Abstract
Rhodococcus fascians is a Gram-positive phytopathogen that induces shooty hyperplasia on its hosts through the secretion of cytokinins. Global transcriptomics using microarrays combined with profiling of primary metabolites on infected Arabidopsis (Arabidopsis thaliana) plants revealed that this actinomycete modulated pathways to convert its host into a niche. The transcript data demonstrated that R. fascians leaves a very characteristic mark on Arabidopsis with a pronounced cytokinin response illustrated by the activation of cytokinin perception, signal transduction, and homeostasis. The microarray data further suggested active suppression of an oxidative burst during the R. fascians pathology, and comparison with publicly available transcript data sets implied a central role for auxin in the prevention of plant defense activation. Gene Ontology categorization of the differentially expressed genes hinted at a significant impact of infection on the primary metabolism of the host, which was confirmed by subsequent metabolite profiling. The much higher levels of sugars and amino acids in infected plants are presumably accessed by the bacteria as carbon and nitrogen sources to support epiphytic and endophytic colonization. Hexoses, accumulating from a significantly increased invertase activity, possibly inhibited the expression of photosynthesis genes and photosynthetic activity in infected leaves. Altogether, these changes are indicative of sink development in symptomatic tissues. The metabolomics data furthermore point to the possible occurrence of secondary signaling during the interaction, which might contribute to symptom development. These data are placed in the context of regulation of bacterial virulence gene expression, suppression of defense, infection phenotype, and niche establishment.
Plants have evolved a remarkable level of developmental plasticity enabling them to deal with changes in their immediate surroundings throughout their life cycle. Environmental stress affects plants in their growth and development by imposing alterations in gene expression and, consequently, in physiology and metabolism. Pathogenic bacteria, fungi, viruses, oomycetes, nematodes, and insects can have a devastating effect on crop plants either at the survival or the yield level. Although the dynamic interaction between pathogen and host is complex and, at first sight, seemingly specific for each plant-pathogen combination, several biotrophic phytopathogens display related mechanisms to convert the plant into a suitable niche (Jameson, 2000).
In contrast to necrotrophic pathogens, biotrophs rely on living tissues for survival and multiplication. To exploit the plant as a source of energy and assimilates, the first requirement is to avoid preformed and suppress induced defense mechanisms of the host. Typical defense responses include cell wall strengthening, production of phytoalexins and proteins with antimicrobial properties, and synthesis of stress signaling molecules, such as salicylic acid, jasmonic acid, ethylene, and reactive oxygen species (ROS; Hammond-Kosack and Jones, 1996; Gadjev et al., 2006; Garcia-Brugger et al., 2006; Berger et al., 2007). Bacterial effector molecules have been proposed to suppress or delay defense and, in Gram-negative pathogenic bacteria, are typically secreted via the type III secretion system (Biemelt and Sonnewald, 2006; Jones and Dangl, 2006; Truman et al., 2006). In the next phase toward successful niche establishment, plant (carbohydrate) metabolism is diverted for bacterial nutrition. Indeed, pathogens have proven to be very effective sinks for photosynthate: accumulation of soluble sugars has been reported in maize (Zea mays), wheat (Triticum spp.), tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), and Arabidopsis (Arabidopsis thaliana) infected with tobacco mosaic virus, different Pseudomonas strains, smuts, rusts, and powdery mildews (Wright et al., 1995; Chou et al., 2000; Herbers et al., 2000; Scharte et al., 2005; Doehlemann et al., 2008). Consequently, the induction of sucrolytic cell wall invertase genes upon pathogen attack is now an established marker for source-to-sink transition in leaves (Herbers et al., 2000; Biemelt and Sonnewald, 2006; Swarbrick et al., 2006; Berger et al., 2007, and refs. therein). The reprogramming of the host metabolism and nutrient partitioning, in turn, triggers several signaling cascades. The modified carbohydrate metabolism represses photosynthetic genes and decreases photosynthetic activity, possibly by end-product inhibition (Nielsen et al., 1998; Roitsch, 1999; Smeekens, 2000; Truman et al., 2006; Kocal et al., 2008). The reduction of photosynthesis and assimilatory metabolic pathways allows the initiation of respiration and other processes required for defense. Interestingly, the high sugar levels that result from the induced sink metabolism also induce a number of defense-related genes (Herbers et al., 1996; Ehness et al., 1997; Herbers and Sonnewald, 1998; Berger et al., 2004; Scharte et al., 2005). Indeed, recent data suggest that cell wall invertases play an important role in host defense against pathogen attack (Essmann et al., 2008). Therefore, the hexose accumulation often observed has a dual role: on the one hand, it serves as nutrition for the microbe; on the other hand, it is part of the stress response of the plant aimed at eradicating the invading pathogen.
Upon infection with the Gram-positive, cytokinin-producing phytopathogen Rhodococcus fascians, the architecture of the plant is drastically changed (Crespi et al., 1992, 1994; Vereecke et al., 2000, 2002a, 2002b; Goethals et al., 2001; de O Manes et al., 2004). Remarkably little is known about the molecular basis of the pathologies of Gram-positive phytopathogens: they do not possess type III secretion systems, the typical Gram-negative effectors have not been identified, and no defense suppression mechanisms have been uncovered (Loria et al., 2006; Robert-Seilaniantz et al., 2007; Gartemann et al., 2008). Although it is becoming clear how R. fascians alters the shape of its hosts (Simón-Mateo et al., 2006; Depuydt et al., 2008, 2009), the establishment of an altered metabolic state had not been investigated until now. Therefore, with an integrated approach of genome-wide transcriptomics and primary metabolite profiling, we compared the response of Arabidopsis upon confrontation with two nearly isogenic R. fascians strains: the wild-type D188 that contains a linear virulence plasmid pFiD188, and its plasmid-free nonpathogenic derivative D188-5. Based on the obtained results, we propose a model for the role of the physiological alterations during initiation and maintenance of the interaction between Arabidopsis and R. fascians.
RESULTS
Transcriptome Analysis Underlines the Key Role for Cytokinins in Symptom Development
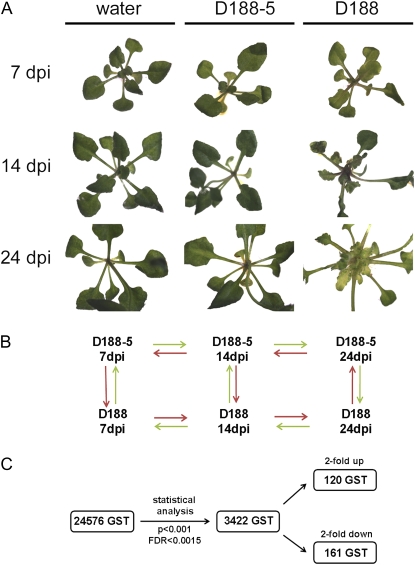
Arabidopsis plants that are infected with the phytopathogenic actinomycete R. fascians display typical phenotypes, such as smaller and serrated leaves, activated axillary and de novo formed meristems, and usually very compact rosettes (Vereecke et al., 2000; de O Manes et al., 2004; Simón-Mateo et al., 2006; Depuydt et al., 2008). To gain a genome-wide view of the processes that are involved in disease establishment, we compared the transcriptome of Arabidopsis plants (ecotype C24) infected with the virulent strain D188 with that of plants infected with the nonpathogenic derivative D188-5 as control samples using two-color CATMA microarrays (Crowe et al., 2003; Hilson et al., 2004). These arrays contain 24,576 gene-specific tags (GST) and probes for small open reading frames and 615 probes tiling the mitochondrial and chloroplastic genomes. For RNA preparation, plant material was harvested at three developmentally distinct time points: at 7 d postinfection (dpi) with the virulent R. fascians D188 strain, no clear symptoms were observed; at 14 dpi, the macroscopic phenotype became apparent; and at 24 dpi, Arabidopsis displayed the typical disease symptoms described above (Fig. 1A). Because the axillary regions are important for colonization, virulence gene expression, and symptom initiation by R. fascians (Cornelis et al., 2002), roots, cotyledons, and leaves were removed during harvesting. We selected a loop design as an experimental setup for the arrays (Fig. 1B) to evaluate gene expression as a function of time (see “Materials and Methods”). The choice of the control samples ensured the exclusion of genes that were merely triggered by the bacterial presence.
Figure 1.
Sampling, setup, and analysis of microarray hybridizations of Arabidopsis plants infected with R. fascians D188-5 and D188. A, Phenotype at the selected time points. Roots, cotyledons, and leaves were removed for sampling. B, Experimental setup in a loop design for the 14 hybridizations. The dye swaps were combined with the two biological replicates. C, Flow chart representing the outcome of the statistical analysis of the microarray data.
Statistical analysis indicated that 3,422 genes showed a significant treatment × time point interaction effect (P < 0.001; false discovery rate [FDR] < 0.0015; see “Materials and Methods”), of which 120 and 161 were more than 2-fold up- or down-regulated, respectively, in at least one time point (Fig. 1C; Tables I and II). The expression patterns obtained with the microarray hybridizations were confirmed for 10 genes randomly taken from the data set using quantitative reverse transcription (qRT)-PCR (Supplemental Fig. S1). The Gene Ontology (GO) annotation of the differentially expressed genes revealed that the major functional categories affected by R. fascians involved metabolism, response to (biotic/abiotic) stress, and transcription activity. Nevertheless, important modulations also occurred in the protein modification, transport, cell organization, and biogenesis categories (Fig. 2).
Table I.
Differentially up-regulated genes upon infection of Arabidopsis with R. fascians D188
| Gene | Function | Codea | Fold Change | ||
|---|---|---|---|---|---|
| 7 dpi | 14 dpi | 24 dpi | |||
| Cell organization and biogenesis | |||||
| At1g26770 | Expansin (putative; EXP10) | u14 | 1.45 | 3.39 | 4.76 |
| At1g31580 | Expressed protein | u101 | 2.89 | 0.86 | 0.95 |
| At1g48920 | Nucleolin (putative) | u82 | 0.95 | 1.32 | 2.13 |
| At1g69530 | Expansin (putative; EXP1) | u5 | 0.73 | 2.50 | 2.69 |
| At2g03090 | Expansin (putative; EXP15) | u92 | 0.81 | 1.69 | 4.22 |
| At2g43590 | Chitinase (putative) | u41 | 1.21 | 2.84 | 1.62 |
| At2g43610 | Glycoside hydrolase family 19 protein | u89 | 1.22 | 2.98 | 1.33 |
| At3g18000 | Phosphoethanolamine N-methyltransferase1 (PEAMT1) | 0.58 | 1.40 | 2.27 | |
| At3g18080 | Glycosyl hydrolase family 1 protein | u94 | 0.70 | 0.88 | 2.24 |
| At4g08950 | Phosphate-responsive protein (putative; EXO) | u54 | 1.49 | 0.62 | 2.29 |
| At4g28250 | β-Expansin (putative; EXPB3) | u11 | 0.60 | 1.86 | 4.30 |
| Development | |||||
| At3g17520 | Late embryogenesis abundant domain-containing protein | u102 | 0.99 | 0.94 | 2.76 |
| At3g51810 | Em-like protein GEA1 (EM1) | u99 | 1.03 | 1.05 | 3.61 |
| At5g03840 | Terminal flower 1 protein (TFL1) | u28 | 1.00 | 0.99 | 3.12 |
| At5g57390 | Ovule development protein (putative) | u36 | 1.23 | 1.46 | 2.01 |
| At5g28640 | SSXT protein-related/Gly-rich protein | u9 | 1.26 | 1.60 | 3.12 |
| Electron transport or energy pathway | |||||
| At1g66540 | Cytochrome P450 (putative) | u57 | 0.80 | 1.07 | 2.20 |
| At4g27080 | Thioredoxin family protein | u69 | 2.64 | 0.71 | 1.29 |
| At5g49730 | Ferric reductase-like transmembrane component family protein | u71 | 0.76 | 0.88 | 2.26 |
| Metabolism | |||||
| At1g06080 | Δ9 desaturase (ADS1) | u76 | 1.13 | 2.50 | 5.59 |
| At1g30820 | CTP synthase (putative)/UTP-ammonia ligase (putative) | u19 | 0.98 | 2.02 | 2.99 |
| At1g56430 | Nicotianamine synthase (putative) | u25 | 2.15 | 2.31 | 3.05 |
| At1g80050 | Adenine phosphoribosyltransferase 2 (APT2) | u77 | 1.57 | 2.06 | 4.44 |
| At1g78580 | Trehalose-6-phosphate synthase (putative) | u93 | 0.93 | 1.46 | 3.31 |
| At3g21420 | Oxidoreductase, 2OG-Fe(II) oxygenase family protein | u21 | 1.04 | 1.21 | 2.32 |
| At3g30775 | Pro oxidase, mitochondrial (POX; PRO1; ERD5) | u91 | 0.99 | 2.21 | 1.20 |
| At3g57010 | Strictosidine synthase family protein | u38 | 0.84 | 0.94 | 2.01 |
| At3g63440 | Cytokinin oxidase family protein | u105 | 0.99 | 1.48 | 2.26 |
| At4g02290 | Glycosyl hydrolase family 9 protein | u90 | 0.97 | 1.37 | 2.63 |
| At4g30140 | GDSL-motif lipase/hydrolase family protein | u87 | 1.45 | 2.93 | 2.06 |
| At4g30290 | Endoxyloglucan transferase (putative) | u40 | 1.65 | 2.48 | 5.88 |
| At5g13930 | Chalcone synthase/naringenin-chalcone synthase | u75 | 3.21 | 1.74 | 3.06 |
| At5g17220 | Glutathione S-transferase (putative) | u84 | 1.22 | 0.79 | 2.27 |
| At5g18670 | β-Amylase (putative; BMY3) | u81 | 0.56 | 1.11 | 2.16 |
| At5g45670 | GDSL-motif lipase/hydrolase family protein | u10 | 1.11 | 1.88 | 2.22 |
| At5g51210 | Gly-rich protein/oleosin | u50 | 1.12 | 0.97 | 2.39 |
| At5g55250 | S-adenosylmethionine:carboxyl methyltransferase family protein | u49 | 1.09 | 1.49 | 2.54 |
| Protein metabolism | |||||
| At2g47020 | Peptide chain release factor (putative) | u27 | 3.24 | 0.99 | 1.61 |
| At4g25740 | 40S ribosomal protein S10 (RPS10A) | u51 | 1.30 | 1.25 | 2.01 |
| At5g19110 | Extracellular dermal glycoprotein (EDGP)-related | u16 | 1.45 | 2.21 | 1.23 |
| At5g53450 | Protein kinase family protein | u60 | 3.61 | 4.52 | 6.27 |
| Response to biotic or abiotic stimulus | |||||
| At1g23130 | Bet v I allergen family protein | u100 | 2.42 | 0.67 | 0.20 |
| At1g56150 | Auxin-responsive family protein | u26 | 2.22 | 1.52 | 0.61 |
| At1g73260 | Trypsin and protease inhibitor/Kunitz family protein | u45 | 2.05 | 1.12 | 0.65 |
| At1g73330 | Protease inhibitor (putative; DR4) | u61 | 2.06 | 0.65 | 0.13 |
| At2g38310 | Expressed protein | u52 | 2.31 | 1.00 | 1.04 |
| At2g38870 | Protease inhibitor (putative) | u35 | 2.62 | 1.61 | 0.50 |
| At2g43510 | Trypsin inhibitor (putative) | u53 | 2.12 | 1.46 | 0.67 |
| At2g43530 | Trypsin inhibitor (putative) | u56 | 2.20 | 0.60 | 0.45 |
| At3g57240 | β-1,3-Glucanase (BG3) | u29 | 1.07 | 2.17 | 1.73 |
| At4g30660 | Low-temperature and salt-responsive protein (putative) | u79 | 2.15 | 0.84 | 1.12 |
| RNA/DNA metabolism | |||||
| At1g02840 | Pre-mRNA splicing factor SF2 (SF2)/SR1 protein | u66 | 1.06 | 1.03 | 2.01 |
| At3g27060 | Ribonucleotide reductase (putative) | u42 | 1.45 | 2.08 | 2.21 |
| Signal transduction | |||||
| At1g10470 | Two-component-response regulator (ARR4) | u73 | 1.09 | 2.20 | 3.20 |
| At1g19050 | Two-component-response regulator (ARR7) | u1 | 1.81 | 3.75 | 8.88 |
| At2g40670 | Two-component-response regulator (ARR16) | u13 | 1.04 | 1.38 | 2.01 |
| At2g41310 | Two-component-response regulator (ARR8) | u8 | 1.01 | 1.37 | 2.24 |
| At2g46070 | Mitogen-activated protein kinase MAPK (putative; MPK12) | u86 | 0.96 | 1.00 | 2.31 |
| At3g57040 | Two-component-response regulator (ARR9) | u15 | 0.98 | 1.26 | 3.06 |
| At5g15230 | Gibberellin-regulated protein 4 (GASA4)/GA-responsive protein 4 | u39 | 1.68 | 2.37 | 2.49 |
| At5g62920 | Two-component-response regulator (ARR6) | u104 | 1.65 | 2.80 | 6.57 |
| Transcription factor | |||||
| At1g04250 | Auxin-responsive protein/IAA-induced protein 17 (IAA17) | u55 | 1.37 | 1.58 | 2.26 |
| At1g56170 | Transcription factor (putative) | u78 | 1.51 | 1.07 | 2.01 |
| At1g67260 | Pseudogene (putative) cycloidea cyc4 protein | 1.00 | 1.48 | 2.50 | |
| At1g68360 | Zinc finger protein-related | u17 | 1.13 | 1.43 | 2.65 |
| At1g71030 | Myb family transcription factor | u23 | 0.56 | 0.73 | 2.35 |
| At2g28510 | Dof-type zinc finger domain-containing protein | u80 | 0.79 | 1.73 | 2.86 |
| At3g15540 | Auxin-responsive protein/IAA-induced protein 19 (IAA19) | u48 | 1.32 | 1.49 | 2.72 |
| At3g16770 | AP2 domain-containing protein RAP2.3 (RAP2.3) | u74 | 0.85 | 1.06 | 2.15 |
| At3g25710 | Basic helix-loop-helix (bHLH) family protein | u62 | 0.97 | 1.27 | 2.27 |
| At4g37750 | Ovule development protein aintegumenta (ANT) | u63 | 1.14 | 1.54 | 2.97 |
| At5g25190 | Ethylene-responsive element-binding protein (putative) | u58 | 1.28 | 2.15 | 4.11 |
| At5g47370 | Homeobox-Leu zipper protein 2 (HAT2)/HD-ZIP protein 2 | u7 | 1.55 | 1.59 | 3.15 |
| At5g50010 | Expressed protein/similar to SAC51 transcription factor | u10 | 0.99 | 1.33 | 2.32 |
| Transport | |||||
| At1g19450 | Integral membrane protein (putative)/sugar transporter family protein | u103 | 0.99 | 1.49 | 2.09 |
| At1g59740 | Proton-dependent oligopeptide transport (POT) family protein | u24 | 1.29 | 1.51 | 3.09 |
| At2g45180 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | u97 | 2.20 | 0.95 | 0.38 |
| At3g25620 | ABC transporter family protein | 0.97 | 1.03 | 3.30 | |
| At3g25620 | ABC transporter family protein | u46 | 0.86 | 0.88 | 5.88 |
| At4g22520 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | 3.10 | 4.91 | 3.10 | |
| At4g27780 | Acyl-CoA binding protein 2 (ACBP2) | u109 | 1.03 | 1.11 | 2.02 |
| At5g13740 | Sugar transporter family protein | u83 | 1.51 | 1.67 | 2.41 |
| At5g50200 | Wound-responsive gene | u20 | 1.30 | 2.84 | 1.18 |
| Others | |||||
| At1g02205 | CER1 protein | 1.08 | 1.06 | 2.33 | |
| At1g03870 | Fasciclin-like arabinogalactan protein (FLA9) | u106 | 2.91 | 0.78 | 0.63 |
| At1g48310 | SNF2 domain-containing protein/helicase domain protein | u67 | 1.18 | 0.95 | 2.39 |
| At1g66100 | Thionin (putative) | u4 | 1.60 | 0.92 | 2.62 |
| At1g70560 | Alliinase C-terminal domain-containing protein | u33 | 1.17 | 1.90 | 2.60 |
| At2g34700 | Pollen Ole e 1 allergen and extensin family protein | u64 | 3.02 | 7.24 | 7.94 |
| At3g09390 | Metallothionein protein (putative; MT2A) | u22 | 2.24 | 0.79 | 1.01 |
| At3g13520 | Arabinogalactan protein (AGP12) | u31 | 2.47 | 0.82 | 0.89 |
| At3g16640 | Translationally controlled tumor family protein | u68 | 0.98 | 0.94 | 2.08 |
| At4g04810 | SeIR domain-containing protein | 2.40 | 0.62 | 0.38 | |
| At4g04830 | SeIR domain-containing protein | u111 | 2.94 | 0.65 | 0.30 |
| At4g24780 | Pectate lyase family protein | u85 | 0.71 | 1.24 | 2.10 |
| At4g31290 | ChaC-like family protein | u70 | 1.43 | 1.28 | 2.92 |
| At5g48850 | Male sterility MS5 family protein | u72 | 2.58 | 0.94 | 0.62 |
| Unknown | |||||
| At1g15270 | Expressed protein | u59 | 2.09 | 1.05 | 1.12 |
| At1g29980 | Expressed protein | u34 | 1.15 | 1.65 | 2.16 |
| At1g64980 | Expressed protein | u88 | 1.39 | 2.27 | 2.42 |
| At1g68250 | Expressed protein | u43 | 0.90 | 0.87 | 2.71 |
| At2g27385 | Expressed protein | 1.24 | 1.11 | 2.12 | |
| At2g30760 | Hypothetical protein | u44 | 1.28 | 1.50 | 3.03 |
| At2g32280 | Expressed protein | u18 | 1.27 | 1.56 | 2.03 |
| At2g32560 | F-box family protein | 1.18 | 1.26 | 2.49 | |
| At2g34160 | Expressed protein | u47 | 2.06 | 1.19 | 1.04 |
| At2g39870 | Expressed protein | u12 | 0.96 | 1.11 | 2.32 |
| At3g05730 | Expressed protein | u96 | 1.26 | 2.25 | 1.89 |
| At3g08030 | Expressed protein | u30 | 0.93 | 1.36 | 2.73 |
| At3g11720 | Expressed protein | 1.00 | 1.39 | 2.14 | |
| At3g16660 | Expressed protein | u110 | 1.10 | 2.63 | 1.67 |
| At3g19200 | Hypothetical protein | u107 | 1.10 | 1.54 | 3.59 |
| At3g56360 | Expressed protein | u65 | 1.07 | 1.25 | 2.85 |
| At4g09840 | Expressed protein | u95 | 1.07 | 0.97 | 2.07 |
| At4g30410 | Expressed protein | u2 | 1.20 | 1.58 | 2.94 |
| At5g03545 | Expressed protein | u37 | 3.21 | 1.07 | 0.25 |
| At5g05250 | Expressed protein | u32 | 1.32 | 1.72 | 2.78 |
| At5g19260 | Expressed protein | u3 | 1.21 | 1.76 | 3.98 |
| At5g24660 | Expressed protein | u98 | 3.40 | 1.14 | 0.63 |
| At5g52900 | Expressed protein | u6 | 1.17 | 1.64 | 3.22 |
Refers to genes in the left panel of Figure 4.
Table II.
Differentially down-regulated genes upon infection of Arabidopsis with R. fascians D188
| Gene | Function | Codea | Fold Change | ||
|---|---|---|---|---|---|
| 7 dpi | 14 dpi | 24 dpi | |||
| Biotic/abiotic stress | |||||
| At1g23130 | Bet v I allergen family protein | d131 | 2.42 | 0.67 | 0.20 |
| At1g54040 | Kelch repeat-containing protein | d129 | 0.98 | 0.63 | 0.46 |
| At1g70850 | Bet v I allergen family protein | d94 | 0.96 | 0.59 | 0.25 |
| At1g70880 | Bet v I allergen family protein | d61 | 1.09 | 0.73 | 0.48 |
| At2g15490 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | d31 | 1.27 | 0.98 | 0.48 |
| At2g18420 | Gibberellin-responsive protein (putative) | d73 | 1.10 | 0.79 | 0.27 |
| At2g35980 | HIN1 family protein | d85 | 1.39 | 1.00 | 0.45 |
| At2g45210 | Auxin-responsive protein-related | d109 | 0.97 | 0.74 | 0.42 |
| At3g28930 | avrRpt2-induced AIG2 protein (AIG2) | d70 | 1.13 | 0.90 | 0.47 |
| At5g14920 | Gibberellin-regulated family protein | d133 | 1.45 | 0.99 | 0.49 |
| At5g53160 | Expressed protein | d110 | 0.87 | 1.02 | 0.42 |
| Cell organization and biogenesis | |||||
| At4g36380 | Cytochrome P450 90C1 (CYP90C1)/rotundifolia3 (ROT3) | d107 | 0.92 | 0.81 | 0.44 |
| At5g66540 | Expressed protein | d36 | 1.31 | 0.72 | 0.41 |
| Development | |||||
| At1g02820 | Late embryogenesis abundant 3 (LEA3) family protein | d145 | 1.74 | 0.76 | 0.26 |
| At4g02380 | Late embryogenesis abundant 3 (LEA3) family protein | d140 | 1.72 | 0.78 | 0.26 |
| DNA and RNA metabolism | |||||
| At5g64200 | Arg/Ser-rich splicing factor SC35 | d91 | 1.41 | 0.64 | 0.45 |
| Electron transport or energy pathway | |||||
| At1g26410 | FAD-binding domain-containing protein | d144 | 1.48 | 0.98 | 0.40 |
| At2g44790 | Uclacyanin II | d86 | 0.84 | 0.91 | 0.35 |
| At3g22370 | Alternative oxidase 1a, mitochondrial (AOX1A) | d84 | 1.13 | 0.90 | 0.43 |
| At3g56060 | Glc-methanol-choline (GMC) oxidoreductase family protein | d87 | 0.81 | 0.67 | 0.32 |
| At4g15760 | Monooxygenase (putative; MO1) | 1.03 | 0.82 | 0.50 | |
| At4g15765 | Monooxygenase family protein | 0.85 | 0.75 | 0.40 | |
| At4g38540 | Monooxygenase (putative; MO2) | d46 | 1.16 | 0.87 | 0.47 |
| At5g36220 | Cytochrome P450 81D1 (CYP81D1; CYP91A1) | d138 | 1.12 | 0.97 | 0.48 |
| Metabolism | |||||
| At1g05010 | Ethylene-forming enzyme (ACO; EAT1) | d111 | 1.02 | 1.15 | 0.48 |
| At1g21440 | Mutase family protein | d5 | 0.44 | 0.50 | 0.77 |
| At1g24100 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | d34 | 0.45 | 0.69 | 0.83 |
| At1g64660 | Cys/Met metabolism pyridoxal-phosphate-dependent enzyme | d113 | 1.30 | 0.83 | 0.28 |
| At2g25450 | 2-Oxoglutarate-dependent dioxygenase (putative) | d55 | 0.54 | 0.44 | 0.22 |
| At2g26560 | Patatin (putative) | d108 | 1.40 | 0.66 | 0.41 |
| At3g08860 | β-Ala-pyruvate aminotransferase (putative)/AGT (putative) | d142 | 1.01 | 0.69 | 0.43 |
| At3g09260 | Glycosyl hydrolase family 1 protein | d96 | 1.14 | 1.16 | 0.48 |
| At3g09270 | Glutathione S-transferase (putative) | d77 | 1.02 | 0.96 | 0.50 |
| At3g13450 | Branched chain α-keto acid dehydrogenase E1 β-subunit (DIN4) | d143 | 1.01 | 0.83 | 0.47 |
| At3g14990 | 4-Methyl-5(β-hydroxyethyl)-thiazole-monophosphate bios (putative) | d75 | 0.96 | 0.83 | 0.44 |
| At3g19710 | Branched chain amino acid aminotransferase (putative; BCAT4) | d16 | 0.46 | 0.57 | 1.25 |
| At3g29250 | Short-chain dehydrogenase/reductase (SDR) family protein | d83 | 1.06 | 0.89 | 0.46 |
| At3g44300 | Nitrilase 2 (NIT2) | d150 | 1.43 | 0.72 | 0.10 |
| At3g48990 | AMP-dependent synthetase and ligase family protein | d25 | 0.93 | 0.73 | 0.47 |
| At3g58990 | Aconitase C-terminal domain-containing protein | d20 | 0.40 | 0.53 | 1.45 |
| At3g60130 | Glycosyl hydrolase family 1/β-glucosidase (putative; YLS1) | d28 | 1.02 | 0.87 | 0.36 |
| At4g13250 | Short-chain dehydrogenase/reductase (SDR) family protein | d54 | 0.37 | 0.48 | 0.31 |
| At4g13430 | Aconitase family protein/aconitate hydratase family protein | d130 | 0.40 | 0.49 | 0.72 |
| At4g15530 | Pyruvate phosphate dikinase family protein | d63 | 0.87 | 0.78 | 0.47 |
| At4g28780 | GDSL-motif lipase/hydrolase family protein | d103 | 1.04 | 0.71 | 0.30 |
| At5g07440 | Glu dehydrogenase 2 (GDH2) | d148 | 1.13 | 0.95 | 0.38 |
| At5g02780 | In2-1 protein (putative) | d73 | 1.89 | 0.91 | 0.31 |
| At5g09440 | Phosphate-responsive protein (putative) | d56 | 1.32 | 0.50 | 0.32 |
| At5g09530 | Hyp-rich glycoprotein family protein | d22 | 1.25 | 0.88 | 0.46 |
| At5g17380 | Pyruvate decarboxylase family protein | d53 | 0.78 | 0.91 | 0.49 |
| At5g23020 | 2-Isopropylmalate synthase 2 (IMS2) | d11 | 0.64 | 0.66 | 0.34 |
| At5g23660 | Nodulin MtN3 family protein | d68 | 1.86 | 0.63 | 0.47 |
| At5g44020 | Acid phosphatase class B family protein | d122 | 1.50 | 0.81 | 0.27 |
| At5g44930 | Exostosin family protein | d127 | 1.06 | 0.64 | 0.36 |
| At5g49360 | Glycosyl hydrolase family 3 protein | d152 | 1.04 | 0.53 | 0.38 |
| At5g51970 | Sorbitol dehydrogenase (putative)/l-iditol 2-dehydrogenase (putative) | d39 | 0.98 | 0.70 | 0.48 |
| At5g59530 | 2-Oxoglutarate-dependent dioxygenase (putative) | d37 | 1.23 | 0.99 | 0.47 |
| At5g65010 | Asn synthetase 2 (ASN2) | d88 | 1.03 | 0.91 | 0.44 |
| Protein modification | |||||
| At1g72070 | DNAJ heat shock N-terminal domain-containing protein | d80 | 1.86 | 0.65 | 0.32 |
| At3g01290 | Band 7 family protein | d134 | 1.18 | 0.91 | 0.45 |
| At4g16563 | Aspartyl protease family protein | d93 | 1.31 | 0.67 | 0.48 |
| At4g38690 | 1-Phosphatidylinositol phosphodiesterase-related | d2 | 1.06 | 0.64 | 0.50 |
| At5g23210 | Ser carboxypeptidase S10 family protein | d146 | 1.26 | 0.71 | 0.48 |
| AtCg00670 | ATP-dependent Clp protease proteolytic subunit | 1.05 | 0.86 | 0.47 | |
| Response to stress | |||||
| At1g07890 | l-Ascorbate peroxidase 1, cytosolic (APX1) | d42 | 0.59 | 0.66 | 0.40 |
| At1g08830 | Copper/zinc superoxide dismutase (CSD1) | d92 | 0.38 | 1.43 | 0.86 |
| At1g09560 | Germin-like protein (GLP4; GLP5) | d44 | 1.23 | 0.78 | 0.36 |
| At1g45145 | Thioredoxin H-type 5 (TRX-h5; TOUL) | d102 | 1.37 | 0.69 | 0.41 |
| At1g73330 | Protease inhibitor (putative; DR4) | d69 | 2.06 | 0.65 | 0.13 |
| At2g23680 | Stress-responsive protein (putative) | d71 | 1.36 | 0.76 | 0.37 |
| At2g28190 | Copper/zinc superoxide dismutase (CSD2) | d106 | 0.44 | 1.18 | 1.12 |
| At2g37130 | Peroxidase 21 (PER21; P21; PRXR5) | d8 | 0.74 | 0.52 | 0.31 |
| At2g37180 | Water stress-induced tonoplast intrinsic protein (RD28) | d49 | 0.84 | 0.84 | 0.46 |
| At2g38870 | Protease inhibitor (putative) | d45 | 2.62 | 1.61 | 0.50 |
| At2g39800 | Δ1-Pyrroline-5-carboxylate synthetase A/P5CS A (P5CS1) | d95 | 0.48 | 0.70 | 1.94 |
| At3g50970 | Dehydrin xero2 (XERO2)/low-temperature-induced protein LTI30 | d67 | 1.27 | 0.47 | 0.45 |
| At4g02520 | Glutathione S-transferase (putative) | d119 | 1.10 | 0.73 | 0.21 |
| At4g08390 | l-Ascorbate peroxidase, stromal (sAPX) | d149 | 0.43 | 0.68 | 0.38 |
| At4g25100 | Superoxide dismutase (Fe), chloroplast (SODB; FSD1) | d120 | 0.67 | 0.41 | 0.24 |
| At4g35770 | Senescence-associated protein (SEN1) | d38 | 1.37 | 0.81 | 0.42 |
| At4g37530 | Peroxidase 50 (PER50; PRXR50) | d132 | 0.68 | 0.75 | 0.42 |
| At5g14780 | Formate dehydrogenase (FDH) | d79 | 0.74 | 0.80 | 0.45 |
| Signal transduction | |||||
| At1g02340 | Reduced phytochrome signaling (REP1; BHLH26) | d29 | 0.86 | 0.55 | 0.27 |
| At4g30270 | MERI-5 protein (MERI-5; MERI5B) | d116 | 1.66 | 0.48 | 0.28 |
| At5g15410 | Cyclic nucleotide-gated channel (CNGC2) | d3 | 0.97 | 0.84 | 0.48 |
| Transcription factor | |||||
| At2g36890 | Myb family transcription factor (MYB38) | d51 | 0.99 | 0.90 | 0.48 |
| At2g43060 | Expressed protein | d59 | 1.17 | 0.72 | 0.39 |
| At2g46970 | Basic helix-loop-helix (bHLH) protein (putative) | 1.18 | 0.68 | 0.23 | |
| At3g05690 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | d32 | 0.87 | 0.69 | 0.43 |
| At3g51910 | Heat shock transcription factor family protein | d24 | 1.13 | 0.87 | 0.45 |
| At3g56400 | WRKY family transcription factor | d105 | 0.46 | 1.20 | 1.62 |
| At3g60530 | Zinc finger (GATA type) family protein | d26 | 1.35 | 0.93 | 0.37 |
| At5g07690 | Myb family transcription factor (MYB29) | d7 | 0.33 | 0.53 | 1.05 |
| At5g07700 | Myb family transcription factor (MYB76) | d125 | 0.35 | 0.51 | 1.28 |
| At5g23000 | Myb family transcription factor (MYB37) | d114 | 1.04 | 0.81 | 0.49 |
| At5g53980 | Homeobox-Leu zipper family protein | d136 | 0.87 | 1.07 | 0.49 |
| At5g63790 | No apical meristem (NAM) family protein | d43 | 0.62 | 0.97 | 0.41 |
| Transport | |||||
| At1g08230 | Amino acid transporter family protein | d115 | 1.02 | 0.79 | 0.46 |
| At1g66760 | MATE efflux family protein | d101 | 1.16 | 0.85 | 0.47 |
| At1g78000 | Sulfate transporter (Sultr1;2) | d76 | 1.54 | 0.57 | 0.47 |
| At1g80830 | NRAMP metal ion transporter 1 (NRAMP1) | d82 | 0.68 | 0.67 | 0.37 |
| At2g10940 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | d153 | 0.85 | 0.91 | 0.37 |
| At2g26650 | Potassium channel protein 1 (AKT1) | d104 | 1.15 | 0.74 | 0.44 |
| At2g40300 | Ferritin (putative) | d123 | 0.78 | 0.79 | 0.42 |
| At2g45180 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | d17 | 2.20 | 0.95 | 0.38 |
| At3g48970 | Copper-binding family protein | d97 | 1.15 | 0.85 | 0.48 |
| At3g53980 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | d30 | 1.68 | 0.48 | 1.18 |
| At3g56090 | Ferritin (putative) | d14 | 1.03 | 1.07 | 0.48 |
| At3g56200 | Amino acid transporter family protein | d52 | 1.90 | 0.69 | 0.44 |
| At4g00370 | Sugar transporter family protein | d12 | 1.22 | 0.77 | 0.43 |
| At4g04770 | ATP-binding cassette transporter (ABC1) | d9 | 0.36 | 0.46 | 0.40 |
| At4g12030 | Bile acid:sodium symporter family protein | d1 | 0.38 | 0.48 | 1.26 |
| At4g30110 | ATPase E1-E2-type family protein | d6 | 0.77 | 0.54 | 0.41 |
| At5g10180 | Sulfate transporter | d10 | 0.46 | 0.42 | 0.14 |
| At5g61520 | Hexose transporter (putative) | d60 | 1.23 | 0.81 | 0.45 |
| At5g62670 | ATPase, plasma membrane-type (putative)/proton pump (putative) | d128 | 0.82 | 0.84 | 0.46 |
| Others | |||||
| At1g02850 | Glycosyl hydrolase family 1 protein | d126 | 1.11 | 1.01 | 0.43 |
| At1g04660 | Gly-rich protein | d117 | 0.99 | 0.61 | 0.47 |
| At1g18590 | Sulfotransferase family protein | d4 | 0.40 | 0.56 | 1.66 |
| At1g49320 | BURP domain-containing protein | d78 | 0.57 | 0.48 | 0.33 |
| At1g76590 | Zinc-binding family protein | d58 | 1.00 | 0.65 | 0.36 |
| At1g76800 | Nodulin (putative) | d13 | 0.88 | 0.63 | 0.30 |
| At1g78830 | Curculin-like (Man-binding) lectin family protein | d48 | 1.24 | 0.72 | 0.34 |
| At2g03890 | Phosphatidylinositol 3- and 4-kinase family protein | d139 | 1.22 | 0.96 | 0.45 |
| At2g05440 | Gly-rich protein | d135 | 1.39 | 0.54 | 0.09 |
| At2g05540 | Gly-rich protein | d74 | 0.84 | 0.78 | 0.39 |
| At2g32870 | MATH domain-containing protein | d141 | 0.77 | 0.48 | 0.62 |
| At2g39030 | GCN5-related N-acetyltransferase (GNAT) family protein | d50 | 1.32 | 0.81 | 0.37 |
| At2g39310 | Jacalin lectin family protein | d100 | 1.05 | 0.80 | 0.48 |
| At2g39330 | Jacalin lectin family protein | d35 | 0.78 | 0.46 | 0.41 |
| At2g41380 | Embryo-abundant protein-related | d147 | 1.33 | 0.90 | 0.47 |
| At2g43530 | Trypsin inhibitor (putative) | d65 | 2.20 | 0.60 | 0.45 |
| At3g04720 | Hevein-like protein (HEL) | d90 | 1.50 | 0.73 | 0.13 |
| At3g15356 | Legume lectin family protein | 1.16 | 0.71 | 0.21 | |
| At3g22740 | Homocysteine S-methyltransferase 3 (HMT-3) | d18 | 0.34 | 0.60 | 1.09 |
| At3g48390 | MA3 domain-containing protein | 1.01 | 0.77 | 0.47 | |
| At3g50440 | Hydrolase, α/β-fold family protein | d15 | 1.00 | 0.61 | 0.35 |
| At3g50440 | Hydrolase, α/β-fold family protein | 0.99 | 0.70 | 0.45 | |
| At3g54600 | DJ-1 family protein | 0.46 | 0.58 | 1.02 | |
| At4g00780 | MATH domain-containing protein | d64 | 0.84 | 0.49 | 0.97 |
| At4g01440 | Nodulin MtN21 family protein | d121 | 0.93 | 0.83 | 0.50 |
| At4g04810 | SeIR domain-containing protein | 2.40 | 0.62 | 0.38 | |
| At4g04830 | SeIR domain-containing protein | d21 | 2.94 | 0.65 | 0.30 |
| At4g15610 | Integral membrane family protein | d137 | 1.48 | 0.74 | 0.24 |
| At4g28050 | Senescence-associated protein (putative) | d99 | 1.10 | 0.57 | 0.25 |
| At4g36850 | PQ-loop repeat family protein/transmembrane family protein | 0.99 | 0.65 | 0.46 | |
| At4g38080 | Hyp-rich glycoprotein family protein | d98 | 1.58 | 0.80 | 0.33 |
| At4g39940 | Adenylylsulfate kinase 2 (AKN2) | d112 | 0.39 | 0.59 | 1.13 |
| Unknown | |||||
| At1g01430 | Expressed protein | d72 | 0.96 | 0.78 | 0.49 |
| At1g07090 | Expressed protein | d40 | 1.26 | 0.85 | 0.46 |
| At1g10090 | Expressed protein | d27 | 0.92 | 0.60 | 0.37 |
| At1g29050 | Expressed protein | d41 | 1.18 | 0.66 | 0.45 |
| At1g49310 | Expressed protein | d89 | 0.85 | 0.66 | 0.45 |
| At1g54740 | Expressed protein | d62 | 0.78 | 0.66 | 0.48 |
| At1g73810 | Expressed protein | 1.05 | 0.65 | 0.44 | |
| At2g04795 | Expressed protein | d19 | 1.24 | 0.63 | 0.42 |
| At3g19030 | Expressed protein | d124 | 1.61 | 0.71 | 0.49 |
| At3g28320 | Hypothetical protein | d66 | 0.78 | 0.83 | 0.34 |
| At4g31330 | Expressed protein | d118 | 1.01 | 0.70 | 0.49 |
| At4g32480 | Expressed protein | 0.93 | 0.62 | 0.36 | |
| At5g01740 | Expressed protein | d23 | 1.43 | 0.75 | 0.28 |
| At5g03545 | Expressed protein | d47 | 3.21 | 1.07 | 0.25 |
| At5g40450 | Expressed protein | 0.87 | 0.88 | 0.49 | |
| At5g40450 | Expressed protein | d151 | 1.46 | 0.86 | 0.47 |
| At5g45410 | Expressed protein | d81 | 0.62 | 0.74 | 0.47 |
| At5g64090 | Expressed protein | d57 | 1.49 | 0.49 | 0.52 |
Refers to genes in the right panel of Figure 4.
Figure 2.
GO annotation of the 2-fold differentially up- and down-regulated genes upon R. fascians infection.
We examined the expression of all genes involved in cytokinin metabolism and the canonical cytokinin signaling pathway (Fig. 3). The expression of six A-type ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, which are transcriptionally induced by cytokinins and mediate a feedback regulation of the cytokinin response (D'Agostino et al., 2000), and of the cytokinin receptor AHK4 was activated, albeit at different levels. Moreover, two CYTOKININ OXIDASE/DEHYDROGENASE (CKX) genes were up-regulated and ISOPENTENYL TRANSFERASE3 (IPT3) was down-regulated, illustrating the occurrence of cytokinin homeostasis and confirming previous data (Depuydt et al., 2008). IPT8, unlike IPT3 not negatively regulated by cytokinins (Miyawaki et al., 2004), showed a minor up-regulation throughout infection. BiNGO analysis (Maere et al., 2005) of all up-regulated genes indicated a clear response toward a cytokinin stimulus, further underlining the central position of cytokinins in the R. fascians pathology (Supplemental Fig. S2).
Figure 3.
Differentially expressed cytokinin-associated genes during the R. fascians plant interaction at 7, 14, and 24 dpi detected via microarray hybridizations. Fold changes between R. fascians D188 and D188-5 (control) are presented.
The Genome-Wide Host Response toward R. fascians Infection Suggests a Highly Specialized Interaction
As the data presented above confirm the central role of cytokinins, we wondered how conserved the global transcriptional response upon R. fascians infection was when compared with cytokinin treatment and upon challenge with other pathogens. Therefore, biclustering analyses were done with the Genevestigator tool (Zimmermann et al., 2004) on the genes listed in Tables I and II in relevant publicly available data sets (zeatin treatment; infection with bacteria [Agrobacterium tumefaciens and Pseudomonas syringae], gray mold [Botrytis cinerea], two powdery mildews [Erysiphe cichoracearum and Erysiphe orontii], and an oomycete [Phytophthora infestans]). Generally, the transcriptional profiles of both the up- and down-regulated genes overlapped a little (Fig. 4). Approximately 12% of the down-regulated and 30% of the up-regulated transcripts were common between cytokinin treatment and R. fascians infection. Among the latter were cell wall-loosening expansins that have been implicated in cytokinin-induced cell proliferation (Downes and Crowell, 1998; Rashotte et al., 2003; indicated by u5, u11, and u14 in Fig. 4) and four ARR genes (indicated by u1, u8, u15, and u104 in Fig. 4). However, many of the transcriptional changes induced upon cytokinin treatment were not activated upon infection. For instance, Bet v 1 genes, encoding putative cytokinin-detoxifying proteins, were induced by cytokinins (Mogensen et al., 2002; Brenner et al., 2005), but three family members were down-regulated upon R. fascians infection (Table II). Interestingly, the transcriptional overlap with A. tumefaciens was considerably higher, 38% and 45% of the down- and up-regulated genes, respectively, implying extensive similarities in the host response toward infection with both gall-forming bacteria. The more conserved up-regulation of a wound-responsive protein, a putative protease inhibitor, a putative chitinase, and two trypsin inhibitors (u20, u35, u41, u45, and u53 in Fig. 4) in many different treatments suggested that plant defense was activated during these biotic interactions. Intriguingly, in the R. fascians interaction, at 24 dpi most of these genes were down-regulated (Table I), hinting at an active down-regulation of defense.
Figure 4.

Output of Genevestigator bicluster analyses comparing R. fascians infection with hormone treatment and biotic interactions. Rows represent the up-regulated (left) and down-regulated (right) genes, and the coded identities of the genes are given in Tables I and II, respectively. Black squares mark matching expression profiles for R. fascians-infected samples and the considered treatment (www.genevestigator.ethz.ch). [See online article for color version of this figure.]
R. fascians Actively Suppresses Defense
Detailed symptom analysis in tobacco and Arabidopsis did not point to a defense reaction of the plant in response to infection (de O Manes et al., 2001, 2004). As ROS are typically involved in restraining pathogen spread (Hammond-Kosack and Jones, 1996; Dempsey et al., 1999), we analyzed the microarray data set for genes related to ROS production or ROS scavenging. Five out of seven differential peroxidases (PER21, PER33, PER50, PERATP23A, and a putative peroxidase), together with a putative copper amino oxidase (At1g62810) that catalyzes the oxidative deamination of polyamines resulting in the release of hydrogen peroxide (H2O2), were significantly down-regulated upon infection with strain D188 (Table II; Supplemental Table S1), suggesting that R. fascians D188 colonization did not trigger H2O2 production. Indeed, no differential accumulation of H2O2 could be visualized by 3,3′-diaminobenzidine (DAB) staining of infected Arabidopsis plants at different time points during the interaction (data not shown), and the oxidative stress marker thioredoxin H5 (TRX-h5; Laloi et al., 2004) was severely repressed (Table II). Moreover, the expression of (stromal) l-ascorbate peroxidase (APX1 and sAPX), glutathione peroxidase (GPX5 and GPX6), copper-zinc superoxide dismutase (CSD1 and CSD2), and catalase (CAT1 and CAT2) genes, known to be ROS scavengers (Mittler et al., 2004), was also down-regulated upon infection with strain D188 (Table II; Supplemental Table S1). Furthermore, dehydroascorbate reductase (DHAR3) and monodehydroascorbate reductase 4 (MDAR4) and MDAR5, which convert dehydroascorbate to the antioxidant ascorbate, were repressed (Table II; Supplemental Table S1). The latter transcript data were supported by the strong reduction in ascorbate levels and the accumulation of its precursor dehydroascorbate in infected tissues (Supplemental Fig. S3). Finally, the genes encoding ferritin 2 and 3, hypothesized to protect cells against ROS by regulating the amount of Fe(II) and Fe(III) (Murgia et al., 2001), were also severely repressed (Table II). Altogether, these data indicated that infection with R. fascians does not cause an oxidative burst in the plant.
Next, we analyzed the microarray data set for genes encoding (putative) pathogenesis-related proteins, such as (endo)glucanases, chitinases, proteinase inhibitor proteins, thionins, glutathione S-transferases, lipid transfer proteins, Phe ammonia lyases, and chalcone synthases known to function in defense (Jwa et al., 2006). Although a few genes were up-regulated throughout R. fascians infection, most were down-regulated in the later stages (Supplemental Table S1), among them established defense markers, such as Phe ammonia lyases, suggesting active defense suppression by R. fascians D188.
To get a clue on which bacterial signal could be involved in defense suppression, we evaluated the expression of the differentially expressed stress-related genes upon D188 infection (Fig. 5A for Table II, Fig. 5B for Supplemental Table S1) in data sets obtained from other biotic interactions and cytokinin and auxin treatments. Interestingly, except for A. tumefaciens, expression of most genes was up-regulated in the selected biotic interactions (Fig. 5A). Unexpectedly, zeatin treatment activated the expression of almost all analyzed genes (Fig. 5), while auxin had either no effect or a repressing effect, implying that auxin secretion by R. fascians might be involved in defense avoidance.
Figure 5.
Genevestigator study of the stress- and defense-related genes upon R. fascians infection. A, Down-regulated stress-related genes of Table II and their response upon different biotic interactions and hormone treatments. B, Responses of all differentially regulated defense-related genes after R. fascians infection (Supplemental Table S1) and their responses upon hormone treatment. Genes that are down-regulated in at least one time point upon R. fascians infection are not marked, while asterisks indicate up-regulated genes throughout R. fascians infection. Green and red represent genes that are down- and up-regulated, respectively. Black indicates no significant changes. IAA, Indole-3-acetic acid; NAA, naphthaleneacetic acid.
R. fascians Reprograms the Host Primary Metabolism
GO annotation revealed that genes involved in metabolism constituted 11% and 12% of the down- and up-regulated genes, respectively. Moreover, BiNGO analysis of the down-regulated genes implied an impact on tricarboxylic acid (TCA) cycle intermediate metabolism and on (branched chain) amino acid and derivative catabolism (Supplemental Fig. S3). These transcript data suggested that infection might significantly modulate the primary metabolism of the plant. For a more comprehensive view, profiles of 38 primary metabolites were analyzed by gas chromatography-mass spectrometry (GC-MS) of extracts prepared from complete Arabidopsis shoots at 4, 7, 14, and 24 dpi treated as follows: mock inoculation with water, control infection with strain D188-5, or infection with strain D188. Although this study is focused on the differences between D188-5 and D188 infections, to correlate metabolic shifts with disease development, we noticed that for some metabolites D188-5 infection caused notable changes compared with mock-inoculated controls (indicated by # in Figs. 6, 10, and 11 and Supplemental Fig. S4), which could be correlated with a general reaction to bacterial colonization or with early flowering, which is a conserved developmental response on bacterial infection (Korves and Bergelson, 2003). Generally, almost 66% of the tested metabolites exhibited similar profiles over time for the three treatments, although in 50% of these cases the final concentration differed significantly from that of the controls. For the remaining 34% of the metabolites, the profiles were completely dissimilar (Figs. 6, 10, and 11; Supplemental Fig. S4).
Figure 6.
Effect of R. fascians infection on carbon metabolism in planta. Inoculation with water, R. fascians D188-5, and D188 at 4, 7, 14, and 24 dpi. Symbols (# and *) indicate statistically significant differences between D188-5 and mock-infected samples (P < 0.01) and between D188-5 and D188 samples (P < 0.01), respectively. The metabolite abundances are expressed relative to the internal standard ribitol at each time point and normalized to the fresh weight. Error bars represent sd (n = 6). N.D., Not determined.
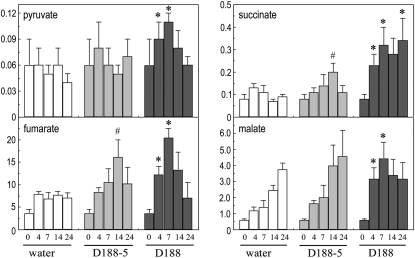
Figure 10.
Responses of pyruvate and TCA cycle intermediates upon infection with R. fascians. Mock, R. fascians D188-5, and D188 infection at 4, 7, 14, and 24 dpi. Symbols (# and *) indicate statistically significant differences between D188-5 and mock samples (P < 0.01) and between D188-5 and D188 samples (P < 0.01), respectively. The metabolite abundances are expressed relative to the internal standard ribitol at each time point and normalized to the fresh weight. Error bars represent sd (n = 6).
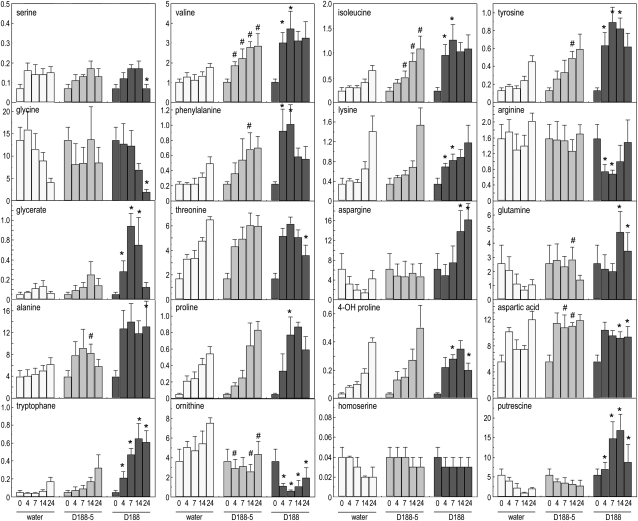
Figure 11.
Effect of R. fascians infection on amino acids, putrescine, and glycerate levels. Mock, R. fascians D188-5, and D188 infection at 4, 7, 14, and 24 dpi. Symbols (# and *) indicate statistically significant changes between D188-5 and mock-infected samples (P < 0.01) and between D188-5 and D188 samples (P < 0.01), respectively. The metabolite abundances are expressed relative to the internal standard ribitol at each time point and normalized to the fresh weight. Error bars represent sd (n = 6).
R. fascians Infection Triggers Sink Development in Symptomatic Tissues
During plant development, but also upon pathogen infections, source-to-sink transitions occur in tissues, with the conversion from a sugar-producing and -exporting status to a sugar-importing and -accumulating status as a result. Interestingly, throughout the experiment, the total sugar content of shoots was between 2.5- and 2.9-fold higher upon D188 infection than that of the mock-infected control. While Suc levels did not differ from 4 dpi onward, the concentrations of the hexoses Glc, Fru, and sorbose as well as of the disaccharide maltose and of the trisaccharide raffinose increased strongly (Fig. 6). For the sugar alcohols, galactinol accumulated in both D188- and D188-5-infected tissues, albeit with different kinetics, whereas erythritol levels only increased upon D188-5 infection. The profiles for glycerol and glycerol-3-phosphate were comparable for the three treatments, whereas the concentration of myoinositol was somewhat higher upon infection. The most striking difference was observed for the nonreducing disaccharide trehalose, which accumulated upon infection with strain D188-5 and strain D188 at 4 dpi between 64- and 42-fold, respectively. At 24 dpi, the level of trehalose strongly decreased in D188-5-infected tissue and was 5-fold higher in the D188-infected plants. The microarray data indeed showed that a trehalose phosphate synthetase gene, TPS1, encoding the first step of trehalose biosynthesis, was differentially up-regulated from 14 dpi onward (Table I). No differential expression was measured for trehalose-6-phosphate-phosphatase genes that mediate the final dephosphorylation step to trehalose.
The specific increase in the hexose-Suc ratio only upon R. fascians D188 infection hinted at a possible involvement of invertases. In the microarray data set, no differentially expressed invertase genes were present; however, it could not be ruled out that this was caused by the different sampling for metabolome and transcriptome analysis. Therefore, we examined the expression pattern of cell wall (FRUCT1), cytoplasmic (FRUCT3 and FRUCT4), and vacuolar and chloroplastic invertase (INV-H and INV-E, respectively) genes in complete shoot tissues at 4, 7, 14, and 24 d after treatment by RT-PCR (Fig. 7A). The expression of the cell wall invertase gene was already induced at 4 dpi by both bacterial infections (D188-5 and D188), but the induction was stronger and only persisted throughout the experiment upon infection with strain D188. A comparable, although less marked, pattern was obtained for FRUCT3, while FRUCT4 was not differentially expressed. For INV-H, no clear pattern was observed, although at 24 dpi the expression was highest in D188-infected plants. Finally, the INV-E expression profile resembled that of FRUCT1. These transcript profiles and the hexose metabolite data were confirmed by measuring invertase enzyme activities in infected and control shoot material. Indeed, for all three invertase types, the enzyme activities were significantly higher upon D188 infection. In agreement with the RT-PCR data, a differential activity could also be measured for the cell wall-bound invertase in D188-5-infected tissue (Fig. 7B).
Figure 7.
Invertase gene expression and activities upon R. fascians infection. RT-PCR profiling of five invertase genes (A) and enzymatic activities of the cell wall, vacuolar, and cytoplasmic invertases (B) after inoculation with water, R. fascians D188-5, and D188. FW, Fresh weight.
Photosynthesis upon D188 Infection: Reduced Activity and Enhanced Competition
Hexoses presumably down-regulate photosynthetic activity and photosynthesis gene expression and thus influence the source/sink status of the plant tissues (Biemelt and Sonnewald, 2006). Given the accumulation of hexoses and the typical sink-associated invertase activity in D188-infected plants, we investigated the effect of infection on photosynthesis. The pale green color of symptomatic Arabidopsis plants (Fig. 1A) already hinted at an altered photosynthetic capacity. The loss of chlorophyll during symptom establishment was indeed confirmed by spectrophotometric analysis (Fig. 8A). Among the down-regulated genes identified in the microarray (Table II), 5% was classified in the GO category “Electron transport/energy pathway” (Fig. 2). Interestingly, At2g28000 and At1g55490, encoding the α- and β-subunits of chaperonin-60, a molecular chaperone involved in correct Rubisco folding and, thus, activity (Salvucci, 2008), were found to be significantly down-regulated in our data set, although by less than 2-fold (data not shown). Since leaves were not harvested for the microarray experiment, the expression of the photosynthesis marker genes RbcS and CAB2 was investigated with RT-PCR on RNA extracted from complete shoots. Generally, D188-5 infection had no or little effect on the age-dependent decline of RbcS and CAB2 expression levels. In contrast, upon D188 infection, both RbcS and CAB2 were significantly down-regulated already at 4 dpi (Fig. 8B). The photosynthetic capacity of infected plants was further assessed by chlorophyll fluorescence imaging (Fig. 8, C–F). Chlorophyll fluorescence is indicative of the overall rate of the photosynthetic function and can be used to determine the efficiency of PSII photochemistry under biotic and abiotic stress conditions (Maxwell and Johnson, 2000; Quilliam et al., 2006). The effective quantum yield of PSII [Y(II)], the maximum quantum yield of PSII (Fv/Fm), and the nonphotochemical quenching parameters NPQ and qN (see “Materials and Methods”) were measured during R. fascians infection. Figure 8C shows a false-color-coded image of Y(II) that represents the proportion of absorbed light that will be used for electron transport through PSII. Whereas Y(II) was not significantly altered by infection of Arabidopsis with strain D188-5, it was markedly reduced by D188 infection. The decrease in Y(II) started at 7 dpi at the leaf edges and, with time, spread throughout the whole leaf. Interestingly, R. fascians colonization starts with the formation of microcolonies at leaf edges (Fig. 9A; Cornelis et al., 2001), suggesting that local secretion of morphogens by the bacteria directly affects photosynthesis. Infected ARR5:GUS lines indeed showed a patchy expression pattern in leaves, which is mostly confined to the margins (Fig. 9B). CKX genes are instrumental for cytokinin degradation and homeostasis upon infection of Arabidopsis with R. fascians (Depuydt et al., 2008; Pertry et al., 2009). Spot inoculation on leaves of CKX5:GUS and CKX6:GUS marker lines revealed a colocalization of the cytokinin-secreting R. fascians colonies and GUS staining (Fig. 9C), further supporting the localized action of the bacterial signals.
Figure 8.
Decrease in photosynthesis after R. fascians infection. A, Chlorophyll content in plants inoculated with water, R. fascians D188-5, and D188 at 24 dpi. B, RT-PCR profiling of the RbcS and CAB2 genes upon mock, R. fascians D188-5, and D188 infections. C, False-color-coded image of Y(II) upon R. fascians D188-5 and D188 infection at 4, 7, 14, and 24 dpi. D, Fv/Fm values upon water, R. fascians D188-5, and D188 infection. E, False color-coded image of qN after R. fascians D188-5 and D188 infection at 4, 7, 14, and 24 dpi. F, Distribution of Y(II), Y(NPQ), and Y(NO) upon mock, R. fascians D188-5, and D188 infection at 4, 7, 14, and 24 dpi. Error bars represent sd (n > 15). Asterisks mark significant differences (P < 0.05). FW, Fresh weight.
Figure 9.
Colocalization of R. fascians microcolonies with cytokinin marker gene expression. A, Electron micrographs showing microcolonies of R. fascians at leaf edges. B, Symptomatic leaves of infected ARR5:GUS marker lines showing GUS staining at leaf edges. C, Spot inoculation of CKX5:GUS and CKX6:GUS lines with GUS staining at the inoculation sites.
The Fv/Fm, a measure of the intrinsic efficiency of PSII, was also significantly lower upon D188 infection (Fig. 8D), indicating photoinhibition. The qN was specifically higher when Y(II) was reduced (Fig. 8E). Y(NPQ) reflects nonphotochemical quenching by heat (thermal) dissipation of excitation energy in the antenna system. Y(NO), representing nonregulated energy dissipation due to PSII inactivity, Y(NPQ), and Y(II) added up to unity, and their distribution during the different treatments is given in Figure 8F. The decrease of Y(II) upon D188 infection was largely paralleled by an increase in Y(NO), indicating inhibition of photosynthesis.
The TCA cycle in the mitochondria is considered to be an integral part of the photosynthetic metabolism (Fernie et al., 2004; Nunes-Nesi et al., 2007; Noguchi and Yoshida, 2008). Among the 2-fold down-regulated genes (Table II), putative aconitases, pyruvate decarboxylases, and pyruvate phosphate dikinase, all involved in the TCA cycle, were identified. Moreover, all of the TCA cycle genes in the list of 3,422 differentially regulated genes were down-regulated at 24 dpi (Supplemental Table S2), further suggesting reduced functioning of the TCA cycle upon infection with R. fascians D188. This observation was partly supported by the metabolite profiles (Fig. 10). Although infection with strain D188 initially led to an accumulation of pyruvate, succinate, fumarate, and malate, at 24 dpi, when symptoms were fully established, the levels of all except succinate dropped below those measured in tissues infected with D188-5.
The bifunctional Rubisco enzyme also functions in the photorespiration process that is in competition with photosynthesis at the level of enzyme substrate (either O2 or CO2). In the complete differential transcript data set, photorespiration-related genes were initially down-regulated, but at 24 dpi, the expression was activated by strain D188, suggesting that photorespiration might be enhanced (Supplemental Table S2). At the metabolite level, Gly and Ser are intermediates in the formation of glycerate that will ultimately be redirected to the chloroplasts, where it is phosphorylated to 3-phosphoglycerate to reenter the Calvin cycle. Upon infection with D188, especially at the later time points of the interaction, Gly and Ser levels were significantly lower than those of the controls (Fig. 11); in contrast, glycerate levels were up to 7-fold higher upon D188 infection, implying a flux toward this metabolite and illustrating a possible enhancement of photorespiration.
R. fascians Infection Stimulates Amino Acid and Polyamine Biosynthesis
Besides a photosynthesis-related function, the TCA cycle provides carbon skeletons for the biosynthesis of several other metabolites, such as amino acids and polyamines. The profiles of the detected amino acids indicated that the bacterial presence had a stimulating effect on the metabolic pathways, resulting in amino acid biosynthesis (Fig. 11). Nevertheless, infection with D188 generally resulted in a faster and stronger increase. The most pronounced differential accumulation was measured for Asn, Trp, Tyr, and Ala, whereas the Arg and Orn contents dropped considerably in D188-infected tissues. Interestingly, the latter are intermediates for polyamine biosynthesis. Indeed, putrescine levels strongly increased upon R. fascians D188 infection already at 4 dpi (Fig. 11).
DISCUSSION
The plant pathogenic actinomycete R. fascians is rather unique among the hyperplasia-inducing bacteria because it induces the formation of differentiated galls upon infection of its many hosts (Putnam and Miller, 2007). Typical symptoms observed in Arabidopsis are small serrated leaves, early flowering, inhibition of floral stalk elongation, activation of axillary meristems, de novo meristem formation in the axillary regions of the rosette, and delayed senescence. These symptoms are triggered by cytokinins secreted by the colonizing bacteria, which set off a signaling cascade leading to activation of mitotic cell divisions, prevention of endoreduplication, and ectopic expression of meristem-specific KNOX genes (Crespi et al., 1992, 1994; Depuydt et al., 2008, 2009). Consequently, infection results in the amplification of young tissues that do not mature. Here, we addressed the questions of how and why symptomatic tissues differ from normal tissues and how the eventual changes could contribute to the establishment of the bacterial population.
Through transcriptome analysis, we investigated the genome-wide molecular basis of symptom development. The overrepresentation of genes involved in cytokinin perception, signal transduction, and homeostasis supported the central role of these hormones in the pathology. However, the data set of genes 2-fold differentially expressed upon infection overlapped only minimally with publicly available microarray data sets that dealt with hormone treatments or biotic interactions. In other words, the R. fascians interaction leaves a very specific transcriptome fingerprint on Arabidopsis. The expression profiles of both up- and down-regulated genes resembled most those obtained from the interaction of A. tumefaciens and Arabidopsis. Interestingly, both bacteria induce galls on their host through elevated levels of cytokinins and auxins. Nevertheless, the mechanisms resulting in the hormone imbalance in the infected plant are completely different, and generally A. tumefaciens-induced tumors consist only of undifferentiated cells (Johnson et al., 1974).
Remarkably, the expression of genes encoding several peroxidases, a copper amino oxidase, and catalase, superoxide dismutase, and l-ascorbate peroxidases was significantly down-regulated upon infection with the pathogenic strain D188. These proteins are either ROS-producing or ROS-scavenging enzymes, and they play a key role in plant defense against pathogen attack by mediating an oxidative burst (Lamb and Dixon, 1997; Mittler et al., 2004). The observed down-regulation implies that during the interaction with R. fascians D188, no oxidative burst is initiated, which is supported by the down-regulated expression of TRX-h5, a marker for oxidative stress, and the reduced level of ascorbate, a major H2O2-scavenging antioxidant, in infected tissues. Moreover, differential accumulation of H2O2 could not be visualized with DAB staining and localized necrosis or cell death was not observed in Arabidopsis tissues infected with the pathogenic or the nonpathogenic R. fascians strains (data not shown). ROS is not only involved in structural aspects related to plant defense, such as restraining pathogen spread, modulation of plant cell wall architecture, programmed plant cell death, and the hypersensitive response (Hammond-Kosack and Jones, 1996; Dempsey et al., 1999; Torres et al., 2006) but also has a significant signaling function during these processes (Gechev et al., 2006; Torres et al., 2006). Consequently, the apparently active suppression of ROS formation by R. fascians possibly avoids defense onset in general. Indeed, phenolic compounds do not accumulate at the infection site (Vereecke et al., 2000; Cornelis et al., 2001), and many defense-related genes are down-regulated upon D188 infection. Intriguingly, comparison with other data sets revealed that, while these genes were up-regulated during other biotic interactions and by cytokinin treatment, generally their expression was down-regulated during the interaction with A. tumefaciens and by auxin treatment. Auxin has been implicated in defense suppression, and bacterial auxin biosynthesis by A. tumefaciens and P. syringae pv savastanoi is crucial for repression of the hypersensitive response (Robinette and Matthysse, 1990; Navarro et al., 2004, 2006; Robert-Seilaniantz et al., 2007). Interestingly, auxins accumulate in symptomatic tissues induced by R. fascians (Vereecke et al., 2000; de O Manes et al., 2001) and the bacterium has been demonstrated to secrete auxins (Vandeputte et al., 2005), altogether suggesting that this bacterial morphogen might be of central importance for defense suppression.
GO annotation of the differentially expressed genes clearly pointed toward a change in the primary metabolism, a finding that was confirmed by metabolic profiling of tissues infected with R. fascians D188 and D188-5. A schematic overview of these metabolic changes is given in Supplemental Figure S5. Intriguingly, bacterial infection in general had a measurable impact on the primary metabolism of the plant, although the alterations induced by the pathogenic strain were much stronger. Evaluation of the carbohydrates revealed that, whereas Suc levels did not change, Glc, Fru, raffinose, maltose, and sorbose accumulated from 4 dpi onward. Importantly, upon infection, invertase transcripts and activities were strongly enhanced, probably accounting for the observed increase in the hexose-Suc ratio and illustrating the establishment of a sink (Smeekens, 2000; Roitsch and González, 2004; Rolland et al., 2006; Berger et al., 2007). Cytokinins are known to induce invertase activity (Chou et al., 2000; Roitsch et al., 2003; Walters and McRoberts, 2006), but high sugar contents have similar effects, implying that the sink strength could be augmented by this amplified signaling. Moreover, cytokinin-induced cell wall invertase activity has been shown to increase sugar uptake by plant cells (Roitsch and Ehneß, 2000). Several genes encoding sugar transporters were indeed differentially regulated upon R. fascians infection (data not shown), which might contribute to the sink strength. Finally, cytokinins and sugars both control the expression of CYCLIND3 genes, which have been shown to be instrumental for the G1-to-S transition of the cell cycle (Riou-Khamlichi et al., 2000) and for symptom development upon R. fascians infection (Depuydt et al., 2009). Therefore, the sink characteristics are probably correlated with symptom establishment that is an energy-demanding process, on the one hand, and with nutrition of the pathogen, on the other hand. Indeed, the accumulating carbohydrates are good carbon sources for R. fascians (Temmerman et al., 2000), and conversion of infected tissues into a sink has been demonstrated in several other plant-pathogen interactions (Wright et al., 1995; Chou et al., 2000; Herbers et al., 2000; Scharte et al., 2005).
Typically, source-to-sink transitions are accompanied by changes in photosynthetic capacity (Scholes et al., 1994; Chou et al., 2000; Walters and McRoberts, 2006). Gene expression data, determination of chlorophyll content, and chlorophyll fluorescence imaging altogether supported the occurrence of photosynthesis inhibition upon infection with R. fascians. Similarly, transcript and metabolite data indicated that the flux through the TCA cycle was also reduced when the disease became fully established, although the concentration of succinate remained high throughout the interaction and pyruvate levels were elevated at the early stages of infection. Interestingly, virulence gene expression in R. fascians is induced by an autoregulatory compound and the induction levels are significantly increased in the presence of succinate and pyruvate (Temmerman et al., 2000; Maes et al., 2001). The decreasing amounts of Ser and Gly, together with the increase in glycerate levels, may point to photorespiratory Gly decarboxylation that would add to the reduced photosynthetic and TCA cycle activities (Atkin et al., 2000; Raghavendra and Padmasree, 2003; Nunes-Nesi et al., 2007). Intriguingly, photorespiration had been hypothesized to play a potential role in providing specialized nutrients to the bacterium in support of endophytic colonization (Vereecke et al., 2002a, 2002b; Vandeputte et al., 2007).
Amino acid levels had generally increased during symptom development, suggesting that R. fascians might use them as nitrogen sources. In vitro, auxin production by the bacterium is significantly induced when excess Trp is added to the medium (Vandeputte et al., 2005). In this context, it is interesting that the augmentation of Trp was one of the highest measured. Production of the autoregulatory compound by the att locus involves Arg biosynthesis, and expression of the att genes is inhibited by Orn and Arg (Maes et al., 2001). Again, it is remarkable that the concentrations of these amino acids were lower than those of the controls at the onset of the interaction.
Orn and Arg are also precursors of polyamine biosynthesis, and the polyamine putrescine accumulates to high levels during infection. This accumulation could be interpreted as a sign of reduced catabolism; polyamine degradation has recently been described as an important source for H2O2 production (Walters, 2003; Cona et al., 2006; Jubault et al., 2008; Kusano et al., 2008, and refs. therein), further supporting the absence of an oxidative burst upon R. fascians infection. Putrescine and other polyamines have also been associated with young, metabolically active healthy and diseased tissues (Walters and Shuttleton, 1985; Galston and Sawhney, 1990). Moreover, polyamine biosynthesis is induced by cytokinins and other hormone treatments, and they are considered to be a separate class of plant hormones that control plant development (Kusano et al., 2008). The accumulation of putrescine might thus be part of the (secondary) signaling cascade that mediates symptom establishment. Similarly, trehalose, a disaccharide known as an osmoprotectant (Wingler, 2002; Penna, 2003; Luo et al., 2008), has also been attributed a signaling function in plant development (Müller et al., 2001; Vogel et al., 2001; Wingler, 2002). Trehalose concentrations in Arabidopsis are generally low through the action of trehalase. During different plant-microbe interactions, however, trehalose accumulates strongly and is believed to serve as a major carbon source for bacteria (Müller et al., 1995; Brodmann et al., 2002; Boboye, 2004; Rolland et al., 2006). Trehalose levels increased upon infection with both R. fascians strains, although the effect of D188 was much stronger, and interestingly, trehalose biosynthesis has been reported to be activated by cytokinin application (Brenner et al., 2005). In accordance, transcript data showed an up-regulation of TPS1 but no differential expression of the trehalase gene.
In conclusion, the transcript and metabolite data presented here allow us to propose a model for the initiation of the interaction between R. fascians and Arabidopsis and the subsequent niche establishment. Epiphytic colonization triggers minor metabolic changes in the plant through the production of very low levels of morphogens and, eventually, other bacterial effectors. Importantly, early Trp accumulation might feed into the auxin biosynthetic pathway of R. fascians, which might down-regulate plant defense, permitting elaborate epiphytic and endophytic colonization. The metabolic modifications include a decrease in Arg and Orn and an increase in pyruvate and succinate levels, together allowing the synthesis of the bacterial autoregulatory compound. Consequently, bacterial cytokinin biosynthesis is strongly triggered, which significantly affects the transcriptome and metabolome of the plant. Secondary signals, such as polyamines and trehalose, involved in plant development accumulate and amplify the developmental alterations that are initiated by the bacterial cytokinins. The symptomatic tissue converts into a sink through the activation of invertases, leading to an increase in carbon source levels. The simultaneous buildup of amino acids and eventually of specialized photorespiration-derived metabolites ultimately results in the establishment of a rich niche for R. fascians.
MATERIALS AND METHODS
Plant Material, Sampling, and Infection Conditions
Arabidopsis (Arabidopsis thaliana ecotype C24) was used throughout and was obtained from the European Arabidopsis Stock Centre (N906). ARR5:GUS, CKX5:GUS, and CKX6:GUS lines were obtained from T. Schmülling (Freie Universität Berlin). The seeds were sterilized by submergence for 2 min in 70% (v/v) ethanol, subsequently for 12 min in 5% (w/v) NaOCl supplemented with 0.1% (v/v) polyoxyethylenesorbitan 20, and rinsed at least five times with sterile water. The seeds were germinated and grown on half-strength Murashige and Skoog medium in a growth chamber under a 16-h/8-h light/dark photoperiod at 21°C ± 2°C. GUS staining was done as described previously (Depuydt et al., 2008).
The Rhodococcus fascians strains used were the pathogenic strain D188, containing the linear virulence plasmid pFiD188, and its plasmid-free nonpathogenic derivative D188-5 (Desomer et al., 1988). These strains were grown in liquid yeast extract broth for 2 d at 28°C under gentle agitation until late exponential phase. Prior to infection, the cultures were washed and concentrated four times by resuspending the bacterial pellets in sterile distilled water. At 16 d after germination, Arabidopsis plants were infected at the 1.05 stage (Boyes et al., 2001) by local application of a drop of bacterial culture to the shoot apical meristem. For genome-wide expression profiling, plants were sampled at 7, 14, and 24 dpi. Roots, cotyledons, and leaves were removed, yielding samples enriched in the meristematic regions, and were immediately snap frozen in liquid nitrogen. Two independent replicates of each time point-treatment combination were harvested, for a total of 12 samples. For the qRT-PCR experiments and the metabolomics analysis, complete shoots were harvested at 4, 7, 14, and 24 dpi. For spot inoculations, 5 μL of a washed bacterial culture was applied to the upper side of the leaves of CKX5:GUS or CKX6:GUS plants, incubated for 7 d, and subsequently stained for GUS activity, as described (Depuydt et al., 2008).
RNA Extraction and Labeling
RNA was extracted using the RNease Plant Mini Kit (Qiagen) according to the manufacturer's instructions. For each time point-treatment combination, RNA was extracted from a pool of 50 plants. These RNA preparations were DNase treated and purified through NH4Ac (5 m) precipitation. Samples were quality controlled and quantified with a NanoDrop Spectrophotometer (Isogen). Samples were labeled with either Cy3 or Cy5 dye.
Hybridization and Preprocessing
Samples were hybridized to the CATMA Arabidopsis arrays (Crowe et al., 2003; Hilson et al., 2004), which are two-color arrays containing 24,576 GSTs and 384 controls. Hybridization, washing, and scanning of the arrays were carried out at the Plant Genomics Research Unit, as described previously (Lurin et al., 2004). Quality of the hybridization was assessed using control spots.
The raw expression data, comprising the logarithm of median feature pixel intensity at wavelengths 653 nm (Cy5) and 532 nm (Cy3), were uploaded into GenStat (Payne and Arnold, 2002) for further processing. The Loess procedure was applied for global intensity-dependent normalization on the log base 2 foreground intensities to correct for dye bias. No background correction was made. Each of the 14 hybridization samples was subjected to the linear mixed normalization model of the form (random effects in italics; Wolfinger et al., 2001): response = μ + array + residual, where the response variable represents the Loess-corrected log2-transformed Cy3 and Cy5 fluorescence intensity measurements of the 24,576 GSTs. Next, the estimated residuals from the normalization mixed model were subjected to a gene-specific mixed ANOVA model of the form (random effects in italics): residuals = μ + dye + treatment + time point + treatment × time point + array + error. The dye term models the fixed dye and replicate effects, as both effects are confounded; the array term models the effects for each spot and equals the gene × array interaction effect. Mixed models were fitted to the data by restricted maximum likelihood. Wald statistics were calculated, and significance was assigned to the interaction term treatment × time point, which identified genes that showed a differential expression pattern for both parameters (time point and treatment). An FDR was calculated for the treatment × time point interaction effect by modeling the adjusted P values as a two-component mixture of uniform and beta densities, as implemented in GenStat; default parameter settings were used to estimate π0, the proportion of features that are truly null. A P value cutoff of 0.001 was chosen, corresponding to a FDR of 0.15% for the treatment × time point interaction effect.
All procedures comply with the MIAME (for minimum information about a microarray experiment) standards for array data (Brazma et al., 2001). The profiling data have been submitted to ArrayExpress (accession no. E-MEXP-1893).
RT-PCR Primers and Conditions
For qRT-PCR analysis, 2 μg of RNA was reverse transcribed into cDNA synthesis with the SuperScript Reverse Transcriptase Kit (Invitrogen), subsequently diluted 50 times, and stored at −20°C until further use. RT-PCR was done on cDNA derived from complete shoot material to investigate invertase and photosynthetic gene expression. The cell wall (FRUCT1 and FRUCT2) and vacuolar (FRUCT3 and FRUCT4) invertases were amplified in 25 cycles with primers described by Tymowska-Lalanne and Kreis (1998). The expression profile of the cytoplasmic/neutral invertases was examined with primers developed by Vargas et al. (2008) in 22 amplification cycles. The RbcS primer set was obtained from Laval et al. (2002), and CAB2 was amplified with primers described by Delessert et al. (2004), both in 18 amplification cycles.
Metabolomics Analysis
The primary metabolite profiles of complete shoots of mock-inoculated controls and plants infected with R. fascians D188-5 and D188 were compared in six independent biological replicates at 0, 4, 7, 14, and 24 dpi. To extract soluble metabolites for GC-MS analysis, 100 mg of ground shoot material was extracted in 1.4 mL of 100% (v/v) methanol together with 60 μL of an internal standard (0.2% [w/v] ribitol in water). The mixture was heated for 15 min at 70°C with vigorous mixing. After centrifugation, 750 μL of chloroform and 1.5 mL of water were added to the supernatant and vortexed for 30 s. The phases were separated by centrifugation, and aliquots of the methanol/water phase (containing the polar metabolites) were taken and reduced to dryness in a SpeedVac concentrator. Samples were dissolved in 40 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine for 2 h at 37°C to protect the carbonyl moieties. Next, 10 μL of a retention time standard mixture (0.029% [v/v] n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotracontane, and n-hexatriacontane dissolved in pyridine) was added. Acidic protons were derivatized by treatment with 70 μL of N-methyl-N-(trimethylsilyl)trifluoracetamide for 30 min at 37°C. The GC-MS profiling method was largely as described previously (Lisec et al., 2006). Samples in each batch were analyzed in random order. GC-MS spectra were obtained with an AS 2000 autosampler and a GC8000 gas chromatograph coupled to a Thermo Finnigan Voyager quadrupole-type mass spectrometer, operated by MassLab software (ThermoQuest; Thermo Fisher Life Scientific). GC-MS hardware, settings, and data analysis were as described (Lisec et al., 2006; Schauer et al., 2006).
Chlorophyll Fluorescence Imaging
Chlorophyll fluorescence parameters were measured with an Imaging-PAM Chlorophyll Fluorometer (Walz). Mock-inoculated controls and R. fascians-infected plants (strains D188 and D188-5) were compared in their photosynthetic capacity at 4, 7, 14, and 21 dpi. At least 15 areas of interest, over which the values of the selected fluorescence parameters were averaged, were marked for each condition of each parameter, and their average was used for downstream analysis. With the Imaging-PAM, the current fluorescence yield (Ft) was continuously measured. After determining the dark-level fluorescence yield (Ft = F0), for which the plants were dark adapted for 15 min, an 800-ms saturating pulse was applied to determine the maximum fluorescence (Fm). The maximum quantum yield of PSII photochemistry, Fv/Fm = (Fm − F0)/Fm, was then automatically calculated by the ImagingWin software (Walz). In the presence of actinic illumination, the current fluorescence yield (Ft = F) and the maximum light-adapted fluorescence (Fm′) were determined, from which the effective PSII quantum yield [Y(II) = (Fm′ − Ft)/Fm′] was automatically derived. The coefficient of nonphotochemical quenching, qN = (Fm − Fm′)/(Fm′ − F0′), with F0′ estimated according to Oxborough and Baker (1997), was calculated by the ImagingWin software, as were the parameters of the effective PSII quantum yields of regulated nonphotochemical energy dissipation, Y(NPQ), and nonregulated energy dissipation, Y(NO), which are complementary with the photochemical quantum yield [i.e. Y(II) + Y(NPQ) + Y(NO) = 1]. Y(II) represents the proportion of absorbed light that is used for photosynthetic electron transport through PSII and is thus essentially the operating efficiency of PSII measured in the light. Fv/Fm is the maximum efficiency of PSII and is a useful measure for photoinhibition, and Y(NPQ) describes the efficiency with which energy is dissipated nonphotochemically as heat. Images of the fluorescence parameters were displayed with the help of a false-color code ranging from 0 (black) to 1 (purple) as implemented in the ImagingWin software.
Scanning Electron Microscopy
The samples were analyzed by scanning electron microscopy with a table-top microscope TM-1000 (Hitachi) without sample processing. Images were taken with the accompanying software.
Invertase Activity Tests
To assay the activities of the cytosolic, vacuolar, and cell wall invertases, the extraction was carried out as described (Wright et al., 1998), with approximately 150 mg of ground shoot tissue of each infection condition supplemented with 1.2 mL of extraction buffer (50 mm HEPES-KOH, pH 8.0, 5 mm MgCl2, 2 mm EDTA, 1 mm CaCl2, 1 mm benzamidine, 1 mm dithiothreitol, and 0.1 mm phenylmethylsulfonyl fluoride). The mixture was incubated on ice for 10 min. After centrifugation (13,000g, 10 min, 4°C), the supernatant (containing cytosolic and vacuolar invertases) was transferred to a new tube. For determination of cell wall invertase activities, the pellet was washed three times and finally resuspended in 1.2 mL of extraction buffer. Extracts were used without dialysis. The invertase reactions were carried out in 0.08 m citrate/phosphate buffers with 0.1 m Suc as a substrate at pH 4.5, 5.5, and 7.5 for cell wall, vacuolar, and cytoplasmic invertases, respectively (Scholes et al., 1994). Of the extracts, 100 μL was added to the reaction mixtures in a final volume of 300 μL. After 45 min of incubation at 37°C, the reaction was stopped by adding 0.1 mL of 1 m Tris-HCl and incubated for 4 min at 95°C. The amounts of Glc and Fru liberated in the reaction were determined by an enzyme-linked assay (Scholes et al., 1994): 30 μL of the invertase reaction was added to 200 μL of a buffer cocktail (1 m imidazole-HCl, pH 6.9, 2 m MgCl2, 100 mm NADP, 100 mm ATP, and 400 units mL−1 Glc-6-P dehydrogenase) on a multiwell plate. Next, 5 μL of hexokinase (300 units mL−1) was added, and the minimum and maximum absorbance were recorded at 340 nm. Subsequently, 5 μL of phosphoglucoisomerase (140 units mL−1) was added, and the minimum and maximum optical densities were recorded to calculate the enzymatic activity of the samples. Each reaction was done in triplicate.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. qRT-PCR confirmation of 10 differentially expressed genes randomly taken from the microarray data set.
Supplemental Figure S2. BiNGO analysis of all up-regulated genes throughout R. fascians infection.
Supplemental Figure S3. BiNGO analysis of all down-regulated genes throughout R. fascians infection.
Supplemental Figure S4. Effect of R. fascians infection on ascorbate and dehydroascorbate levels.
Supplemental Figure S5. Overview of significant changes in the primary metabolism upon infection with R. fascians D188 when compared with D188-5 infection.
Supplemental Table S1. Differential expression of putative defense genes upon R. fascians D188 infection.
Supplemental Table S2. TCA cycle and photorespiration genes differentially expressed upon R. fascians D188 infection.
Supplementary Material
Acknowledgments
We thank Koen Goethals and Rosemary Loria for fruitful discussions, Thomas Schmülling for ARR5:GUS, CKX5:GUS, and CKX6:GUS seeds, Elisabeth Stes for DAB staining, Annick De Keyser for technical assistance, and Martine De Cock for help in preparing the manuscript.
This work was supported by the Bijzonder Onderzoeksfonds of Ghent University and the European Molecular Biology Organization (predoctoral and short-term fellowships, respectively, to S.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Danny Vereecke (danny.vereecke@psb.ugent.be).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Atkin OK, Millar AH, Gardeström P, Day DA (2000) Photosynthesis, carbohydrate metabolism and respiration in leaves of higher plants. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Photosynthesis: Physiology and Metabolism, Advances in Phytosynthesis and Respiration, Vol 9. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 153–175
- Berger S, Papadopoulos M, Schreiber U, Kaiser W, Roitsch T (2004) Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol Plant 122 419–428 [Google Scholar]
- Berger S, Sinha AK, Roitsch T (2007) Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J Exp Bot 58 4019–4026 [DOI] [PubMed] [Google Scholar]
- Biemelt S, Sonnewald U (2006) Plant-microbe interactions to probe regulation of plant carbon metabolism. J Plant Physiol 163 307–318 [DOI] [PubMed] [Google Scholar]
- Boboye B (2004) Degradation of trehalose by rhizobia and characteristics of a trehalose-degrading enzyme isolated from Rhizobium species NGR234. J Appl Microbiol 97 256–261 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbusch J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29 365–371 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44 314–333 [DOI] [PubMed] [Google Scholar]
- Brodmann D, Schuller A, Ludwig-Müller J, Aeschbacher RA, Wiemken A, Boller T, Wingler A (2002) Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol Plant Microbe Interact 15 693–700 [DOI] [PubMed] [Google Scholar]
- Chou H-M, Bundock N, Rolfe SA, Scholes JD (2000) Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Mol Plant Pathol 1 99–113 [DOI] [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11 80–88 [DOI] [PubMed] [Google Scholar]
- Cornelis K, Maes T, Jaziri M, Holsters M, Goethals K (2002) Virulence genes of the phytopathogen Rhodococcus fascians show specific spatial and temporal expression patterns during plant infection. Mol Plant Microbe Interact 15 398–403 [DOI] [PubMed] [Google Scholar]
- Cornelis K, Ritsema T, Nijsse J, Holsters M, Goethals K, Jaziri M (2001) The plant pathogen Rhodococcus fascians colonizes the exterior and interior of the aerial parts of plants. Mol Plant Microbe Interact 14 599–608 [DOI] [PubMed] [Google Scholar]
- Crespi M, Messens E, Caplan AB, Van Montagu M, Desomer J (1992) Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J 11 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Vereecke D, Temmerman W, Van Montagu M, Desomer J (1994) The fas operon of Rhodococcus fascians encodes new genes required for efficient fasciation of host plants. J Bacteriol 176 2492–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouzé P, Hilson P, Beynon J, Weisbeek P, Van Hummelen P, Reymond P, et al (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R (2004) Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 55 165–181 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Shah J, Klessig DF (1999) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18 547–575 [Google Scholar]
- de O Manes C-L, Beeckman T, Ritsema T, Van Montagu M, Goethals K, Holsters M (2004) Phenotypic alterations in Arabidopsis thaliana plants caused by Rhodococcus fascians infection. J Plant Res 117 139–145 [DOI] [PubMed] [Google Scholar]
- de O Manes C-L, Van Montagu M, Prinsen E, Goethals K, Holsters M (2001) De novo cortical cell division triggered by the phytopathogen Rhodococcus fascians in tobacco. Mol Plant Microbe Interact 14 189–195 [DOI] [PubMed] [Google Scholar]
- Depuydt S, De Veylder L, Holsters M, Vereecke D (2009) Eternal youth, the fate of developing Arabidopsis leaves upon Rhodococcus fascians infection. Plant Physiol 149 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Doležal K, Van Lijsebettens M, Moritz T, Holsters M, Vereecke D (2008) Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiol 146 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desomer J, Dhaese P, Van Montagu M (1988) Conjugative transfer of cadmium resistance plasmids in Rhodococcus fascians strains. J Bacteriol 170 2401–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Wahl R, Vranes M, de Vries RP, Kämper J, Kahmann R (2008) Establishment of compatibility in the Ustilago maydis/maize pathosystem. J Plant Physiol 165 29–40 [DOI] [PubMed] [Google Scholar]
- Downes BP, Crowell DN (1998) Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol 37 437–444 [DOI] [PubMed] [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T (1997) Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9 1825–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann J, Schmitz-Thom I, Schön H, Sonnewald S, Weis E, Scharte J (2008) RNA interference-mediated repression of cell wall invertase impairs defense in source leaves of tobacco. Plant Physiol 147 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7 254–261 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galston AW, Sawhney RK (1990) Polyamines in plant physiology. Plant Physiol 94 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, Wendehenne D, Pugin A (2006) Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact 19 711–724 [DOI] [PubMed] [Google Scholar]
- Gartemann K-H, Abt B, Bekel T, Burger A, Engemann J, Flügel M, Gaigalat L, Goesmann A, Gräfen I, Kalinowski J, et al (2008) The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J Bacteriol 190 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28 1091–1101 [DOI] [PubMed] [Google Scholar]
- Goethals K, Vereecke D, Jaziri M, Van Montagu M, Holsters M (2001) Leafy gall formation by Rhodococcus fascians. Annu Rev Phytopathol 39 27–52 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense responses. Plant Cell 8 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Meuwly P, Frommer WB, Métraux J-P, Sonnewald U (1996) Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell 8 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Sonnewald U (1998) Molecular determinants of sink strength. Curr Opin Plant Biol 1 207–216 [DOI] [PubMed] [Google Scholar]
- Herbers K, Takahata Y, Melzer M, Mock H-P, Hajirezaei M, Sonnewald U (2000) Regulation of carbohydrate partitioning during the interaction of potato virus Y with tobacco. Mol Plant Pathol 1 51–59 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson P (2000) Cytokinins and auxins in plant-pathogen interactions—an overview. Plant Growth Regul 32 369–380 [Google Scholar]
- Johnson R, Guderian RH, Eden F, Chilton M-D, Gordon MP, Nester EW (1974) Detection and quantitation of octopine in normal plant tissue and in crown gall tumors. Proc Natl Acad Sci USA 71 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Jubault M, Hamon C, Gravot A, Lariagon C, Delourme R, Bouchereau A, Manzanares-Dauleux MJ (2008) Differential regulation of root arginine catabolism and polyamine metabolism in clubroot-susceptible and partially resistant Arabidopsis genotypes. Plant Physiol 146 2008–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jwa N-S, Agrawal GK, Tamogami S, Yonekura M, Han O, Iwahashi H, Rakwal R (2006) Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol Biochem 44 261–273 [DOI] [PubMed] [Google Scholar]
- Kocal N, Sonnewald U, Sonnewald S (2008) Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol 148 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, Bergelson J (2003) A developmental response to pathogen infection in Arabidopsis. Plant Physiol 133 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano T, Berberich T, Tateda C, Takahashi Y (2008) Polyamines: essential factors for growth and survival. Planta 228 367–381 [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld J-P (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48 251–275 [DOI] [PubMed] [Google Scholar]
- Laval V, Koroleva OA, Murphy E, Lu C, Milner JJ, Hooks MA, Tomos AD (2002) Distribution of actin gene isoforms in the Arabidopsis leaf measured in microsamples from intact individual cells. Planta 215 287–292 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1 387–396 [DOI] [PubMed] [Google Scholar]
- Loria R, Kers J, Joshi M (2006) Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44 467–487 [DOI] [PubMed] [Google Scholar]
- Luo Y, Li W-M, Wang W (2008) Trehalose: protector of antioxidant enzymes or reactive oxygen species scavenger under heat stress? Environ Exp Bot 63 378–384 [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess enrichment of Gene Ontology categories in Biological Networks. Bioinformatics 21 3448–3449 [DOI] [PubMed] [Google Scholar]
- Maes T, Vereecke D, Ritsema T, Cornelis K, Ngo Thi Thu H, Van Montagu M, Holsters M, Goethals K (2001) The att locus of Rhodococcus fascians strain D188 is essential for full virulence on tobacco through the production of an autoregulatory compound. Mol Microbiol 42 13–28 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51 659–668 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network in plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37 128–138 [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Wimmer R, Larsen JN, Spangfort MD, Otzen DE (2002) The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J Biol Chem 277 23684–23692 [DOI] [PubMed] [Google Scholar]
- Müller J, Aeschbacher RA, Wingler A, Boller T, Wiemken A (2001) Trehalose and trehalase in Arabidopsis. Plant Physiol 125 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Boller T, Wiemken A (1995) Trehalose and trehalase in plants: recent developments. Plant Sci 112 1–9 [Google Scholar]
- Murgia I, Briat JF, Tarantino D, Soave C (2001) Plant ferritin accumulates in response to photoinhibition but its ectopic overexpression does not protect against photoinhibition. Plant Physiol Biochem 39 797–805 [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JDG (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Krapp A, Röper-Schwarz U, Stitt M (1998) The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP glucose pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ 21 443–454 [Google Scholar]
- Noguchi K, Yoshida K (2008) Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8 87–99 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Sweetlove LJ, Fernie AR (2007) Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol Plant 129 45–56 [Google Scholar]
- Oxborough K, Baker NR (1997) An instrument capable of imaging chlorophyll a fluorescence from intact leaves at very low irradiance and at cellular and subcellular levels of organization. Plant Cell Environ 20 1473–1483 [Google Scholar]
- Payne RW, Arnold GM (2002) GenStat Release 6.1 Reference Manual Part 3: Procedure Library PL14. VSN International, Hemel Hempstead, UK
- Penna S (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8 355–357 [DOI] [PubMed] [Google Scholar]
- Pertry I, Václavíková K, Depuydt S, Galuszka P, Spíchal L, Temmerman W, Stes E, Schmülling T, Kakimoto T, Van Montagu M, et al (2009) Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc Natl Acad Sci USA 106 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam ML, Miller ML (2007) Rhodococcus fascians in herbaceous perennials. Plant Dis 91 1064–1076 [DOI] [PubMed] [Google Scholar]
- Quilliam RS, Swarbrick PJ, Scholes JD, Rolfe SA (2006) Imaging photosynthesis in wounded leaves of Arabidopsis thaliana. J Exp Bot 57 55–69 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8 546–553 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JMS, Murray JAH (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10 372–379 [DOI] [PubMed] [Google Scholar]
- Robinette D, Matthysse AG (1990) Inhibition by Agrobacterium tumefaciens and Pseudomonas savastanoi of development of the hypersensitive response elicited by Pseudomonas syringae pv. phaseolicola. J Bacteriol 172 5742–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2 198–206 [DOI] [PubMed] [Google Scholar]
- Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinha AK (2003) Extracellular invertase: key metabolic enzyme and PR protein. J Exp Bot 54 513–524 [DOI] [PubMed] [Google Scholar]
- Roitsch T, Ehneß R (2000) Regulation of source/sink relations by cytokinins. Plant Growth Regul 32 359–367 [Google Scholar]
- Roitsch T, González M-C (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9 606–613 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Salvucci ME (2008) Association of Rubisco activase with chaperonin-60β: a possible mechanism for protecting photosynthesis during heat stress. J Exp Bot 59 1923–1933 [DOI] [PubMed] [Google Scholar]
- Scharte J, Schön H, Weis E (2005) Photosynthesis and carbohydrate metabolism in tobacco leaves during an incompatible interaction with Phytophthora nicotianae. Plant Cell Environ 28 1421–1435 [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24 447–454 [DOI] [PubMed] [Google Scholar]
- Scholes JD, Lee PJ, Horton P, Lewis DH (1994) Invertase: understanding changes in the photosynthetic and carbohydrate metabolism of barley leaves infected with powdery mildew. New Phytol 126 213–222 [Google Scholar]
- Simón-Mateo C, Depuydt S, de Oliveira Manes CL, Cnudde F, Holsters M, Goethals K, Vereecke D (2006) The phytopathogen Rhodococcus fascians breaks apical dominance and activates axillary meristems by inducing plant genes involved in hormone metabolism. Mol Plant Pathol 7 103–112 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51 49–81 [DOI] [PubMed] [Google Scholar]
- Swarbrick PJ, Schulze-Lefert P, Scholes J (2006) Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ 29 1061–1076 [DOI] [PubMed] [Google Scholar]
- Temmerman W, Vereecke D, Dreesen R, Van Montagu M, Holsters M, Goethals K (2000) Leafy gall formation is controlled by fasR, an AraC-type regulatory gene, in Rhodococcus fascians. J Bacteriol 182 5832–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Torres de Zabala M, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46 14–33 [DOI] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M (1998) Expression of the Arabidopsis thaliana invertase gene family. Planta 207 259–265 [DOI] [PubMed] [Google Scholar]
- Vandeputte O, Öden S, Mol A, Vereecke D, Goethals K, El Jaziri M, Prinsen E (2005) Biosynthesis of auxin by the Gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl Environ Microbiol 71 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte O, Vereecke D, Mol A, Lenjou M, Van Bockstaele D, El Jaziri M, Baucher M (2007) Rhodococcus fascians infection accelerates progression of tobacco BY-2 cells into mitosis through rapid changes in plant gene expression. New Phytol 175 140–154 [DOI] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL (2008) New insights on sucrose metabolism: evidence for an active A/N-Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta 227 795–807 [DOI] [PubMed] [Google Scholar]
- Vereecke D, Burssens S, Simón-Mateo C, Inzé D, Van Montagu M, Goethals K, Jaziri M (2000) The Rhodococcus fascians-plant interaction: morphological traits and biotechnological applications. Planta 210 241–251 [DOI] [PubMed] [Google Scholar]
- Vereecke D, Cornelis K, Temmerman W, Holsters M, Goethals K (2002. a) Versatile persistence pathways for pathogens of animals and plants. Trends Microbiol 10 485–488 [DOI] [PubMed] [Google Scholar]
- Vereecke D, Cornelis K, Temmerman W, Jaziri M, Van Montagu M, Holsters M, Goethals K (2002. b) Chromosomal locus that affects the pathogenicity of Rhodococcus fascians. J Bacteriol 184 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Fiehn O, Jean-Richard-dit-Bressel L, Boller T, Wiemken A, Aeschbacher RA, Wingler A (2001) Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J Exp Bot 52 1817–1826 [DOI] [PubMed] [Google Scholar]
- Walters DR (2003) Polyamines and plant disease. Phytochemistry 64 97–107 [DOI] [PubMed] [Google Scholar]
- Walters DR, McRoberts N (2006) Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci 11 581–586 [DOI] [PubMed] [Google Scholar]
- Walters DR, Shuttleton MA (1985) Polyamines in the roots of turnip infected with Plasmodiophora brassicae Wor. New Phytol 100 209–214 [Google Scholar]
- Wingler A (2002) The function of trehalose biosynthesis in plants. Phytochemistry 60 437–440 [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Wright DP, Baldwin BC, Shephard MC, Scholes JD (1995) Source-sink relationships in wheat leaves infected with powdery mildew. I. Alterations in carbohydrate metabolism. Physiol Mol Plant Pathol 47 237–253 [Google Scholar]
- Wright DP, Read DJ, Scholes JD (1998) Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ 21 881–891 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.