 3.8% of profiled genes, with known molecular markers of Arabidopsis defense ranked very highly by the multivariate empirical Bayes statistic (T2 statistic). Functional analyses of T2-selected genes identified statistically significant PM-impacted processes, including photosynthesis, cell wall modification, and alkaloid metabolism, that are ICS1 independent. ICS1-impacted processes include redox, vacuolar transport/secretion, and signaling. Our data also support a role for ICS1 (SA) in iron and calcium homeostasis and identify components of SA cross talk with other phytohormones. Through our analysis, 39 novel PM-impacted transcriptional regulators were identified. Insertion mutants in one of these regulators, PUX2 (for plant ubiquitin regulatory X domain-containing protein 2), results in significantly reduced reproduction of the PM in a cell death-independent manner. Although little is known about PUX2, PUX1 acts as a negative regulator of Arabidopsis CDC48, an essential AAA-ATPase chaperone that mediates diverse cellular activities, including homotypic fusion of endoplasmic reticulum and Golgi membranes, endoplasmic reticulum-associated protein degradation, cell cycle progression, and apoptosis. Future work will elucidate the functional role of the novel regulator PUX2 in PM resistance.
The obligate biotrophic powdery mildew (PM) Golovinomyces orontii is virulent on Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0). It exclusively infects epidermal cells with a defined progression of infection, including germination of the conidia (1–2 h post inoculation [hpi]), penetration of the epidermal cell (approximately 5 hpi), and development of the haustorial complex, the feeding structure (14–24 hpi). This is followed by the further growth and reproduction of the fungus from 1 to 7 d post inoculation (dpi) with formation of the reproductive structures known as conidiophores that contain the conidia (Plotnikova et al., 1998). As part of this intimate interaction, the PM must extract sufficient nutrients (carbon, nitrogen, etc.) from the plant host while maintaining viable plant tissue and limiting host defense responses, including cell death (for a recent review, see Micali et al., 2008).
Plant cell architecture is altered to allow fungal entry and development of the haustorial complex, composed of both fungus- and plant-derived components. In addition, specific alteration of plant cell metabolism is needed to provide the fungus with requisite nutrients. Plant factors involved in these processes are often referred to as compatibility factors. These include the seven-transmembrane, calmodulin-binding proteins AtMLO2, -6, and -12 that mediate fungal entry into the Arabidopsis epidermal cell (Collins et al., 2003; Consonni et al., 2006). In addition, PM-resistant mutants such as pmr5 and pmr6 that are specifically resistant to virulent PMs such as G. orontii and Golovinomyces cichoracearum likely contain mutations in compatibility factors (Vogel et al., 2002, 2004). Recessive mutations in the genes encoding AtPMR5 (a novel protein) and AtPMR6 (glycosylphosphatidylinositol-anchored pectate lyase-like) result in altered cell wall composition, smaller cells, and a significant reduction in PM growth and reproduction. In general, resistance conferred by the loss of compatibility factors has been found to be SA independent (Vogel et al., 2002, 2004; Consonni et al., 2006).
G. orontii induces SA-dependent defense responses in infected Arabidopsis leaves, with mutants in SA biosynthesis and signal transduction exhibiting enhanced susceptibility to the fungus (Reuber et al., 1998; Dewdney et al., 2000). This indicates that the plant is mounting active defenses in response to virulent PMs that limit the extent of infection. These induced responses, however, do not include cell death or cell death-associated responses. Indeed, Arabidopsis mutants with enhanced resistance to virulent PMs often exhibit constitutive or PM-induced cell death. For example, the Arabidopsis enhanced disease resistance mutants edr1, edr2, and edr3, which contain recessive mutations in genes resulting in loss of function of a mitogen-activated protein kinase kinase kinase (Frye et al., 2001), a novel protein with pleckstrin and steroidogenic acute regulatory protein-related lipid transfer domains (Vorwerk et al., 2007), and a dynamin-related protein 1E (Tang et al., 2006) exhibit enhanced cell death at the site of PM infection and are more resistant to G. cichoracearum. Similarly, the recessive pen3 Arabidopsis mutant exhibits PM-induced lesions and enhanced resistance to G. cichoracearum (Stein et al., 2006). These PM-induced lesions and associated plant defense responses are SA dependent (Frye et al., 2001; Tang et al., 2006; Vorwerk et al., 2007), as is mildew-induced lesions4-mediated resistance (S. Somerville, personal communication).
Arabidopsis transcriptional profiling in response to virulent PMs has focused on early phases of the PM interaction (0–24 hpi), including penetration and formation of the haustorial complex (Zimmerli et al., 2004; Fabro et al., 2008). Here, we focus on the later phase of the PM infection (1–7 dpi), the growth and reproduction phase. In this phase, we would expect to see expression changes associated with continued nutrient acquisition by the fungus, delayed senescence of local tissue to support the continued growth and reproduction of this obligate biotroph, suppression of cell death and some host plant defenses, and continued host plant defenses that limit the extent of infection. Previous studies have shown that SA biosynthesis and signal transduction pathways mediate this later phase of PM infection, with mutants in ICS1 (for isochorismate synthase 1), EDS5 (for enhanced disease susceptibility 5), NPR1 (for nonexpresser of PR genes 1), and PAD4 (for phytoalexin-deficient 4) exhibiting enhanced growth of PM on infected leaves, assessed visually at 7+ dpi (Reuber et al., 1998; Dewdney et al., 2000). In addition, npr1 and pad4 mutants exhibit an increased number of conidiophores per germinated conidium (Reuber et al., 1998). We established that this is also the case for the ics1-2 mutant (8.6 ± 3.9 conidiophores per conidium for ics1-2 [16 colonies] compared with 5.8 ± 3.4 conidiophores per conidium for Col-0 [51 colonies]; P < 0.006). These differences in conidiophores per conidium provide further evidence that SA plays a role in limiting PM reproduction. However, a mechanistic understanding of how SA limits PM growth and reproduction is unknown.
Therefore, to examine the later growth and reproduction phase of a virulent PM infection and to specifically examine the impact of SA on this phase of infection, we performed replicated global expression profiling using identically treated wild-type and ics1-2 mutant plants. By acquiring replicated progressive time series data with six time points and by employing the multivariate empirical Bayes statistic (T2 statistic) specifically designed for the analysis of replicated time series data (Tai and Speed, 2006), we can ascertain distinct patterns of host response and can resolve genes with more subtle responses. For example, genes with mean expression changes of less than 2-fold at any given time point but with a clear pattern of response across the time series can be identified. Analysis of this T2-selected data then allows us to identify known and novel processes and components of the PM-Arabidopsis interaction. Very few plant transcription factors mediating PM growth and reproduction have been reported (Micali et al., 2008). As described below, our analyses identified four of the five known transcription factors as well as 39 novel regulators with no previously published role in plant disease resistance or abiotic stress response. We then established a functional role in PM resistance/susceptibility for one of these novel regulators.
RESULTS AND DISCUSSION
We performed replicated global expression profiling with the Affymetrix ATH1 array using RNA extracted from mature whole leaves harvested at 0 (just prior to infection), 0.25, 1, 3, 5, and 7 dpi with G. orontii and 7-d uninfected (UI) leaves from parallel, identically treated wild-type and ics1-2 mutant plants. We then applied the T2 statistic to select genes based on differences in their expression profiles in (1) wild-type plants in response to PM and (2) ics1 versus wild-type plants in response to PM to assess the impact of SA on the PM-induced transcriptional response. The T2 statistic is specifically designed for use with replicated temporal expression data of a longitudinal nature. It tests the null hypothesis that there is no difference in the vector of a gene's mean expression levels (e.g. between mutant and the wild type), taking into account correlations among observations across time, replication, and moderation (Tai and Speed, 2006). Thus, genes with a more dramatic change in their pattern of expression (e.g. in the mutant versus the wild type) have a higher T2 statistic independent of the pattern or sign of their difference in expression.
All additional analyses were performed using the T2-selected gene sets, which are provided with ranking, annotation, and cluster designation in Supplemental Worksheet S1, with expression profiles plotted in Supplemental Figures S1 and S2. To confirm our ATH1 data, we performed real-time quantitative reverse transcription-PCR (qPCR) for 11 genes: ICS1 (rank 30), PR1 (rank 1), PR2 (rank 33), the lupeol synthase At1g66960 (rank 28), PDF1.2a (rank 21), PUX2 (rank 381), PAD4 (rank 418), ACA2 (rank 469), ethylene-responsive transcription factor (ERF) At1g06160 (rank 545), NPR1 (rank 851), and the control UBQ5. Genes with different temporal expression patterns, differing magnitudes of expression change, and different levels of expression were selected. In addition, the chosen genes are near the top, middle, and bottom of those selected by the T2 statistic. For example, NPR1 (rank 851 of 870) is very near the bottom of our selection cutoff; it exhibits a visually discernible difference in its ATH1 expression patterns for the wild type compared with the ics1 mutant but less than 2-fold change in mean expression at any time point. As shown in Supplemental Figure S3, temporal patterns of gene expression for wild-type and ics1 mutant samples obtained by qPCR paralleled those obtained with ATH1 GeneChips.
Global Transcriptional Response to PM Infection
Nine hundred seventy-three genes exhibited altered expression in response to PM in wild-type plants over the course of infection. Of the 973 PM-responsive genes, 38% (373) are ICS1 impacted, with 90% of the ICS1-impacted genes also induced by exogenous treatment with the SA analog benzo(1,2,3) thiadiazole-7-carbothioc acid S-methyl ester (BTH), as reported by Wang et al. (2006; Fig. 1A; Supplemental Worksheet S2). The set of 154 genes that are both ICS1 independent and unresponsive to BTH are likely to exist in pathways upstream of SA biosynthesis or independent of SA. This set of genes includes peroxidases, which may be involved in generating hydrogen peroxide, nine DNA-binding regulatory factors, and three type A response regulators (ARRs): ARR6, ARR7, and ARR16.
Figure 1.
Venn diagrams showing genes with differential expression in wild-type Arabidopsis (PM) or in the ics1 mutant versus the wild type (PM:ICS1-impacted) over the time course of PM infection or in response to exogenous BTH (data from Wang et al., 2006). A, ATH1 genes. B, Transcription factors. PM and PM:ICS1-impacted genes were selected with log10(T2) ≥ 2.5. BTH-responsive genes exhibited significantly altered expression in wild-type versus BTH-treated samples, selected using ANOVA with P < 0.05 and 2-fold change cutoff (Wang et al., 2006).
Four clusters represent the dominant patterns of expression of wild-type plants in response to PM (Fig. 2A). Genes in wild-type clusters 1 and 2, representing 57% of the T2-selected genes, exhibit increased expression over the time course of PM infection, with cluster 1 genes tending to exhibit a higher magnitude of response. Wild-type cluster 1 genes include those involved in (1) plant defense, such as the pathogenesis-related proteins PR1 (At2g14610), PR2 (At3g57260), and PR5 (At1g75040); (2) nutrient reallocation and acquisition, such as the Suc-proton symporter SUC1 (At1g71880) and the putative mannitol transporter At4g36670; (3) PM compatibility/resistance, such as HR4 (At3g50480), a susceptible homolog of the PM resistance gene RPW8 (Xiao et al., 2001, 2004); and (4) cell death suppression, such as the Arabidopsis BI-1 gene (At5g47120; Hückelhoven et al., 2003; Kawai-Yamada et al., 2004). Similarly, wild-type cluster 2 genes include genes involved in SA biosynthesis and signal transduction (such as ICS1, EDS5, and PAD4), nutrient transporters, and genes mediating PM compatibility/resistance (i.e. MLO12 [At2g39200]). The remaining 43% of the PM-impacted genes exhibit reduced expression over the time course of PM infection, with wild-type clusters 3 and 4 differing in their magnitude of response. They include genes involved in processes impacting nutrient reallocation/acquisition, such as photosynthesis, chlorophyll synthesis, chloroplast development, and carbohydrate metabolism (e.g. vacuolar invertase β-FRUCTOSIDASE4 [BFRUCT4; At1g12240]). Wild-type cluster 4 also contains four type A AARs (ARR5, -6, -7, and -16) that act as negative regulators of cytokinin (CK) signaling (Muller and Sheen, 2007). Down-regulation of these negative regulators could result in enhanced CK-induced responses. This is consistent with previous reports of CK accumulation at PM infection sites and roles for CK in prolonged leaf longevity and cell death suppression (Walters and McRoberts, 2006).
Figure 2.
Expression profile plots for clusters of genes selected based on differences in temporal expression for wild-type (WT) T2-selected data (A) or ics1 versus wild-type T2-selected data (B). For wild-type clusters, log2 expression values for 0, 1, 3, 5, and 7 dpi (I) and 7-d UI are shown. For ICS1 clusters, log2 expression values for 0, 0.25, 1, 3, 5, and 7 dpi for wild-type and ics1 are displayed. Data for biological replicates are shown sequentially.
Global Transcriptional Impact of ICS1 Mutation
SA is a phytohormone and signaling molecule mediating response to (a)biotic stress (Dempsey et al., 1999; Durrant and Dong, 2004). Here, we identify 870 probe sets (3.8% of those on the ATH1 array) with differential expression in the SA biosynthetic mutant ics1 compared with the wild type over the time course of PM infection. Three hundred seventy-three of these 870 probe sets also show significantly altered expression in wild-type plants in response to PM (Fig. 1). Seventy-five percent of the 870 ICS1-impacted genes are also BTH responsive, consistent with the role of ICS1 in SA synthesis. In addition, 177 genes were only identified in the PM:ICS1-impacted set. The majority of these genes exhibit slightly altered expression in wild-type plants over the course of PM infection but fall below the T2 selection threshold. The marked change in PM-induced expression in the ics1 mutant compared with the wild type resulted in their selection in the PM:ICS1-impacted data set. This is the case for NPR1. Another set of genes identified only in the PM:ICS1-impacted set were those in which gene expression did not change in wild-type plants infected with PM but did change in the ics1 plants, as is the case for PDF1.2a, a marker of ethylene (ET)/jasmonate (JA) signaling (Glazebrook, 2005).
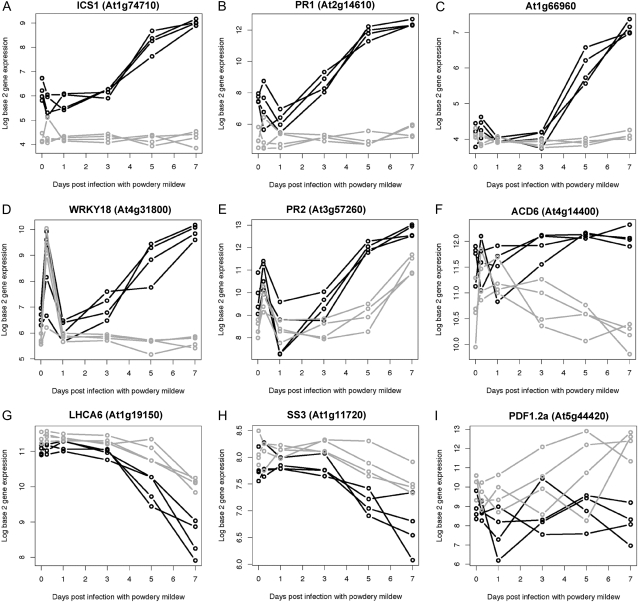
Six clusters represent the dominant patterns of ICS1-impacted expression (Fig. 2B). Figure 3 shows the expression profile for a selected gene of interest for each cluster, with additional subprofiles shown for ICS1 cluster 1. Displayed genes were selected either because they were the top T2-ranked gene within that cluster and/or they were of particular biological interest. Seventy-six percent of the 870 PM:ICS1-impacted genes exhibit induced expression in response to PM over the progression of infection, with this induced expression reduced in the ics1 mutant. Those genes exhibiting a dramatic difference in expression between the wild type and ics1 are ICS1 cluster 1 members, whereas those with more moderate expression differences are typically found in cluster 2. ICS1 clusters 1 and 2 include genes involved in pathogen defense and disease resistance, redox, secretion and trafficking, nutrient acquisition, protein modification, and calcium signaling. Cluster 1 includes ICS1 (At1g74710), which exhibits lower basal expression and lack of induction by PM in the ics1-2 mutant, consistent with the large deletion in ICS1 caused by fast-neutron mutagenesis (Wildermuth et al., 2001; S.K. Marr and M.C. Wildermuth, unpublished data). The highest ranked member of this PM:ICS1-impacted set is PR1. PR1 (At2g14610), which encodes a small secreted peptide, has been associated specifically with SA-dependent induced defense responses including systemic acquired resistance (Shah, 2003). The expression of PR1, which increases 37-fold over the course of infection, is completely abolished in the ics1 mutant (Fig. 3B). At1g66960 (Fig. 3C) and WRKY18 (Fig. 3D) represent additional patterns of response within ICS1 cluster 1. At1g66960 exhibits delayed induction in response to PM, with a substantial increase in expression evident at 5 dpi. This delayed induction may reflect processes specifically associated with PM reproduction and sustained growth. In the ics1 mutant, we observe genes in which this delayed expression is abrogated, as is the case for At1g66960 and others with reduced expression. At1g66960 (rank 28) is the most highly T2-ranked gene with the delayed SA-dependent paired expression profile. This gene encodes lupeol synthase, which is likely involved in the synthesis of the antimicrobial pentacyclic triterpenoid lupeol in Arabidopsis (Herrera et al., 1998; Husselstein-Muller et al., 2001; Ohyama et al., 2007). Genes with the partially SA-dependent delayed induction pattern include the cell wall invertase BFRUCT1 (At3g13790), which catalyzes the breakdown of Suc to Glc and Fru, hexoses that can be taken up by the fungus. In addition, WRKY75 (At5g13080) and WRKY30 (At5g24110) transcription factors exhibit this delayed pattern of response. ICS1 cluster 1 member WRKY18 (rank 14) is representative of genes that exhibit a distinct early peak of expression (6 h) that may in some cases be uncoupled from the later response. The 6-h induced response of WRKY18 (At4g31800) is not significantly impacted by the ics1 mutation, whereas the later response is abolished in the mutant (Fig. 3D). This suggests that the initial response (at 6 h) is ICS1 independent, whereas subsequent responses are impacted by the ICS1 mutation. One explanation for this type of pattern is that the unaltered change in expression at 6 h is due to circadian effects that are SA independent. All time points, excepting the 6-h time point, were collected at the same time of day. Alternatively, this initial peak in expression may be pathogen dependent but independent of SA. This is likely the case for WRKY18, as others have reported similar results using other pathosystems. For example, in response to an avirulent bacterial pathogen, WRKY18 exhibits rapid (≤2 h) SA-independent induction, with SA required for full expression at 24 hpi (Dong et al., 2003).
Figure 3.
Patterns of ICS1-impacted transcriptional responses. Gene expression profiles for the wild type (black) and ics1 mutant (gray) over the time course of PM infection from 0 (UI) to 7 dpi. Expression profiles for four paired (wild-type and mutant) biological replicates are shown for members of each ICS1 cluster: cluster 1 (A–D), cluster 2 (E), cluster 3 (F), cluster 4 (G), cluster 5 (H), and cluster 6 (I).
ICS1 cluster 2 members often exhibit a partial ICS1 dependence, with reduced PM-induced expression over the progression of infection, as shown for PR2 (Fig. 3E). PR2 (At3g57260; rank 33) encodes an acidic β-glucanase whose expression is generally associated with SA-dependent induced defense responses; however, a component of its expression has been reported to be SA independent (Dewdney et al., 2000; Wildermuth et al., 2001), as we observe. Other ICS1 cluster 2 members include the thaumatin-like protein PR5 (At1g75040) and the SA-pathway lipase-like regulator PAD4 (At3g52430). Genes in ICS1 clusters 4 and 5 exhibit reduced expression in wild-type plants in response to PM, with less of a reduction in the ics1 mutant. Genes in clusters 4 and 5 include genes involved in photosynthesis (e.g. PSI light-harvesting complex gene 6 [LHCA.6; At1g19150]; Fig. 3G), carbohydrate metabolism (e.g. starch synthase AtSS3 [At1g11720]; Fig. 3H), and iron homeostasis (AtFER1, -3, and -4). Genes in ICS1 clusters 3 and 6 do not exhibit altered expression in wild-type plants but have either reduced (cluster 3) or enhanced (cluster 6) expression in the ics1 mutant over the course of PM growth and reproduction. Cluster 3 genes include the ankyrin repeat-containing proteins ACD6 (At4g14400; Fig. 3F), an SA-dependent activator of cell death (Lu et al., 2003), and At5g54610. Expression of these genes may be subject to phytohormone cross talk, in part controlled by SA (Lu et al., 2003; Blanco et al., 2005). ICS1 cluster 6 genes, such as the defensin PDF1.2a (At5g44420; rank 21), are not typically induced in response to PM but are induced in the ics1 mutant (Fig. 3I). PDF1.2a is the highest T2-ranked gene in this cluster, with 18-fold higher expression in ics1 compared with the wild type at 7 dpi, similar to our previous results by northern blot (Dewdney et al., 2000). In addition to PDF1.2a, a marker of ET/JA-dependent induced defense responses, other ET/JA-associated genes are in this cluster, suggesting that this cluster may represent ET/JA-impacted genes that are typically repressed by SA in response to PM. Unlike the temporal expression profiles of most genes exhibiting differential expression, the temporal expression profile of PDF1.2a is quite variable for independent biological samples. This variability in PDF1.2a expression is not due to limits on sensitivity in detecting PDF1.2a but reflects the underlying biology. Many of the other genes in cluster 6 exhibit similar variability, suggestive of shared factors mediating this variable expression pattern. The ability of the T2 statistic to highly rank this gene and to identify other genes with a similar pattern and replicate variability is unusual and very powerful.
Identification of PM-Impacted Functional Processes and Protein Families
By looking at PM-impacted genes independent of sign or pattern of differential expression as selected using the T2 statistic, we can obtain an integrated overview of PM-impacted functional processes and protein families. We did this because select genes involved in a process may be down-regulated (e.g. a negative regulator), while others, such as positive regulators or targets, may be up-regulated. In addition, the observed temporal patterns of expression are not all exclusively up or down throughout the progression of infection. Furthermore, altered expression of genes at earlier time points may impact later transcriptional responses. For our analysis, we used MapMan (version 2.1.1; Thimm et al., 2004) to identify functional processes that were impacted significantly (P ≤ 0.05) in the wild-type PM and PM:ICS1-impacted T2-selected gene sets (Table I; Supplemental Worksheets S3 and S4). We also looked for significantly enriched PFAM domains (Finn et al., 2008) in the proteins encoded by our selected genes compared with those of the ATH1 array (Table II; Supplemental Worksheet S5). We then discuss the sign and pattern of gene expression for genes identified as part of an enriched functional process or encoding proteins whose PFAM domains are enriched in the T2 data sets.
Table I.
PM-impacted processes
MapMan analysis (Thimm et al., 2004) was performed using T2-selected genes to identify statistically enriched processes. MapMan bins with P ≤ 0.05 are shown. Specific genes in MapMan bins are provided in Supplemental Worksheets S3 and S4.
| MapMan Bin | Process | Wild Type
|
ics1 versus Wild Type
|
||
|---|---|---|---|---|---|
| P | Genes | P | Genes | ||
| ICS1 independent | |||||
| 1 | Photosynthesis | 0.02 | 20 | ||
| 10 | Cell wall | 0.03 | 39 | ||
| 10.7 | Cell wall modification | 0.01 | 9 | ||
| 11.1.5 | Lipid metabolism, fatty acids | 0.05 | 2 | ||
| 11.3 | Lipid metabolism, phospholipid synthesis | 0.03 | 3 | ||
| 16.4 | Secondary metabolism, nitrogen miscellaneous | 0.01 | 13 | ||
| 16.4.1 | Nitrogen miscellaneous, alkaloid-like | 0.01 | 13 | ||
| 29.2 | Protein synthesis | <0.01 | 28 | ||
| 29.2.1 | Chloroplast/mitochondrial plastid ribosomal | 0.00 | 18 | ||
| 29.5 | Protein degradation | 0.02 | 31 | ||
| 29.5.11 | Ubiquitin | 0.02 | 14 | ||
| 29.5.11.4 | Ubiquitin E3 | 0.02 | 14 | ||
| 26.21 | Miscellaneous protease inhibitor/seed storage/lipid transfer | 0.02 | 8 | ||
| 26.19 | Miscellaneous plastocyanin-like | 0.02 | 2 | ||
| ICS1 impacted | |||||
| 2.1 | Major carbohydrate metabolism, degradation, synthesis | 0.05 | 3 | ||
| 17.4 | Hormone metabolism, cytokinin | 0.04 | 3 | ||
| 20 | Stress | 0.04 | 61 | <0.01 | 63 |
| 20.1 | Stress, biotic | <0.01 | 47 | ||
| 21.4 | Redox, glutaredoxins | 0.05 | 2 | ||
| 27.2 | RNA transcription | 0.02 | 4 | ||
| 27.3.32 | RNA regulation of transcription, WRKY domain | 0.02 | 13 | 0.01 | 13 |
| 29.3.4.3 | Protein targeting, secretory pathway, vacuole | 0.02 | 3 | ||
| 30 | Signaling | 0.01 | 96 | <0.01 | 91 |
| 30.2 | Receptor kinases | 0.01 | 50 | <0.01 | 62 |
| 30.2.1 | LRRI | 0.03 | 3 | ||
| 30.2.17 | DUF26 | 0.04 | 14 | 0.04 | 15 |
| 30.2.24 | S-locus glycoprotein-like | 0.03 | 3 | ||
| 30.2.25 | Wall-associated kinase | 0.03 | 2 | 0.01 | 4 |
| 31.1 | Cell organization | <0.01 | 12 | ||
| 33 | Development | 0.05 | 21 | ||
| 33.99 | Development, unspecified | 0.01 | 18 | ||
Table II.
PFAM protein domains enriched in response to PM
Genes encoding proteins with enriched PFAM domains (Finn et al., 2008) are provided in Supplemental Worksheet S5.
| PFAM Domain | Enrichment | Adjusted P | No. in T2 Set | No. in ATH1 |
|---|---|---|---|---|
| ICS1 independent | ||||
| Chitin recognition protein (PF00187) | 16.3 | 3.00E-04 | 6 | 8 |
| Dimerization domain (PF08100) | 9.1 | 5.00E-02 | 5 | 12 |
| Patatin-like phospholipase (PF01734) | 7.8 | 9.00E-02 | 5 | 14 |
| Berberine and berberine-like (PF08031) | 5.8 | 5.00E-02 | 7 | 26 |
| ICS1 impacted | ||||
| Ferritin-like domain (PF00210) | 18.5 | 7.00E-02 | 3 | 4 |
| Haloacid dehalogenase-like hydrolase (PF08282) | 13.7 | 6.00E-03 | 5 | 9 |
| Calreticulin family (PF00262) | 12.3 | 6.00E-02 | 4 | 8 |
| Calcium-binding EGF domain (PF07645) | 12.3 | 6.00E-02 | 4 | 8 |
| UAA transporter family (PF08449) | 11.0 | 7.00E-02 | 4 | 9 |
| Chitinase class I (PF00182)a | 8.2 | 7.00E-02 | 5 | 15 |
| S-locus glycoprotein family (PF00954) | 5.1 | 8.00E-02 | 7 | 34 |
| WRKY DNA-binding domain (PF03106)a | 5.0 | 1.00E-03 | 13 | 64 |
| Domain of unknown function DUF26 (PF01657)a | 4.6 | 2.00E-03 | 13 | 70 |
| LRR N-terminal domain (PF08263)a | 2.8 | 2.00E-03 | 24 | 209 |
| LRR (PF00560)a | 2.7 | 1.00E-07 | 53 | 488 |
| Protein kinase domain (PF00069)a | 2.4 | 1.00E-10 | 86 | 888 |
In both T2 data sets, with values shown for the ics1 versus the wild-type T2 set.
Genes involved in photosynthesis, cell wall modification, secondary metabolism, and protein synthesis and degradation are enriched exclusively in the wild-type data set and thus appear to be predominantly ICS1 independent (Table I). The photosynthesis bin is dominated by light reaction-associated and Calvin cycle-associated genes. Of the 20 photosynthesis-related genes, 19 exhibit decreased temporal expression in response to PM in wild-type plants, with 7 dpi versus UI mean ratios of 0.2 to 0.7. Photosynthesis has been shown to be progressively reduced in virulent PM-infected leaves when compared with uninoculated leaves, with this lower rate of photosynthesis associated with an increase in invertase (e.g. BFRUCT1) activity, an accumulation of hexoses, and down-regulation of photosynthetic gene expression (Swarbrick et al., 2006). The cell wall modification bin includes genes encoding pectin-degrading enzymes (e.g. pectate lyases, pectinesterases, β-galactosidases, and a polygalacturonase) and cell wall-loosening enzymes (e.g. expansins and xyloglucan endotransglycosylases) with reduced expression in response to PM. Down-regulation of sets of genes involved in cell wall modification might be required for maintenance of the haustorial complex or limitation of defense signaling via cell wall components. In contrast, two pectinesterases (At3g90410 and At4g02330) show increased temporal expression in response to infection. These genes might play a role in increasing nutrient availability to the fungus during the growth and reproduction phase. Within protein degradation processes, the ubiquitin E3 bin exhibits enriched altered expression. This is significant as ubiquitin-dependent proteolysis of repressors has been shown to play an important role in phytohormone- and (a)biotic stress-induced responses (Stone and Callis, 2007; Chico et al., 2008). The C3HC4-type RING finger member At4g11370, which exhibits very strong similarity to RHA1a, is present; RHA1 mediates vacuolar trafficking of soluble cargo proteins (Sohn et al., 2003). In addition, ATL6 (At3g05200) and ATL6-like (At5g27420) C3HC4-type RING fingers reside in this bin. LeATL6 appears to contribute to fungal elicitor-activated defense responses in tomato (Solanum lycopersicum; Hondo et al., 2007), and both of these Arabidopsis genes exhibit chitin-induced expression (Libault et al., 2007). Chitin is a fungal wall component that is hydrolyzed by plant chitinases, with chitin oligomers inducing plant defense responses. Loss-of-function mutations in three chitin rapidly responsive genes resulted in increased susceptibility to the PM G. cichoracearum, supporting a role for this plant response pathway in inducing plant defense responses that limit PM growth and reproduction (Ramonell et al., 2005). Indeed, chitin recognition proteins and chitinase protein domains are enriched in our data sets, with these genes exhibiting enhanced expression in response to PM (Table II). In addition, the o-methyl transferase dimerization and berberine domains are both enriched exclusively in the wild-type data set, with all but one gene in each set exhibiting enhanced expression in response to PM (Supplemental Worksheet S5). Enzymes with these domains are involved in the formation of plant alkaloids (Kutchan and Dittrich, 1995; Zubieta et al., 2001) known to inhibit fungal sporulation and growth (Singh et al., 2000). PM-induced changes in alkaloid metabolism are also identified as being statistically significant using MapMan (Table I).
Processes exclusively enriched in the ICS1-impacted data set include redox, carbohydrate metabolism, CK metabolism, vacuolar protein secretion/targeting, stress, and signaling (Table I). Glutaredoxins exhibit statistically significantly altered expression in wild-type plants in response to PM, with 8- and 4.4-fold mean increases in expression at 7 dpi versus UI for At1g03850 and At1g28480, respectively. This increased expression from 1 to 7 dpi is completely abrogated in the ics1 mutant. Glutaredoxins activate thioredoxins such as TRX-H5 (At1g4515; Reichheld et al., 2007), a cytosolic thioredoxin that facilitates the formation of the NPR1 monomer (Tada et al., 2008). We observe PM-induced expression of TRX-H5 with reduced expression in the ics1 mutant as well as similar expression profiles for other genes involved in redox homoeostasis (see ICS1 clusters 1 and 2). As SA is required for the redox-mediated formation of the NPR1 monomer that allows NPR1 to translocate to the nucleus and activate the expression of genes involved in defense, protein targeting, and secretion (Mou et al., 2003), much of the observed transcriptional impact of the ics1 mutation may be through its impact on redox status. Subsets of genes involved in carbohydrate metabolism also display altered expression in the ics1 mutant compared with the wild type. Most of these genes are involved in starch metabolism (e.g. AtSS3) and are predominantly members of ICS1 clusters 4 and 5, exhibiting reduced gene expression in wild-type plants over the time course of infection with less of a reduction in the ics1 mutant. Although negative regulators of CK signaling were down-regulated in response to PM, this was ICS1 independent. However, as shown in Table I, a subset of CK genes with minimal change in expression in response to PM do exhibit slightly altered expression in the ics1 mutant, suggestive of cross talk between SA and CK signaling and response pathways that allows for fine-tuning of phytohormone signaling. These consist of the cytokinin oxidase CKX4, the His-containing phosphotransfer protein AHP5, and the cytokinin His kinase AHK4.
The vacuolar secretion/transport-associated genes impacted by the ics1 mutation include three Atbp80 vacuolar sorting receptors, SEC14 cytosolic factor (At1g22180), and the SEC61β family member At3g60540. These genes exhibit increased expression in response to PM, which is reduced in the ics1 mutant. In parallel, expression of a number of PM-induced defense-associated genes that are targeted to the vacuole, including chitinases and 1,3-β-glucanases, is down-regulated in the ics1 mutant. Our findings support the established role of SA as inducing both expression of plant defensive products and the genes required for the trafficking and secretion of these products (Wang et al., 2005). In addition, we identify additional “novel” genes associated with SA-impacted vacuolar secretion and transport (Supplemental Worksheets S3 and S4).
Furthermore, the ICS1-impacted data set is enriched in genes involved in signaling, including specific classes of receptor kinases, such as leucine-rich repeat (LRR) kinases (Table I). This is mirrored by an enrichment in protein kinase and LRR PFAM domains (Table II). The kinases identified as differentially regulated include mitogen-activated protein kinase cascade components (such as AtMKK1, -2, -4, and -5), LRR kinases, cell wall-associated kinases (e.g. AtWAK1 to -3), and receptor-like kinases (including AtRLK4 to -6). Of the 86 genes with a protein kinase domain identified in the ics1 versus wild-type T2 gene set, 70 also exhibit altered expression in response to the SA analog BTH (Supplemental Worksheet S2). Thus, altered expression of these kinases in the SA ics1 biosynthetic mutant further confirms their role in SA-associated processes; however, their modes of action and functional roles are largely unknown. The two most highly T2-ranked LRR protein kinases in the PM:ICS1-impacted data set are At3g11010 (rank 11) and At1g35710 (rank 20). These kinases, both of which are induced by the SA analog BTH, are induced 6.3- and 4.8-fold, respectively, at 7 dpi with PM compared with 7-d UI, with dramatically reduced expression in the ics1 mutant compared with the wild type (mean 7-dpi expression ratios of 0.1 and 0.2 for ics1 versus the wild type, respectively). In addition, they both exhibit NPR1-dependent expression in response to BTH and are repressed by SN1 (for suppressor of NPR1-inducible; Mosher et al., 2006). ICS1 expression is also repressed by SN1 (Mosher et al., 2006), which leads us to speculate that repression of these LRR kinases may be mediated by SA.
Enriched SA-impacted protein families also include those involved in calcium signaling and iron storage/homeostasis (Table II). Ca2+ plays an important role in mediating SA and hydrogen peroxide signal transduction (Yang and Poovaiah, 2003), and we observe enrichment of the calreticulin and calcium-binding EGF domains specifically in the ICS1-impacted data set. Calreticulin is best known as a high-capacity Ca2+-binding endoplasmic reticulum-resident chaperone involved in calcium homoeostasis, quality control, and proper folding of newly synthesized (glyco)proteins (Gelebart et al., 2005), although it appears to play additional roles in varied locales (Sharma et al., 2004). In plants, direct interactions between calreticulin and the endoplasmic reticulum chaperone BiP and between calreticulin and SNAREs have been reported (Crofts et al., 1998; Laporte et al., 2003). Calcium-binding EFG domains modulate Ca2+-dependent conformational changes, including that of the putative vacuolar sorting receptor PV72 (Watanabe et al., 2002). In addition, calcium mediates the function of the calmodulin-binding MLO compatibility factors (Kim et al., 2002). Further support for a link between calcium signaling and SA in the PM interaction is provided by our previous identification of enriched CGCG box elements in the promoters of genes whose expression increases in response to PM, with decreased expression in the ics1 mutant (Zhang et al., 2008). The CGCG box is recognized by Arabidopsis SR proteins, Ca2+/calmodulin-binding, DNA-binding proteins that are induced in response to a variety of plant phytohormones and stresses (Yang and Poovaiah, 2003).
PFAM ferritin-like domain members are also enriched in the ICS1-dependent data set. Ferritin is the major nonheme iron storage protein in animals, plants, and microorganisms (Theil, 1987, 2003). Iron sequestration, release, and transport are highly integrated and regulated; thus, a decrease in SA production could result in enhanced ferritin expression, as observed in our data set. The sample-to-sample variability in temporal ferritin expression for the ics1 mutant (Supplemental Fig. S2) may result from a threshold-dependent transcriptional response, as is common for iron-regulated pathways (e.g. siderophore biosynthesis) in bacteria (Ratledge and Dover, 2000). Although host limitation of available iron is well defined in mammal-pathogen interactions, our knowledge of its role in plant-pathogen interactions is much more limited (Boughammoura et al., 2007; Johnson, 2008). In response to Blumeria graminis f. sp. tritici PM, it appears that reactive Fe3+ is deposited at wheat cell wall appositions, where it accumulates and mediates the oxidative burst while cytosolic Fe is depleted in response to B. graminis f. sp. tritici (Liu et al., 2007). In that study, ferritin gene expression was down-regulated at 24 hpi, while PR genes were up-regulated (Liu et al., 2007). Our data suggest that SA might be a component of this response in the G. orontii-Arabidopsis system. SA may act by impacting reactive oxygen species status either chemically, via the Fenton reaction, or by binding to and inhibiting antioxidant enzymes such as catalase and ascorbate peroxidase (Dempsey et al., 1999), via its impact on cellular redox status or via the intracellular transport of iron within the cell (Ratledge and Dover, 2000).
cis-Acting Regulatory Elements Associated with Patterns of PM Transcriptional Response
The 1-kb promoters of genes in each wild-type and ICS1 cluster were analyzed using The Arabidopsis Information Resource (TAIR) Statistical Motif Analysis (Rhee et al., 2003) and MDScan (Liu et al., 2002; Supplemental Fig. S4) to identify known and novel cis-acting regulatory motifs. Known motifs strongly enriched in the promoters of genes in wild-type clusters 1 and 2 and ICS1 clusters 1 and 2, which exhibit enhanced expression in response to PM, include the W box, bound by WRKY family transcription factors, the ocs element, bound by OBF transcription factors, and the OBP-1 site, which enhances the ability of OBF proteins to bind ocs elements (Lescot et al., 2002). The ocs and OBP-1 sites were also enriched in the promoters of cluster B genes induced in response to Blumeria graminis f. sp. hordei or G. cichoracearum assessed at 8, 18, and 24 hpi (Zimmerli et al., 2004). Promoters of genes in wild-type cluster 4 and ICS1 cluster 5, which exhibit decreased expression in response to PM, are enriched in the bHLH MYC-binding motif, consistent with previous reports of MYCs mediating Arabidopsis defense (Boter et al., 2004). Promoters of genes in ICS1 cluster 6 are enriched in the GCC box, bound by ERFs, in accordance with our previous analysis using an adaptive model-building procedure (Zhang et al., 2008). Additional significant putative cis-acting regulatory elements of interest include the cell cycle 1b motif, a transactivator in cell cycle-dependent transcription, and an MSA-like motif, a regulatory element involved in cell cycle regulation (Lescot et al., 2002).
Transcriptional Factors Mediating the PM Interaction
As discussed below, there are only five known transcription factors (WRKY18, WRKY40, WRKY70, ATAF1, and AtERF1) mediating Arabidopsis susceptibility to PM. Of these, only two single transcription factor knockouts result in altered PM resistance/susceptibility: wrky70-1 (Li et al., 2006) and ataf1-1 (Jensen et al., 2007). This paucity of known transcription factors impacting PM resistance/susceptibility is likely due to partial functional redundancy and the lack of assessment of PM resistance/susceptibility as part of standard disease characterizations (i.e. compared with Pseudomonas syringae). In addition, in order to resolve the impact of a single knockout on PM resistance/susceptibility, macroscopic whole leaf PM disease scores may be insufficient. Microscopic analyses such as penetration success rate or conidiophores per colony may be required.
We found 47 transcription factors with significantly altered expression in wild-type plants in response to G. orontii and 51 with altered expression in ics1 versus the wild type (Fig. 1B; Table III). Here, we defined transcription factors as genes with a DNA-binding domain, even if a role for that DNA-binding domain has not yet been established. Of the 81 total transcription factors with altered expression in response to PM, 35 are both ICS1 impacted and responsive to BTH, consistent with role for SA in regulating the expression of these genes in planta. Thirty-nine of the 81 transcription factors have not been previously reported to play a role in plant defense or abiotic stress response, as assessed by mutation in or overexpression of the gene. WRKY transcription factors are the dominant family impacted, with several NAC family members, but not ATAF subfamily members, exhibiting altered expression. In accordance with our cis-acting regulatory element analysis, we also identified ERF factors and OBF2. However, no MYC transcription factors or R1R2R3 MYB factors, shown to bind the MSA element (Haga et al., 2007), were identified.
Table III.
PM-impacted transcription factors
Transcription factors are defined as containing a DNA-binding domain (Davuluri et al., 2003; Rhee et al., 2003; Palaniswamy et al., 2006). Altered expression was determined using T2-selected gene sets for the wild type and ics1 versus the wild type. Locus numbers in boldface represent novel transcription factors with no known role in (a)biotic stress, defined by mutation or overexpression of the transcription factor.
| Family/Domain | Wild Type Only | Both | ics1 versus Wild Type Only |
|---|---|---|---|
| NAC | At3g10500, At3g12910 | At2g43000, At2g17040 | At1g01010, At1g25580, At1g34180, At1g69490 |
| AP2 | At1g25560 | ||
| ERF | At4g17500 (ERF1) | At5g47220 (ERF2) | At5g25190 At3g50260 (ERF11) At1g06160 (ORA59) |
| bHLH | At2g18300, At4g01460, At5g15160, At5g46690, At5g39860 (PRE1) At1g73830 (BEE3) | At3g05800 | At4g00050 (UNE10) At3g56980 (ORG3) At5g04150 |
| MYB | At1g18570 (MYB51) | At5g05090 At4g17695 (KAN3) | |
| WRKY | At3g01970 (WRKY45) At2g46400 (WRKY46) At1g62300 (WRKY6) At1g29860 (WRKY71) | At4g31800 (WRKY18) At5g24110 (WRKY30) At2g38470 (WRKY33) At5g22570 (WRKY38) At1g80840 (WRKY40) At4g01720 (WRKY47) At4g23810 (WRKY53) At2g25000 (WRKY60) At5g13080 (WRKY75) | At2g30250 (WRKY25) At2g40750 (WRKY54) At3g01080 (WRKY58) At3g56400 (WRKY70) |
| Zinc fingers | |||
| CCCH-type | At3g55980 At2g40140 (ZFAR1) | At3g21810 | |
| DHHC-type | At1g69420 | ||
| GATA-type | At5g66320 | ||
| C2H2-type | At3g13810 (IDD11) At1g27730 (STZ) At5g59820 (RHL41) At4g17810 | At5g67450 (AZF1) | At3g45260 At3g57480 At2g01650 (PUX2) |
| bZIP | At3g58120 | At2g46270 (GBF3) | |
| CCR4-NOT | At5g22250 | ||
| Homeobox | At4g17460 (HAT1) | At4g16780 | |
| Heat shock factor | At4g36990 (HSF4) | At4g18880, At1g67970 | |
| B3 domain | At1g68840 (RAV2) | At1g13260 (RAV1) | At2g24650, At3g53310 |
| Transcription initiation | At1g75510 (TFII-B) At1g17440 (TAF12B) | ||
| Miscellaneous | At4g17600 (LIL3.1) At1g18330 (EPR1) At1g22590 (AGL87) At5g08330 | At4g14560 (IAA1) At1g75080 (BZR1) At5g63080 At1g07640 (OBP2) At1g33420 At5g13730 (SIG4) |
The WRKY family, consisting of 72 expressed members, was the only statistically enriched transcription factor family domain (Tables I and II), consistent with their role in SA signaling and response to (a)biotic stress (for review, see Ulker and Somssich, 2004). The three WRKYs (WRKY70, WRKY18, and WRKY40) shown to play direct roles in PM resistance are present. All 13 of the WRKYs in the PM:ICS1-impacted gene set are also induced by BTH, consistent with their role in SA-mediated defense responses. Six (WRKY38, WKRY54, WRKY70, WRKY18, WRKY53, and WRKY58) are direct targets of NPR1 (Wang et al., 2006). For each WRKY in the PM:ICS1-impacted gene set, its expression in the ics1 mutant was lower than in the wild-type plants; however, the patterns of expression varied, with subsets of WRKYs exhibiting similar expression profiles (Supplemental Table S1). One could postulate that those WRKY members with similar expression profiles might interact with each other or be partially functionally redundant. For example, we would predict that this might be the case for WRKY54 and WRKY70. Indeed, single and double mutant analyses suggest that WRKY54 and WRKY70 act in concert as positive regulators of SA signaling (Wang et al., 2006), although remarkably, the single wrky70 knockout exhibited altered resistance to PM (Li et al., 2006). In addition, we would predict that WRKY18 and WRKY40 among others might have complementary functions; this is supported by the finding that the wrky18wrky40 double mutant has altered resistance to PM while the single mutants do not exhibit altered resistance at the macroscopic level (Shen et al., 2007). An additional example includes WRKY30 and WRKY75. Although these two WRKYs are members of different subfamilies, they exhibit very similar temporal patterns of expression, with a delayed induction of expression in the wild type that is reduced in the ics1 mutant. Although no role for these WRKYs in plant defense has been reported, WRKY75 and WRKY30 are induced by reactive oxygen species such as hydrogen peroxide and/or superoxide anions (Gechev et al., 2005; Scarpeci et al., 2008). Superoxide generation at the site of penetration has been detected during the establishment of B. graminis f. sp. hordei haustoria in a compatible interaction, but not a resistant interaction (Hückelhoven and Kogel, 1998). As it is also a component of defense signaling pathways, it would be interesting to evaluate single and double mutants in WRKY75 and WRKY30 for altered PM resistance.
A number of ERFs are present in our data set, with AtERF1 and AtERF2 exhibiting increased expression from 1 to 7 dpi in response to G. orontii in wild-type plants, whereas statistically significant differences in expression of the other three ERFs is only observed in the ics1 versus wild-type data set. Expression of AtERF1 and AtERF2 may act to limit the extent of PM infection, as overexpression of AtERF1 and its tomato ortholog PTI4 in Arabidopsis resulted in enhanced resistance to G. orontii (Gu et al., 2002; M.C. Wildermuth, unpublished data). On the other hand, ERFs selected exclusively in the ics1 versus wild-type data set may mediate cross talk between SA and ET/JA. SA-impacted repression of a subset of ET/JA signaling in the later stages of PM infection (1–7 dpi) is suggested by the ICS1-dependent repression of genes known to be regulated by ET/JA, including PDF1.2a (Fig. 3I), PDF1.2b, a chitinase, and the regulators ERF At1g06160 and ORG3, all members of ICS1 cluster 6. As the GCC box motif was the top identified cis-acting regulatory element in promoters of genes in cluster 6, it is likely that an ERF transcription factor regulates this set of genes. ERF At1g06160 is contained within ICS1 cluster 6 and exhibits similar replicate variance to PDF1.2a and other members of this cluster. Comparatively little is known about ERF At1g06160. It is a member of the B-3cii subfamily of ERF transcription factors and is most similar to AtERF15 (At2g31230; Gutterson and Reuber, 2004). In addition, it does not contain the EAR domain associated with ERF repressors (Ohta et al., 2001). In a study examining JA signaling and response, At1g06160 was found to be up-regulated by ERF2 and down-regulated by WRKY33 (Dombrecht et al., 2007). Both ERF2 and WRKY33 are induced by PM, with reduced expression in the ics1 mutant from 3 to 7 dpi. Furthermore, the promoter of ERF At1g06160 contains both the GCC box and W boxes, suggesting that it may be autoregulated and/or regulated by ERF2 and WRKY33. The competing effects of ERF2 and WRKY33 on ERF At1g06160 expression could be one explanation for its replicate variance. Therefore, ERF At1g06160 is a good candidate regulator of our observed SA-dependent repression of a subset of ET/JA-responsive genes.
pux2 Mutants Exhibit Reduced PM Reproduction
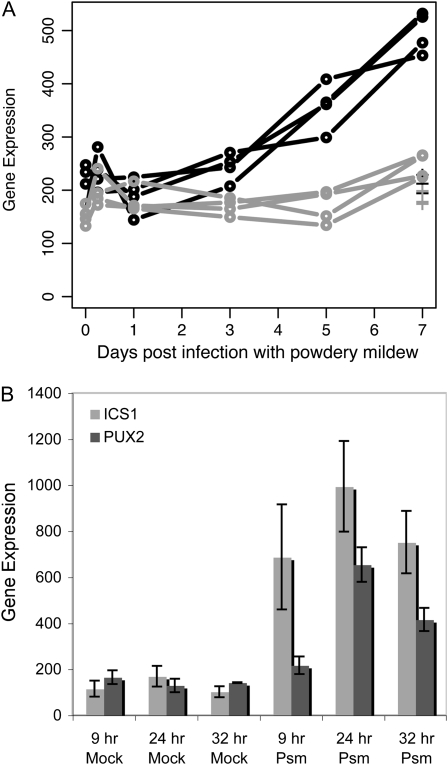
We identified PUX2 (At2g01650), predicted to contain a C2H2 zinc finger DNA-binding domain (Davuluri et al., 2003), as a novel transcriptional regulator with no previously published role in plant defense or abiotic stress response. Although it was exclusively selected in the ics1 versus wild-type T2-data set, PUX2 expression is induced in response to PM in wild-type plants, with 2.35-fold mean induction at 7 dpi/7-d UI (Fig. 4A) and a pattern of expression similar to that of ICS1 (Fig. 3A), although of reduced magnitude. PM-induced PUX2 expression is ICS1 dependent. Consistent with this result, PUX2 expression is also induced by BTH (Supplemental Worksheet S2). A histogram of the frequency of expression over the range of PUX2 expression for the ATH1 GeneChips in the NASCArray database (Craigon et al., 2004) indicates that PUX2 is a lowly expressed gene. The experimental conditions in which PUX2 exhibited maximal expression include infection with Pseudomonas syringae pv maculicola ES4326 (experiment 414). PUX2 expression increases 3.7-fold in response to P. syringae pv maculicola ES3426 in this experiment, paralleling the expression of ICS1 (Fig. 4B).
Figure 4.
PUX2 expression in response to pathogens. A, In response to G. orontii. The wild type is shown in black, and ics1 is shown in gray. Crosses indicate UI 7-d samples. B, In response to P. syringae pv maculicola ES4326 (Psm) or mock treatment (5 mm MgSO4). Data are from the NASCArray database (Craigon et al., 2004), experiment 414 (submitted by R. Mitra and J. Glazebrook).
To determine whether PUX2 plays a role in PM reproduction and growth, we performed macroscopic and microscopic analyses using two independent homozygous T-DNA insertion lines in PUX2. At the macroscopic level, the extent of visible PM infection on mature infected leaves is significantly reduced on the pux2 mutants compared with wild-type plants (Fig. 5A). Microscopic analysis indicates that the number of conidiophores per colony is also significantly decreased in the pux2 mutants compared with the wild type (Fig. 5B). Mutants with enhanced disease resistance to PM often exhibit cell death in the mesophyll cells underlying the infection site (e.g. EDR1 [Frye et al., 2001] and PMR4 [Nishimura et al., 2003]). Therefore, we determined whether macrolesions or microlesions were present in mature leaves of UI or infected pux2 mutant plants using trypan blue staining to identify dead cells. Enhanced cell death was not observed in the pux2 mutants near the site of infection (Fig. 5B) or in UI leaves (data not shown). In addition, we did not observe any enhanced accumulation of autofluorescent compounds (data not shown). These autofluorescent compounds, suggestive of phenolic compounds cross-linked into the cell wall, are often observed at these sites of cell death. Under standard growth conditions, the pux2 mutants are similar in size (assessed via mean largest rosette diameter at 5 weeks) to wild-type plants. In addition, pux2 leaves are similar in color (greenness) to wild-type leaves, indicative of no gross alterations in chlorophyll (or anthocyanin) content. This suggests that the observed PM resistance of the pux2 mutants is not due to a pleiotropic reduction in photosynthetic capacity or growth.
Figure 5.
PUX2 T-DNA insertion mutants exhibit reduced PM reproduction. A, Macroscopic phenotype. Wild-type (Col-0), pux2-1 (SALK_148507), and pux2-2 (SALK_145175) UI whole plants were photographed at 6 weeks. G. orontii-infected leaves were rated and photographed at 11 dpi. Number of plants with disease rating is indicated as described by Reuber et al. (1998) as follows: 0, no visible symptoms of infection; 4, approximately 100% coverage of infected leaves. Bars = 1 cm. B, Microscopic phenotype. Leaves were harvested at 5 dpi and stained with trypan blue for visualization of fungus or cell death. c, Initial germinated conidia; cp, conidiophore. Bars = 100 μm. Independent experiments showed similar results.
PUX2 is one of 15 Arabidopsis plant UBX domain-containing proteins. The UBX domain (PF00789) is present in ubiquitin-regulatory proteins and serves as a general CDC48-interacting module (Bateman et al., 2004). CDC48 is an essential AAA-ATPase chaperone recruited by cofactors/adaptors to modulate cellular pathways, including homotypic fusion of endoplasmic reticulum and Golgi membranes, endoplasmic reticulum-associated protein degradation, cell cycle progression, and apoptosis (Lupas and Martin, 2002; Woodman, 2003). In addition to the C2H2 zinc finger domain and the C-terminal UBX domain, PUX2 contains a PUB domain (PF09409) located between these two other domains. The PUB domain is also known to function as an AAA-ATPase-binding domain (Bateman et al., 2004). Although none of the other Arabidopsis PUX proteins share this domain architecture, we found PUX2 orthologs in a number of other plants, including rice (Oryza sativa; accession no. NM_00106957.1; E value = 2e-84), grape (Vitis vinifera; accession no. AM_481795.2; E value = 3e-71), lotus (Lotus corniculatus; accession no. AP004536.1; E value = 1e-71), and Medicago truncatula (accession no. AC150979.15; E value = 2e-68), suggesting conserved function preceding the divergence of monocots and dicots. Little is known about PUX2. PUX1, the most similar Arabidopsis PUX, with 29% identity (45% similarity) over the full-length protein, acts as a negative regulator of Arabidopsis CDC48 by promoting disassembly of the AAA-ATPase hexamer (Rancour et al., 2004; Park et al., 2007). PUX2 can also interact with Arabidopsis CDC48 in vitro and, like PUX1, can be detected as part of a complex with CDC48 and the syntaxin SYP31 (Rancour et al., 2004). However, the top 300 genes exhibiting correlated expression with PUX1 using curated publicly available data (Obayashi et al., 2009) are very different from PUX2, with an overlap of only three genes. This suggests that PUX1 and PUX2 play distinct roles. The dominant CDC48 isoform CDC48a (At3g09840) is in the top 300 genes correlated with PUX1. However, neither it nor the other two CDC48 isoforms are present in the top 300 genes correlated with PUX2. Furthermore, PUX1 and the CDC48 isoforms do not exhibit significantly altered expression in our T2 data sets. On the other hand, of the 300 top genes correlated with PUX2 (Obayashi et al., 2009), 136 also exhibit altered expression in our data sets. These genes include an AAA-ATPase (At2g46620), genes involved in ubiquitin-mediated processes, including At1g19270, SUM3 (At5g55170), and the F box protein At5g18780, the cell death inhibitor AtBI-1, and numerous genes mediating redox status, including TRX2 and TRX5.
PUX2 appears to act as a negative regulator mediating the PM-Arabidopsis interaction, with the enhanced resistance observed in the pux2 mutants being independent of cell death. The pmr5 and pmr6 mutants also exhibit enhanced resistance to a virulent PM without macroscopic cell death or PM-induced cell death at the site of infection (Vogel et al., 2002, 2004). However, unlike the pmr5 and pmr6 mutants, pux2 mutants do not exhibit microlesions along veins in a subset of the oldest leaves, nor do they exhibit differences in leaf size or shape compared with wild-type plants. In addition, the pux2 phenotype is less dramatic than those of the pmr5 and pmr6 mutants, which exhibit a 10-fold reduction in conidiophores per germinated conidium (Vogel et al., 2002, 2004). Finally, in contrast to PUX2, we did not observe altered PMR5 or PMR6 expression in response to PM. While PMR5 and PMR6 appear to be PM compatibility factors, with resistance in the pmr5 and pmr6 mutants limited to PM pathogens, we do not yet know whether the resistance conferred by insertion mutations in PUX2 is specific to PMs. Further characterization of the pux2 mutants, including assessing disease resistance in response to a diverse suite of pathogens and dissection of the mechanism of action of PUX2 in PM resistance, is in progress.
In general, understanding mechanisms limiting the extent of growth and reproduction of virulent PMs in Arabidopsis could allow for a multipronged approach to limiting the growth and reproduction of PM in important agricultural and ornamental species, including grape, strawberry (Fragaria species), barley (Hordeum vulgare), and roses (Rosa species). This is particularly true for a gene like PUX2 that (1) has orthologs in species including grape and rice and (2) has no apparent yield penalty associated with enhanced PM resistance, unlike many PM-resistant mutants. By limiting PM reproduction, not only is the initial infection limited but also fewer conidia are available for subsequent infection of other leaves and plants. Targeting a set of such host genes could result in dramatically reduced PM growth and reproduction that is less likely to be rapidly overcome by pathogen counterevolution than would be the case if resistance were conferred by a dominant resistance gene.
MATERIALS AND METHODS
Plant Growth and PM Infection for Microarray
To minimize plant-to-plant variation, Arabidopsis (Arabidopsis thaliana ecotype Col-0; wild type) and the ics1-2 (eds16-1) mutant in the Col-0 background (Dewdney et al., 2000; Wildermuth et al., 2001) were grown using a template to ensure even spacing of plants within each box (of nine plants) and using a design that alternates the placement of wild-type and mutant plants. Seeds were stratified in 0.1% agarose for 3 to 5 d to facilitate and synchronize germination. Flats were bottom watered and fertilized (Hoagland's solution at 1/4×) once at 2.5 weeks (Metromix 200; Scotts Sierra Horticultural Products). Plants were grown in Percival AR66L growth chambers at 22°C, 80% relative humidity, and 14-h photoperiod from 6 am to 8 pm with photosynthetically active radiation of 150 μE m−2 s−1. At 4 weeks, a subset of boxes was infected with Golovinomyces orontii (MGH isolate; Plotnikova et al., 1998) using a moderately heavy inoculum (conidia from two to three fully infected leaves at 10–14 dpi per box) using a settling tower as described by Reuber et al. (1998). PM infections were performed using a settling tower and mesh screen for each box to enhance reproducibility from experiment to experiment and to minimize plant-to-plant variation. Boxes containing G. orontii-infected plants were placed in a separate Percival AR66L growth chamber under the same conditions as those used for UI samples. Infections were performed at 2:30 pm to standardize experiments and minimize circadian effects. Samples were harvested at 0 dpi (just prior to infection), 6 hpi, 1 dpi, 3 dpi, 5 dpi, and 7 dpi. A 7-d UI sample was also collected. With the exception of the 6-hpi time point, all samples were harvested at 2:30 pm. For each sample, three to four fully expanded mature leaves were excised per plant, with leaves from two randomly selected plants (approximately 200 mg fresh weight total) used to prepare a single RNA sample; triplicate samples (designated -1, -2, and -3) were collected for each data point. Plants were not resampled once used. Leaves were immediately frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. Three independent experiments (designated 1, 2, and 3) were performed. At 7 dpi, symptoms of infection were visible on leaves of wild-type Col-0 plants, with enhanced symptoms for the ics1-2 mutant.
Tissue Processing, Hybridization, and Scanning
Total RNA was extracted from each sample using the RNeasy Plant RNA Miniprep kit (Qiagen); samples were split in two before homogenization and repooled before loading on the RNA-binding column. RNA quality was assessed by determining the A260/A280 ratio of RNA in Tris buffer and by checking the integrity of RNA on an Agilent 2100 Bioanalyzer (Agilent Technologies). Target labeling and microarray hybridizations to Affymetrix Arabidopsis ATH1 GeneChips were performed according to the protocol given in the Affymetrix GeneChip Expression Analysis Technical Manual 701025 rev 1. Arrays were scanned using an Affymetrix GeneArray 2500 Scanner at the Harvard University Bauer Center for Genomics Research.
Data Preprocessing
Expression values (log2) were extracted using robust multiarray analysis (Irizarry et al., 2003a, 2003b) implemented in the function rma() in the Bioconductor software package affy (Gautier et al., 2004). Background correction was performed by correcting perfect match only, to reduce noise from the signal intensity. To make different experiments comparable, quantile normalization was performed to make the distributions of signal intensities uniform across chips (Bolstad et al., 2003). Quality assessment was done using the weights outputted from the linear model-fitting function fitPLM() in the R package AffyExtensions. One sample set was processed from each experiment, with experiment 1 denoted 1-3, experiment 2 denoted 2-1, and experiment 3 denoted 3-1. In addition, to compare the variation in gene expression from samples in one experiment with that between different experiments, we processed an additional sample from experiment 2, denoted 2-2. Therefore, in total, 56 ATH1 GeneChips that passed our quality assessment were utilized. As an additional control, we performed a technical replicate (same complementary RNA target, separate hybridizations to ATH1 GeneChips) for one sample (experiment 2-2, Col-0, 0 dpi); signal intensity was highly correlated (weighted value = 0.99).
To determine whether any experimental data point was an outlier, we assessed individual probe-level plots and time profile plots for the four replicates. For each probe set, the median log2 intensity of the four wild-type or mutant replicates at each time point was calculated and subtracted from the log2 intensities of these four replicate samples, yielding log2 ratios of these four replicates relative to their median. Box plots for these log ratios were plotted for the four replicates for comparison. This analysis was performed using all probe sets and T2-selected probe sets (1,470 probe sets; see below). The Col-0 3-dpi data point from experiment 1 was the only outlier; thus, for this data point only, we estimated the log2 expression value at this single time point from all other Col-0 time point samples using the median polish algorithm, implemented in the function medpolish() in the software R (R Development Core Team, 2005), and we removed this sample when performing clustering. We also performed pair-wise correlation analyses for each time point (the wild type and ics1 mutant) for the 1-3, 2-1, 2-2, and 3-1 samples, excluding the 1-3 wild-type 3-dpi sample. For all genes, the correlation of gene expression between the 2-1 and 2-2 data sets (Pearson's r = 0.99 ± 0.003) was similar to that for all paired data sets (Pearson's r = 0.99 ± 0.004). This was also the case for T2-selected genes (1,470 probe sets; see below) with Pearson's r = 0.97 ± 0.01 for the 2-1 versus 2-2 data sets and r = 0.96 ± 0.02 for all paired data set comparisons. As our analysis indicated that both intraexperimental and interexperimental correlations were similar and very high, for our statistical analyses below, we treated the 1-3, 2-1, 2-2, and 3-1 data sets as independent biological replicates and describe our data set as composed of four biological replicates for simplicity.
Gene Selection by T2 Statistic
Following the preprocessing step described above, genes of interest were selected using these background-adjusted, normalized log2 intensities. The calculation of the T2 statistics (Tai and Speed, 2006) was performed using the mb.long() function in the R software package time course with the default parameters (Tai, 2007). For the ics1 versus wild-type analysis, four biological replicates for paired ics1 and wild-type samples at 0, 0.25, 1, 3, 5, and 7 dpi were utilized. For the wild-type data, wild-type samples at 0, 1, 3, 5, and 7 dpi were utilized in the T2 analysis. The 0.25-dpi sample was not included, so as not to introduce possible circadian effects on expression. The ATH1 probe sets were then ordered by their T2 statistics, and genes were selected based on a log10(T2) cutoff of 2.5. This cutoff was chosen as it included genes of biological interest near the selection cutoff, such as NPR1 and RIN4, that exhibit clear differences in temporal gene expression and a false discovery rate of less than 5%. For the wild-type data, the 7-d UI data were used to cull T2-selected genes without an appreciable difference in expression at 7 dpi versus 7 UI, as appropriate given the observed pattern of expression.
Validation of ATH1 Gene Expression Data by qPCR
RNA from wild-type and ics1 mutant samples was treated with Ambion DNA-free, and first-strand cDNA was synthesized using iScript reverse transcriptase (Bio-Rad) according to the manufacturer's instructions. Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa RR041A) in an ABI 7300 instrument according to the manufacturer's instructions. Three technical replicates were performed for each sample analyzed. Following activation of Taq polymerase for 10 s at 95°C, 40 cycles of amplification were performed consisting of denaturation for 5 s at 95°C followed by annealing at 60°C for 31 s. The absence of primer-dimer formation was confirmed by performing a melting curve. The specificity of the product was confirmed by analyzing the amplified product on a 2% agarose gel and sequencing the amplified products from one well. Raw fluorescence data were converted into Ro values using DART-PCR 1.0 (Peirson et al., 2003), and expression of analyzed genes was given relative to UBQ5 Ro values.
Primers were designed using Amplify (http://engels.genetics.wisc.edu/amplify). The specificity of primers was verified by performing National Center for Biotechnology Information BLAST searches and using Amplify to test the ability of primer combinations to amplify closely related gene family members. Primers used in amplifying ICS1 (At1g74710), PR1 (At2g14610), PR2 (At3g57260), PAD4 (At3g52430), NPR1 (At1g64280), the lupeol synthase At1g66960, PDF1.2 (At5g44420), ERF At1g06160, ACA2 (At4g37640), PUX2 (At2g01650), and UBQ5 (At3g62250) are shown in Supplemental Table S2. All primer sets used showed more than 95% efficiency when the slopes of their amplification plots were analyzed using DART-PCR software.
Analysis of Expression Data
TAIR8 (April 2008 update) gene functional descriptions were employed (Rhee et al., 2003). MapMan was used to identify enriched processes in the T2-ranked data sets (Thimm et al., 2004). Enriched PFAM (Bateman et al., 2004; Finn et al., 2008) domains (PFAM version 22.0) were determined for nucleus-encoded genes in the T2 data sets versus the ATH1 GeneChip, and P values were computed using a hypergeometric distribution. The resulting P values were adjusted for multiple hypothesis testing as described by Benjamini and Hochberg (1995), with the false discovery rate controlled to be less than 10%. We wrote Python scripts to parse the PFAM domain data and developed code in R to perform the enrichment analysis (provided in Supplemental Text S1).
Clustering of the top T2-ranked probe sets was performed using Partek Genomics Suite (Partek). For ICS1 clusters, hierarchical clustering with Euclidian distances was performed, with manual curation, using paired replicate samples for 0 dpi (UI), 6 hpi, 1 dpi, 3 dpi, 5 dpi, and 7 dpi for wild-type and ics1 plants. For wild-type clusters, partition clustering with subclustering using Max Value was optimal. Wild-type clustering used replicate samples for 0 dpi (UI), 1 dpi, 3 dpi, 5 dpi, 7 dpi, and 7-d UI, excluding the experiment 1-3 7-d UI sample. The 6-h time point was excluded to minimize circadian effects. cis-Acting regulatory element analysis was performed on 1-kb sequences upstream of the start codon using TAIR Statistical Motif Analysis (Rhee et al., 2003) and on 1-kb upstream sequences, downloaded from TAIR8, for MDScan with a motif width of 6 (Liu et al., 2002).
pux2 Genotyping
Seeds for pux2-1 (SALK_148507) and pux2-2 (SALK_145175) T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center at Ohio State University (Alonso et al., 2003). To obtain homozygous lines for the pux2 mutants, genomic DNA from pux2-1 and pux2-2 leaves was used for PCR-based genotyping. The left and right primers for pux2-1 were 5′-AATCAAAATTATTGCGCAACG-3′ and 5′-TCATCCGCCGAGAGACTATAG-3′, and those for pux2-2 were 5′-CAGTCTCGCCTGTGAGATCTC-3′ and 5′-CGGAGTCACAGCTTTTGATTC-3′, respectively. The left border primer (LBb1.3) for T-DNA was 5′-ATTTTGCCGATTTCGGAAC-3′. Homozygous pux2-1 and pux2-2 lines were used in all experiments.
pux2 Growth Conditions, PM Inoculations, and Microscopy
Arabidopsis Col-0 and homozygous pux2-1 and pux2-2 plants were grown in Metromix 200 in a Percival AR66L growth chamber at 22°C, 80% relative humidity, and 12-h photoperiod with photosynthetically active radiation of 180 μE m−2 s−1. For each box, a template was used to provide even spacing of plants with Col-0 and pux2 mutants alternated. Four-week-old plants were infected with a moderate inoculum of G. orontii (two half-infected leaves with 10- to 14-dpi conidia per box) using a settling tower. A total of 12 mature, fully expanded leaves from four plants were scored at 11 dpi. Whole leaf PM scores were assessed as described (Reuber et al., 1998). To visualize fungal structures and cell death, infected leaves (5 dpi) were cleared in 95% ethanol for 2 h, stained with 250 μg mL−1 trypan blue in a solution of lactic acid, glycerol, and water (1:1:1:1) for 15 min, and mounted in 50% glycerol. For cell death, an additional step, overnight clearing in chloral hydrate (2.5 g dissolved in 1 mL of water), was used to minimize background staining. Well-separated fungal colonies were chosen for analysis, and the number of conidiophores was counted per colony. A QImaging camera attached to a Zeiss Axioimager microscope was used for photographs. All experiments were repeated at least twice with similar results, with results shown for one experiment.
The ATH1 GeneChip expression data are deposited in Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (GEO accession no. GSE13739).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Wild-type T2 gene expression plots.
Supplemental Figure S2. ics1 versus wild-type T2 gene expression plots.
Supplemental Figure S3. qPCR plots.
Supplemental Figure S4. cis-Acting RE analysis: TAIR hexamer and MDScan.
Supplemental Table S1. WRKY table.
Supplemental Table S2. qPCR primers.
Supplemental Text S1. Code in R used to perform PFAM enrichment analysis.
Supplemental Worksheet S1. T2-selected probe sets with T2 values, cluster designation, and gene description: sheet 1 = the wild type and sheet 2 = ics1 versus the wild type.
Supplemental Worksheet S2. Venn diagram subsets.
Supplemental Worksheet S3. MapMan analysis: wild-type genes in enriched bins.
Supplemental Worksheet S4. MapMan analysis: ics1 versus wild-type genes in enriched bins.
Supplemental Worksheet S5. PFAM analysis: genes containing enriched PFAM domains.
Supplementary Material
Acknowledgments
We thank Dr. Shauna Somerville and Greg Wiberg for comments on the manuscript.
This work was supported by the National Science Foundation (Arabidopsis 2010 grant nos. MCB–0420267 to M.C.W. and T.P.S. and DBI–0114783 to X. Dong, S. Somerville, and F.M.A.) and the Winkler Family Foundation (grant to M.C.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mary C. Wildermuth (wildermuth@nature.berkeley.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al (2004) The Pfam protein families database. Nucleic Acids Res 32 D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B Methodological 57 289–300 [Google Scholar]
- Blanco F, Garreton V, Frey N, Dominguez C, Perez-Acle T, Van der Straeten D, Jordana X, Holuigue L (2005) Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59 927–944 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193 [DOI] [PubMed] [Google Scholar]
- Boter M, Ruiz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughammoura A, Franza T, Dellagi A, Roux C, Matzanke-Markstein B, Expert D (2007) Ferritins, bacterial virulence and plant defence. Biometals 20 347–353 [DOI] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 11 486–494 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977 [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, et al (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38 716–720 [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Leborgne-Castel N, Pesca M, Vitale A, Denecke J (1998) BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell 10 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DMA, Shah J, Klessig DF (1999) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18 547–575 [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24 205–218 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51 21–37 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Fabro G, Di Rienzo JA, Voigt CA, Sevchenko T, Dehesh K, Somerville S, Alvarez ME (2008) Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol 146 1421–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al (2008) The Pfam protein families database. Nucleic Acids Res 36 D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 307–315 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J (2005) Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57 181–188 [DOI] [PubMed] [Google Scholar]
- Gelebart P, Opas M, Michalak M (2005) Calreticulin, a Ca2+-binding chaperone of the endoplasmic reticulum. Int J Biochem Cell Biol 37 260–266 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7 465–471 [DOI] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Voss U, Jurgens G, et al (2007) R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134 1101–1110 [DOI] [PubMed] [Google Scholar]
- Herrera JB, Bartel B, Wilson WK, Matsuda SP (1998) Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry 49 1905–1911 [DOI] [PubMed] [Google Scholar]
- Hondo D, Hase S, Kanayama Y, Yoshikawa N, Takenaka S, Takahashi H (2007) The LeATL6-associated ubiquitin/proteasome system may contribute to fungal elicitor-activated defense response via the jasmonic acid-dependent signaling pathway in tomato. Mol Plant Microbe Interact 20 72–81 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Dechert C, Kogel KH (2003) Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. Proc Natl Acad Sci USA 100 5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Kogel KH (1998) Tissue-specific superoxide generation at interaction sites in resistant and susceptible near-isogenic barley lines attacked by the powdery mildew fungus (Erysiphe graminis f. sp. hordei). Mol Plant Microbe Interact 11 292–300 [Google Scholar]
- Husselstein-Muller T, Schaller H, Benveniste P (2001) Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol Biol 45 75–92 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, Joehnk N, Lyngkjaer MF, Collinge DB (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65 137–150 [DOI] [PubMed] [Google Scholar]
- Johnson L (2008) Iron and siderophores in fungal-host interactions. Mycol Res 112 170–183 [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M, Ohori Y, Uchimiya H (2004) Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416 447–451 [DOI] [PubMed] [Google Scholar]
- Kutchan TM, Dittrich H (1995) Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem 270 24475–24481 [DOI] [PubMed] [Google Scholar]
- Laporte C, Vetter G, Loudes AM, Robinson DG, Hillmer S, Stussi-Garaud C, Ritzenthaler C (2003) Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15 2058–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46 477–491 [DOI] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20 900–911 [DOI] [PubMed] [Google Scholar]
- Liu G, Greenshields DL, Sammynaiken R, Hirji RN, Selvaraj G, Wei Y (2007) Targeted alterations in iron homeostasis underlie plant defense responses. J Cell Sci 120 596–605 [DOI] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20 835–839 [DOI] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12 746–753 [DOI] [PubMed] [Google Scholar]
- Micali C, Göllner K, Humphry M, Consonni C, Panstruga R (2008) The powdery mildew disease of Arabidopsis: a paradigm for the interaction between plants and biotrophic fungi. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Muller B, Sheen J (2007) Arabidopsis cytokinin signaling pathway. Sci STKE 2007 cm5. [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37 D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Suzuki M, Masuda K, Yoshida S, Muranaka T (2007) Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem Pharm Bull (Tokyo) 55 1518–1521 [DOI] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY (2007) Protein domain-domain interactions and requirements for the negative regulation of Arabidopsis CDC48/p97 by the plant ubiquitin regulatory X (UBX) domain-containing protein, PUX1. J Biol Chem 282 5217–5224 [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31 e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova JM, Reuber TL, Ausubel FM (1998) Powdery mildew pathogenesis of Arabidopsis thaliana. Mycologia 90 1009–1016 [Google Scholar]
- R Development Core Team (2005) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Park S, Knight SD, Bednarek SY (2004) Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. J Biol Chem 279 54264–54274 [DOI] [PubMed] [Google Scholar]
- Ratledge C, Dover LG (2000) Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54 881–941 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J 16 473–485 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM (2008) Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol 66 361–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6 365–371 [DOI] [PubMed] [Google Scholar]
- Sharma A, Isogai M, Yamamoto T, Sakaguchi K, Hashimoto J, Komatsu S (2004) A novel interaction between calreticulin and ubiquitin-like nuclear protein in rice. Plant Cell Physiol 45 684–692 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103 [DOI] [PubMed] [Google Scholar]
- Singh UP, Sarma BK, Mishra PK, Ray AB (2000) Antifungal activity of venenatine, an indole alkaloid isolated from Alstonia venenata. Folia Microbiol (Praha) 45 173–176 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Callis J (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10 624–632 [DOI] [PubMed] [Google Scholar]
- Swarbrick PJ, Schulze-Lefert P, Scholes JD (2006) Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ 29 1061–1076 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai YC (2007) Timecourse: statistical analysis for developmental microarray time course data. In R Package Version 1.10.0. University of California, Berkeley, CA
- Tai YC, Speed TP (2006) A multivariate empirical Bayes statistic for replicated microarray time course data. Annals of Statistics 34 2387–2412 [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW (2006) A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J 47 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil EC (1987) Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem 56 289–315 [DOI] [PubMed] [Google Scholar]
- Theil EC (2003) Ferritin: at the crossroads of iron and oxygen metabolism. J Nutr 133 1549S–1553S [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40 968–978 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, Vogel J, Somerville C, Somerville S (2007) EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol 7 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DR, McRoberts N (2006) Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci 11 581–586 [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 1036–1040 [DOI] [PubMed] [Google Scholar]
- Watanabe E, Shimada T, Kuroyanagi M, Nishimura M, Hara-Nishimura I (2002) Calcium-mediated association of a putative vacuolar sorting receptor PV72 with a propeptide of 2S albumin. J Biol Chem 277 8708–8715 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565 [DOI] [PubMed] [Google Scholar]
- Woodman PG (2003) p97, a protein coping with multiple identities. J Cell Sci 116 4283–4290 [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291 118–120 [DOI] [PubMed] [Google Scholar]
- Xiao S, Emerson B, Ratanasut K, Patrick E, O'Neill C, Bancroft I, Turner JG (2004) Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol Biol Evol 21 1661–1672 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8 505–512 [DOI] [PubMed] [Google Scholar]
- Zhang NR, Wildermuth MC, Speed TP (2008) Transcription factor binding site prediction with multivariate gene expression data. Ann Appl Stat 2 332–365 [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40 633–646 [DOI] [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8 271–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.