Abstract
While the presence of a complete shikimate pathway within plant plastids is definitively established, the existence of a cytosolic postchorismate portion of the pathway is still debated. This question is alimented by the presence of a chorismate mutase (CM) within the cytosol. Until now, the only known destiny of prephenate, the product of CM, is incorporation into tyrosine (Tyr) and/or phenylalanine (Phe). Therefore, the presence of a cytosolic CM suggests that enzymes involved downstream of CM in Tyr or Phe biosynthesis could be present within the cytosol of plant cells. It was thus of particular interest to clarify the subcellular localization of arogenate dehydrogenases (TYRAs) and arogenate dehydratases (ADTs), which catalyze the ultimate steps in Tyr and Phe biosynthesis, respectively. The aim of this study was to address this question in Arabidopsis (Arabidopsis thaliana) by analysis of the subcellular localization of the two TYRAAts and the six AtADTs. This article excludes the occurrence of a spliced TYRAAt1 transcript encoding a cytosolic TYRA protein. Transient expression analyses of TYRA- and ADT-green fluorescent protein fusions reveal that the two Arabidopsis TYRA proteins and the six ADT proteins are all targeted within the plastid. Accordingly, TYRA and ADT proteins were both immunodetected in the chloroplast soluble protein fraction (stroma) of Arabidopsis. No TYRA or ADT proteins were immunodetected in the cytosol of Arabidopsis cells. Taken together, all our data exclude the possibility of Tyr and/or Phe synthesis within the cytosol, at least in green leaves and Arabidopsis cultured cells.
A complete plastid-localized biosynthesis pathway of the three aromatic amino acids has been clearly demonstrated (Bickel et al., 1978; Schulze-Siebert et al., 1984). However, among all of the enzymes involved, chorismate mutase (CM; Fig. 1), which catalyzes the first committed step in the Phe and Tyr biosynthesis pathway, is the only one known to be present in both the plastid and the cytosol. The presence of a cytosolic form of CM has been clearly detected both at the enzymatic and molecular levels in several plant species, such as Arabidopsis (Arabidopsis thaliana), Nicotiana sylvestris, and Papaver somniferum (Benesova and Bode, 1992; d'Amato et al., 1992; Eberhard et al., 1996). The physiological significance of this cytosolic CM is still an enigma. In contrast to the two plastid isoforms (AtCM1 and AtCM3), the cytosolic CM (AtCM2) is not elicitor and pathogen inducible and is not allosterically regulated by Trp, Phe, and Tyr (Eberhard et al., 1996). The contribution of the cytosolic CM to Tyr and/or Phe and phenylpropanoids thus remained questionable. Nevertheless, to date, the only known destiny of prephenate, the product of the CM reaction, is incorporation into Phe or Tyr. Therefore, if this cytosolic CM participates in the cytosolic postchorismate biosynthesis of Tyr and/or Phe, other enzymes involved in downstream Tyr or Phe biosynthesis would also be present within the cytosol.
Figure 1.
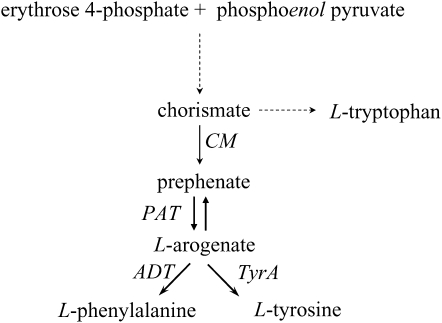
Biosynthesis pathway leading to Trp, Tyr, and Phe. Abbreviation not defined in the text: PAT, prephenate aminotransferase.
Whereas the upper part of this pathway, from 3-deoxy-d-arabino-heptulosonate-7-P synthase to CM, has been extensively studied, both at the molecular and the biochemical level (for review, see Schmid and Amrhein, 1995, 1999; Herrmann and Weaver, 1999), its postchorismate branch, from prephenate to Tyr or Phe, has received much less attention. In most plants, prephenate is first transaminated into arogenate by an aromatic amino acid transaminase and is then decarboxylated into Tyr by arogenate dehydrogenase (TYRA; Hall et al., 1982) or into Phe by arogenate dehydratase (ADT; Jung et al., 1986; Siehl and Conn, 1988; Cho et al., 2007; Yamada et al., 2008; Fig. 1). In the genome of the model plant Arabidopsis, two genes encoding TYRA proteins were identified and named TYRAAt1 and TYRAAt2 (AGI nos. At5g34930 and At1g15710, respectively; Rippert and Matringe, 2002a, 2002b). Both proteins possess a putative plastid transit peptide. However, TYRAAt1, the product of the gene TYRAAt1 (At5g34930), has a very peculiar structure because it is constituted by three exons encoding a single polypeptide chain housing two highly similar TYRA domains (TYRAAt1-D1 and TYRAAt1-D2; Fig. 2A; Supplemental Fig. S1). We previously shown that separate overexpression of each of the two domains in Escherichia coli sustained TYRA activity with catalytic properties very similar to those of the entire TYRAAt1 protein (Rippert and Matringe, 2002a). The second isoform, TYRAAt2, the product of the gene TYRAAt2, is composed of only one TYRA protein domain (Rippert and Matringe, 2002a, 2002b).
Figure 2.
Analysis of the TYRAAt1 transcript. A, Schematic representation of the intron/exon organization of the TYRAAt1 gene. Position of primers used in this study is shown. B, Northern-blot analysis of TYRAAt1 transcript in young Arabidopsis rosette leaves. Total RNAs were isolated from young Arabidopsis rosette leaves using the RNeasy plant mini kit isolation system according to the manufacturer's instructions (Qiagen). Total RNAs (10 μg) were denatured for 1 h at 50°C in 10 mm NaH2PO4 (pH 7), 2% (v/v) dimethyl sulfoxide, 1.08 m glyoxal, and separated by 1% (w/v) agarose gel electrophoresis. It was then transferred to nylon membrane (Nitran). The resulting blot was subjected to hybridization with the corresponding 32P-labeled DNA probes matching exon 3 of the Arabidopsis TYRAAt1 gene. A single transcript of 1.9 kb corresponding to the complete TYRAAt1 mRNA was hybridized.
Analyses of available genomic databases reveal the presence of an ortholog of TYRAAt1 in other plant species. Indeed, sequence similarity searching using National Center for Biotechnology Information (NCBI) BLAST allowed us to identify genomic regions in Medicago truncatula (AC151744; region 56,869–59,500) and Lotus corniculatus (AP006375; region 47,076–49,436), respectively, which encode putative TYRA proteins with two highly similar TYRA catalytic domains (Supplemental Fig. S1). These two putative TYRA genes are formed by two large exons, each of them encoding an open reading frame (ORF) corresponding to one of the two catalytic domains of these two TYRA proteins. The Arabidopsis TYRAAt1 gene (At5g34930) presents the same organization, except for the presence of an additional very short 29-bp exon at the 5′-terminus (Fig. 2A; Supplemental Fig. S1). In addition, a partial genomic sequence from Brassica oleracea (BH495674), encoding a polypeptide sequence encompassing two putative TYRA catalytic domains, was also identified (Supplemental Fig. S1). This partial genomic region is apparently constituted by only one exon encoding both protein domains. In contrast, analysis of the rice (Oryza sativa) genome database revealed the presence of three putative TYRA isoforms, all of which have only one catalytic domain. The presence of putative orthologs of the TYRAAt1 gene in several dicotyledonous species raises the possibility that the function of this unusual organization is to generate a TYRA protein devoid of plastid transit peptide. This could occur either by alternative splicing of exon 1 and/or exon 2 or by proteolytic cleavage of the entire TYRAAt1 protein between its two catalytic domains.
Concerning the last step of Phe biosynthesis, which in most plants is catalyzed by ADT (Jung et al., 1986; Siehl and Conn, 1988; Cho et al., 2007; Yamada et al., 2008), not less than six genes encoding ADT were identified in the Arabidopsis genome (ADT1 [At1g11790]; ADT2 [At3g07630]; ADT3 [At2g27820]; ADT4 [At3g44720]; ADT5 [At5g22630]; and ADT6 [At1g08250]; Ehlting et al., 2005; Cho et al., 2007). They all present an N-terminal extension when compared with bacterial enzymes. However, one of them (ADT1) presents an N-terminal extension that was not clearly recognized as putative plastid transit peptide by protein-targeting prediction software like ChloroP (http://www.cbs.dtu.dk/services/ChloroP) or TargetP (http://www.cbs.dtu.dk/services/TargetP). ADT1 could then be a cytosolic protein.
It is thus of great importance to clarify the subcellular localization of TYRA and ADT in plants. This study aimed to address this question by analyzing the possible presence of TYRA and/or ADT proteins within the cytosol of Arabidopsis cells. The expression patterns of the two TYRA genes and the six ADT genes in different organs were also analyzed by means of quantitative real-time PCR experiments to gain insight into their physiological functions in planta.
RESULTS AND DISCUSSION
TYRAAt1 mRNA Is Not Modified by a Posttranscriptional Mechanism
In contrast to TYRAAt2, which is formed by only one exon, TYRAAt1 contains three exons, encoding two TYRA catalytic domains (Fig. 2A). This particular structure opens the possibility of generating a cytosolic TYRA protein. Indeed, three potential ATG initiation codons in-frame with the TYRAAt1 ORF are present in the 5′-region of TYRAAt1; the first one (ATG1) in the first exon, the second one (ATG2) in the first intron, and the third one (ATG3) at the beginning of the second exon (Fig. 2A). Prediction software ChloroP (http://www.cbs.dtu.dk/services/ChloroP) or TargetP (http://www.cbs.dtu.dk/services/TargetP) reveal that the first two ATG codons, ATG1 and ATG2, would initiate TYRAAT1 ORFs containing putative plastid transit peptide, but (ATG3) would initiate a TYRAAt1 protein devoid of plastid transit peptide. Identification of the TYRAAt1 transcription start site was carried out by 5′-RACE experiments. All of the clones obtained by RACE included exon 1, confirming that ATG1 was the sole transcription start of TYRAAt1. Splicing of exon 2 could in theory initiate a TYRAAt1 transcript encoding a TYRA protein corresponding to TYRAAt1-D2 devoid of plastid transit peptide (Fig. 2A). However, no in-frame ATG codon, initiating this TYRAAt1-D2 protein by a splicing event respecting the GT/AG rules of intron borders, could be identified in the nucleotide sequence of the TYRAAt1 gene. Absence of exon 2 splicing events was confirmed by 5′-RACE and northern-blot experiments. All of the clones obtained by RACE using a reverse primer matching exon 3 revealed correct splicing of the second intron and the presence of exon 2. Accordingly, northern-blot analysis of TYRAAt1 transcripts from young Arabidopsis seedlings, by hybridization of total RNAs with a 32P-radiolabeled probe matching exon 3, reveals the presence of a single transcript of 1.9 to 2 kb, corresponding to the expected size of the full-length TYRAAt1 mRNA (Fig. 2B). No signal corresponding to a spliced mRNA constituted by exons 1 and 3 could be detected. To exclude any other posttranscriptional mechanism, we also conducted in vitro transcription/translation studies of the full-length TYRAAt1 mRNA. Indeed, two in-frame AUG codons, in positions 1,015 and 1,018 in the TYRAAt1 ORF, are present just upstream of the sequence encoding the second polypeptide domain of TYRAAt1. Although very unlikely, due to their distance from the first AUG codon, alternative utilization of one of these two AUG codons as a translation start would give rise to the catalytic TYRAAt1-D2 protein. In vitro transcription/translation experiments revealed that the full-length mRNA of TYRAAt1 was always translated entirely and always gave rise to a 66-kD protein corresponding to the complete TYRAAt1 protein (Supplemental Fig. S2).
Taken together, these data exclude the presence of posttranscriptional events leading to the production of mRNA encoding a TYRAAt1 protein devoid of plastid transit peptide. We thus decided to confirm this situation by carrying out biochemical and immunological analyses of TYRAAt1 and TYRAAt2 in planta.
Analysis of TYRA Activity from Arabidopsis Leaves
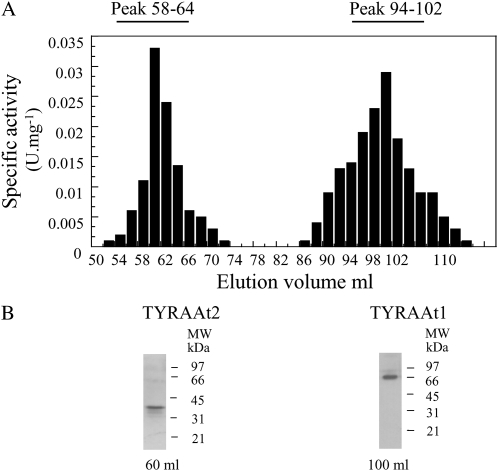
The presence of the two TYRA isoforms and their respective contribution to total TYRA activity from total soluble protein extract of Arabidopsis leaves was analyzed. TYRA activity could hardly be detected in this extract. We thus partially purified the soluble extract by ammonium sulfate precipitation followed by a gel filtration purification step. Total TYRA activity was recovered in the 20% to 60% fraction of an ammonium sulfate fractionation, which was then loaded onto a S200 gel filtration column. As shown in Figure 3A, total TYRA activity was recovered in two separate peaks of activity from the S200 gel filtration column. The first peak (54–74 mL of elution) corresponds to a protein eluted in the void volume of the gel filtration column, indicating that the corresponding TYRA protein behaves as an oligomer with an apparent molecular mass higher than 600 kD. The second TYRA peak of activity (86–110 mL of elution) is consistent with a TYRA protein of an apparent molecular mass of 65 to 67 kD. These results are in agreement with our previous biochemical studies carried out on purified recombinant TYRAAt1 and TYRAAt2 overproduced in E. coli cells (Rippert and Matringe, 2002b). In this previous study, the purified recombinant TYRAAt2 eluted in the void volume of the S200 gel filtration column, whereas the purified recombinant TYRAAt1 eluted with a mobility consistent with a protein of 66 kD. Therefore, in this article, the first peak of activity was attributed to TYRAAt2 and the second one to TYRAAt1. This was further confirmed by a western-blot analysis of the two peaks using polyclonal antibodies raised against the recombinant TYRAAt1 and TYRAAt2 proteins. Under our assay conditions, affinity-purified TYRAAt1 antibody used in this study was specific to TYRAAt1 proteins and did not cross-react with recombinant TYRAAt2 (Supplemental Fig. S3A).Western-blot analyses carried out on the first peak of activity (54–74 mL of elution) by antibodies raised against TYRAAt2 revealed the presence of only one peptide of 38 kD (Fig. 3B). No peptide was detected using antibodies raised against TYRAAt1, confirming that the first peak of activity corresponds to TYRAAt2. A western-blot analysis carried out on the second peak of activity (86–110 mL of elution) using antibodies raised against TYRAAt1 revealed only one peptide of 65 kD (Fig. 3B). No peptide of 38 kD corresponding to TYRAAt2 was detected by TYRAAt2 antibodies, confirming that the second peak of activity corresponds to TYRAAt1 (Fig. 3B). The apparent mobility on the SDS-PAGE of this TYRAAt1 protein is consistent with an entire TYRAAt1 protein formed by the association of its two catalytic domains TYRAAt1-D1 and TYRAAt1-D2. No traces of proteolytic cleavage of TYRAAt1 protein leading to TYRAAt1-D1 and TYRAAt1-D2 could be detected in any fraction eluted from the S200 gel filtration column using TYRAAT1 antibodies, which were able to immunodetect recombinant TYRAAT1-D2 protein (Supplemental Fig. S3B). This biochemical analysis revealed that in Arabidopsis leaves both TYRAAt1 and TYRAAt2 are present and functional. Their elution into two separate peaks of activity allowed us to calculate their respective contribution to the total TYRA activity recovered from Arabidopsis leaves. Total TYRAAt1 activity was found twice as high as TYRAAt2 activity. These data also confirm the absence of a TYRA protein corresponding to TYRAAt1-D2.
Figure 3.
TYRA activity (A) and western-blot analysis (B) of Arabidopsis protein soluble extract separated on a S200 gel filtration column. A, Total soluble extracts were obtained from Arabidopsis plants as described in “Materials and Methods” and separated on a S200 gel filtration column. TYRA activity was determined using 300 μm arogenate and 1 mm NADP+ in 50 mm Tris-HCl, pH 7.5. B, Fifty micrograms of protein extract resulting from the gel filtration column was separated by SDS-PAGE using 12% acrylamide gels, transferred to nitrocellulose membranes, and probed with the antibodies raised against TYRAAt2 (left) or TYRAAt1 (right). Molecular masses are indicated on the right (in kD).
Analysis of the Subcellular Localization of the Two TYRAs
Chloroplastic TYRA activity was previously reported (Byng et al., 1981; Siehl et al., 1986); however, none of these studies examined its possible presence within the cytosol. Sequence comparison with the Synechocystis enzyme (accession no. NP-441290) shows that both TYRAAt1 and TYRAAt2 have N-terminal extensions. Analysis of these N-terminal extensions by prediction programs such as TargetP (http://www.cbs.dtu.dk/services/TargetP) or ChloroP (http://www.cbs.dtu.dk/services/ChloroP) revealed that both TYRAAt2 and TYRAAt1 N-terminal extensions have characteristics of putative plastid transit peptides. Furthermore, the apparent molecular mass of the peptides revealed by western-blot immunodetection in Arabidopsis leaf extracts (Fig. 3B) is consistent with processed proteins.
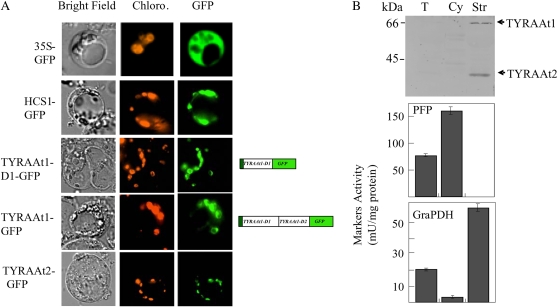
To confirm these predictions, and to definitively clarify the subcellular localization of the two TYRA proteins in Arabidopsis, we conducted transient expression of the two TYRA proteins fused to the GFP in Arabidopsis protoplasts. As shown in Figure 4A, the expression of TYRAAt1-GFP, TYRAAt1-D1-GFP, and TYRAAt2-GFP resulted in punctuate patterns of green fluorescence colocalized with the red autofluorescence of chlorophyll. This pattern is similar to the one observed with GFP fused to the plastid transit peptide sequences of the biotin holocarboxylase synthetase from Arabidopsis (hcs-1 gene; GenBank accession no. U41369; Tissot et al., 1997; Puyaubert et al., 2008), but is clearly distinct from the green fluorescence spread throughout the cytoplasm that is observed with GFP alone. This analysis revealed that the two TYRA proteins are both exclusively targeted within the plastid. Furthermore, the transient expression of the fusion protein TYRAAt1-GFP, which gives rise only to chloroplastic punctuated pattern of GFP fluorescence, confirmed the absence of any proteolytic cleavage leading to a cytosolic TYRAAt1-D2 protein.
Figure 4.
Subcellular localization of the two isoforms of Arabidopsis TYRA. A, Transient expression of TYRAAt1 and TYRAAt2 fused to GFP in Arabidopsis protoplasts. Constructs encoding GFP alone (35S-GFP), GFP fused to the N terminus of HOLOCARBOXYLASE SYNTHETASE1 (HCS1-GFP; a chloroplastic marker), GFP fused to TYRAAt1 (TYRAAt1-GFP and TYRAAt1-D1-GFP), and GFP fused to TYRAAt2 (TYRAAt2-GFP) were introduced into Arabidopsis protoplasts. Images are optical photomicrographs (Bright Field), chlorophyll fluorescence (Chloro.; red pseudocolor), and GFP fluorescence (GFP; green pseudocolor). B, Immunolocalization of TYRAAt1 and TYRAAt2 in Arabidopsis. Proteins, 100 μg for total soluble extract of Arabidopsis rosette leaves (T), 100 μg for the cytosolic enriched fraction (Cy), and 100 μg for the chloroplast stroma (Str), were separated by SDS-PAGE and analyzed by western blotting using affinity-purified antibodies raised against TYRAAt1 and TYRAAt2. Specific activities of cytosolic (pyrophosphate:Fru-6-P 1-phosphotransferase; PFP) and chloroplast stroma (NADP-dependent glyceraldehyde-3-P dehydrogenase; GraPDH) markers were measured (Tissot et al., 1997) to analyze cross-contaminations between subfractions. Data are means ± sd of three replicates (bottom).
As a complementary approach, we investigated the subcellular distribution of TYRA proteins in Arabidopsis by western-blot immunodetection using polyclonal antibodies raised against purified recombinant Arabidopsis TYRA proteins. To overcome the difficulty in immunodetection of endogenous proteins due to their low abundance, a sensitive chemifluorescent detection system, was used. Intact chloroplasts from Arabidopsis rosette leaves were purified on Percoll density gradients, providing chloroplasts devoid of contamination from the other compartments (Fig. 4B, bottom). A highly enriched cytosolic fraction was also prepared with <6% of stromal contamination based on enzymatic markers (Fig. 4B, bottom). The total soluble protein fraction, the soluble protein fraction from purified chloroplasts (stroma), and the cytosolic-enriched protein fractions were then analyzed with a mixture of affinity-purified antibodies raised against the recombinant TYRAAt1 and TYRAAt2 proteins. As shown in Figure 4B, antibodies raised against the recombinant TYRAAt1 and TYRAAt2 proteins revealed two labeled bands of about 66 and 38 kD in the stroma fraction of Arabidopsis chloroplasts, corresponding to the TYRAAt1 and TYRAAt2 proteins, respectively. These two proteins are slightly detectable in the total soluble protein extract in good agreement with the low activity detected in the total soluble protein extract. As expected for plastid proteins, the signal in the stroma fraction was 3 to 4 times as high as in the crude extract. In contrast, no immunological signal could be detected in the cytosolic-enriched fraction (Fig. 4B). These results confirmed that the two TYRA proteins are plastid proteins in Arabidopsis. Our data thus definitively exclude the presence of a cytosolic TYRA protein and consequently of any postchorismate synthesis of Tyr within the cytosol.
Analyses of the Pattern of Expression of the TYRAAt1 and TYRAAt2 Genes in Different Organs by Quantitative RT-PCR
The presence of several isoforms is a common feature of enzymes involved in the shikimate pathway (for reviews, see Schmid and Amrhein, 1999; Herrmann and Weaver, 1999). To complete our knowledge on the two Arabidopsis TYRAs, we evaluated the relative abundance of TYRAAt1 and TYRAAt2 transcripts in various plant organs by means of real-time quantitative reverse transcription (RT)-PCR experiments (Fig. 5). Each TYRA gene is expressed in all organs tested at similar levels, except in seeds, where both transcripts are more abundant. The relative amount of TYRAAt2 mRNA was about 2 times higher than that of TYRAAt1 mRNA in all organs examined. A very similar pattern was also observed for the Arabidopsis ecotype Columbia (Col-0) using the freely available microarray database in Genevestigator (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004). Taken together, our data revealed that the two Arabidopsis TYRA genes are constitutively expressed and encode plastid proteins, which both contribute to the overall Arabidopsis TYRA activity.
Figure 5.
Expression profiles of TYRAAt1 and TYRAAt2 in different organs. Steady-state levels of TYRAAt1 and TYRAAt2 mRNAs were measured by quantitative real-time RT-PCR on total RNA from Arabidopsis organs (flowers [Flow.]; stems, siliques [Siliq.]; rosette leaves [Leav.]; roots; and dry mature seeds [Seeds]) using isoform-specific primers (Supplemental Table S1) as described in “Materials and Methods.” Data are the mean ± sd of three independent experiments performed. The amplification of actin cDNA has been used as internal standard of RNA integrity and cDNA preparation.
Analysis of the Subcellular Localization of the Six ADTs
As for TYRAs, previous biochemical studies on ADTs report activities within the chloroplast (Jung et al., 1986; Siehl and Conn, 1988); however, here, again, none of these studies examined their possible presence within the cytosol. Among the six Arabidopsis ADTs, only ADT1 N-terminal extension is not clearly recognized as a putative plastid transit peptide by protein-targeting prediction software like ChloroP or TargetP. Furthermore, in a recent study, Warpeha et al. (2006) report that AtADT3 activity (named PDT1 in their study), was regulated by blue light via protein-protein interaction with the G-protein α-subunit GPA1, which is likely to be involved in many processes critical for plant development, including blue light response (Ma, 2001). This protein-protein interaction implies that AtADT3 (PDT1) should be present within the cytosol because GPA1 is a plasma membrane protein (Warpeha et al., 1991). Accordingly, in their study, the authors present AtADT3 (PDT1) as a cytosolic protein, which catalyzes prephenate dehydratase activity. This subcellular assumption, based only on an in silico analysis of the N-terminal sequence, was quite surprising because the AtADT3 N-terminal sequence is predicted to contain a putative plastid transit peptide by ChloroP and TargetP.
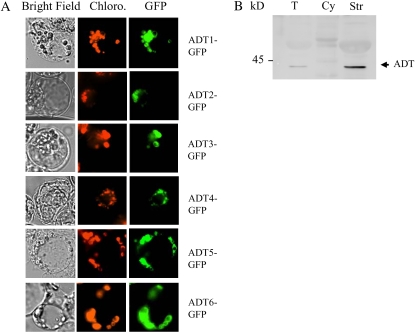
To clarify the subcellular localization of the six ADTs in Arabidopsis, we conducted transient expression of the six ADT proteins fused to the N terminus of the GFP in Arabidopsis protoplasts. As shown in Figure 6A, in all cases, the expression of the six AtADT-GFP fusions resulted in punctuate patterns of green fluorescence colocalized with the red autofluorescence of chlorophyll, but clearly distinct from the green fluorescence spread throughout the cytoplasm that is observed with GFP alone (Fig. 4A). All six Arabidopsis ADTs are thus targeted within the plastid of Arabidopsis cells.
Figure 6.
Subcellular localization of the six isoforms of Arabidopsis ADT. A, Transient expression of AtADT1 to AtADT6 fused to GFP in Arabidopsis protoplasts. Constructs encoding GFP fused to AtADT1 to AtADT6 (AtADT1–6-GFP) were introduced into Arabidopsis protoplasts. Images are optical photomicrographs (Bright Field), chlorophyll fluorescence (Chloro.; red pseudocolor), and GFP fluorescence (GFP; green pseudocolor). B, Immunolocalization of ADT in Arabidopsis. Proteins, 100 μg for total soluble extract of rosette leaves (T), 100 μg for the cytosolic-enriched fraction (Cy), and 100 μg for the chloroplast stroma (Str), were separated by SDS-PAGE and analyzed by western blotting using affinity-purified antibodies raised against AtADT1.
As a complementary approach, western-blot immunodetection of ADT proteins was conducted using total soluble protein extract, chloroplast soluble protein extract (stroma), and highly enriched cytosolic fraction from Arabidopsis cells (see “Materials and Methods”). Arabidopsis subfractions were analyzed with affinity-purified antibody raised against purified recombinant AtADT1 protein, which cross-reacts with the six recombinant isoforms (Supplemental Fig. S5). As shown in Figure 6B, immunopurified antibody revealed only one labeled band of about 40 to 42 kD in the total soluble protein extract and in the stroma fraction of Arabidopsis chloroplasts (Fig. 6B). As expected for plastid proteins, the signal in the stroma fraction was 3 to 4 times as high as in the crude extract. Furthermore, the molecular mass of the labeled band corresponds well to the expected size of mature ADTs, when it could be predicted (see Supplemental Fig. S5). No signal could be detected in the cytosolic-enriched fraction, revealing the absence of any ADT within the cytosol of Arabidopsis cultured cells. The presence of only one labeled band in the stroma could either indicate that all six ADT mature proteins have similar molecular mass and could not be distinguished on SDS-PAGE (see Supplemental Fig. S5) and/or that not all ADT are present in sufficient quantity in chloroplast stroma to be detected. Nevertheless, the absence of any cytosolic signal, and the presence of a strong chloroplastic signal, confirm the ADT-GFP fusion data indicating that the all six ADT proteins are plastid proteins. Taken together, our data definitively exclude any postchorismate synthesis of Phe within the cytosol in Arabidopsis, at least in green leaves and cultured cells.
These results are in apparent contradiction with the work reported by Warpeha et al. (2006) revealing a protein-protein interaction between AtADT3 and GPA1 leading to the activation of the prephenate dehydratase activity sustained by AtADT3. This prephenate dehydratase activity seems difficult to reconcile with the biochemical characterization of AtADT3 activity as strictly arogenate dependent reported by Cho et al. (2007). The protein-protein interaction between AtADT3 and GPA1 is a very interesting observation, which, according to our current knowledge, is difficult to interpret. It remains possible that the different models used (e.g. green leaves and cultured Arabidopsis cells in this study), versus etiolated Arabidopsis seedlings submitted to blue light in their study, could explain this discrepancy in both activity and subcellular localization.
Analyses of the Pattern of Expression of the Six AtADT Genes in Different Organs by Quantitative RT-PCR
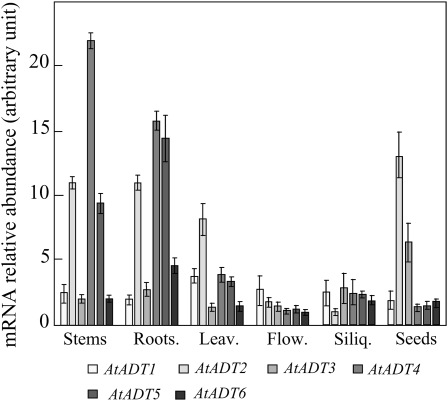
Quantitative PCR data confirm that, as observed by Cho et al. (2007) using northern-blot analyses, the six isoforms are all expressed at the mRNA level in all Arabidopsis organs. However, our analysis (Fig. 7) reveals that the pattern of expression of the six AtADTs differs greatly depending on the organ considered. AtADT2 is the more abundant isoform in both leaves and seeds. This isoform is also well represented in stems and roots, but less expressed in flowers and siliques. This result is in good agreement with a recent study of the Arabidopsis plastid proteome, revealing that AtADT2 is the sole isoform of ADT clearly identified by master peptides (N. Roland, personal communication). This ADT isoform could have a specific role in maintaining the housekeeping needs of Phe in Arabidopsis. Interestingly, AtADT4 and AtADT5 mRNAs, which belong to the same subgroup (Cho et al., 2007), are clearly more abundantly expressed in stems and roots compared to leaves, flowers, siliques, and seeds. A previous study aimed at the identification of candidate genes triggering lignin biosynthesis and modification also highlighted that the gene encoding AtADT5 is strongly up-regulated during stem elongation (Ehlting et al., 2005). Analyses of freely available microarray databases (https://www.genevestigator.ethz.ch) are in good agreement with our quantitative RT-PCR analyses, revealing that genes encoding AtADT4 and AtADT5 are up-regulated during stem and root development. Interestingly, these microarray data revealed that they are both specifically down-regulated by treatment with the herbicide isoxaben, an inhibitor of cellulose biosynthesis. These two genes are also strongly up-regulated during biotic stress. All these data suggest coordinated expression of the genes encoding AtADT5 and AtADT4, together with other genes of the shikimate pathway and the phenylpropanoid pathway for the synthesis of lignin and plant defense phenylpropanoids.
Figure 7.
Expression profiles of AtADT1 to AtADT6 in different organs. Steady-state levels of transcript of the six AtADT genes were measured by quantitative real-time RT-PCR on total RNA from Arabidopsis organs (flowers [Flow.]; stems, siliques [Siliq.]; rosette leaves [Leav.]; roots; and dry mature seeds [Seeds]) using isoform-specific primers (Supplemental Table S1) as described in “Materials and Methods.” Data are the mean ± sd of three independent experiments performed. The amplification of actin cDNA has been used as internal standard of RNA integrity and cDNA preparation.
CONCLUSION
The possible presence of a cytosolic postchorismate Phe and/or Tyr biosynthesis remained an open question raised by the presence of a cytosolic CM; Benesova and Bode, 1992; d'Amato et al., 1992; Eberhard et al., 1996). However, among the three Arabidopsis CM isoforms, only the two chloroplastic CMs (AtCM1 and AtCM3) are elicitor and pathogen inducible and allosterically regulated by Phe and Tyr (Eberhard et al., 1996). The contribution of the cytosolic CM (AtCM2) to phenylpropanoids, or metabolites deriving from Tyr, thus remained questionable, and would necessitate the presence of enzymes catalyzing the final steps of Tyr and/or Phe synthesis within the cytosol. Until now, very little information was available on these enzymes in higher plants, particularly concerning their subcellular localization. All the data presented herein aimed to address the question of the possible presence of cytosolic TYRA and/or ADT proteins in Arabidopsis revealed that the two TYRA proteins and the six AtADT proteins are all plastid proteins. Our data exclude any cytosolic postchorismate synthesis of Tyr and/or Phe in Arabidopsis green leaves and cultured cells. Because the only known destiny of prephenate is to be processed into Tyr or Phe, the physiological reason for the presence of a cytosolic CM still remains an open and exciting question.
MATERIALS AND METHODS
Materials
All chemicals were obtained from Sigma. Arogenate was synthesized enzymatically from prephenate and Asp as previously described (Rippert and Matringe, 2002b).
Plant Material
Arabidopsis (Arabidopsis thaliana ecotype Wassilewskija) plants were grown in soil under greenhouse conditions (23°C with a 16-h photoperiod and a light intensity of 200 μmol of photons m−2 s−1) until harvested for analysis. Arabidopsis (ecotype Col-0) cell suspension cultures were grown under continuous white light (40 μE m−2 s−1) at 23°C with rotary agitation at 125 rpm in Gamborg's B5 medium supplemented with 1 μm 2-naphthalene acetic acid and 1.5% (w/v) Suc.
Preparation of Crude Extract of Arabidopsis Rosette Leaves and Fractionation on a Gel Filtration S200 Column
Young Arabidopsis rosette leaves were frozen in liquid nitrogen and finely ground using a mortar and pestle. The powder was then homogenized in buffer A (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, pH 8.0, 1 mm dithiothreitol [DTT], 1 mm benzamidine-HCl, 5 mm amino caproic acid, 0.2 mm phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 40,000g for 30 min at 6°C. The supernatant classified as the crude extract was used immediately for ammonium sulfate fractionation.
The soluble protein extract was subjected to ammonium sulfate clarification by addition of solid (NH4)2SO4 (20% saturation) at 4°C. After 20 min of stirring, the mixture was centrifuged at 40,000g for 20 min and the supernatant was brought to 60% saturation with solid (NH4)2SO4 at 4°C. The precipitate was recovered by centrifugation and resuspended in a minimal volume of buffer A. The resulting protein extract was applied to a Hiload Superdex S200 (3.2 × 60 cm; Pharmacia), column connected to an FPLC system (Pharmacia) previously equilibrated in 200 mL of buffer B (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, pH 8.0, 1 mm DTT, 150 mm NaCl). Two milliliters of eluted fractions were collected at 1.5 mL/min and tested for TYRA activity.
Preparation of a Cytosolic-Enriched Fraction
Arabidopsis protoplasts were prepared by enzymatic digestion of 6-d-old cell suspension cultures, using the procedure described by Tissot et al. (1997). Protoplasts were gently ruptured by passing through a 20-μm nylon mesh and subsequently through a 10-μm nylon mesh. The protoplast lysate was centrifuged successively at 100g for 5 min, 900g for 5 min, and 13,000g for 20 min. The 13,000g supernatant fraction was centrifuged further at 100,000g for 1 h to remove membranes. Pellets were pooled and used, together with the 13,000g supernatant fraction, to measure marker enzyme activities (Tissot et al., 1997). Most of the chloroplast (94% ± 2%, mean of three independent experiments ± se) and mitochondria (96 ± 2%) marker activities were recovered in the pooled pellets, whereas the 13,000g supernatant fraction contained 72% ± 3% of the cytosolic marker activity. This cytosolic-enriched fraction contained a small proportion of mitochondrial (4% ± 2%) and chloroplastic (6% ± 2%) marker enzymes.
Purification of Chloroplasts
Chloroplasts were purified from 4-week-old Arabidopsis leaves as described by Ravanel et al. (2004). Intact chloroplasts were purified on preformed Percoll gradients and lysed in 20 mm MOPS, pH 7.5, 4 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine-HCl, and 5 mm ɛ-amino caproic acid. Chloroplast subfractions were separated on a step gradient of 0.93-0.6-0.33 m Suc in 10 mm MOPS, pH 7.5, by centrifugation at 70,000g for 1 h. The soluble fraction (stroma) was collected at the top of the 0.33 m Suc layer.
ADT cDNA Isolation and Expression
Full-length ADT cDNAs were obtained by PCR amplification of an Arabidopsis (var. Col-0) cDNA library constructed in pYES (Elledge et al., 1991). Primers were designed on the basis of the predicted genomic sequences of the six ADTs and generated appropriate restriction sites for the cloning of a complete ORF into pET-28b (Novagen) expression vectors (see Supplemental Table S1). The PCR fragments were first cloned into pPCR-Script (Stratagene) for maintenance and sequenced in both strands (Genome Express). The cloned sequence encoding Arabidopsis ADT was designated AtADT1 (At1g11790), AtADT2 (At3g07630), AtADT3 (At2g27820), AtADT4 (At3g44720), AtADT5 (At5g22630), and AtADT6 (At1g08250) according to the nomenclature of Ehlting et al. (2005). For overproduction, the six ADT ORFs were inserted between the NdeI and EcoRI or EcoRV sites of pET-28b vector (Novagen); this procedure added an N-terminal hexa-His tag. The resulting pET-ADT constructs were introduced first into Escherichia coli DH5α, then into E. coli Rosetta (DE3) cells (Stratagene).
Overexpression and Purification of Recombinant AtADTs
E. coli Rosetta cells transformed with the pET-AtADT constructs were grown at 37°C in Luria-Bertani medium with the appropriate antibiotics until A600 was 0.6, at which point isopropylthio-β-galactoside was added to a final concentration of 0.4 mm. Incubation was continued for 16 h at 28°C. Pelleted cells from 2-L cultures were resuspended in 15 mL of buffer A (20 mm Tris-HCl, pH 8) and disrupted by sonication with a Vibra-Cell disrupter. The six AtADT recombinant proteins were all recovered as inclusion bodies. AtADT1 and AtADT5, which belong to the two subgroups of ADT, were purified from inclusion bodies. Inclusion bodies were washed twice in 20 mm Tris-HCL, pH 8, 0.5% triton, and then solubilized in 20 mm Tris-HCl, pH 8, 0.5 m NaCl, 8 m urea, and purified through a nickel nitrilotriacetic acid agarose column (1 × 3 cm) in denaturing conditions according to the manufacturer (Qiagen).
Protein Determination, Antibody Production, and Immunoblot Analysis
Proteins were measured by the Bradford method (Bradford, 1976) using Bio-Rad protein assay reagent, with γ-globulin as a standard. The purified recombinant TYRAAt1, TYRAAt2, AtADT1, and AtADT5 were purified further by SDS-PAGE and injected into rabbits to raise antibodies (Elevage Scientifique des Dombes). Total soluble proteins from Arabidopsis cells were extracted by grinding powdered samples in 20 mm MOPS, pH 7.5, 5% (w/v) glycerol, 1 mm DTT, 1 mm EDTA, 1 mm benzamidine-HCl, and 5 mm ɛ-amino caproic acid. Samples were centrifuged at 130,000g for 20 min at 4°C, and the supernatant was used as a source of soluble proteins. Proteins from Arabidopsis cell subfractions were resolved by SDS-PAGE and electroblotted to nitrocellulose membrane. The blots were probed by using the affinity-purified antibodies (dilution 1:5,000), horseradish peroxidase-conjugated rabbit IgGs, and detection was achieved by ECL+ on a Typhoon 9400 according to the manufacturer (GE Healthcare Europe).
Enzyme Activities
TYRA activities were assayed at 25°C according to Bonner and Jensen (1987) by following the formation of NADPH at 340 nm in 50 mm Tris-HCl, pH 7.5, 300 μm arogenate, and 1 mm NADP+ in a total volume of 200 μL as previously described (Rippert and Matringe, 2002a, 2002b).
Marker enzyme activities pyrophosphate:Fru-6-P 1-phosphotransferase and NADP-dependent glyceraldehyde-3-P dehydrogenase were measured as described by Tissot et al. (1997).
Expression of TYRAAt1 and TYRAAt2 and the Six AtADTs in Different Organs of Arabidopsis
Preparation of Total RNAs
Total RNA from different organs (rosette leaves, stems, roots, flowers, siliques, dry mature seeds) of Arabidopsis was prepared using the RNeasy plant mini kit isolation system, according to the manufacturer's instructions (Qiagen). First-strand cDNA was synthesized from 1 μg of total RNAs in a final volume of 20 μL using oligo(dT)20 primers (Thermoscript RT-PCR system; Invitrogen).
Identification of TYRAAt1 Transcription Start Site
The transcription start site of TYRAAt1 was determined by 5′-RACE using Arabidopsis leaf cDNAs (GeneRacer kit; Invitrogen). Two sequential PCRs were performed with two pairs of nested adaptor primers and gene-specific primers in exon 2 (TYRAAt1 E2) and exon 3 (TYRAAt1 E3) of TYRAAt1. The resulting PCR products were cloned into pPCR-Script and sequenced.
Quantitative Real-Time PCR Experiments
Relative quantification experiments of mRNA were done by real-time PCR using the LightCycler system (Roche Biomolecular Biochemicals) and the LightCycler-Faststart DNA Master SYBR Green I kit. For each measurement, 5 μL of cDNA preparation was used as a template in a standard 1-μL LightCycler PCR reaction with appropriate primers (used at a final concentration of 1 μm) and 3 mm MgCl2. Primers used during real-time PCR experiments are given in Supplemental Table S1.
Amplification and detection were performed using the following program: 95°C for 8 min followed by 40 cycles of 95°C for 10 s, 66°C for 15 s, and 72°C for 8 s. Data were analyzed with LightCycler Relative Quantification software (Roche) using dilutions of linear plasmid pPCR-Script-TYRAAt1, pPCR-Script-TYRAAt2, and pPCR-Script-AtADTs as standards for quantification and actin cDNA (accession no. U39449; primers listed in Supplemental Table S1) as a reference. The specificity of the reaction was verified by melting curve analysis obtained by increasing temperature from 55°C to 95°C (0.1°C/s).
In Vitro Transcription/Translation Experiments
pET21-TYRAAt1, pET21-TYRAAt1-D1, and pET21-TYRAAt1-D2 constructs (Rippert and Matringe, 2002a) were linearized and transcribed with T7 RNA polymerase from mMessage mMachine kit (Ambion) in accordance with the instructions of the manufacturer. Capped TYRA mRNAs produced were then translated using a reticulocyte lysate from Ambion kit (Retic lysate IVT) and [35S]Met, according to the manufacturer's instructions. A control reaction with empty plasmid was performed under the same conditions. Radiolabeled proteins were separated by SDS-PAGE and analyzed by phosphor imaging analysis using a Typhoon 9400 scanner (GE Healthcare Europe).
Transient Expression of GFP Fusion Proteins in Arabidopsis Protoplasts
A pUC18 plasmid expressing a modified version of GFP (mGFP4), under the control of the cauliflower mosaic virus 35S promoter (Haseloff et al., 1997), was used for transient expression experiments in Arabidopsis protoplasts. The first 1,130 nucleotides of TYRAAt1, the first 250 to 300 nucleotides of TYRAAt1-D1 and TYRAAt2, and the six AtADT ORFs were fused upstream and in-frame with mGFP4. The transit peptide sequences of the biotin holocarboxylase synthetase from Arabidopsis (HCS1 gene; GenBank accession no. U41369) were used as a control for the targeting of GFP to plastids (Tissot et al., 1997).
TYRAAt1, TYRAAt2, and the six AtADTs were PCR amplified using Pfu DNA polymerase and specific flanking primers containing restriction sites (underlined) listed in Supplemental Table S1 from pPCR-Script-TYRAAt1, pPCR-Script-TYRAAt2, and pPCR-Script-AtADTs plasmids. The PCR products were digested with Xba1 and BamH1 and inserted between the Xba1 and BamH1 sites of the GFP-containing plasmid to give plasmids pGFP-TYRAAt1, pGFP-TYRAAt1-D1, pGFP-TYRAAt2, and pGFP-AtADTs. Arabidopsis protoplasts prepared from a 4-d-old cell suspension culture were transformed essentially as described by Abel and Theologis (1994). For each transformation, 300 μL of protoplasts (2 × 106 cells) were mixed with 20 to 30 μg of plasmid DNA, 50 μg of salmon sperm carrier DNA, and 350 μL of polyethylene glycol solution (0.4 m mannitol, 0.1 m CaNO3, 40% [w/v] PEG-4000) and incubated at room temperature for 30 min. Protoplasts were cultured in 3 mL of medium (0.4 m Suc; Murashige and Skoog basal medium; 4 mm CaCl2, and 250 mg L−1 Xyl) under dim light at 22°C for up to 48 h. Samples were analyzed by fluorescence microscopy using a Zeiss Axioplan2 fluorescence microscope and the images were captured with a digital CCD camera (Hamamatsu). The filter sets used were Zeiss filter set 13, 488013-0000 (exciter BP 470/20; beam splitter FT 493; emitter BP 505–530), and Zeiss filter set 15, 488015-0000 (exciter BP 546/12, beam splitter FT 580, emitter LP 590) for GFP and chlorophyll fluorescence, respectively.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure S1. Orthologs of the TYRAAt1 gene.
Supplemental Figure S2. Alignment of the deduced amino acid sequence from the six Arabidopsis ADT genes using the Multalin program.
Supplemental Figure S3. Analysis of the nature of the translation product of TYRAAt1 mRNA by in vitro transcription/translation experiments.
Supplemental Figure S4. Specificity of antibodies directed against TYRAAt1 and TYRAAt2.
Supplemental Figure S5. All six AtADT isoforms are immunodetected by polyclonal antibodies raised against AtADT1.
Supplemental Table S1. Synthetic oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We are grateful to Dr. Claude Alban, Gilles Curien, and Renaud Dumas for helpful discussions.
This work was supported by the Institut National de la Recherche Agronomique, the Centre National de la Recherche Scientifique, the Commissariat à l'Energie Atomique, and Grenoble I University.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michel Matringe (michel.matringe@cea.fr).
The online version of this article contains Web-only data.
References
- Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5 421–427 [DOI] [PubMed] [Google Scholar]
- Benesova M, Bode R (1992) Chorismate mutase isoforms from seeds and seedlings of Papaver somniferum. Phytochemistry 31 2983–2987 [Google Scholar]
- Bickel H, Palme L, Schultz G (1978) Incorporation of shikimate and other precursors into aromatic acids and prenylquinones of isolated spinach chloroplasts. Phytochemistry 17 119–124 [Google Scholar]
- Bonner CA, Jensen RA (1987) Arogenate dehydrogenase. Methods Enzymol 142 488–494 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of dye-binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Byng GS, Whitaker RJ, Flick C, Jensen RA (1981) Enzymology of l-tyrosine biosynthesis in corn (Zea mays). Phytochemistry 20 1289–1292 [Google Scholar]
- Cho MH, Corea ORA, Yang H, Bedgar DL, Laskar DD, Anterola AM, Moog-Anterola FA, Hood RL, Kohalmi SE, Bernards MA, et al (2007) Phenylalanine biosynthesis in Arabidopsis thaliana: identification and characterization of arogenate dehydratases. J Biol Chem 282 30827–30835 [DOI] [PubMed] [Google Scholar]
- d'Amato TA, Ganson RJ, Gaines CG, Jensen RA (1992) Subcellular localization of chorismate mutase isoenzymes in protoplasts from mesophyll and suspension-cultured cells of Nicotiana silvestris. Planta 162 104–108 [DOI] [PubMed] [Google Scholar]
- Eberhard J, Ehrler TT, Epple P, Felix G, Raesecke HR, Amrhein N, Schmid J (1996) Cytosolic and plastidic chorismate mutase isozymes from Arabidopsis thaliana: molecular characterization and enzymatic properties. Plant J 10 815–821 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42 618–640 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW (1991) Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA 88 1731–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall GC, Flick MB, Gherna RL, Jensen RA (1982) Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J Bacteriol 149 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50 473–503 [DOI] [PubMed] [Google Scholar]
- Jung E, Zamir LO, Jensen RA (1986) Chloroplasts of higher plants synthesize L-phenylalanine via arogenate. Proc Natl Acad Sci USA 83 7231–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H (2001) Plant G proteins: the different faces of GPA1. Curr Biol 11 869–871 [DOI] [PubMed] [Google Scholar]
- Puyaubert J, Denis L, Alban C (2008) Dual-targeting of Arabidopsis thaliana Holocarboxylase Synthetase 1: a small upstream open reading frame (uORF) regulates translation initiation and protein targeting. Plant Physiol 146 478–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279 22548–22557 [DOI] [PubMed] [Google Scholar]
- Rippert P, Matringe M (2002. a) Molecular and biochemical characterisation of an A. thaliana arogenate dehydrogenase with two highly similar and active domains. Plant Mol Biol 48 361–368 [DOI] [PubMed] [Google Scholar]
- Rippert P, Matringe M (2002. b) Purification and kinetic analysis of the two recombinant arogenate dehdyrogenase isoforms of Arabidopsis thaliana. Eur J Biochem 269 4753–4761 [DOI] [PubMed] [Google Scholar]
- Schmid J, Amrhein N (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39 737–749 [Google Scholar]
- Schmid J, Amrhein N (1999) The shikimate pathway. In B Singh, ed, Plant Amino Acids. Marcel Dekker, New York, pp 147–169
- Schulze-Siebert D, Heineke D, Scharf H, Schultz G (1984) Pyruvate-derived amino acids in spinach chloroplasts. Plant Physiol 76 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehl DL, Conn EE (1988) Kinetic and regulatory properties of arogenate dehydratase in seedlings of Sorghum bicolor (L.) Moench. Arch Biochem Biophys 260 822–829 [DOI] [PubMed] [Google Scholar]
- Siehl DL, Connelly JA, Conn EE (1986) Tyrosine biosynthesis in Sorghum bicolor: characteristics of prephenate aminotransferase. Z Naturforsch [C] 41 79–86 [DOI] [PubMed] [Google Scholar]
- Tissot G, Douce R, Alban C (1997) Evidence for multiple forms of biotin holocarboxylase synthetase in pea (Pisum sativum) and Arabidopsis thaliana: subcellular fractionation studies and isolation of a cDNA clone. Biochem J 323 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Hamm HE, Rasenick MM, Kaufman LS (1991) A blue-light-activated GTP-binding protein in the plasma membranes of etiolated peas. Proc Natl Acad Sci USA 88 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee BS, Kaufman LS (2006) G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol 140 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Matsuda F, Kasai K, Fukuoka S, Kitamura K, Tozawa Y, Miyagawa H, Wakasa K (2008) Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell 20 1316–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.