Abstract
The Signal Peptide Peptidases (SPP) are members of the Intramembrane Cleaving Proteases, which are involved in an array of protein-processing and intracellular signaling events in animals. Arabidopsis (Arabidopsis thaliana) has six genes encoding SPP-like proteins, the physiological functions of which are unknown. As a first step in defining the roles of the SPPs in plants, we examined the distribution and activities of Arabidopsis SPP (AtSPP; accession no. At2g03120), the SPP-like gene with the highest degree of similarity to human SPP. The protease is expressed at low levels throughout the plant, with the highest levels in emerging leaves, roots, and floral tissues. Homozygous plants carrying a T-DNA insertion mutation in AtSPP, spp-2, could not be recovered, and transmission of the mutant allele through pollen was reduced to less than 2% in reciprocal cross experiments. Although viable, pollen from spp-2 heterozygous plants exhibited a 50% reduction in germination rate and a disruption in male germ unit organization. These data demonstrate that AtSPP is required for male gametophyte development and pollen maturation in Arabidopsis.
Intramembrane Cleaving Proteases (I-CLiPs) are a family of integral membrane proteases involved in a wide array of protein-processing and intracellular signaling events (Golde and Eckman, 2003; Weihofen and Martoglio, 2003). The I-CLiPs include aspartyl proteases (e.g. presenilin), metalloproteases (e.g. site-2 protease), and Ser proteases (e.g. Rhomboid). Regulated intramembrane proteolysis involving I-CLiPs is an essential step in many cellular and developmental processes in animals, including cholesterol homeostasis, Notch signaling, the unfolded protein response, and the pathogenesis of Alzheimer's disease (Brown et al., 2000). Although the functions of several I-CLiPs are well established in animal systems, their roles in plants have not been extensively investigated (Kanaoka et al., 2005).
One class of I-CLiPs is exemplified by Signal Peptide Peptidase (SPP; also PSH and IMPAS), an endoplasmic reticulum (ER)-resident protease that cleaves certain signal peptides after they have been released from proteins entering the secretory pathway (Grigorenko et al., 2002; Weihofen et al., 2002). Since the signal peptide fragments are then released from the ER membrane, one function of SPP simply may be to prevent the accumulation of signal peptides in the ER membrane. However, a growing body of evidence implicates SPP family members in regulated intramembrane proteolysis, because their proteolytic products have downstream functions in signaling and protein trafficking. For example, cleavage by SPP is a required step in generating human lymphocyte antigen E epitopes from the signal peptide of major histocompatibility complex class I proteins (Lemberg et al., 2001). Two other SPP-like proteins, SPPL2a and SPPL2b, promote cleavage of tumor necrosis factor-α at the cell surface (Fluhrer et al., 2006; Friedmann et al., 2006). Thus, the SPP family plays essential roles in immune surveillance and response. SPP also appears to be involved in protein processing and quality control at the ER. SPP is exploited by hepatitis C virus to process an internal signal peptide and release the viral coat protein to the cytosol (McLauchlan et al., 2002). A recent cross-linking study showed an association between SPP and unassembled opsin fragments, suggesting a role for SPP in quality control of polytopic membrane proteins (Crawshaw et al., 2004). Other recent data implicate SPP in US2-mediated dislocation and degradation of class I major histocompatibility complex heavy chains from the ER membrane (Loureiro et al., 2006), suggesting a role for SPP in ER-associated protein degradation.
Human SPP is an integral membrane protein with seven transmembrane segments, oriented such that its N terminus is lumenal and its C terminus is cytosolic. SPP is an aspartic protease and, consistent with the enzyme's ability to catalyze intramembrane cleavage, the two active site Asp residues are located in the middle of adjacent transmembrane domains. The active site of SPP is similar to that of presenilin, and the two enzymes have overlapping inhibitor sensitivities. Numerous SPP-like proteins have been identified in humans and other animals, and they all share certain structural features: (1) they are predicted to be polytopic integral membrane proteins; (2) they contain the conserved active site motifs YD and LGLGD; and (3) they contain a conserved sequence of unknown function, QPALLYxxP (Weihofen et al., 2002).
Several recent studies have used genetic approaches to examine the physiological functions of SPP in various animal models. Knockdown of SPP in zebrafish embryos led to cell death within the nervous system (Krawitz et al., 2005). RNA interference of the SPP ortholog in Caenorhabditis elegans resulted in a set of developmental defects similar to those caused by disruption of lipid steroid homeostasis and signaling (Grigorenko et al., 2004). An SPP ortholog is also necessary for normal development in Drosophila melanogaster (Casso et al., 2005). Although in each case loss of SPP led to relatively specific defects in various organs and processes in these diverse animals, the specific biochemical pathways or SPP substrates involved have not yet been defined.
The SPP family is conserved across all multicellular eukaryotes examined, including plants. Arabidopsis (Arabidopsis thaliana) contains six potential genes encoding SPP-like proteins (Ponting et al., 2002; Friedmann et al., 2004; Tamura et al., 2008). A recent study showed that AtSPP, the Arabidopsis protein most closely related to human SPP, and five other SPP-like genes are differentially expressed in Arabidopsis and that their proteins are differentially localized in subcellular organelles (Tamura et al., 2008). However, the roles of these proteases in cell signaling and development in plants are unknown. In this report, we provide evidence that one member of this family, AtSPP, is highly expressed in pollen, developing ovules, endosperm, and developing vegetative tissues. Consistent with the expression profile, we provide genetic evidence that AtSPP activity is essential for pollen maturation and germination. These data demonstrate a critical role for AtSPP in pollen germination and reveal important roles for the SPP family of proteases in plant reproduction.
RESULTS
Expression Profile of AtSPP
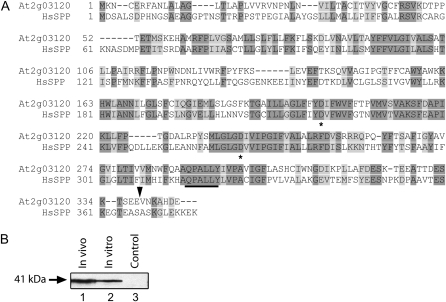
As a first step in defining the functions of the SPP-like genes in Arabidopsis, we focused our attention on the gene most closely related to human SPP (Fig. 1A; Weihofen et al., 2002). This gene, At2g03120, was previously designated AtSPP by Tamura et al. (2008). AtSPP is predicted to encode an integral membrane protein of the ER with an N-terminal signal sequence. Subcellular fractionation and fluorescence localization data have confirmed its ER localization (Tamura et al., 2008). To examine the processing and localization of AtSPP in vivo, we transformed wild-type Arabidopsis plants with a gene expressing AtSPP fused to two tandem Flag epitope tags (Hopp et al., 1988) inserted upstream of the putative ER localization signal (Fig. 1A; AtSPP-Flag, 41 kD). This construct was placed under the control of the putative AtSPP native promoter (i.e. a 1-kb fragment of intragenic sequence upstream of the At2g03120 transcriptional start site). To determine whether AtSPP is processed in vivo, we compared the size of AtSPP-Flag expressed in vivo in transgenic plants with that of in vitro translated AtSPP-Flag (Fig. 1B) by immunoblotting with anti-Flag antibodies. Both proteins exhibited the same mobility on SDS-PAGE, supporting the conclusion that AtSPP is not processed upon insertion into the ER membrane. It should be noted that, unlike human SPP, AtSPP lacks apparent consensus N-linked glycosylation sites (Asn-Xaa-Ser/Thr triplet) within its primary structure (Ronin et al., 1978a, 1978b).
Figure 1.
Primary sequence and processing of AtSPP. A, Alignment of the predicted primary sequence of AtSPP (At2g03120) with human SPP (HsSPP). Identical residues are highlighted in dark gray, and similar residues are highlighted in light gray. The conserved active site Asp residues are marked with asterisks, and the conserved QPALLY sequence is underlined. The black arrowhead indicates the point of disruption of the primary sequence by the T-DNA insertion in the spp-2 mutant. B, Comparison of in vivo expressed and in vitro translated AtSPP-Flag. Protein extracted from AtSPP-Flag transgenic plants (lane 1) or translated in an in vitro reticulocyte lysate (lane 2) was resolved by SDS-PAGE and immunoblotted with anti-Flag antibody. Protein extract from untransformed plants was used as a negative control (lane 3).
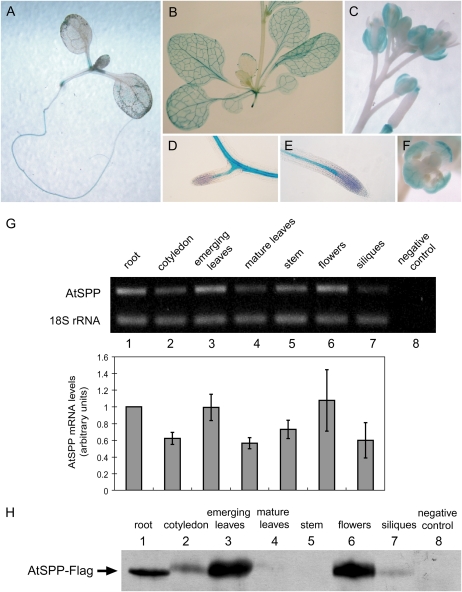
To determine the tissue distribution of AtSPP expression, we created transgenic lines carrying the GUS gene fused to the putative promoter region of AtSPP. Faint GUS staining was observed in most tissues throughout the plant, with somewhat stronger staining in emerging leaves and especially intense staining in the stipules, roots, flowers, and vasculature of leaves (Fig. 2, A–F). To confirm the GUS staining results, we estimated the relative abundance of transcript from the endogenous AtSPP gene in various organs using comparative reverse transcription (RT)-PCR with 18S rRNA as our internal control (Fig. 2G). Root, cotyledon, and emerging leaves were dissected from 7-d-old seedlings. Mature leaves, stems, flowers, and siliques were collected from mature plants at least 3 weeks old. AtSPP transcript levels were highest in emerging leaves, flowers, and roots, with moderate levels detected in other tissues. These results are generally consistent with microarray data available online (https://www.genevestigator.ethz.ch/). For example, developmental data from the AtGenExpress project (Schmid et al., 2005) also showed that AtSPP transcript levels were highest in root, shoot apex, carpel, and stamen/pollen as well as in stage 6 and 7 seeds.
Figure 2.
Expression pattern of AtSPP. A to F, Expression of a transcriptional fusion of the GUS reporter gene to the promoter of AtSPP. A, One-week-old seedling. B, Rosette and stem of a 3-week-old plant. C, Inflorescence. D, Lateral root of a 1-week-old plant. E, Primary root tip of a 1-week-old plant. F, Open flower. G, Semiquantitative RT-PCR analysis of AtSPP transcript abundance. The ratio of AtSPP transcript over 18S rRNA in RT-PCR of each tissue was normalized against the average ratio in root samples and plotted using arbitrary units. Each bar represents the mean and sd of three replicates. H, Expression pattern of AtSPP-Flag in Col-0 plants transformed with AtSPP-Flag expressed from its native promoter. Total protein (20 μg per lane) was extracted from various organs of a single-copy transgenic line (n = 2) and subjected to immunoblot analysis using an anti-Flag monoclonal antibody. An extract from flower buds of untransformed plants (wild type) was used as the negative control (lane 8).
To examine the distribution of AtSPP protein, we immunoblotted total protein extracts from wild-type plants expressing AtSPP-Flag with anti-Flag antibody. Plant material was harvested at a similar stage of plant development as that used for RT-PCR. The AtSPP signals were strongest in emerging leaves and flowers, with lower levels of protein detected in roots of 7-d-old seedlings (Fig. 2H). A faint signal also was seen in cotyledons, mature leaves, and siliques, with no detectable protein in stem extracts. These results were in general agreement with the mRNA levels, although the protein expression data suggested a more significant difference in AtSPP expression in different tissues.
T-DNA Insertion Mutants in AtSPP
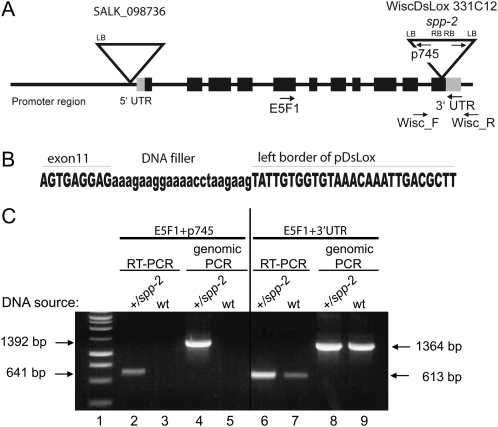
The tissue expression profile of AtSPP suggested that this gene might play a specialized role in roots and developing tissues, such as emerging leaves and reproductive tissues. To further explore the function of AtSPP, we characterized two AtSPP T-DNA insertion lines in the Columbia (Col-0) ecotype, obtained from the Arabidopsis Biological Resource Center (ABRC). The first T-DNA line, WiscDsLox 331C12 (http://www.hort.wisc.edu/krysan/DS-Lox/; http://signal.salk.edu/cgi-bin/tdnaexpress) or spp-2, was identified by a BLAST search of available T-DNA border sequences using the AtSPP genomic sequence. Although the annotation of this insertion does not reference the At2g03120 locus, our analysis indicated that it contains tandem repeated copies of the T-DNA inserted in the last exon of the gene (Fig. 3, A and B). Therefore, this line appears to be misannotated in the databases. This line was backcrossed with Col-0 wild-type plants to eliminate any additional T-DNA insertions that are unrelated to the T-DNA insertion in At2g03120.
Figure 3.
T-DNA insertion alleles of AtSPP. A, Map of the two T-DNA insertions (spp-1 and spp-2) in AtSPP (At2g03120). LB, T-DNA left border; RB, T-DNA right border; UTR, untranslated region. B, Detailed sequence of the pDsLox331C12 (spp-2) insertion site into the AtSPP locus. Extra nucleotides are apparent between the AtSPP and T-DNA sequences, resulting in the misannotation of this T-DNA line. C, RT-PCR detection of the aberrant AtSPP transcript in cDNA from +/spp-2 floral tissue. Genomic PCR was performed to verify the genotype of plants used to extract RNA. PCR was performed using cDNA or genomic DNA and the E5F1 (exon-specific primer) and p745 (T-DNA left border-specific primer) primer set (lanes 2–5) or the E5F1 and 3′ UTR-R (3′ UTR-specific primer; lanes 6–9) primer set as indicated. The positions of the primers are indicated in A.
Genomic PCR with gene-specific and T-DNA-specific primers confirmed the presence of normal and T-DNA-disrupted alleles (Fig. 3C, compare lanes 4 and 8) in spp-2 heterozygous plants. RT-PCR demonstrated that the spp-2 heterozygotes produce, in addition to the normal AtSPP transcript (Fig. 3C, lane 6), an aberrant transcript that includes most of the coding region of AtSPP and extends into the T-DNA (Fig. 3C, lane 2). If translated, this transcript would produce a protein that is missing the last seven amino acids of AtSPP, including a putative ER retention signal (Fig. 1A), and has an additional 29 amino acids derived from the T-DNA sequence.
The second T-DNA insertion line, SALK_098736 (Alonso et al., 2003), contains a T-DNA insertion in the putative promoter region of AtSPP, 40 bp upstream of the predicted transcriptional start site (Fig. 3A). The expected PCR product was amplified from DNA extracted from SALK_098736 heterozygous plants using a primer for the left border of the T-DNA and a gene-specific primer upstream of the insertion site (data not shown). However, no product was generated from the right border side of the T-DNA insertion in this line. Southern-blot analysis indicated the likelihood of a deletion or chromosomal rearrangement in this region of the inserted allele (data not shown; Bonhomme et al., 1998; Tax and Vernon, 2001). Analysis of SALK_098736 indicated that it exhibited fertility defects, and homozygous SALK_098736 plants were not recovered (data not shown). However, we were unable to complement these phenotypes by transgenic expression of AtSPP using constructs that complemented spp-2 phenotypes (data not shown; see below). These observations suggested that the phenotypes resulting from the T-DNA insertion in SALK_098736 could not be attributed solely to AtSPP. As a result, we did not further characterize this insertion line.
T-DNA Insertion Causes a Male Gametophyte Defect in spp-2
Heterozygous spp-2 individuals (+/spp-2) appeared normal, but no homozygotes were recovered based on PCR genotyping (Table I), indicating that very few, if any, homozygous embryos were viable (Drews et al., 1998). Segregation analysis of the spp-2 allele (conferring basta resistance) indicated that 52.4% of the progeny of heterozygous plants were basta resistant, compared with the expected 75% resistance expected for a nonlethal allele or the 66.6% resistance expected for an embryo-lethal allele. These data suggested a potential gametophytic defect in the T-DNA mutant line (Drews and Yadegari, 2002). To determine whether the defect in the spp-2 mutant was associated with the male or female gametophytes, or both, we performed reciprocal crosses of spp-2 heterozygotes with wild-type plants (Table I). When spp-2 heterozygotes served as the male parent, the transmission efficiency was only 1.6%. The progeny of outcrosses using spp-2 heterozygotes as the female recipient showed a transmission efficiency of 95.2%, indicating no significant female gametophytic defect. This result indicated that pollen carrying the mutant allele was nearly completely defective. Thus, the inability to recover spp-2 homozygous plants appeared to be due to a severe defect in male gametophyte development or function. The fact that spp-2 homozygotes cannot be recovered suggested that the aberrant transcript generated in the T-DNA line (Fig. 3C) is not translated or generates a mislocalized or dysfunctional protein.
Table I.
Segregation of the spp-2 mutant allele in the progeny of self-crossed and reciprocal outcrossed plants
BastaR, Basta resistant; BastaS, basta sensitive; wt, wild type.
| Self-Crosses | No. of Progeny | Genotypes of Progeny (PCR)
|
||
|---|---|---|---|---|
| +/+ (%) | +/spp-2 (%) | spp-2/spp-2 | ||
| Parental genotype | ||||
| +/spp-2 | 81 | 44 (54.3)a | 37 (45.7)a | 0 |
| Resistance to Basta
|
||||
| BastaS (%)
|
BastaR (%)
|
|||
| +/spp-2 | 643 | 306 (47.6)b | 337 (52.4)b | |
| Reciprocal Outcrosses | No. of Progeny | Resistance to Basta
|
Transmission Efficiencyc | |
| BastaS (+/+) | BastaR (+/spp-2) | |||
| Recipient × donor | ||||
| wt × +/spp-2 | 376 | 370 | 6 | 1.6 |
| +/spp-2 × wt | 123 | 63 | 60 | 95.2 |
Significantly different from the expected 1:2:1 ratio for normal Mendelian segregation (χ2 = 49.2; P < 0.001). Not significantly different from the 1:1:0 ratio for a gametophytic defect (χ2 = 0.605; P > 0.05).
Significantly different from the expected 1:2:1 ratio for normal Mendelian segregation (χ2 = 174.0; P < 0.001). Not significantly different from the 1:1:0 ratio for a gametophytic defect (χ2 = 0.222; P > 0.05).
Transmission efficiency = (mutant/wild type) × 100 (%).
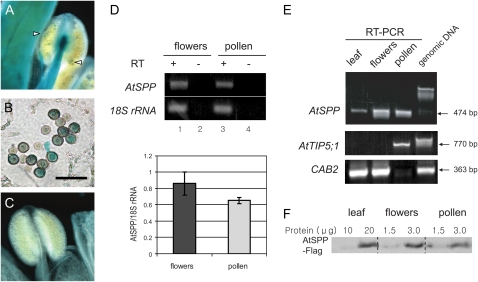
More detailed studies of AtSPP expression in male gametophytic tissues were consistent with a role for the protease in pollen development and function. Analysis of expression of the AtSPP promoter-GUS transcriptional fusion in the anthers of transgenic plants demonstrated expression in intact (Fig. 4A) and sectioned (Fig. 4B) pollen grains. Comparative RT-PCR analysis of RNA isolated from whole flowers and purified pollen from wild-type plants indicated that AtSPP mRNA levels are similar in both samples (Fig. 4D). mRNA levels in these tissues are significantly higher than in leaf tissue (Fig. 4E, top), but AtSPP expression is less tissue-specific than that of the pollen-specific marker, AtTIP5;1, or a photosynthesis gene not expressed in pollen, CAB2. The high levels of expression of AtSPP in flowers and pollen relative to leaf tissues were confirmed by immunoblotting of protein extracts from plants expressing AtSPP-Flag under the control of the AtSPP promoter (Fig. 4F).
Figure 4.
Expression of AtSPP in male gametophyte tissues. A to C, Photomicrographs of anthers from stage 12 to 13 (Smyth et al., 1990) flowers of wild-type (C) or AtSPP-GUS (A and B; white arrowheads) plants. A cross-section image of a paraffin-embedded anther of the AtSPP-GUS plant is shown in B. Bar = 50 μm. D, Semiquantitative RT-PCR analysis of AtSPP transcript abundance in flowers or pure pollen from wild-type plants at the same stage of flower development. Transcript abundance was normalized by measuring the ratio of AtSPP transcript to 18S rRNA. Each bar represents the mean and sd of three replicates. E, RT-PCR analysis of AtSPP transcript abundance in leaf, flower, and pollen relative to the pollen-specific transcript, AtTIP5;1, and a transcript not expressed in pollen, CAB2. F, Comparison of the expression levels of AtSPP-Flag in leaves, flowers, and pure pollen of transgenic plants. Two-fold serial dilutions of total protein extracts from leaves, flowers, and pure pollen were immunoblotted using an anti-Flag monoclonal antibody. Note that the amount of protein loaded for the leaf samples is 7-fold higher than that of the corresponding samples from flowers and pollen.
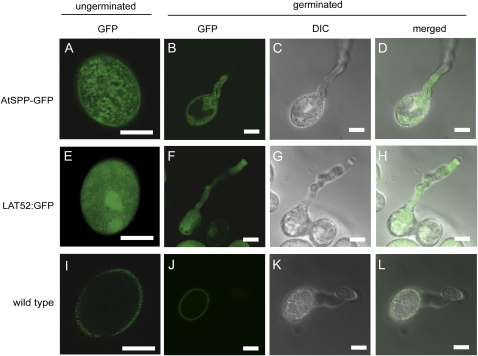
We transformed plants with an AtSPP construct containing the GFP (AtSPP-GFP) inserted just upstream of the putative ER retention signal to examine the subcellular distribution of AtSPP in pollen. GFP fluorescence in pollen from AtSPP-GFP plants exhibited a reticular pattern (Fig. 5A). The pattern was distinct from the diffuse cytoplasmic and nuclear signal when GFP was expressed under the control of the LAT52 pollen-specific promoter (Fig. 5E). Furthermore, the reticular AtSPP-GFP signal appears to move from the pollen body into the growing pollen tube (Fig. 5, B–D), whereas GFP alone is distributed evenly throughout the body and tube of germinated pollen (Fig. 5, F–H). Taken together, the results in Figures 4 and 5 were consistent with the high expression levels and ER localization of AtSPP in pollen.
Figure 5.
Localization of AtSPP-GFP in pollen. Confocal microscopy images of GFP fluorescence, differential interference contrast (DIC), and merged images are shown as indicated. A to D, AtSPP-GFP fluorescence in ungerminated (A) and germinated (B–D) pollen of AtSPP-GFP plants. E to H, GFP fluorescence in ungerminated (E) and germinated (F–H) pollen of LAT52:GFP plants. I to L, Autofluorescence signal in wild-type ungerminated (I) and germinated (J–L) pollen. Bars = 10 μm.
Pollen Produced by spp-2 Heterozygotes Showed Poor Germination
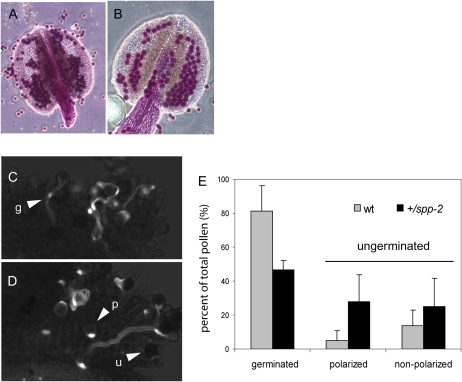
To determine the basis of the male defect conferred by the spp-2 allele, we first examined the viability and morphology of pollen shed from heterozygous plants. Col-0 wild-type plants and wild-type siblings from the spp-2 heterozygous line were used as controls. Using Alexander stain to score pollen viability, there was little difference between spp-2 heterozygote pollen and that from the wild-type controls (Fig. 6, A and B; 98.7% and 99.0% viability, respectively). A detached pistil assay (Lalanne et al., 2004) was used to measure the germination rate of pollen from spp-2 heterozygotes (Fig. 6, C and D). Despite the normal appearance of the mutant pollen, they showed a significantly lower rate of germination (47%) compared with that of wild-type pollen (81%) within the 2 h of the assay (P < 0.01; Fig. 6, C–E). Assuming that half of the pollen from the heterozygote was wild type and germinated at the normal rate, these data implied that the mutant pollen accounts for the approximately 50% reduction in germination rate in +/spp-2 plants (Fig. 6E). A large proportion of ungerminated pollen grains from spp-2 heterozygotes polarized but failed to germinate, showing intense aniline blue fluorescence at one side of the grain (Fig. 6D, arrowheads). This result suggested that many of the ungerminated pollen grains of the mutant were viable and able to respond to stigma signals but were retarded or unable to proceed further in the germination process within the 2 h of the assay. The germination defect is likely to account for the extremely low transmission efficiency of the spp-2 allele (Table I).
Figure 6.
Pollen viability and germination in +/spp-2 plants compared with wild-type plants. A and B, Alexander-stained anthers from Col-0 wild-type (A) and +/spp-2 (B) plants. C and D, Micrographs of pollen germination assays of wild-type (C) and spp-2 heterozygote (D) pollen on detached wild-type pistils. Examples of germinated (g), polarized (p), and ungerminated (u) pollen grains are indicated. E, Proportion of pollen grains that germinated after 2 h at room temperature for wild-type (wt) siblings (n = 420; gray bars) and heterozygotes (n = 330; black bars) of spp-2. Ungerminated pollen was subdivided into polarized and nonpolarized.
Genetic Complementation of spp-2 Plants
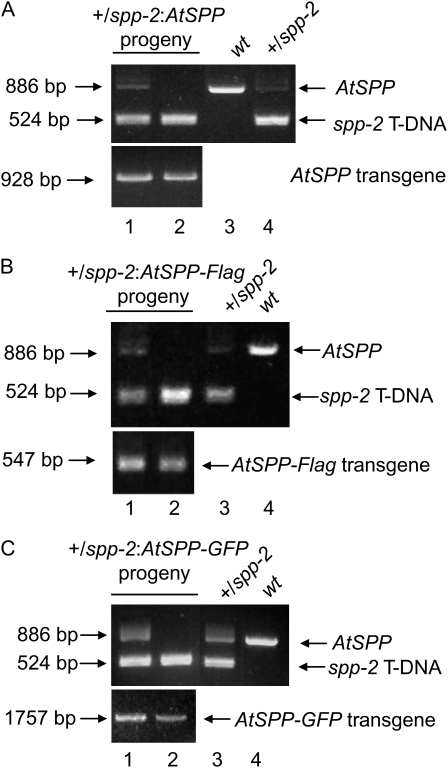
In order to confirm that the male defect in spp-2 is due to disruption of AtSPP, genes encoding AtSPP, AtSPP-Flag (Fig. 2), or AtSPP-GFP (Fig. 5) under the control of the AtSPP native promoter were introduced into +/spp-2 plants by pollinating +/spp-2 with the pollen from transgenic plants expressing each transgene. Segregation analysis of the F2 generation from the crosses demonstrated that AtSPP, AtSPP-Flag, or AtSPP-GFP expression increased the ratio of basta-resistant to basta-sensitive progeny, indicating complementation of the spp-2 allele (Table II). Homozygous spp-2 plants were recovered in each of the three lines, as demonstrated by PCR genotyping of the progeny from transformed +/spp-2 plants (Fig. 7, lane 2). These data confirmed that the male gametophyte defect in the spp-2 lines was due to a defect in AtSPP function. Furthermore, these data validate the expression and localization studies using AtSPP-Flag and AtSPP-GFP lines that are presented in Figures 2, 4, and 5.
Table II.
Complementation of the spp-2 mutant by expression of AtSPP, AtSPP-Flag, or AtSPP-GFP
BastaR, Basta resistant; BastaS, basta sensitive.
| Parental Genotype | No. of Progeny | Resistance to Basta in F2
|
χ2a | P | |
|---|---|---|---|---|---|
| BastaR (%) | BastaS (%) | ||||
| +/spp-2 | 906 | 456 (50.3) | 450 (49.7) | 0.040 | NSb |
| +/spp-2:AtSPP | 414 | 256 (62) | 158 (38) | 23.198 | <0.001 |
| +/spp-2:AtSPP-Flag | 608 | 368 (60.5) | 240 (39.5) | 26.9 | <0.001 |
| +/spp-2:AtSPP-GFP | 764 | 428 (56.0) | 336 (44.0) | 11.1 | <0.001 |
Calculated χ2 values were based on an expected 1:1 ratio.
NS, Not significant (P > 0.05).
Figure 7.
Complementation of spp-2 plants. Genes encoding AtSPP (A), AtSPP-Flag (B), and AtSPP-GFP (C) under the control of the AtSPP native promoter were introduced into +/spp-2 plants. +/spp-2 plants homozygous for each transgene were self-crossed, and their progeny were genotyped using PCR to detect the wild-type AtSPP allele, the spp-2 T-DNA insertion, or the appropriate transgene. PCR genotypes of spp-2 heterozygous (lane 1) and homozygous (lane 2) plants carrying the transgenes are shown. Lanes 3 and 4 contain control PCR genotypes of wild-type (wt) and +/spp-2 plants as indicated.
The Organization of the Male Germ Unit Is Disrupted in +/spp-2
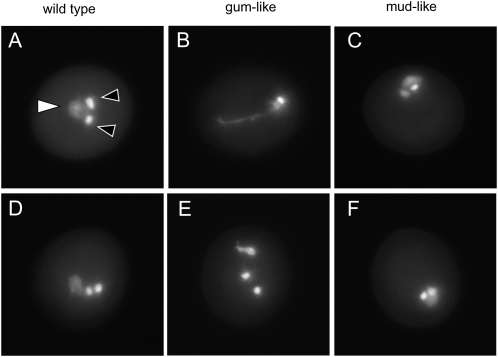
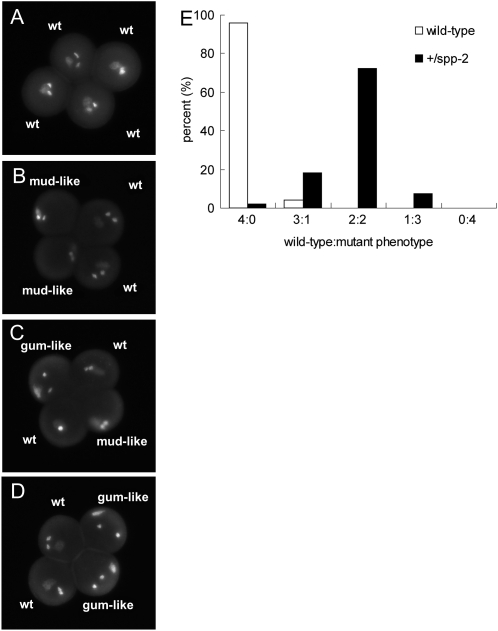
During our analyses of pollen structure and viability in the +/spp-2 plants, we frequently observed abnormal nuclear morphology in pollen stained with 4′,6-diamidino-2-phenylindole (DAPI) solution. In tricellular pollen species such as Arabidopsis, two consecutive mitotic events (pollen mitosis I and II) following meiosis give rise to three cells within each pollen grain. These correspond to a single, round vegetative nucleus and two small sperm cells each containing an additional nucleus. These three structures constitute the male germ unit (MGU; Matthysrochon et al., 1987). Normal pollen exhibit a less densely stained vegetative nucleus and two densely stained sperm nuclei (Fig. 8, A and D), which are centrally located within the grain. Interestingly, the pollen population in +/spp-2 showed a high frequency of abnormal nuclear morphology (Fig. 8, B, C, E, and F). The vegetative nuclei were malformed or stretched, with displacement of the two sperm cells (Fig. 8, B and E), or the entire MGU was mislocalized to the pollen periphery (Fig. 8, C and F). Those phenotypes were similar to the previously reported gum (for germ unit malformed) and mud (for MGU displaced) mutants (Lalanne and Twell, 2002). Consequently, we refer to the spp-2 phenotypes as gum-like and mud-like (Fig. 8, B, C, E, and F; Table III). Quantification of the nuclear defects (Table III) indicates that approximately 50% of pollen grains from +/spp-2 plants were affected, with the majority of abnormal pollen exhibiting a mud-like phenotype (displaced MGU). However, there was a minor difference between the mud-like phenotype of +/spp-2 and the previously described mud mutants. In the case of +/spp-2, the vegetative nucleus in the MGU was positioned closer to the pollen periphery than the sperm cells (Fig. 8, C and F). By contrast, the vegetative nucleus of mud mutants was positioned inward, with the sperm cells more closely apposed to the pollen periphery (Lalanne and Twell, 2002).
Figure 8.
MGU morphology of mature pollen in +/spp-2. The nuclei of mature pollen grains from +/spp-2 plants were visualized by DAPI staining. Wild-type pollen grains (A and D) show a typical arrangement of the MGU, consisting of a well-centered vegetative nucleus (white arrowhead) and two sperm cells (black arrowheads). Pollen from +/spp-2 plants shows an abnormal gum-like (B and E) or mud-like (C and F) MGU morphology characteristic of the gum and mud mutants (Lalanne and Twell, 2002).
Table III.
Comparison of MGU morphology of pollen from +/spp-2 and spp-2 complemented plants with that of wild-type siblings (+/+)
| Genotype | No. of Pollen Grains | Pollen Phenotypes
|
||
|---|---|---|---|---|
| Wild Type | Gum Like | Mud Like | ||
| % | ||||
| +/+ | 489 | 100 | 0 | 0 |
| +/spp-2 | 831 | 53.8 | 18.6 | 27.6 |
| spp-2:AtSPP-Flag | 109 | 97.2 | 2.8 | 0 |
| spp-2:AtSPP-GFP | 147 | 83.7 | 8.1 | 8.2 |
To confirm that the gum-like and mud-like phenotypes were due to the disruption of AtSPP, we tested whether the phenotypes were attenuated in spp-2 plants complemented with AtSPP-Flag or AtSPP-GFP. The frequency of gum-like and mud-like phenotypes was reduced to 2.8% and 16.3% in the spp-2:AtSPP-Flag and spp-2:AtSPP-GFP lines, respectively (Table III). These results demonstrated a direct correlation between AtSPP disruption and the gum-like and mud-like mutant phenotypes in spp-2 pollen.
The approximately 50% incidence of the gum-like and mud-like phenotypes in +/spp-2 pollen suggested a gametophytic rather than a sporophytic defect in pollen development (Table III). To test this, we performed tetrad analysis of +/spp-2 plants that had been crossed with the quartet1 (qrt1) mutant (Preuss et al., 1994; Johnson-Brousseau and McCormick, 2004). In the qrt mutant, all four microspores remain attached during meiosis. If the ratio of normal to affected pollen in each quartet is close to 2:2, the effect must be gametophytic. A biased ratio of defective to normal pollen in the tetrads would indicate a sporophytic defect that affects the development of pollen rather than a pure gametophytic defect (Johnson-Brousseau and McCormick, 2004). After crossing the two lines, we selected plants that were qrt1/qrt1 and +/spp-2. We quantified the mutant phenotype in pollen from these plants. As expected, 72.2% of total intact quartets exhibited the mutant phenotypes, with a 2:2 ratio of normal to abnormal (i.e. mud-like or gum-like) pollen in each tetrad (Fig. 9, B–E). Therefore, the tetrad analysis strongly suggested that the gum-like and mud-like phenotypes resulting from the mutation in AtSPP are the results of a gametophytic defect.
Figure 9.
Tetrad analysis of wild-type and +/spp-2 pollen. A, MGU morphology of a single wild-type (wt) quartet (+/+; qrt1/qrt1). B to D, Examples of MGU morphology in single spp-2 quartets (+/spp-2; qrt1/qrt1). E, Quantification of tetrad analysis of wild-type (white bars; n = 101) and +/spp-2 (black bars; n = 133) pollen. Only intact quartets of spp-2 heterozygous or wild-type plants were counted. Both gum-like and mud-like morphologies were combined and defined as mutant phenotypes in the graph.
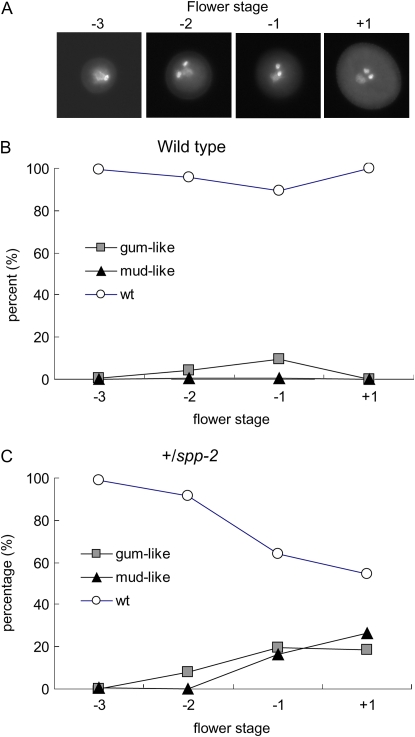
The gum and mud phenotypes began to appear after pollen mitosis II and increase during pollen maturation (Lalanne and Twell, 2002). We examined the frequency of the mutant phenotypes in +/spp-2 (Fig. 10C) or wild-type (Fig. 10B) pollen over the time course of pollen development to determine the stage that the mud-like and gum-like phenotypes appeared in the AtSPP mutant (Fig. 10). The gum-like phenotype first appeared at the early tricellular stage (−2 stage) and increased up to a maximum of approximately 20% of mature pollen (Fig. 10C). The appearance of the mud-like phenotype began at the late tricellular stage (−1 stage) and continued to increase to approximately 25% of mature pollen (+1 stage; Fig. 10C). The onset of each mutant phenotype during pollen maturation is similar to that observed for the gum and mud mutants, suggesting that the AtSPP defect and the mud and gum mutations affected a similar stage in male gametophyte development (Lalanne and Twell, 2002). Wild-type pollen appeared to develop normally, although a low frequency of the gum-like phenotypes appeared transiently at a low frequency (approximately 10%) during the late tricellular stage of pollen development (Fig. 10B). This morphology disappeared at pollen maturation (Fig. 10B).
Figure 10.
Occurrence of the gum-like and mud-like morphologies in +/spp-2 plants during pollen maturation. The frequency of wild-type and mutant phenotypes was determined during pollen maturation after pollen mitosis II by visual inspection of DAPI-stained pollen. A, DAPI-stained nuclei of wild-type pollen to illustrate the bicellular/early tricellular (−3), early tricellular (−2), late tricellular (−1), and mature (+1) pollen stages that were used for developmental analysis (Lalanne and Twell, 2002). B and C, The frequencies of wild-type (wt) and mutant phenotypes were measured in wild-type (B) and +/spp-2 (C) plants. A total of 9.7% of pollen in the wild type showed an extended vegetative nuclei-like, gum-like phenotype before dehiscence; however, no gum-like phenotype was observed in released wild-type pollen. A minimum of 300 pollen grains were scored at each developmental stage. [See online article for color version of this figure.]
DISCUSSION
In this study, we investigated the expression and physiological function of AtSPP in Arabidopsis. There is a family of six related SPP genes in Arabidopsis with similarity to human SPP (Tamura et al., 2008). AtSPP (At2g03120) is the most likely ortholog of human SPP in Arabidopsis; it is the most closely related SPP at the amino acid sequence level and contains a potential ER retention signal (Fig. 1). SPPL (for SPP-like) family members are also present in other plants and appear to fall into three broad classes: (1) those with similarity to HsSPP; (2) those with similarity to HsSPPL3 (e.g. AtSPPL1); and (3) N-terminally extended SPPLs (e.g., AtSPPL2–AtSPPL5; Tamura et al., 2008). In this study, we focused on the characterization of AtSPP in Arabidopsis.
Tamura et al. (2008) demonstrated that AtSPP is an ER protein biochemically and showed that GFP-tagged AtSPP is localized at the ER. Similar to human SPP (Friedmann et al., 2004), the predicted signal peptide of AtSPP does not appear to be processed upon entry to the ER. However, unlike human SPP, AtSPP does not have any predicted sites for N-linked glycosylation, and the similar mobility of in vivo expressed versus in vitro expressed AtSPP-Flag also indicates that the protein is not glycosylated (Fig. 1).
Analysis of the spp-2 T-DNA insertion line indicated that AtSPP performs an essential function in Arabidopsis. The pollen phenotype of spp-2 heterozygotes indicated that AtSPP is specifically required for pollen development and germination. Consistent with a role in post-pollination events such as gametophyte development and pollen tube elongation, a microarray study of gene expression during pollen development showed that AtSPP clustered with other genes expressed late in development (cluster 6; Honys and Twell, 2004). There are many dynamic changes in the cytoplasm of pollen during development (Yamamoto et al., 2003), and germinating pollen and pollen tubes are very active in secretion (Tung et al., 2005). Thus, pollen would be expected to be especially sensitive to disruption of the secretory pathway. spp-2 might be affected in secretory function or in the processing of an essential membrane component, resulting in disruption of pollen germination (Cheung, 1995; Nasrallah, 2000; Swanson et al., 2004). Interestingly, we found that +/spp-2 pollen has a defect in organizing the MGU during pollen development. Although the mechanism of the defect remains unclear, we hypothesize that the spp-2 mutation causes developing pollen to arrest in a late stage of development (Fig. 10). The pollen is viable but fails to mature to a stage that allows proper germination during fertilization.
The MGU defect in spp-2 pollen was very reminiscent of the mud and gum phenotypes described previously by Lalanne and Twell (2002). The genes carrying the mud and gum mutations have not been identified. The mud1 and mud2 mutations were mapped to chromosomes III and II, respectively, and the gum1 and gum2 mutants were mapped to a location between the markers M506-SSLP (21.9 cM) and nga8 (26.6 cM) on chromosome IV of Arabidopsis (Lalanne and Twell, 2002). The AtSPP locus is not located close to any of the identified loci for gum and mud mutants. Therefore, the AtSPP gene does not correspond to the known mud and gum genes, although it may play a role in the same developmental process.
AtSPP shows relatively high expression in certain vegetative organs of Arabidopsis, including stipules, emerging leaves, and the vasculature of roots and leaves, but the lack of homozygous mutant plants prevented an investigation of AtSPP's role in vegetative organs. A recent study in Medicago truncatula showed that an SPP-like gene is coinduced with a large family of secreted nodule-specific Cys-rich polypeptides (Mergaert et al., 2003). These polypeptides share an unusually well-conserved signal peptide, and the authors hypothesize that the signal peptides could be further processed by the nodule-specific SPP to produce oligopeptides with signaling functions (Mergaert et al., 2003). Interestingly, a similar family of Cys-rich polypeptides, also with well-conserved signal peptides, is abundantly expressed in rice pollen (Park et al., 2006). Peptides associated with self-incompatibility also have unusually conserved signal peptides (O'Brien et al., 2002).
The physiological function of AtSPP will not be entirely clear until its substrates are identified. Nevertheless, our results demonstrate a critical role for AtSPP in the development and function of reproductive tissues in Arabidopsis, especially in pollen development. Future advances in the biochemistry of pollen development may provide clues to the exact function of AtSPP in plant reproduction.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana Col-0 ecotype) T-DNA lines SALK_098736, WiscDsLox 331C12, and qrt1 mutant were obtained from the ABRC (www.Arabidopsis.org/abrc/). LAT52:GFP transgenic plants were kindly provided by Dr. Alice Cheung at the University of Massachusetts-Amherst. Plate-grown plants were grown on half-strength Murashige and Skoog salts containing 1% (w/v) Suc, buffered with 5 g L−1 MES, pH 5.7, and solidified with 0.5% (w/v) Phyto Agar (Research Products International). Soil-grown plants were grown on Promix BX (Premier Horticulture) wetted with deionized water containing 1 g L−1 Miracle-Gro (Scotts Miracle-Gro Company) and 5 mL L−1 Gnatrol concentrate (Abbot Laboratories). Plants were grown in a growth chamber at 22°C with a 16-h-light/8-h-dark photoperiod.
For T-DNA lines SALK_098736 and WiscDsLox 331C12 (spp-2), the mutant alleles were verified by PCR genotyping and by sequencing the PCR products. The SALK_098736 insertion allele was detected using the left border primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′; see http://signal.salk.edu/cgi-bin/tdnaexpress) and the gene-specific primer 5′-TCGGACGCGTCTTTGCTACTA-3′. The WiscDsLox 331_C12 allele, spp-2, was detected using the left border primer p745 (5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′) and the gene-specific primers Wisc_F and Wisc_R (5′-ACTGGTTTCAAGCAGCACAGG-3′ and 5′-TCTCCAAATAGCTCCCCAAGC-3′, respectively). For PCR genotyping, DNA was extracted as described (Edwards et al., 1991).
Plasmids and DNA Constructs
The AtSPP promoter:GUS (pProAtSPP:GUS) fusion construct was generated using the Gateway system (Invitrogen). A 1,096-bp fragment, containing the putative promoter and the first three codons of the AtSPP gene, was amplified from genomic DNA by PCR using the forward primer 5′-CACCTTCATTTTTGATGATGTCGTC-3′ (the underlined bases were introduced for directional TOPO cloning) and the reverse primer 5′-ATTCTTCATAATTGCTTCTTGTG-3′ and cloned into pENTR-DTOPO using the pENTR Directional TOPO Cloning kit (Invitrogen). The resulting entry vector, pProAtSPP, was used in a LR recombination reaction with the pDEST G2 destination vector, which encodes the Escherichia coli uidA gene. The resulting binary vector, pProAtSPP:GUS, carried the GUS reporter gene fused in frame to the putative promoter and first three codons of AtSPP, with kanamycin resistance as the plant selectable marker and spectinomycin resistance as the bacterial selectable marker.
AtSPP-Flag and AtSPP-GFP were constructed from TAP0180, a cDNA for AtSPP that is complete except for the first 21 bases of the 5′ untranslated region (UTR) and the last 24 bases of the 3′ UTR (obtained from the ABRC). TAP0180 was first mutagenized by PCR to remove a SpeI site at the junction between the cDNA insert and the multicloning site, resulting in TAP0180-one-SpeI. For pAtSPP-Flag, two complementary oligonucleotides, containing the Flag codons (DYKDDDDK; Hopp et al., 1988) and SpeI-compatible ends (5′-CTAGCGATTACAAGGATGACGACGATAAGT-3′ and 5′-CTAGACTTATCGTCGTCATCCTTGTAATCG-3′), were annealed and ligated into a unique SpeI site just upstream of the putative ER retention signal in TAP0180-one-SpeI (one additional Ser residue was added as a consequence). The resulting construct had two tandem Flag tags inserted in frame into the SpeI site with an additional Ser residue. For pAtSPP-GFP, the GFP coding sequence (excluding the start and stop codons) was amplified from a plasmid encoding a S65T mutant of Aequoria victoria GFP (Heim et al., 1995) using Pfu polymerase (Stratagene) and primers that also introduced SpeI sites near the ends of the PCR product (5′-CGGACTAGTGTGAGCAAGGGCGAG-3′ and 5′-CGGACTAGTCTTGTACAGCTCGTCCATG-3′). This product was cloned into the EcoRV site of pBluescript II SK− (Stratagene), removed using SpeI, and ligated into the unique SpeI site of TAP0180-one-SpeI to produce pAtSPP-GFP.
AtSPP-Flag, AtSPP-GFP, and a 3-kb genomic DNA fragment encoding AtSPP were introduced into the pDEST-NOS binary vector using the Gateway system (Invitrogen). All transgenic plants were generated using the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Transgenic plants were selected on growth medium supplemented with 50 mg L−1 gentamycin or kanamycin or 10 to 50 mg L−1 basta (glufosinate-ammonium; Sigma-Aldrich), as appropriate. For segregation analysis, plants were grown on growth medium containing 10 to 50 mg L−1 basta for spp-2 (WiscDsLox331_C12). When grown on soil, plants were sprayed with a 120 mg L−1 solution of basta (Finale; AgrEvo Environmental Health).
For complementation of spp-2, +/spp-2 plants were pollinated with transgenic plants carrying genes encoding AtSPP, AtSPP-Flag, and AtSPP-GFP under the control of the AtSPP native promoter. In the F1 generation, +/spp-2 plants expressing each transgene were self-crossed and their progeny were genotyped using PCR to detect the wild-type AtSPP allele, the spp-2 T-DNA insertion, or the appropriate transgene.
RNA Extraction and RT-PCR Analysis
For RNA extraction, tissues were frozen in liquid nitrogen and ground with mortar and pestle. Pollen was collected from open flowers on ice as described previously (Honys and Twell, 2003) and was ground with a Dounce homogenizer. Total RNA was extracted from each sample using an RNeasy kit (Qiagen) according to the manufacturer's protocol and then treated with DNaseI (Promega). RT was carried out using the SuperScript First-Strand synthesis system (Invitrogen) with 200 ng of total RNA and random hexamer primers. For each PCR amplification, 1 μL of cDNA was used in a 20-μL reaction. PCR was run for 15 cycles for 18S rRNA and 29 cycles for AtSPP. The PCR cycle number was optimized by testing for linear amplification with serially diluted cDNA. The following primer pairs were used: for 18S rRNA, 5′-AAACGGCTACCACATCCAAG-3′ and 5′-ACTCGAAAGAGCCCGGTATT-3′; for AtSPP, 5′-TTCTTCTGTGCGTGGTATGC-3′ and 5′-GCAGTGAGAAGCCAAGAACC-3′; for AtTIP5;1 (At3g47440), 5′-GGACTAGTGAAATTGATGAGAAGAATGATTCC-3′ and 5′-GGGAATTCAGTCATTACACACCAATGGCATC-3′ (Soto et al., 2008); and for CAB2 (LHCB1.1; At1g29920), 5′-TACTTGGGTCCATTCTCTGGC-3′ and 5′-CGTTGAAGGTTACAGAGTCGCA-3′.
PCR products were resolved on 1% Tris-acetate EDTA agarose gels containing ethidium bromide. Band intensities were analyzed using a Kodak EDAS 290 camera and 1D image analysis software (Eastman Kodak). For each tissue, the ratio of AtSPP to 18S rRNA was normalized to the average AtSPP-18S rRNA ratio obtained for the root sample.
Protein Extraction and Immunoblot Analysis
For immunodetection of in vivo expressed AtSPP-Flag, total protein extracts were prepared by grinding plant tissue samples (100 mg fresh weight) in 100 μL of extraction buffer containing 0.35 m Tris base, 7.5% glycerol, 5% SDS, and protease inhibitor cocktail (Sigma P-9599). Each sample was centrifuged at 12,000g for 5 min, and the supernatant was used as the total protein extract. Protein concentration was measured using the bicinchoninic acid protein assay kit (Pierce). Protein samples were resolved by SDS-PAGE and immunoblotted following standard protocols. Mouse anti-Flag M2 monoclonal antibody (Sigma F3165) was the primary antibody, and horseradish peroxidase-conjugated rabbit anti-mouse IgG (Rockland Immunochemicals) was the secondary antibody. Bound antibody was detected by chemiluminescence detection.
In Vitro Translation
For in vitro translation of AtSPP, PCR was used to amplify the coding region from TAP0180 and pAtSPP-Flag and simultaneously introduce an NdeI site at the start codon for AtSPP. The PCR products were then cloned into pZErO-2 (Invitrogen) by blunt end ligation. Finally, the AtSPP coding region was removed with NdeI and EcoRI and subcloned into pET21a (Novagen) to yield pET21a AtSPP and pET21a AtSPP-Flag. 35S-labeled (Perkin-Elmer Life Sciences) in vitro translation products were generated in 25 μL of a TnT coupled reticulocyte system (Promega) according to the manufacturer's instructions. All samples were resolved by SDS-PAGE and imaged with a Fuji FLA 5000 phosphoimager (Fuji Film Medical Systems) or detected by immunoblotting.
GUS Histochemical Assay
Tissue samples were fixed in ice-cold 10% (v/v) acetone, vacuum infiltrated for 15 min on ice, and incubated at 37°C for 16 or 24 h in a GUS staining solution containing 10 mm EDTA, 2 mm K3Fe(CN6), 2 mm K4Fe(CN6)·3H2O, 0.1% Triton X-100, 100 μg mL−1 chloramphenicol, and 2 mm X-GlcUA (Gold Biotech Technology; dissolved in N,N-dimethylformamide) in 50 mm sodium phosphate buffer, pH 7.0 (Vielle-Calzada et al., 2000; Yadegari et al., 2000). For whole plant samples, the initial vacuum infiltration step was omitted. After staining, samples were cleared with several changes of 70% ethanol.
Microscopy
GUS staining was observed with a SMZ800 stereomicroscope or a E-600 microscope (Nikon Instruments), and GFP fluorescence was observed using an E-600 epifluorescence microscope (Nikon Instruments) equipped with an FITC-HYQ filter set (EX460-500, DM505, BA510-560). A Spot-RT camera system (Diagnostic Instruments) was used for image capture.
Aniline blue staining of pollen tubes and ovules was based on the method of Schiott et al. (2004). Samples were observed using a Nikon Labophot2 fluorescence microscope and photographed with a SPOT camera and software. For the viability test, mature pollen was stained with Alexander stain (Alexander, 1969) with the chloral hydrate omitted (Johnson-Brousseau and McCormick, 2004). To visualize pollen nuclei, the grains were stained with DAPI (Park et al., 1998). Phenotypes were classified as gum like or mud like based on the MGU morphology as defined previously (Lalanne and Twell, 2002). In brief, the gum-like phenotype is defined by vegetative nuclei that are malformed and displaced from the two sperm cells. The mud-like phenotype is defined as a complete displacement of the MGU from its normal position in the pollen center to a position at the pollen periphery. For measuring the frequency of mutant phenotypes in pollen during development, the stage of the flower bud was determined as described previously (Lalanne and Twell, 2002).
For AtSPP-GFP localization, pollen grains collected from AtSPP-GFP, LAT52:GFP, and wild-type flowers were germinated on glass slides in 25 μL of germination medium (Hicks et al., 2004) for approximately 5 to 6 h at 22°C in humidified chambers as described previously (Johnson-Brousseau and McCormick, 2004). Ungerminated and germinated pollen grains were observed with a confocal laser scanning microscope (LSM510 Meta; Carl Zeiss MicroImaging). Samples were excited at 488 nm with an argon laser, and the emission was collected using a band-pass filter set (505/530) to detect the GFP signal. Images were processed using LSM software (Carl Zeiss MicroImaging). Micrograph figures were prepared using Adobe Photoshop CS and Adobe Illustrator CS.
Pollen Germination Assay
Pollen germination rates were determined using an in vivo pollination assay (Lalanne et al., 2004). Briefly, pistils were excised from mature wild-type flower buds prior to anthesis and inserted upright in 1% (w/v) agar in water. Up to 25 pollen grains were applied to each stigma and allowed to germinate for 2 h at room temperature. The pistils were then stained with aniline blue, and the pollen grains were scored with a fluorescence microscope. Pollen grains with a pollen tube longer than the radius of the pollen grain were scored as germinated. Those with no emerging pollen tube were scored as ungerminated, and these were further divided as follows: those with strong localized fluorescence but no tube were scored as polarized, and the rest were scored as nonpolarized.
Tetrad Analysis
WiscDsLox 331C12 (+/spp-2) plants were pollinated with pollen from homozygous qrt1/qrt1 plant lines (Landsberg erecta ecotype; qrt1-1; Preuss et al., 1994). Plants exhibiting the qrt phenotype in the F3 generation were genotyped to select the +/spp-2; qrt1/qrt1 genotype. Mature pollen from those plants was analyzed by DAPI staining. The frequency of incomplete quartets was 4.7% and 11.3% in wild-type and +/spp-2 pollen, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CAD13132.1 (HsSPP), NM_126363 (AtSPP), NM_102732 (CAB2), and NM_114612 (AtTIP5;1).
Acknowledgments
We thank Dr. Peter Hepler, Dr. Tobias Baskin, Dr. Maura Cannon, and Dr. Joe Jerry for the generous use of their microscopes and technical support for histological studies. We thank Dr. Elsbeth Walker for supplying us with plasmids (pDEST G2) for this work, Zeke Nims for assistance with Southern-blot analysis, Dr. Caleb Rounds and Dr. Sook-Young Yoon for assistance in collecting GFP and DAPI confocal images, and Dr. Natasha Raikhel for AtγTIP antisera. We also thank Dr. Alice Cheung for providing the LAT52:GFP plants.
This work was supported by the National Institutes of Health (grant no. R01 GM–61893 to D.J.S.) and by the Central Microscopy Facility of the University of Massachusetts, which is supported by the National Science Foundation (grant no. BBS 8714235).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Danny J. Schnell (dschnell@biochem.umass.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alexander M (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Horlow C, Vezon D, de Laissardiere S, Guyon A, Ferault M, Marchand M, Bechtold N, Pelletier G (1998) T-DNA mediated disruption of essential gametophytic genes in Arabidopsis unexpectedly rare and cannot be inferred from segregation distortion alone. Mol Gen Genet 260 444–452 [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100 391–398 [DOI] [PubMed] [Google Scholar]
- Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB (2005) Drosophila signal peptide peptidase is an essential protease for larval development. Genetics 170 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY (1995) Pollen-pistil interactions in compatible pollination. Proc Natl Acad Sci USA 92 3077–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Crawshaw SG, Martoglio B, Meacock SL, High S (2004) A misassembled transmembrane domain of a polytopic protein associates with signal peptide peptidase. Biochem J 384 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA (1998) Genetic analysis of female gametophyte development and function. Plant Cell 10 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36 99–124 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, Bohland C, Imhof A, Martoglio B, Teplow DB, et al (2006) A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol 8 894–896 [DOI] [PubMed] [Google Scholar]
- Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, Kuhn PH, Stauffer D, Rovelli G, Martoglio B (2006) SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat Cell Biol 8 843–848 [DOI] [PubMed] [Google Scholar]
- Friedmann E, Lemberg MK, Weihofen A, Dev KK, Dengler U, Rovelli G, Martoglio B (2004) Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J Biol Chem 279 50790–50798 [DOI] [PubMed] [Google Scholar]
- Golde TE, Eckman CB (2003) Physiologic and pathologic events mediated by intramembranous and juxtamembranous proteolysis. Sci STKE 2003 RE4. [DOI] [PubMed] [Google Scholar]
- Grigorenko AP, Moliaka YK, Korovaitseva GI, Rogaev EI (2002) Novel class of polytopic proteins with domains associated with putative protease activity. Biochemistry (Mosc) 67 826–835 [DOI] [PubMed] [Google Scholar]
- Grigorenko AP, Moliaka YK, Soto MC, Mello CC, Rogaev EI (2004) The Caenorhabditis elegans IMPAS gene, imp-2, is essential for development and is functionally distinct from related presenilins. Proc Natl Acad Sci USA 101 14955–14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY (1995) Improved green fluorescence. Nature 373 663–664 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV (2004) Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol 134 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. BioTechnology 6 1204–1210 [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39 761–775 [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Urban S, Freeman M, Okada K (2005) An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett 579 5723–5728 [DOI] [PubMed] [Google Scholar]
- Krawitz P, Haffner C, Fluhrer R, Steiner H, Schmid B, Haass C (2005) Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin in homologues SPPL2b and SPPL3. J Biol Chem 280 39515–39523 [DOI] [PubMed] [Google Scholar]
- Lalanne E, Michaelidis C, Moore JM, Gagliano W, Johnson A, Patel R, Howden R, Vielle-Calzada JP, Grossniklaus U, Twell D (2004) Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167 1975–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Twell D (2002) Genetic control of male germ unit organization in Arabidopsis. Plant Physiol 129 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B (2001) Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol 167 6441–6446 [DOI] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL (2006) Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441 894–897 [DOI] [PubMed] [Google Scholar]
- Matthysrochon E, Vergne P, Detchepare S, Dumas C (1987) Male germ unit isolation from three tricellular pollen species: Brassica oleracea, Zea mays, and Triticum aestivum. Plant Physiol 83 464–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J, Lemberg MK, Hope G, Martoglio B (2002) Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J 21 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah JB (2000) Cell-cell signaling in the self-incompatibility response. Curr Opin Plant Biol 3 368–373 [DOI] [PubMed] [Google Scholar]
- O'Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP (2002) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J 32 985–996 [DOI] [PubMed] [Google Scholar]
- Park JI, Hakozaki H, Endo M, Takada Y, Ito H, Uchida M, Okabe T, Watanabe M (2006) Molecular characterization of mature pollen-specific genes encoding novel small cysteine-rich proteins in rice (Oryza sativa L.). Plant Cell Rep 25 466–474 [DOI] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125 3789–3799 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE (2002) Identification of a novel family of presenilin homologues. Hum Mol Genet 11 1037–1044 [DOI] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the Quartet (Qrt) genes. Science 264 1458–1460 [DOI] [PubMed] [Google Scholar]
- Ronin C, Bouchilloux S, Granier C, van Rietschoten J (1978. a) Enzymatic N-glycosylation of synthetic Asn-X-Thr containing peptides. FEBS Lett 96 179–182 [DOI] [PubMed] [Google Scholar]
- Ronin C, Granier C, Van Rietschoten J, Bouchilloux S (1978. b) Enzymatic glycosylation of an Asn-Ala-Thr-containing peptide from oligosaccharide-lipids (proceedings). Arch Int Physiol Biochim 86 885–886 [PubMed] [Google Scholar]
- Schiott M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA 101 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP (2008) AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett 582 4077–4082 [DOI] [PubMed] [Google Scholar]
- Swanson R, Edlund AF, Preuss D (2004) Species specificity in pollen-pistil interactions. Annu Rev Genet 38 793–818 [DOI] [PubMed] [Google Scholar]
- Tamura T, Asakura T, Uemura T, Ueda T, Terauchi K, Misaka T, Abe K (2008) Signal peptide peptidase and its homologs in Arabidopsis thaliana: plant tissue-specific expression and distinct subcellular localization. FEBS J 275 34–43 [DOI] [PubMed] [Google Scholar]
- Tax FE, Vernon DM (2001) T-DNA-associated duplication/translocations in Arabidopsis: implications for mutant analysis and functional genomics. Plant Physiol 126 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB (2005) Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol 138 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U (2000) Delayed activation of the paternal genome during seed development. Nature 404 91–94 [DOI] [PubMed] [Google Scholar]
- Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B (2002) Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296 2215–2218 [DOI] [PubMed] [Google Scholar]
- Weihofen A, Martoglio B (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol 13 71–78 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB, et al (2000) Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Nishimura M, Hara-Nishimura I, Noguchi T (2003) Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol 44 1192–1201 [DOI] [PubMed] [Google Scholar]