Abstract
RNA silencing in plants serves as a potent antiviral defense mechanism through the action of small interfering RNAs (siRNAs), which direct RNA degradation. siRNAs can be derived directly from the viral genome or via the action of host-encoded RNA-dependent RNA polymerases (RDRs). Plant genomes encode multiple RDRs, and it has been demonstrated that plants defective for RDR6 hyperaccumulate several classes of virus. In this study, we compared the effectiveness of virus-induced gene silencing (VIGS) and RNA-directed DNA methylation (RdDM) in wild-type and RDR6-deficient Nicotiana benthamiana plants. For the potexvirus Potato virus X (PVX) and the potyvirus Plum pox virus (PPV), the efficiency of both VIGS and RdDM were compromised in RDR6-defective plants despite accumulating high levels of viral siRNAs similar to infection of wild-type plants. The reduced efficiency of VIGS and RdDM was unrelated to the size class of siRNA produced and, at least for PVX, was not dependent on the presence of the virus-encoded silencing suppressor protein, 25K. We suggest that primary siRNAs produced from PVX and PPV in the absence of RDR6 may not be good effectors of silencing and that RDR6 is required to produce secondary siRNAs that drive a more effective antiviral response.
RNA silencing is a unifying term to describe a sequence-specific RNA degradation process found in eukaryotic organisms. The sequence specificity of this process is determined by 21- to 24-nucleotide (nt) small interfering RNAs (siRNAs), which associate with a multiprotein complex, the RNA-induced silencing complex (RISC). RISC has endonuclease activity and thus is able to direct the cleavage of target RNAs having complementarity to the siRNA associated with it (Baumberger and Baulcombe, 2005; Hammond, 2005; Brodersen and Voinnet, 2006; Vaucheret, 2006). As well as directing RNA degradation, siRNAs can also participate in RNA-directed DNA methylation (RdDM; Wassenegger et al., 1994). Natural roles for RNA silencing include regulation of gene expression and suppression of transposon mobility as well as resistance against viruses (Robert et al., 2004; Voinnet, 2005; Ding and Voinnet, 2007). siRNAs are produced by cleavage of double-stranded RNA (dsRNA) through the action of RNase III-type enzymes known as Dicer (DCR) in animals or Dicer-like (DCL) in plants (Hammond, 2005; Vazquez, 2006). dsRNA can be produced by the action of plant-encoded RNA-dependent RNA polymerases (RDRs) on a variety of RNA templates. For example, RDR6 copies TAS transcripts and transgenic mRNAs to produce dsRNAs, which in turn are cleaved by DCL4 protein to give rise to trans-acting siRNAs and secondary siRNAs, respectively (Vaistij et al., 2002; Peragine et al., 2004; Vazquez et al., 2004; Axtell et al., 2006; Moissiard et al., 2007). However, not all plant transcripts appear to be good templates for RDR6 (Vaistij et al., 2002).
Plant RNA viruses are strong triggers of RNA silencing, and engineered viruses can be used to silence target transcripts in a process termed virus-induced gene silencing (VIGS; Robertson, 2004). Significant levels of siRNAs accumulate during infection, and in theory these could be processed from double-stranded viral RNA replication intermediates, from self-complementary regions of the viral genome, or via the action of RDRs on viral templates. Nicotiana benthamiana plants deficient in RDR6 are hypersusceptible to a subset of viruses, demonstrating the role of this enzyme in antiviral defense and illustrating that not all viruses are templates for RDR6. For example, RDR6-deficient N. benthamiana plants hyperaccumulate Potato virus X (PVX) and Potato virus Y but not Tobacco rattle virus (TRV), Tobacco mosaic virus, or Turnip crinkle virus (Qu et al., 2005; Schwach et al., 2005). In Arabidopsis (Arabidopsis thaliana) DCL2, DCL3, and DCL4 produce virus-derived siRNAs of distinct size classes (Park et al., 2002; Xie et al., 2004, 2005; Borsani et al., 2005; Dunoyer and Voinnet, 2005; Gasciolli et al., 2005): DCL4 and DCL2 act redundantly to produce 21/22-nt siRNAs responsible for antiviral silencing activities (Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Diaz-Pendon et al., 2007), whereas DCL3 gives rise to 24-nt siRNAs that are not active in directing RNA cleavage (Deleris et al., 2006; Fusaro et al., 2006).
In this work, we have investigated the role of RDR6 in RNA silencing and DNA methylation driven by different viral vectors in N. benthamiana plants. These experiments relate to the observations of enhanced virus accumulation in RDR6-deficient plants and the question of why this occurs if virus replication is a strong trigger of RNA silencing. One prediction of enhanced virus accumulation is that there should be high levels of virus-derived siRNAs and that this should lead to self-silencing. In order to investigate why this does not happen, we have undertaken a series of experiments to address the activity of viral siRNAs produced in the absence of RDR6 (primary siRNAs) compared with the secondary siRNAs that are derived via RDR6 activity. We found that both VIGS and RdDM driven by the potexvirus PVX or the potyvirus Plum pox virus (PPV) were compromised in RDR6-deficient plants. However, this was not the case for TRV, whose accumulation is not affected by lack of RDR6. We show that the impairment of PVX- or PPV-driven silencing does not correlate with a significant reduction of virus-derived siRNAs in the RDR6-deficient background and provide evidence that the mechanism underlying this inefficiency appears unrelated to the presence of virus-encoded silencing suppressor proteins encoded by the viral vectors or to the size class of siRNA produced.
RESULTS
PVX-Driven VIGS Is Impaired in RDR6-Deficient Plants
Previous work has demonstrated that PVX hyperaccumulates in RDR6-deficient N. benthamiana plants (Schwach et al., 2005). In order to investigate whether this hyperaccumulation influences the efficiency of VIGS, 35S:GFP transgenic (GFP) or GFP/RDR6-deficient (GFP/rdr6i) N. benthamiana plants were infected with PVX vectors carrying the 3′ 359 nucleotides of the GFP open reading frame (PVX:P) or the PVX empty vector (PVX:00) as a negative control. Plants were then assessed 21 d later for green fluorescence and RNA accumulation. Newly developed leaves of PVX:00-infected plants were green under UV light and accumulated GFP mRNA (Fig. 1, A and B). The levels of GFP transcript accumulation in PVX:00-infected plants were similar to those of noninfected plants regardless of their genetic background (data not shown). As expected, upper leaves of PVX:P-infected GFP plants showed GFP silencing, as visualized under UV light and confirmed by northern-blot analysis (Fig. 1, A and B). In contrast, the upper leaves of PVX:P-infected GFP/rdr6i plants remained mainly green and accumulated GFP mRNA and PVX:P at much higher levels than in equivalent PVX:P-infected GFP plants (Fig. 1, A and B). In order to investigate whether the inefficiency of silencing in the rdr6i background corresponded to a reduction in siRNA accumulation, RNA samples were assessed for antisense (as) siRNAs complementary to the P portion carried by the PVX:P vector. asP-specific siRNAs accumulated in both genetic backgrounds, although the levels were slightly higher in wild-type GFP plants (Fig. 1B). The siRNAs were predominantly of 21 nt, the size class that has been shown to be active in RNA silencing (Deleris et al., 2006; Fusaro et al., 2006).
Figure 1.
RDR6-dependent RNA silencing driven by PVX. A, Leaves of GFP and GFP/rdr6i plants systemically infected with PVX:00 or PVX:P (21 dpi) visualized under UV light. B, Accumulation of GFP mRNA, PVX:P genomic RNA (gRNA), and asP siRNAs assessed by northern-blot analyses using probes specific for the GF and P regions (see Fig. 2C). C, Wild-type (WT) and rdr6i plants systemically infected with PVX:PDS (21 dpi). D, Accumulation of PDS mRNA assessed by semiquantitative RT-PCR followed by Southern blotting with amplification of actin used as a control for quantification. PVX:PDS gRNA and asPDS and sPDS siRNAs were assessed by northern blot analysis. Ethidium bromide-stained rRNAs are shown as loading controls, and positions of 24- and 21-nt size markers are indicated.
In wild-type plants, triggering GFP RNA silencing will lead to RDR6-dependent amplification of silencing via the production of secondary siRNAs (Vaistij et al., 2002; Schwach et al., 2005). As this amplification effect will be reduced in the rdr6i plants, a difference in GFP silencing may have been predicted. Therefore, we assessed PVX-driven silencing of the phytoene desaturase (PDS) gene, which is believed not to be a template for RDR6 (Vaistij et al., 2002). The product of this gene protects leaves from photobleaching; thus, PDS silencing can be easily visualized as areas of white tissue (Kumagai et al., 1995). A PVX vector carrying a fragment of the PDS coding region (PVX:PDS) was used to infect plants, and PDS silencing was assessed by the degree of bleaching and the level of PDS mRNA accumulation. At 21 d post inoculation (dpi), the uppermost leaves of PVX:PDS-infected wild-type plants showed extensive and strong bleaching, whereas the bleaching was only observed at a lower extent and intensity in equivalent leaves of rdr6i plants (Fig. 1C). These observations correlated with a higher accumulation of PDS mRNA in rdr6i compared with wild-type plants (Fig. 1D). PVX:PDS RNA accumulated to higher levels in rdr6i compared with wild-type plants; however, asPDS-specific PVX-derived siRNAs were detected at similar levels in both genetic backgrounds (Fig. 1D). As expected from the DCL processing of the viral dsRNA replication intermediates, sense (s) PDS-specific siRNAs were also present, although at slightly lower levels, in rdr6i plants (Fig. 1D). Taken together, these observations suggest that efficient PVX-driven VIGS requires RDR6 and that the siRNAs produced directly from the virus are compromised for driving RNA degradation.
Impairment of PVX-Driven RdDM in RDR6-Deficient Plants
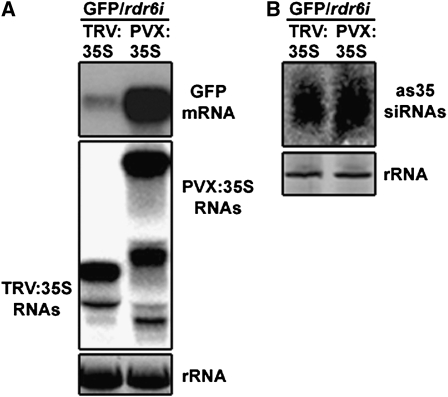
Since PVX-derived siRNAs accumulate in rdr6i plants but transcript cleavage is impaired, we were interested to determine whether RdDM was also affected. If RdDM is targeted to promoter regions, transcriptional gene silencing (TGS) can be achieved (Jones et al., 1999; Mette et al., 2000). Thus, TGS was assessed in GFP and GFP/rdr6i plants by infection with a PVX vector (PVX:35S) carrying the 35S promoter of the GFP transgene (Jones et al., 1999). At 21 dpi, upper leaves of GFP plants fluoresced red under UV light and accumulated GFP mRNA to low levels, indicating that TGS was occurring. In contrast, equivalent PVX:35S-infected GFP/rdr6i leaves remained mainly green and showed no significant reduction in GFP mRNA levels (Fig. 2, A and B). Levels of PVX:35S RNA were higher in GFP/rdr6i compared with GFP plants, and assessment of as35S-specific siRNA levels demonstrated that equivalent levels accumulated in both genetic backgrounds (Fig. 2B).
Figure 2.
Impairment of PVX-driven transcriptional silencing and DNA methylation in rdr6i plants. A, Leaves of GFP and GFP/rdr6i plants systemically infected with PVX:00 or PVX:35S (21 dpi) visualized under UV light. B, Accumulation of GFP mRNA, PVX:35S gRNA, and as35S siRNAs assessed by northern-blot analyses using probes specific for GF and 35S promoter regions. Ethidium bromide-stained rRNAs are shown as loading controls. C, Schematic representation of the 35S:GFP:NOS (NOS, Nopaline Synthase) transgene illustrating the sizes (in kilobases) of the expected fragments after full Sau96I digestion. The 5′ (GF) and 3′ (P) regions are denoted in different shades of gray. D, Southern blot analysis after Sau96I digestion and detection with a probe specific for GF.
DNA samples were prepared from the same tissue used for the RNA analysis, and DNA methylation of the 35S promoter was assessed using the methylation-sensitive restriction enzyme Sau96I, which cuts within the targeted region (Fig. 2C). In PVX:00-infected control plants, a hybridizing band of 0.56 kb was detected, the size expected if the Sau96I site is unmethylated. PVX:35S-infected GFP plants produced an additional band of approximately 0.96 kb, indicative of DNA methylation within the 35S promoter, whereas in PVX:35S-infected GFP/rdr6i plants, the 0.56-kb band was the major band, with only a faint band visible at 0.96 kb (Fig. 2D). Thus, despite having high levels of 35S-specific siRNAs, the rdr6i plants were defective in PVX-driven RdDM.
TRV- But Not PVX-Derived siRNAs Drive Silencing in RDR6-Deficient Plants
Previous work has shown that the strength of TRV-related symptoms is similar in wild-type and rdr6i N. benthamiana plants (Schwach et al., 2005), suggesting that RDR6 does not influence TRV RNA accumulation. By assessing TRV RNA levels, we found that they accumulate to similar amounts in both genetic backgrounds (Supplemental Fig. S1A) and therefore provide molecular support to the prior observations that were based upon severity of symptoms. Additionally, by infecting N. benthamiana with TRV:PDS or TRV:35S, we show that TRV-driven RNA silencing and TGS are unaffected in rdr6i plants (Supplemental Fig. S1, B and C), which is in agreement with data previously obtained in Arabidopsis (Dalmay et al., 2000; Vaistij et al., 2002). We then inoculated GFP/rdr6i plants with TRV:35S or PVX:35S vectors and compared the levels GFP silencing and accumulation of 35S-specific siRNAs (Fig. 3): Infection by either vector produced similar levels of 35S siRNAs; however, only TRV:35S triggered a strong reduction of GFP mRNA accumulation (Fig. 3). Thus, these observations indicate that the difference in efficiency of silencing driven by TRV and PVX in the RDR6-deficient background is not due to quantitative differences in siRNA levels.
Figure 3.
Comparison of TRV:35S- and PVX:35S-driven silencing in GFP/rdr6i plants. A, Accumulation of GFP mRNA and genomic and subgenomic RNAs of TRV:35S and PVX:35S (both at 21 dpi) assessed by northern-blot analyses using GF and 35S probes. B, as35S siRNA accumulation. Ethidium bromide-stained rRNAs are shown as loading controls.
Impairment in PVX-Driven Silencing Is Independent of the PVX-Encoded Suppressor of Silencing
Since PVX accumulates at higher levels in the RDR6-deficient plants, one possible explanation for the inefficiency of VIGS is that a virus-encoded suppressor of silencing is hyperaccumulating. The PVX-encoded suppressor is the 25K protein that also has a role in virus movement (Voinnet et al., 2000; Bayne et al., 2005). PVX vectors deleted for the 25K gene [PVX(Δ25)] are able to replicate, produce 21-nt siRNAs, and trigger silencing but are unable to move out of the leaf veins (Himber et al., 2003). A PVX(Δ25) vector that carries fragments of both the GFP and the Rubisco small subunit (RbcS) genes [PVX(Δ25):GFP/RbcS] was used to infect GFP and GFP/rdr6i plants. At 28 dpi, GFP silencing extended over the whole leaf lamina, whereas areas of chlorosis that are indicative of RbcS silencing were restricted to around leaf veins (Fig. 4A; Himber et al., 2003). This difference in the extent of silencing is attributable to the ability of the GFP transcript (and not the RbcS mRNA) to act as a template for RDR6 to further generate dsRNA and secondary siRNAs and hence achieve silencing amplification. Interestingly, in equivalent leaves of PVX(Δ25):GFP/RbcS-infected GFP/rdr6i plants, no GFP or RbcS silencing was observed (Fig. 4A). Similar results showing a lack of silencing in GFP/rdr6i plants were obtained using a PVX(Δ25):GFP/PDS vector (data not shown). A PVX(Δ25):35S vector also failed to trigger silencing in GFP/rdr6i plants, whereas in GFP plants, GFP silencing was restricted to leaf veins (Fig. 4B). The difference in the extent of GFP silencing triggered by PVX(Δ25):GFP/RbcS or PVX(Δ25):35S in wild-type plants is attributable to the lack of a transcript corresponding to the 35S promoter that can be amplified by RDR6. The possibility that the absence of silencing observed in rdr6i plants is due to lack of infection by the PVX(Δ25) viral vectors was ruled out using the sensitive Rx-based PVX detection assay (Supplemental Fig. S2). Taken together, these observations show that the impairment of silencing in rdr6i plants is not a consequence of overaccumulation of the 25K protein.
Figure 4.
The impairment of PVX-driven silencing in rdr6i plants is independent of the 25K suppressor of silencing. A, Leaves of GFP and GFP/rdr6i plants systemically infected with PVX(Δ25):GFP/RbcS pictured at 28 dpi. For each genetic background, the same leaf was photographed under UV light and daylight. B, Leaves of systemically PVX(Δ25):35S-infected GFP and GFP/rdr6i plants pictured at 28 dpi under UV light.
Silencing Driven by PPV Is Also Impaired in RDR6-Deficient Plants
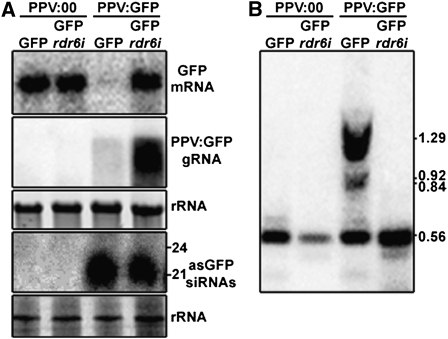
In order to investigate whether the inefficiency of PVX-driven silencing in the RDR6-deficient background also occurs with other viruses, we performed a series of experiments with PPV, a member of the potyvirus family. Initially, we determined that PPV hyperaccumulated in rdr6i plants (Supplemental Fig. S3), as has been reported for another member of the potyvirus family, the Potato virus Y (Schwach et al., 2005). Following these initial studies, a PPV:GFP vector was used to infect GFP and GFP/rdr6i plants in order to assess the efficiency of VIGS. Infections with a PPV:00 empty vector were used as negative controls. At 28 dpi, RNA samples were prepared from upper leaves and assessed for the accumulation of GFP mRNA, PPV:GFP RNA, and asGFP-specific siRNAs. In upper leaves of PPV:GFP-infected GFP plants, GFP mRNA was barely detectable, whereas in PPV:GFP-infected GFP/rdr6i plants, it was present at high levels despite accumulating the viral vector at higher levels than wild-type background plants (Fig. 5A). asGFP-specific siRNAs accumulated to similar levels in rdr6i and wild-type plants and were predominantly of the 21-nt size class in both genetic backgrounds (Fig. 5A).
Figure 5.
RNA degradation and DNA methylation driven by PPV is RDR6 dependent. A, Accumulation of GFP mRNA, PPV:GFP gRNA, and asGFP siRNAs in GFP and GFP/rdr6i plants systemically infected with PPV:00 or PPV:GFP (28 dpi) assessed by northern-blot analyses using probes specific for GFP and the Nopaline Synthase 3′ untranslated region of the transgenic GFP mRNA. Ethidium bromide-stained rRNAs are shown as loading controls. B, Southern-blot analysis of DNA samples extracted from the same tissue used for the RNA analysis. DNAs were digested with Sau96I digestion and analyzed using a GF-specific probe (see Fig. 2C).
RdDM of the GFP coding region was also assessed in PPV:GFP-infected plants using Sau96I digestion. In control PPV:00-infected plants, a major hybridizing band of 0.56 kb was detected. In PPV:GFP-infected GFP plants, higher molecular mass bands of 0.84, 0.92, and 1.29 kb were detected that are indicative of DNA methylation of Sau96I sites within the GFP transgene (Fig. 2C). However these higher molecular mass bands were not detected in PPV:GFP-infected GFP/rdr6i plants, indicating that RdDM was not occurring despite the accumulation of GFP siRNAs (Fig. 5B). Thus, these observations indicate that, in the absence of RDR6, PPV-driven VIGS and RdDM are compromised.
DISCUSSION
In this work, we demonstrate that virus-derived siRNAs can be functionally different in different virus-host systems. We show that for PVX and PPV, both VIGS and RdDM are compromised in RDR6-deficient N. benthamiana plants despite the accumulation of high levels of virus-derived siRNAs. The implication of this is that for some viruses, RDR6 is required to produce siRNAs that are active in directing silencing events and, importantly, that the primary siRNAs derived directly from the replicating virus are inefficient.
RDR6 was originally identified in genetic screens for Arabidopsis mutants defective in RNA silencing of transgenes and has subsequently been shown to be involved in defense against plant viruses and the production of trans-acting siRNAs from endogenous RNA templates (Dalmay et al., 2000; Mourrain et al., 2000; Peragine et al., 2004; Vazquez et al., 2004; Qu et al., 2005; Schwach et al., 2005). Some viruses such as PVX and PPV hyperaccumulate in the absence of RDR6, whereas others such as TRV are unaffected, indicating that RDR6 is only able to use a subset of viral RNAs as templates (Qu et al., 2005; Schwach et al., 2005). In this study, we have shown that efficient RNA silencing and RdDM driven by PVX or PPV were dependent upon RDR6 but those driven by TRV were not. These results confirm that the actual mechanisms of RNA cleavage and DNA methylation do not require RDR6, and we observe that the difference in efficiency of VIGS correlates with the viral RNA accumulation phenotype. A previous study showed that cabbage leaf curl virus (CaLCuV)-driven silencing is compromised in Arabidopsis rdr6 mutant plants (Muangsan et al., 2004). Since CaLCuV is a DNA-based virus belonging to the geminivirus family, it was hypothesized that the requirement for RDR6 was due to the lack of dsRNA intermediates generated during its replication cycle. However, here, we demonstrate that efficient RNA silencing directed by RNA-based viruses, such as PVX and PPV, can also require RDR6.
siRNAs that are produced during infection of wild-type plants can be derived directly from viral replication intermediates through the action of the virus-encoded replicase, from regions of secondary structure or via the copying action of a plant-encoded RDR. It is tempting, therefore, to speculate that the difference between the efficiency of VIGS in wild-type and rdr6i plants reflects the activity of siRNAs derived from distinct sources of dsRNA. The suggestion here is that in the absence of RDR6, PVX- and PPV-derived primary siRNAs are ineffective in driving RISC-mediated target cleavage (and RdDM), which leads to hyperaccumulation of these viruses. In agreement with this, VIGS driven by transgenic PVX amplicons has been shown to be defective in Arabidopsis rdr6 mutant plants despite accumulation of virus-derived siRNAs (Moissiard et al., 2007). The lack of impairment of TRV-driven VIGS in the RDR6-deficient background may be because the TRV-derived primary siRNAs are good effectors of silencing, with the difference between the virus families reflecting differences in viral pathology or interactions with other components of the silencing machinery. Alternatively, an RDR that is distinct from RDR6 may copy TRV RNAs to generate dsRNA and the subsequent effective secondary siRNAs. Interestingly, it was reported recently that, in Arabidopsis, TRV-derived siRNA production is strongly dependent on the combined RDR1, RDR2, and RDR6 functions (Donaire et al., 2008). This opens the possibility that, due to overlapping RNA template specificities, any of these three RDRs may be sufficient to produce TRV dsRNA and consequently secondary siRNAs.
Since both PVX and PPV RNAs accumulate at higher levels in the RDR6-deficient plants, another explanation for the inefficiency of VIGS during infections is the potential hyperaccumulation of virus-encoded silencing suppressor proteins. The PVX-encoded 25K and potyvirus-encoded HcPro proteins have been shown to act as suppressors of RNA silencing (Anandalakshmi et al., 1998; Brigneti et al., 1998; Voinnet et al., 2000; Kasschau and Carrington, 2001). However the lack of silencing by PVX in RDR6-deficient plants was found to be independent of 25K, as demonstrated in the experiments based upon PVX(Δ25). Additionally HcPro has been shown to suppress RNA degradation but not initiation and maintenance of DNA methylation (Mallory et al., 2001). Since, in our experiments, hyperaccumulation of PPV in RDR6-deficient plants failed to trigger both RNA degradation and RdDM, it is unlikely that impairment of silencing is due to the overaccumulation of HcPro. Hyperaccumulation of PVX or PPV causes stunting and leaf deformity, which raises the possibility that impairment of VIGS may be a consequence of the changes in host morphology, physiology, and biochemistry that occur in infected RDR6-deficient plants. However, VIGS and RdDM were also affected in the PVX(Δ25) infections, in which no symptoms of infection are visible.
It has been demonstrated in Arabidopsis that 21/22-nt-long DCL4/DCL2-produced viral siRNAs are active in directing RNA cleavage, whereas 24-nt-long DCL3-produced siRNAs are not (Deleris et al., 2006; Fusaro et al., 2006). A recent study in yeast has shown a physical association between an RDR and a DCR (Colmenares et al., 2007), raising the possibility that different virus-encoded replicases or plant-encoded RDRs may interact with specific DCLs. However, the PVX and PPV siRNAs that were produced in both wild-type and RDR6-deficient plants were predominantly of the 21-nt size class; therefore, we have no evidence to suggest that the difference in silencing is due to a difference in either the size of siRNAs produced or the DCL enzyme responsible for their synthesis. Furthermore, miR163, a 24-nt microRNA produced by DCL1 acting on an endogenous self-complementary dsRNA, leads to RISC-driven degradation of its RNA target (Allen et al., 2004), which shows that the length of the small RNA per se is not the determining feature for silencing activity.
As in plants, RNA silencing in Caenorhabditis elegans can be self-sustaining due to the action of an RDR, which maintains the production of secondary siRNAs following a triggering event driven by primary siRNAs (Sijen et al., 2001). Interestingly, the primary and secondary siRNAs in C. elegans are structurally distinct, with primary siRNAs having a 5′-monophosphate and secondary siRNAs having a 5′-triphosphate (Pak and Fire, 2007; Sijen et al., 2007). In contrast to primary siRNAs, secondary siRNAs are produced independently of DCR by single in-phase events of siRNA synthesis (Sijen et al., 2007). Thus, the biochemical differences between primary and secondary siRNAs may result from the differences in their mode of biosynthesis. Evidence suggests that the secondary siRNAs are more efficient for directing mRNA cleavage and that the difference in 5′-phosphorylation status is responsible for their differential activities (Aoki et al., 2007). In plants, only siRNAs with 5′-monophosphates have been found, and although the sample may be biased due to the methods used so far for cloning small RNAs, the synthesis of secondary siRNAs by host-encoded RDRs is not thought to occur by the same mechanism as in C. elegans. Production of both primary and secondary siRNAs is dependent upon DCL activities; therefore, differences in phosphorylation status between primary and secondary siRNAs in plants appear unlikely.
One possibility to explain the different activities of viral primary and secondary siRNAs is that they may not be incorporated into active effector complexes unless produced via a host-encoded RDR such as RDR6. One biochemical study failed to detect an association between the Argonaute1 (AGO1) RNase component of RISC and virus-derived siRNAs, despite being able to detect associations with other siRNAs (Baumberger and Baulcombe, 2005). Another study showed that only a minority of viral siRNAs are present in protein complexes, suggesting that most do not interact with RISC (Lakatos et al., 2004). However, a third study has demonstrated the incorporation of viral siRNAs into an AGO1/RISC complex (Zhang et al., 2006). Importantly, none of these studies considered whether the siRNAs are primary or secondary in origin. Therefore, it will be interesting to examine the association between viral siRNAs and RISC in RDR6-deficient backgrounds at distinct stages of the infection process of different viruses.
MATERIALS AND METHODS
Plant Material and Viral Inoculations
Plants were grown in a glasshouse with a 16-h supplementary lighting at a temperature of 22°C/20°C. GFP and rdr6i transgenic Nicotiana benthamiana lines (Ruiz et al., 1998; Schwach et al., 2005) as well as PVX(Δ25), PVX:35S, and PPV:GFP viral vectors have been described previously (Jones et al., 1999; Himber et al., 2003; Alamillo et al., 2006). The PVX:P vector was created by inserting the 3′ 359 nucleotides of the GFP coding region (P fragment) into the SmaI site of the pGR107 PVX binary vector in the sense orientation. PVX:PDS was created by introducing a PDS region (amplified using oligonucleotides pdS5' [5′-AATGAGGATGGAAGTGTCAAAT-3′] and pdS3' [5′-CAGGGATCCGGCACTCAACTTTATAAACCC-3′]) into the SmaI site of pGR107. PVX(Δ25):35S was derived from the pGR107-based PVX:35S vector by digestion with Bsu36I and ApaI and blunt end production using T4 DNA polymerase followed by self-ligation. The 35S:GFP and 35S:Rx agroinfiltration vectors were described previously, as was the use of Rx as a tool for detecting PVX infection (Haseloff et al., 1997; Bendahmane et al., 1999; Himber et al., 2003).
Nucleic Acid Extraction and Analysis
Extraction of nucleic acids and transfer for northern and Southern analysis were performed as described previously (Vaistij et al., 2002). 32P-labeled DNA probes were synthesized using a PCR-generated template for detection of high molecular mass viral RNAs and mRNAs following northern blotting or DNA fragments following Southern blotting. Probes used were specific for the 5,453 nucleotides of the GFP open reading frame (GF fragment), the nopaline synthase terminator of the GFP transgene (Vaistij et al., 2002), or the inserts carried by the viral vectors themselves, as indicated in the figure legends. PDS and actin mRNA levels were assessed by semiquantitative reverse transcription-PCR (RT-PCR) using primers PDs5' (5′-GGGATCCGGCACTCAACTTTATAAACC-3′) and PDs3' (5′-CAGCTCGATCTTTTTTATTCGT-3′) and primers actin5' (5′-CGGGAAATTGTTAGGGATGT-3′) and actin3' (5′-GATACGGGGAGCTAATGCAG-3′). Twenty PCR cycles were used, which resulted in visible bands for actin but not for PDS following agarose gel electrophoresis. PCR was continued to 30 cycles to confirm that 20 cycles was below the point of reaction plateau for both actin and PDS. In order to detect the PDS RT-PCR product, Southern blotting was performed with a PDS-specific probe. In vitro synthesized 32P-labeled RNA transcripts using T7 or SP6 RNA polymerases were used to detect small RNA species. Hybridization conditions for all nucleic acid detections were as performed previously (Vaistij et al., 2002), with the modification of using PerfectHyb Plus (Sigma). siRNA size was determined by comparison with 21- and 24-nt-long GFP-specific siRNAs that are produced upon agroinfiltration with a 35S:GFP binary vector (Hamilton et al., 2002).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RDR6-independent silencing driven by TRV.
Supplemental Figure S2. Systemic infection of rdr6i plants inoculated with PVX(Δ25):GFP/RbcS.
Supplemental Figure S3. Accumulation of PPV:00 is RDR6 dependent.
Supplementary Material
Acknowledgments
We thank Dianna Bowles for her generosity and support and Neil Taylor for comments on the manuscript. Biomaterials were kindly supplied by David Baulcombe (transgenic lines and TRV/PVX vectors), Olivier Voinnet [PVX(Δ25) vectors], and Juan-Antonio García (PPV vectors). Work with recombinant viruses was performed under license from the Department for Environment, Food and Rural Affairs (PHL 198A/555).
This work was supported by the Garfield Weston Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Fabián E. Vaistij (fv4@york.ac.uk).
The online version of this article contains Web-only data.
References
- Alamillo JM, Saenz P, Garcia JA (2006) Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J 48 217–227 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36 1282–1290 [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95 13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H (2007) In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, Rakitina DV, Morozov SY, Baulcombe DC (2005) Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J 44 471–482 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27 449–461 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553 [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68–71 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Matínez-García B, Matínez-Priego L, Pagán I, Llave C (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol 82 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Voinnet O (2005) The complex interplay between plant viruses and host RNA-silencing pathways. Curr Opin Plant Biol 8 415–423 [DOI] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15 1494–1500 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM (2005) Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579 5822–5829 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC (1999) RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11 2291–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285 71–81 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyan J (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J 23 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance VB (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Parizotto EA, Himber C, Voinnet O (2007) Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542 [DOI] [PubMed] [Google Scholar]
- Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38 1004–1014 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241–244 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ (2005) RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79 15209–15217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert VJ, Vastenhouw NL, Plasterk RH (2004) RNA interference, transposon silencing, and cosuppression in the Caenorhabditis elegans germ line: similarities and differences. Cold Spring Harb Symp Quant Biol 69 397–402 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55 495–519 [DOI] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476 [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH (2007) Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244–247 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20 759–771 [DOI] [PubMed] [Google Scholar]
- Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11 460–468 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 6 206–220 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167 [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL (1994) RNA-directed de novo methylation of genomic sequences in plants. Cell 76 567–576 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.