Abstract
Transient genetic modification of plant protoplasts is a straightforward and rapid technique for the study of numerous aspects of plant biology. Recent studies in metazoan systems have utilized cell-based assays to interrogate signal transduction pathways using high-throughput methods. Plant biologists could benefit from new tools that expand the use of cell culture for large-scale analysis of gene function. We have developed a system that employs fluorescent positive selection in combination with flow cytometric analysis and fluorescence-activated cell sorting to isolate responses in the transformed protoplasts exclusively. The system overcomes the drawback that transfected protoplast suspensions are often a heterogeneous mix of cells that have and have not been successfully transformed. This Gateway-compatible system enables high-throughput screening of genetic circuitry using overexpression. The incorporation of a red fluorescent protein selection marker enables combined utilization with widely available green fluorescent protein (GFP) tools. For instance, such a dual labeling approach allows cytometric analysis of GFP reporter gene activation expressly in the transformed cells or fluorescence-activated cell sorting-mediated isolation and downstream examination of overexpression effects in a specific GFP-marked cell population. Here, as an example, novel uses of this system are applied to the study of auxin signaling, exploiting the red fluorescent protein/GFP dual labeling capability. In response to manipulation of the auxin response network through overexpression of dominant negative auxin signaling components, we quantify effects on auxin-responsive DR5∷GFP reporter gene activation as well as profile genome-wide transcriptional changes specifically in cells expressing a root epidermal marker.
It has been demonstrated that flow cytometric analysis and fluorescence-activated cell sorting (FACS) of plant protoplasts is practicable; moreover, this technique has yielded valuable results in a number of different fields of research (Harkins and Galbraith, 1984; Galbraith et al., 1995; Sheen et al., 1995). For instance, FACS of protoplasts from Arabidopsis (Arabidopsis thaliana) plants expressing tissue-specific fluorescent protein markers has been used to examine both basal and environmentally stimulated transcriptional profiles in particular cell types (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008; Gifford et al., 2008), and flow cytometry has been employed to analyze reactive oxygen species production and programmed cell death in tobacco (Nicotiana tabacum) protoplasts (Lin et al., 2006). A broad selection of fluorescence tools is available to study a plethora of physiological parameters in plants, for example, cis-regulatory elements fused to fluorescent proteins (Haseloff and Siemering, 2006), genetically encoded molecular sensors (Looger et al., 2005), or dye-based sensors (Haugland, 2002), can be used in combination with cytometry to measure diverse biological processes.
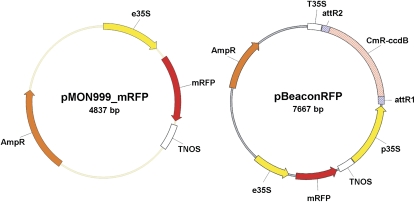
Here, we document the development of a protoplast transfection system that employs cytometry and a transient transformation vector harboring a fluorescent positive selection marker (pBeaconRFP; Fig. 1). The notable advantage of this system is that it allows for the exclusive analysis of the transformed cells and facilitates high-throughput dual-color analysis. The new vector for use in this system is designed in such a way that it not only expresses a gene of interest but also expresses monomeric red fluorescent protein (mRFP). Furthermore, it is compatible with the Gateway recombinase-mediated cloning system, permitting fast and easy cloning. Because of its red emission spectrum, the mRFP marker can easily be used in combination with the commonly utilized GFP. We present two examples of this system's use in the analysis of an important signal transduction cascade involved in many aspects of plant development, namely the auxin perception pathway (Fig. 2; Guilfoyle and Hagen, 2007). Promising alternative uses of the system are further discussed.
Figure 1.
The pBeaconRFP transient transformation system. A schematic representation of the control vector pMON999_mRFP and pBeaconRFP, a high-copy-number plasmid containing a 35S-driven mRFP positive marker and a Gateway cassette. [See online article for color version of this figure.]
Figure 2.
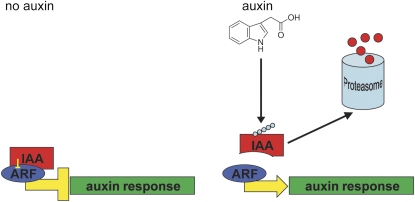
The ARF-Aux/IAA auxin response pathway. In the absence of auxin, Aux/IAAs repress the activity of ARF transcription factors. Upon perception of auxin, Aux/IAAs are ubiquitinated and degraded in a proteasome-dependent manner. Dominant negative mutant isoforms of Aux/IAAs (e.g. IAA7mII and IAA19mII) can no longer be ubiquitinated and effect a stable repression of ARF function. [See online article for color version of this figure.]
Transient transformation of protoplasts is a widely utilized tool in plant research that is swift and unproblematic. The technique can be used, for example, to monitor the regulation of promoter elements, to analyze gene expression or enzymatic activity in response to a variety of stimuli, to examine the roles of transcription factors or signal transduction cascade components, or to study the subcellular localization of proteins (Sheen, 2001; Yoo et al., 2007). As opposed to stable transformation of plants (Arabidopsis being the most commonly used platform), which generally takes months and requires the use of a transfecting agent (usually Agrobacterium tumefaciens), transfection of protoplasts can be achieved in just 1 d and entails only raw DNA and either a chemical- or electroporation-based transfection method. Additionally, transient transformation analyses can overcome problems encountered with stable overexpression, such as pleiotropic developmental effects or nonviability, when a cell-based assay is appropriate. However, due to the fact that protoplast transformation efficiency is never 100%, the results can be convoluted by the nontransformed cells.
Transformation efficiencies are often low and variable (Cummins et al., 2007; <10%) and depend on the employed method as well as properties of the protoplasts and DNA used. We usually get efficiencies ranging from 5% to 20% using Arabidopsis root protoplasts. Others in the field, however, have reported efficiencies of up to 90% using Arabidopsis mesophyll protoplasts (Sheen, 2001). Nonetheless, even a relatively small contamination with nontransformed cells can obscure effects and lead to a misinterpretation of the results. For example, supposing one wanted to measure the ability of a dominant negative signaling component to inhibit the activation of downstream targets and one still sees a level of activation after transfection of the protoplasts. Although significantly reduced as compared with a control, is the remaining activity due to a partial inhibition or is it only present in the nontransformed cells? If a way were to be found to select for successfully transformed cells, a much more precise measurement of the parameter of interest could be obtained.
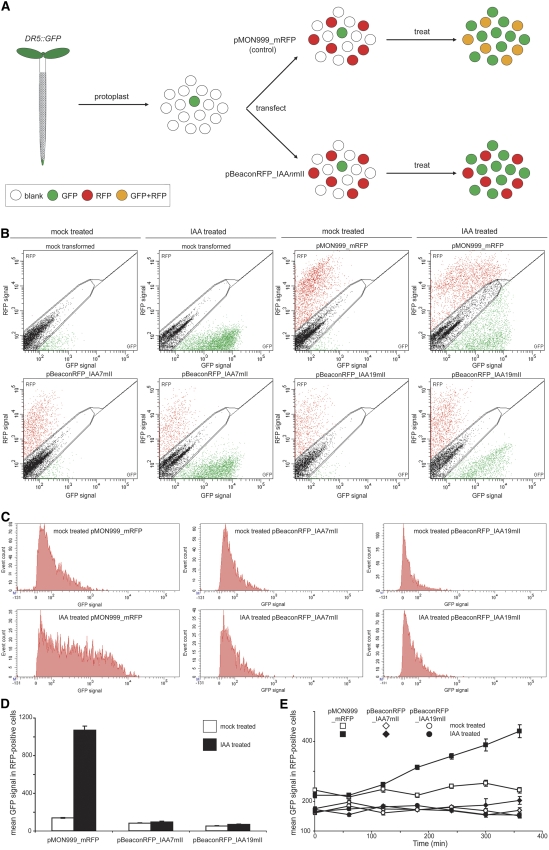
As a first example, we used the pBeaconRFP transient transformation system for the rapid analysis of a regulatory circuit by means of reporter gene readout. We overexpress dominant negative mutant isoforms of the Aux/IAA transcription factors (IAAnmII; Fig. 2; Tiwari et al., 2001) in protoplasts derived from the roots of Arabidopsis seedlings stably transformed with the auxin-sensitive reporter DR5∷GFP (Fig. 3A; Ottenschläger et al., 2003; the pBeaconRFP system is also usable in mesophyll cells, as mRFP is readily distinguishable from chlorophyll autofluorescence cytometrically [data not shown]). In this first experiment, we validate the system using the elegant experiments pioneered in the Guilfoyle laboratory (Ulmasov et al., 1997). A repeat of experiments performed previously (Tiwari et al., 2001), now using pBeaconRFP and flow cytometry, demonstrates that it is possible to assess auxin-induced DR5 promoter activity exclusively in transformed protoplasts by measuring GFP signal intensity.
Figure 3.
Repression of DR5∷GFP activation by IAA7mII and IAA19mII. A, A schematic representation of the experiment. Protoplasts derived from the roots of 1-week-old DR5∷GFP Arabidopsis seedlings were transfected with either pMON999_mRFP, expressing only mRFP, or pBeaconRFP, expressing IAA7mII or IAA19mII. After an overnight incubation, protoplast suspensions were treated for 10 h with 5 μm IAA or solvent alone. B, Flow cytometric analysis of transfected and treated protoplast suspensions. The GFP and RFP intensities for individual protoplasts were recorded and are represented in dot plots; 10,000 events are displayed in each plot. Gates were defined to separate blank, GFP-positive, and RFP-positive events. C, A frequency distribution of the GFP signal of events falling within the RFP gate in B. D, Quantification of the mean GFP signal in RFP-positive cells for auxin- and mock-treated protoplasts transformed with the control vector or pBeaconRFP with IAA7mII or IAA19mII. Data are presented in a histogram ± se (n = 798–2,447). E, A 6-h time course of GFP quantification in an independent experiment. Data are presented in a line graph ± se (n = 579–2,390).
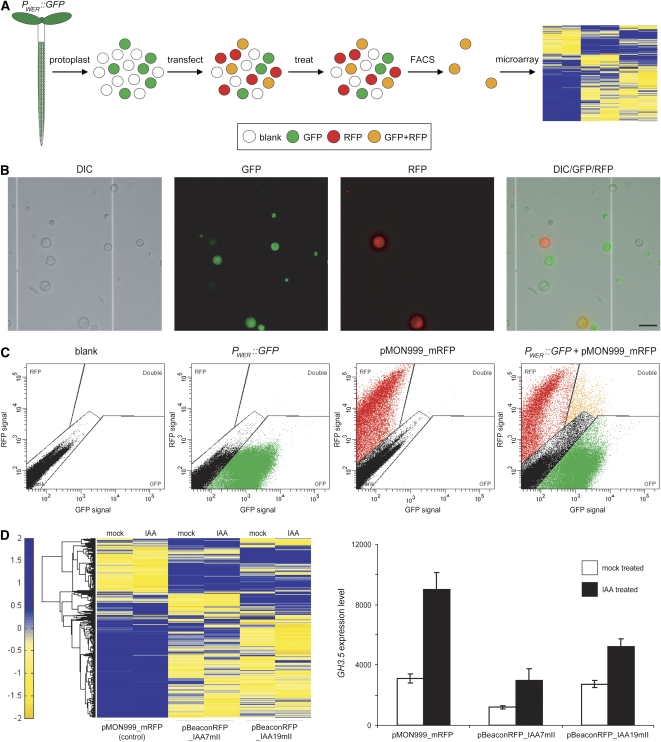
The second example of the application of the pBeaconRFP positive selection marker system involves examination of the transcriptional effects of the expression of IAAnmIIs in a specifically marked cell population (Fig. 4A). We make use of protoplasts derived from a cell type-specific GFP marker line (PWER∷GFP, the WEREWOLF promoter fused to GFP), which expresses primarily in atrichoblasts (Lee and Schiefelbein, 1999), transfected with two different IAAnmII isoforms. Subsequent genome-wide transcriptional profiling of auxin-treated and mock-treated IAAnmII-expressing cells makes it possible to distinguish distinctive patterns of gene expression regulated by the different mutant Aux/IAA isoforms.
Figure 4.
Transcriptional analysis of cell type-specific IAA7mII and IAA19mII expression. A, A schematic representation of the experiment. Protoplasts derived from the roots of 1-week-old PWER∷GFP seedlings were transfected with either pMON999_mRFP, expressing only mRFP, or pBeaconRFP, expressing IAA7mII or IAA19mII. After an overnight incubation, protoplast suspensions were treated for 3 h with 5 μm IAA or solvent alone. Dual-labeled protoplasts were isolated by FACS and used for microarray analysis. B, Microscopic examination of protoplasts derived from the roots of 1-week-old PWER∷GFP seedlings that were transfected with pMON999_mRFP. Bar = 50 μm. C, FACS of the transfected protoplast suspensions. Dot plots are shown depicting the controls used to set up the gates: an untransfected protoplast suspension derived from wild-type roots (blank), an untransfected protoplast suspension derived from PWER∷GFP roots, and a protoplast suspension derived from wild-type roots transfected with pMON999_mRFP. In addition, a dot plot depicting a protoplast suspension derived from PWER∷GFP roots transfected with pMON999_mRFP is shown; protoplasts falling within the gate marked “double” were sorted and used for microarray analysis. A total of 100,000 events are displayed in each dot plot. D, Transcriptional analysis of sorted protoplasts. A log-scale heat map and a histogram quantifying the differences in gene expression between the various collected protoplasts are shown. The heat map displays all of the genes that exhibit any significant difference between mock and auxin treatment, between transformation with the different vectors, and by interaction level (see “Materials and Methods”) as measured by microarray analysis; rows represent genes and columns represent treatment and transformation vectors (n = 3). The histogram presents the difference in GH3.5 expression (as measured by microarray) between the various collected protoplasts ± sd (n = 3).
The use of high-throughput cell-based screening methods in the study of regulatory networks has become a conventional and effective approach in animal systems (Müller et al., 2005; Palmer et al., 2006). Cytometric and FACS-based analyses have also been much more widespread and prolific in animal or microbiology research than in plant research. The combination of a selectable protoplast transformation system along with the use of cytometry now allows us to take these powerful techniques to a new level in plant research.
RESULTS
Modification of Reporter Gene Activation by Transient Overexpression
In order to demonstrate the use of the pBeaconRFP system to study signal transduction, we took advantage of the auxin-sensitive DR5∷GFP reporter gene. DR5 is a highly active synthetic auxin response element created by Ulmasov and coworkers (1997) and derived from the soybean (Glycine max) GH3 indole-3-acetic acid amido synthetase promoter. Upon treatment of seedlings or protoplasts harboring a DR5 reporter gene with auxin, the reporter is activated throughout the plant or protoplast suspension (Ulmasov et al., 1997).
In the original experiments (Ulmasov et al., 1997), carrot (Daucus carota) protoplast suspensions were transfected with three different plasmids: (1) a reporter containing DR5∷GUS, (2) an effector expressing an Aux/IAA, and (3) a transformation efficiency control expressing luciferase. Measurement of auxin-induced GUS activity relative to luciferase activity showed a reduced induction of GUS activity in protoplast suspensions transfected with the Aux/IAA effector plasmid as compared with those transfected with a control vector, indicative of the repressive effect on auxin responses of this family of transcription factors. A drawback of this initial system is that GUS activity induced in protoplasts that have been transformed with the reporter and not the effector will also be measured. The relative amount of protoplasts transformed with fewer than all three of the applied vectors will vary from experiment to experiment and among different effector plasmids. An improved version of this system, in which mesophyll protoplasts from a stably transformed DR5∷GUS Arabidopsis line were utilized, avoided the need for the cotransfection with the reporter and allowed for an analysis of the reporter in a more natural chromatin environment but did not address the issue of measuring the response only in transformed cells (Tiwari et al., 2006).
It has been demonstrated previously that stabilizing mutations in domain II of Aux/IAA proteins lead to a repression of auxin-responsive reporter gene activation (Fig. 2; Tiwari et al., 2001). These authors used the carrot protoplast system described above and presented results indicating that overexpression of these dominant negative mutant isoforms caused a marked reduction in reporter gene activation, although it appeared that the inhibition was incomplete.
Here, we have constructed an mRFP-positive marker containing a Gateway-compatible transient transformation vector, pBeaconRFP (Fig. 1), and have cloned the dominant negative Aux/IAA isoforms IAA7mII and IAA19mII, provided by the Guilfoyle laboratory, into this vector. pMON999_mRFP was utilized as a control vector, expressing only mRFP. These vectors were used to transfect protoplasts derived from the roots of 1-week-old DR5∷GFP Arabidopsis seedlings (Fig. 3A). After an overnight incubation, giving the transformed protoplasts the opportunity to start expressing the IAAnmIIs and mRFP, protoplast suspensions were treated with 5 μm indole-3-acetic acid (IAA) or mock treated with solvent and monitored cytometrically. Figure 3B shows the acquired cytometric data in a dot-plot format. Mock-transfected protoplast suspensions (suspensions that encountered the polyethylene glycol (PEG) transfection procedure without the addition of plasmid; see “Materials and Methods”) displayed only a minor population expressing GFP when treated with solvent alone; this population likely represents protoplasts derived from the natural DR5∷GFP-expressing auxin maxima of the root (i.e. the root tip and lateral root primordia). These suspensions exhibited a sizeable induction of GFP expression when treated with auxin, as expected. In suspensions transfected with the control vector, the induction of GFP expression was clearly apparent in both the nontransformed and RFP-positive, transformed protoplasts. In stark contrast, the auxin-induced GFP expression in suspensions overexpressing either IAA7mII or IAA19mII was only evident in the nontransformed cells and not perceptible in the RFP-positive protoplasts. Quantification of the GFP signal in RFP-positive cells (Fig. 3, C and D) demonstrates that there was an approximately 8-fold increase of GFP signal in protoplasts transformed with the control vector, whereas the protoplasts transformed with the dominant negative Aux/IAA isoforms exhibited no obvious induction. Interestingly, the quantification also showed that the GFP signal in mock-treated IAA7mII- and IAA19mII-expressing protoplasts was already less intense than in the protoplasts transformed with the control vector, a 1.7- and 2.6-fold repression, respectively (Fig. 3D). An independent experiment is presented, showing a time course analysis of GFP induction (Fig. 3E), reiterating the previous result and allowing examination of the kinetics of reporter gene activation.
These results corroborate previous results (Tiwari et al., 2001) and validate the pBeaconRFP system. Furthermore, they demonstrate that we were able to measure reporter gene activation specifically in the transformed cells and indicate that both IAA7mII and IAA19mII effectively repress auxin-induced DR5∷GFP expression. This system permits a highly quantitative live analysis and has the potential for large-scale screening of candidate genes for effects on reporter gene activation.
Transcriptional Analysis of Cell Type-Specific Transient Overexpression
In order to demonstrate an entirely novel use for the system, we used pBeaconRFP in combination with a cell type-specific GFP marker to isolate dual-labeled cells by FACS and to analyze the effects of overexpression in a particular cell population. An expansive collection of cell type-specific fluorescent markers is available to the plant research community (Lee et al., 2006; http://www.plantsci.cam.ac.uk/Haseloff/construction/catalogFrame.html, http://enhancertraps.bio.upenn.edu/default.html). Furthermore, transcriptional changes in specific cell types in response to several environmental stimuli have been scrutinized, and these studies have demonstrated that distinct cell types respond differentially to external cues (Dinneny et al., 2008; Gifford et al., 2008). Auxin responses are also expected to diverge between different cell types; this can be deduced from, among other evidence, the cell type-specific expression of the different isoforms of the ARF-Aux/IAA auxin perception pathway (Weijers and Jürgens, 2004).
We have used pBeaconRFP to transiently express IAAnmIIs in protoplasts derived from the roots of PWER∷GFP Arabidopsis seedlings. Following overnight incubation and a 3-h treatment with IAA or solvent alone, dual-labeled protoplasts were separated using FACS and transcriptionally profiled by means of microarray analysis (Fig. 4A). Protoplast suspensions were transfected with the pMON999_mRFP control vector, pBeaconRFP_IAA7mII, or pBeaconRFP_IAA19mII. Figure 4B shows microscopic images of a PWER∷GFP protoplast suspension transfected with the control vector, demonstrating that there are protoplasts present in all four expected categories: blank, PWER∷GFP alone, pMON999_RFP alone, and dual labeled. Untransfected wild-type and PWER∷GFP protoplast suspensions as well as a wild-type protoplast suspension transfected with the control vector were employed to conservatively set up sorting gates in such a way that exclusively the dual-labeled protoplasts would be sorted (Fig. 4C). The experiment was performed in triplicate; nine separate transfections, 18 treatments, sorts, and microarrays. In corroboration with known auxin responses and our own data, the expression of Arabidopsis GH3.5, as measured by microarray, resembles the DR5∷GFP expression measured in the previous experiment, displaying a drastically reduced auxin-induced increase in expression level and a basal repression of expression in protoplasts transformed with the IAAnmIIs (Figs. 3, D and E, and 4D). Furthermore, genes displaying a response to auxin in the protoplasts transformed with the control vector generally exhibited a dampened response in the protoplasts expressing IAAnmIIs (Table I). Analysis of the data as a whole showed that the protoplasts transformed with the IAAnmIIs were fundamentally already very different compared with the protoplasts transformed with the control vector. Interestingly, although they were more similar to each other than to the control, there was also a substantial number of statistically significant gene expression differences between protoplasts expressing IAA7mII and IAA19mII (Fig. 4D; Table I).
Table I.
Transcriptional changes induced by auxin treatment and IAAnmII overexpression
The number of genes with statistically significant differences (see “Materials and Methods”) in expression between all vectors and treatments are given. The average fold change in expression of the 809 genes responsive to auxin treatment in the pMON999_mRFP control vector is presented for protoplasts transformed with the control vector as well as protoplasts transformed with the IAA7mII and IAA19mII overexpressors.
| Comparison of Treatments | No. of Genes with Significant Changes in Expression
|
Average Fold Change in Expression of pMON999_mRFP Auxin-Responsive Genes | ||||||
|---|---|---|---|---|---|---|---|---|
| pMON999_mRFP
|
pBeaconRFP_IAA7mII
|
pBeaconRFP_IAA19mII
|
||||||
| Mock | IAA | Mock | IAA | Mock | IAA | |||
| pMON999_mRFP | Mock | 0 | 809 | 3,663 | 3,859 | 4,036 | 4,340 | 2.7 ± 0.2a |
| IAA | 0 | 4,182 | 4,231 | 4,513 | 4,677 | |||
| pBeaconRFP_IAA7mII | Mock | 0 | 0 | 2,139 | 2,369 | 1.6 ± 0.1ab | ||
| IAA | 0 | 2,300 | 2,378 | |||||
| pBeaconRFP_IAA19mII | Mock | 0 | 620 | 1.6 ± 0.2ab | ||||
| IAA | 0 | |||||||
Average values are given ± se (n = 809).
Statistically significant difference compared with pMON999_mRFP as determined by a two-tailed unpaired t test (P < 10−4).
These results provide a proof of concept for the feasibility of transcriptional profiling after transient protoplast transformation. This is now possible due to the fact that the system eliminates any contaminating effects of nontransformed cells. Furthermore, the dual-color cell-sorting approach makes it possible to analyze the effect of overexpression in a specific population of cells. In this case, the system allowed us to compare the outcome of expression of two highly homologous signal transduction cascade components, and the results indicate that IAA7 and IAA19 have both overlapping and unique downstream consequences in protoplasts derived from the Arabidopsis root epidermis. These results can be pursued to investigate mechanisms that lead to the specificity of auxin signal transduction. This demonstrates how the pBeaconRFP system can be used as a tool for rapid and high-throughput as well as in-depth analysis of genetic circuitry.
DISCUSSION
The system described here, making use of the pBeaconRFP positive selection marker vector in combination with flow cytometry and FACS, has several advantages over traditional protoplast transient transformation techniques. (1) The vectors containing a fluorescent positive marker make it possible to examine effects exclusively in the transformed protoplasts, thereby avoiding confounding of the results by nontransformed cells. (2) Functional data can be obtained even with protoplasts from tissues or species with intrinsically low transformation efficiencies. (3) The use of RFP as a positive marker allows cytometric analysis of transient gene expression in combination with systems employing any distinguishable fluorophores as readout. (4) FACS-based collection of transformed protoplasts also enables accurate use of any other nonfluorometric downstream analyses. (5) The system is Gateway compatible, making it quick and easy to clone genes of interest and amenable to high-throughput approaches (De Sutter et al., 2005).
Making use of the pBeaconRFP vector and FACS-based collection of cells permits analysis not only of effects on GFP-reporter gene activation or transcriptional profiles, as demonstrated here, but also of any other measurable parameters, such as enzymatic activities and metabolite levels. In combination with cell identity markers, this system now also makes it possible to quickly analyze overexpression effects in a cell type-specific manner. Additionally, measuring effects in a particular cell population, as opposed to a heterogeneous mix of protoplasts, allows for a more defined and specific analysis. Moreover, there is the potential of measuring multiple parameters at once; for instance, one could measure the effect of the manipulation of upstream signal transduction elements on both mitogen-activated protein kinase activation and its ultimate downstream transcriptional responses. Of course, this system does not have to be used exclusively with flow cytometry or FACS; for example, it could also be used to select transformed protoplasts for individual analyses such as patch clamping or subcellular protein localization studies. Alternatively, a use in combination with more basic fluorometric analyses could be envisioned, such as microscopic analyses or assays performed with plate readers. Lastly, the system is conceptually well suited for high-throughput screening purposes (e.g. looking for genes that activate or inhibit the activation of a favorite reporter gene or complementation screens in mutant backgrounds). In conclusion, the technique described here opens up a wide field of possibilities not previously feasible in plant research.
Further development and enhancement of this system is ongoing. A transient silencing vector containing a positive selection marker will allow for RNA interference manipulations. Enhancement with glucocorticoid receptor protein fusion or a transcriptionally inducible system will make it possible to time the activation or overexpression of one's gene of interest (Moore et al., 2006). A Gateway-compatible multicolor protein-tagging set will allow high-throughput protein localization studies as well as protein interaction screens. Additionally, vectors with alternative positive selection markers, such as GFP or other fluorescent proteins, will permit analysis of protoplasts transformed with multiple effectors. Lastly, the development of low stress-eliciting protoplast transfection procedures will allow the examination of protoplasts that more closely resemble their natural state.
The pBeaconRFP vector will be made available through the Flanders Institute of Biotechnology (http://www.psb.ugent.be/gateway/), where the backbone originated. The microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE13783.
MATERIALS AND METHODS
Plant Materials and Treatment
Seeds from wild-type (ecotype Columbia [Col-0]), DR5∷GFP (Col-0; obtained from the Arabidopsis Biological Resource Center; stock no. CS9361), and PWER∷GFP (Col-0; obtained from Dr. John Schiefelbein, University of Michigan) Arabidopsis (Arabidopsis thaliana) plants were sterilized by 5-min incubation with 96% ethanol followed by 20-min incubation with 50% household bleach and rinsing with sterile water. Seeds were plated on square 10-cm × 10-cm plates (Fisher Scientific) with MS agar (2.2 g L−1 Murashige and Skoog salts [Sigma-Aldrich], 1% [w/v] Suc, 1% [w/v] agar, 0.5 g L−1 MES hydrate [Sigma-Aldrich], pH 5.7, with KOH) on top of a sterile nylon mesh (NITEX 03-100/47; Sefar Filtration) to facilitate harvesting of the roots. Seeds were plated in two rows of approximately 150 seeds. Plates were vernalized for 2 d at 4°C in the dark and placed vertically in an Advanced Intellus environmental controller (Percival) set to 35 μmol m−2 s−1 and 22°C with an 18-h-light/6-h-dark regime. Protoplast suspensions were treated with 5 μm IAA (Sigma-Aldrich) or mock treated with solvent alone. A 20 mm IAA stock was dissolved in ethanol and stored at −20°C.
Vector Construction
pMON999_mRFP was obtained from Dr. Joop Vermeer (Universiteit van Amsterdam). pBeaconRFP was constructed by PCR amplification of the 35S∷mRFP∷TNOS cassette from pMON999_mRFP with primers mRFP_F2 (5′-GAATTGCATATGCGTTCAAGCTTCTGCAGG-3′) and mRFP_R (5′-TTAATACATATGCCCGGGGATCGATCC-3′), both with an NdeI restriction site (in boldface), using Phusion polymerase (New England Biolabs). The PCR product was ligated into the NdeI site of p2GW7.0 (http://www.psb.ugent.be/gateway/). The orientation of the insert was checked by PCR.
A pZP211 plasmid containing 35S∷HA-IAA7mII and a pUC18 plasmid containing PIAA19∷HA-IAA19mII were obtained from Dr. Thomas Guilfoyle (University of Missouri). IAA7mII and IAA19mII were PCR amplified with primers IAA7_AttB1 (5′-AAAAAGCAGGCTATGATCGGCCAACTTATGAAC-3′), IAA7AttB2 (5′-AGAAAGCTGGGTTCAAGATCTGTTCTTGCAG-3′), IAA19AttB1 (5′-AAAAAGCAGGCTATGGAGAAGGAAGGACTC-3′), and IAA19AttB2 (5′-AGAAAGCTGGGTTCACTCGTCTACTCCTCTAG-3′) and subsequently reamplified with primers AttB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and AttB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) using Phusion polymerase. The PCR products were recombined into pDONR221 using BP clonase and subsequently shuttled into pBeaconRFP with LR clonase (Invitrogen).
Protoplast Isolation and Transfection
Protoplast isolation and PEG-mediated transfection were performed basically as described by the Sheen laboratory (http://genetics.mgh.harvard.edu/sheenweb/). Roots of 1-week-old seedlings were harvested with a scalpel and placed into a gently shaking flask with 100 mL of protoplasting solution for 3 h. Protoplasting solution was prepared as follows: 1.25% (w/v) cellulase (Yakult), 0.3% (w/v) macerozyme (Yakult), 0.4 m mannitol, 20 mm MES, 20 mm KCl, pH 5.7, with Tris-HCl, pH 7.5; heat for 10 min at 55°C, cool to room temperature; 0.1% (w/v) bovine serum albumin, 10 mm CaCl2, and 5 mm β-mercaptoethanol. The protoplast solution was filtered through a 40-μm cell strainer (BD Falcon), divided over 15-mL conical tubes, and centrifuged for 10 min at 150g. Pellets were washed once with transfection solution (0.4 m mannitol, 15 mm MgCl2 hexahydrate, 4 mm MES, pH 5.7, with KOH), centrifuged again, and resuspended in transfection solution with a final concentration of 4 × 106 protoplasts mL−1 (generally, we obtain between 8 × 103 and 1 × 104 protoplasts from 20 plates). Conical tubes (15 mL) were prepared for each transfection with 50 μg of plasmid DNA (10–20 μL) and 250 μL of protoplasts in transfection solution. PEG solution (250 μL; 40% [w/v] PEG 4000, 0.4 m mannitol, and 0.1 m CaCl2) was added, and the suspension was mixed by flicking the tube repeatedly. Suspensions were incubated for 15 min, after which the protoplasts were washed with 15 mL of incubation solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm MES, pH 5.7, with KOH), centrifuged, and resuspended in 1 mL of incubation solution. Protoplast suspensions were incubated overnight on 24-well plates in the dark.
Flow Cytometry and FACS
Protoplast suspensions were cytometrically analyzed and sorted with a FACSAria (BD Biosciences) fitted with a 100-μm nozzle and using phosphate-buffered saline as a sheath fluid. The sheath pressure was set at 20 psi, and the defection plate voltage was set at 5,000 V (default “low” setting). A 488-nm Coherent Sapphire Solid State laser was used for excitation, and emission was measured at 530/30 nm for GFP and 610/20 nm for RFP. The photomultiplier tube voltage was set at 60 V for forward scatter, 159 V for side scatter, 350 V for GFP, and 335 V for RFP. The threshold value for event detection was set at 8,835 on forward scattering. The drop drive frequency was set to approximately 30 kHz, and the amplitude was set to approximately 45 V; the drop delay value was approximately 27 (these settings will vary slightly with day-to-day operation of the FACSAria). Data were processed using the FACSDiva 5.0.2 software (BD Biosciences). Compensation constraints were set to adjust for spectral overlap between GFP and RFP (GFP, minus 0.50% RFP; RFP, minus 17.91% GFP). Gates for sorting dual-labeled protoplasts were set up using blank (wild type), RFP-only (pMON999_mRFP-transfected wild type), and GFP-only (PWER∷GFP) protoplast suspensions in such a way that the sorted dual-labeled protoplasts in the “double” gate would not be contaminated by blank, RFP-only, or GFP-only protoplasts (Fig. 4C).
Microscopy
Microscopic images of protoplasts mounted in a Bright-Line Hemacytometer (Hausser Scientific) were obtained with differential interference contrast, GFP, and Texas Red filters on an Eclipse 90i microscope (Nikon) running on Metamorph software (Molecular Devices).
Microarray Analysis
Protoplasts were sorted directly into RNA extraction buffer, and RNA was extracted using an RNeasy Micro Kit with RNase-free DNase set according to the manufacturer's instructions (Qiagen). RNA was quantified with a Bioanalyzer (Agilent Technologies) and amplified and labeled with the WT-Ovation Pico RNA Amplification System and FL-Ovation cDNA Biotin Module V2, respectively (NuGEN). The labeled cDNA was hybridized, washed, and stained on an ATH-121501 Arabidopsis full-genome microarray using a Hybridization Control Kit, a GeneChip Hybridization, Wash, and Stain Kit, a GeneChip Fluidics Station 450, and a GeneChip Scanner (Affymetrix).
Data were normalized using the MAS 5.0 method with a scaling factor of 250. Statistical analysis was performed as follows. We first filtered genes that showed expression below noise levels by removing genes whose average expression signal (among three replicates) never exceeded a threshold of 75 in any experiment. The data were subjected to two-way ANOVA (treatment × transient genetic background), and all genes that showed a significant effect at P < 0.05 at any level, including the interaction level, were taken as showing some response to experimental conditions (n = 7,145). These genes are shown on the heat map, which was generated with a log2 transformation of the data followed by row normalization. The heat map was generated in Matlab 7.6.0 (Mathworks). To test for gene expression differences in individual comparisons between the different treatments, we used a procedure that accounts for multiple testing (significance analysis of microarrays, two-class unpaired test, Wilcoxon statistic; q < 10% false discovery rate). In order to assess the effects of IAAnmII expression on auxin responses, the transcripts that showed a significant difference between mock-treated control vector and IAA-treated control vector (basal auxin response; n = 809) were then tested for their fold change response in experiments in which protoplasts were transiently transformed with pBeaconRFP_IAAnmII and mock treated or treated with auxin. Increases and decreases in average expression were converted to an absolute fold change to measure the overall effect of the overexpression on the basal auxin response. IAA7 and IAA19 were removed from analysis in their respective overexpressor samples.
Acknowledgments
We thank Joop Vermeer for the pMON999_mRFP plasmid, Tom Guilfoyle for the 35S∷HA-IAA7mII and PIAA19∷HA-IAA19mII plasmids, and John Schiefelbein for the PWER∷GFP line.
This work was supported by the National Science Foundation (grant no. DBI 0519984).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kenneth D. Birnbaum (ken.birnbaum@nyu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Cummins I, Steel PG, Edwards R (2007) Identification of a carboxylesterase expressed in protoplasts using fluorescence-activated cell sorting. Plant Biotechnol J 5 354–359 [DOI] [PubMed] [Google Scholar]
- De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, Karimi M, Inzé D, Goossens A, Hilson P (2005) Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J 44 1065–1076 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320 942–945 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Lambert GM, Grebenok RJ, Sheen J (1995) Flow cytometric analysis of transgene expression in higher plants: green-fluorescent protein. Methods Cell Biol 50 3–14 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10 453–460 [DOI] [PubMed] [Google Scholar]
- Harkins KR, Galbraith DW (1984) Flow sorting and culture of plant protoplasts. Physiol Plant 60 43–52 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR (2006) The uses of green fluorescent protein in plants. Methods Biochem Anal 47 259–284 [PubMed] [Google Scholar]
- Haugland RP (2002) Handbook of Fluorescent Probes and Research Products. Molecular Probes, Eugene, OR
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483 [DOI] [PubMed] [Google Scholar]
- Lin J, Wang Y, Wang G (2006) Salt stress-induced programmed cell death in tobacco protoplasts is mediated by reactive oxygen species and mitochondrial permeability transition pore status. J Plant Physiol 163 731–739 [DOI] [PubMed] [Google Scholar]
- Looger LL, Lalonde S, Frommer WB (2005) Genetically encoded FRET sensors for visualizing metabolites with subcellular resolution in living cells. Plant Physiol 138 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Samalova M, Kurup S (2006) Transactivated and chemically inducible gene expression in plants. Plant J 45 651–683 [DOI] [PubMed] [Google Scholar]
- Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436 871–875 [DOI] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer EL, Miller AD, Freeman TC (2006) Identification and characterisation of human apoptosis inducing proteins using cell-based transfection microarrays and expression analysis. BMC Genomics 7 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8 777–784 [DOI] [PubMed] [Google Scholar]
- Tiwari S, Wang S, Hagen G, Guilfoyle TJ (2006) Transfection assays with protoplasts containing integrated reporter genes. Methods Mol Biol 323 237–244 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 12 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Jürgens G (2004) Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol 7 687–693 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocols 2 1565–1572 [DOI] [PubMed] [Google Scholar]