Summary
Chitin is an essential component of the fungal cell wall and its synthesis is under tight spatial and temporal regulation. The fungal human pathogen Candida albicans has a four member chitin synthase gene family comprising of CHS1 (class II), CHS2 (class I), CHS3 (class IV) and CHS8 (class I). LacZ reporters were fused to each CHS promoter to examine the transcriptional regulation of chitin synthesis. Each CHS promoter had a unique regulatory profile and responded to the addition of cell wall damaging agents, to mutations in specific CHS genes and exogenous Ca2+. The regulation of both CHS gene expression and chitin synthesis was co-ordinated by the PKC, HOG MAP kinase and Ca2+/calcineurin signalling pathways. Activation of these pathways also resulted in increased chitin synthase activity in vitro and elevated cell wall chitin content. Combinations of treatments that activated multiple pathways resulted in synergistic increases in CHS expression and in cell wall chitin content. Therefore, at least three pathways co-ordinately regulate chitin synthesis and activation of chitin synthesis operates at both transcriptional and post-transcriptional levels.
Introduction
The fungal cell wall is a dynamic structure whose composition and structural organization is regulated during the cell cycle and in response to changing environmental conditions, imposed stresses and mutations in cell wall biosynthetic processes (reviewed in Klis et al., 2006; Ruiz-Herrera et al., 2006). Chitin and β(1–3)-d-glucan, represent the main structural components of the fungal cell wall. These polysaccharides oppose the positive turgor pressure within the cell and ultimately determine the morphology of the cell (Munro and Gow, 2001; Klis et al., 2002; Roncero, 2002). Chitin and glucan synthesis therefore play fundamental roles in maintaining fungal cell integrity during growth and morphogenesis and in adaptation to stress (Cabib, 1987; Wessels, 1990; Shaw et al., 1991; Sietsma and Wessels, 1994; Gooday, 1995). Because these structural polysaccharides do not occur in mammals and are essential for fungi, there is considerable potential for cell wall synthesis as a target for antifungal drugs (Munro and Gow, 1995; Munro et al., 2001; Odds et al., 2003). New generation echinocandins that target the synthesis of cell wall β(1–3)-d-glucan are proving effective agents in the treatment of opportunistic fungal pathogens such as Candida albicans (Denning, 2003). Chitin synthase inhibitors have not yet been discovered that have clinical use in the treatment of fungal infections (Odds et al., 2003).
Regulation of chitin synthesis occurs both at the transcriptional and post-translational levels and is dependent on precise targeting and activation of chitin synthases to specific locations in the plasma membrane, and the provision of adequate substrate (Munro and Gow, 1995). All fungi examined to date have multiple genes encoding chitin synthase families (Munro and Gow, 2001; Roncero, 2002; Ruiz-Herrera and San-Blas, 2003). Individual chitin synthase enzymes perform distinct functions at specific stages of the cell cycle. Saccharomyces cerevisiae has three chitin synthase enzymes – Chs1p (Class I), Chs2p (Class II) and Chs3p (Class IV) while C. albicans has four chitin synthases – two class I enzymes – CaChs2p and CaChs8p, CaChs3p (Class IV) and CaChs1p (a class II enzyme which is the orthologue of ScChs2p). Relatively little is known about the transcriptional regulation of chitin synthase genes in fungi but considerable attention has been focused on post-transcriptional regulation by Chs4–7, which influences Chs3p chitin synthase activation and localization in S. cerevisiae and C. albicans. ScChs7p controls exit of ScChs3p from the ER, ScChs5p and ScChs6p regulate its exit from the trans-Golgi network (Ziman et al., 1996; Santos and Snyder, 1997; Santos et al., 1997; Ziman et al., 1998; Trilla et al., 1999). ScChs4p tethers ScChs3p to the septins at the mother-bud neck via ScBni4p (Demarini et al., 1997; Trilla et al., 1997). Chitin synthesis is therefore influenced by endogenous and exogenous factors that directly and indirectly regulate the chitin synthase catalytic proteins.
Disruption of genes in cell wall biosynthetic pathways of S. cerevisiae and C. albicans often results in alteration and redistribution of chitin and β(1–3)-d-glucan in the cell wall, the synthesis of new cell wall proteins and changes in their cross-linking to cell wall polysaccharides (reviewed in Popolo et al., 2001; Klis et al., 2002; Klis et al., 2006). Defects in cell wall integrity are sensed by the transmembrane proteins of the Mid2p and the Wscp family, which signal via the Rom2p guanine nucleotide exchanger leading to activation of the Rho1p GTPase. Rho1p has many downstream targets including protein kinase C and the β(1–3)-d-glucan synthase subunits Fks1p and Fks2p (Popolo et al., 2001). In S. cerevisiae this ‘cell wall salvage’ or ‘cell wall compensatory’ pathway is activated in response to cell wall perturbing agents such as Calcofluor white (CFW), Congo Red (CR), caffeine, β-glucanases and cell wall mutations and is mediated primarily through the PKC cell integrity MAP kinase cascade and its downstream target the transcription factor Rlm1p (Lagorce et al., 2003; Boorsma et al., 2004; Garcia et al., 2004). In S. cerevisiae, elevation of chitin levels in response to activation of the salvage pathway is largely dependent upon ScChs3p (Valdivieso et al., 2000; Carotti et al., 2002). Several studies have highlighted the importance of signalling systems in co-ordinating this regulation. A higher proportion of ScChs3p localized to the plasma membrane in heat-stressed cells (Valdivia and Schekman, 2003). This mobilization of ScChs3p was dependent upon activation of Rho1p and Pkc1p, and the phosphorylation of ScChs3p by Pkc1p.
A second MAP kinase cascade, the high osmolarity glycerol response (HOG) pathway, has also been suggested to play a role in regulating cell wall architecture in S. cerevisiae (Garcia-Rodriguez et al., 2000; Kapteyn et al., 2001) and in C. albicans (Eisman et al., 2006). In S. cerevisiae, the HOG pathway is required for the response to CFW and mutants in several components of the pathway are resistant to CFW (Garcia-Rodriguez et al., 2000). In addition, changes in osmotic pressure have been shown to regulate chitin synthase activity in the dimorphic fungus Benjaminiella poitrasii suggesting the HOG pathway is involved in chitin regulation (Deshpande et al., 1997).
Transcript profiling studies have implicated Ca2+ in the regulation of ScCHS1 (Yoshimoto et al., 2002). In addition, sequences recognized by the Ca2+/calcineurin-dependent transcription factor Crz1p/Tcn1p have been identified upstream of a number of genes that are upregulated in cell wall mutants that activate the cell wall salvage pathway (Lagorce et al., 2003; Boorsma et al., 2004; Garcia et al., 2004; Karababa et al., 2006). These studies directed us towards examining the role of Ca2+ signalling in the regulation of chitin synthesis in C. albicans.
Each of the four C. albicans Chs enzymes plays a distinct role in cellular growth. CaChs1p synthesizes the septal chitin and contributes to chitin in the lateral cell wall and is essential for viability in both the yeast and hyphal forms (Munro et al., 2001). CaChs2p encodes the major chitin synthase activity in vitro, and chs2Δ null mutants have fractionally less chitin in hyphal cells (Gow et al., 1994; Munro et al., 1998). CaChs3p synthesizes the majority of the chitin in the lateral cell wall and the ring of chitin at the site where a new bud emerges (Bulawa et al., 1995; Mio et al., 1996). CaChs8p and CaChs2p account for almost all the measurable in vitro chitin synthase activity in membrane preparations but are non-essential for growth (Munro et al., 2003). In C. albicans, northern analyses suggested that CaCHS2 and CaCHS3 are upregulated shortly after induction of hyphal formation while CaCHS1 is expressed at low but constant levels in both yeast and hyphae (Chen-Wu et al., 1992; Munro et al., 1998). Hyphal formation in C. albicans is accompanied by a three to fivefold increase in the chitin content of the cell wall (Chattaway et al., 1968; Sullivan et al., 1983; Munro et al., 1998).
Here we examine the regulation of chitin synthesis of C. albicans and describe the signalling pathways that co-ordinate this process. We used a lacZ reporter gene fused to the putative promoters of each of the C. albicans CHS genes to test hypotheses about the expression of CHS genes when cells are challenged with cell wall perturbing agents or subjected to environmental stresses. We show that transcriptional regulation of the CHS genes is stimulated via at least three pathways – the PKC and HOG MAP kinase cascades and the Ca2+/calcineurin pathway. Each of the four chitin synthase promoters was regulated differentially, but all were activated by exogenous Ca2+ in a calcineurin and Crz1p-dependent manner. In addition, hyper-stimulation of CHS gene expression was observed when multiple signalling pathways were activated simultaneously and this resulted in greatly elevated cell wall chitin levels.
Results
Endogenous CHS promoter activity in wild-type cells and chsΔ mutants
Transcriptional activity of the four chitin synthase genes of C. albicans was characterized using a lacZ reporter system. Plasmid placpoly 6 containing URA3 and RPS1 was used to create a fusion between the promoter of each C. albicans chitin synthase gene and the Streptococcus thermophilus lacZ open reading frame (ORF). A 1 kb region upstream from the ATG start codon of CHS1, CHS2, CHS3 and CHS8 was cloned into placpoly 6 generating, respectively, plasmids pCHS1plac, pCHS2plac, pCHS3plac and pCHS8plac. Ura− C. albicans cells were transformed with each linearized plasmid, homologous recombination resulted in integration of the plasmid at the RPS1 locus and Ura+ transformants were selected. Transformants were screened by Southern blot analysis and those with a single copy integration of pCHSplac (strains NGY210-NGY213, Table 1) were analysed further. The CHS2 and CHS3 promoters had the highest and lowest level of expression, respectively, for growth in YPD medium (P < 0.05) (Fig. 1). Real-time quantitative polymerase chain reaction (PCR) confirmed these results (data not shown).
Table 1.
Candida albicans strains used in this study.
| Strain | Parental strain | Genotypea | Source or reference |
|---|---|---|---|
| CAF2-1 | SC5314 | URA3/ura3Δ::λimm434 | Fonzi and Irwin (1993) |
| CAI-4 | CAF2-1 | ura3Δ::λimm434/ura3Δ::λimm434 | Fonzi and Irwin (1993) |
| NGY210 | CAI-4 | RPS1/rps1Δ::pCHS1plac | This study |
| NGY211 | CAI-4 | RPS1/rps1Δ::pCHS2plac | This study |
| NGY212 | CAI-4 | RPS1/rps1Δ::pCHS3plac | This study |
| NGY213 | CAI-4 | RPS1/rps1Δ::pCHS8plac | This study |
| C155 | C154 | chs2Δ::hisG/chs2Δ::hisG | Mio et al. (1996) |
| Myco3 | Myco4 | chs3Δ::hisG/chs3Δ::hisG | Bulawa et al. (1995) |
| NGY128 | CAI-4 | chs8Δ::hisG/chs8Δ::hisG | Munro etal. (2003) |
| C157 | C155 |
chs2Δ::hisG/chs2Δ::hisG; chs3Δ::hisG/chs3Δ::hisG |
Mio et al. (1996) |
| NGY138 | CAI-4 |
chs2Δ::hisG/chs2Δ::hisG; chs8Δ::hisG/chs8Δ::hisG |
Munro etal. (2003) |
| CM1613c | CAI-4 | mkc1Δ::hisG/mkc1Δ::hisG | Navarro-Garcia et al. (1995) |
| DSY2842 | CAI-4 | crz1Δ::hisG/crz1Δ::hisG | Karababa et al. (2006) |
| DSY2101 | CAI-4 | cna1Δ::hisG/cna1Δ::hisG | Sanglard etal. (2003) |
| CNC15 | RIM1000 | hog1Δ::hisG/hog1Δ::hisG | Alonso-Monge et al. (1999) |
| NGY258 | C155 | chs2Δ::hisG/chs2Δ::hisG; RPS1/rps1Δ::pCHS1plac | This study |
| NGY260 | C155 | chs2Δ::hisG/chs2Δ::hisG, RPS1/rps1Δ::pCHS3plac | This study |
| NGY261 | C155 | chs2Δ::hisG/chs2Δ::hisG, RPS1/rps1Δ::pCHS8plac | This study |
| NGY262 | Myco3 | chs3Δ::hisG/chs3Δ::hisG, RPS1/rps1Δ::pCHS1plac | This study |
| NGY263 | Myco3 | chs3Δ::hisG/chs3Δ::hisG, RPS1/rps1Δ::pCHS2plac | This study |
| NGY265 | Myco3 | chs3Δ::hisG/chs3Δ::hisG, RPS1/rps1Δ::pCHS8plac | This study |
| NGY290 | NGY128 | chs8Δ::hisG/chs8Δ::hisG, RPS1/rps1Δ::pCHS1plac | This study |
| NGY291 | NGY128 | chs8Δ::hisG/chs8Δ::hisG, RPS1/rps1Δ::pCHS2plac | This study |
| NGY292 | NGY128 | chs8Δ::hisG/chs8Δ::hisG, RPS1/rps1Δ::pCHS3plac | This study |
| NGY270 | NGY138 |
chs2Δ::hisG/chs2Δ::hisG; chs8Δ::hisG/chs8Δ::hisG; RPS1/rps1Δ::pCHS1plac |
This study |
| NGY272 | NGY138 |
chs2Δ::hisG/chs2Δ::hisG; chs8Δ::hisG/chs8Δ::hisG; RPS1/rps1Δ::pCHS3plac |
This study |
| NGY266 | C157 |
chs2Δ::hisG/chs2Δ::hisG; chs3Δ::hisG/chs3Δ::hisG; RPS1/rps1Δ::pCHS1plac |
This study |
| NGY269 | C157 |
chs2Δ::hisG/chs2Δ::hisG; chs3Δ::hisG/chs3Δ::hisG; RPS1/rps1Δ::pCHS8plac |
This study |
| NGY294 | DSY2101 | cna1Δ::hisG/cna1Δ::hisG; RPS1/rps1Δ::pCHS1plac | This study |
| NGY295 | DSY2101 | cna1Δ::hisG/cna1Δ::hisG; RPS1/rps1Δ::pCHS2plac | This study |
| NGY296 | DSY2101 | cna1Δ::hisG/cna1Δ::hisG; RPS1/rps1Δ::pCHS3plac | This study |
| NGY297 | DSY2101 | cna1Δ::hisG/cna1Δ::hisG; RPS1/rps1Δ::pCHS4plac | This study |
| NGY282 | CM1613c | mkc1Δ::hisG/mkc1Δ::hisG; RPS1/rps1Δ::pCHS1plac | This study |
| NGY283 | CM1613c | mkc1Δ::hisG/mkc1Δ::hisG; RPS1/rps1Δ::pCHS2plac | This study |
| NGY284 | CM1613c | mkc1Δ::hisG/mkc1Δ::hisG; RPS1/rps1Δ::pCHS3plac | This study |
| NGY285 | CM1613c | mkc1Δ::hisG/mkc1Δ::hisG; RPS1/rps1Δ::pCHS8plac | This study |
| NGY314 | DSY2842 | crz1Δ::hisG/crz1Δ::hisG; RPS1/rps1Δ::pCHS1plac | This study |
| NGY315 | DSY2842 | crz1Δ::hisG/crz1Δ::hisG; RPS1/rps1Δ::pCHS2plac | This study |
| NGY316 | DSY2842 | crz1Δ::hisG/crz1Δ::hisG; RPS1/rps1Δ::pCHS3plac | This study |
| NGY317 | DSY2842 | crz1Δ::hisG/crz1Δ::hisG; RPS1/rps1Δ::pCHS8plac | This study |
| NGY321 | CNC15 | hog1Δ::hisG/hog1Δ::hisG; RPS1/rps1Δ::pCHS1plac | This study |
| NGY322 | CNC15 | hog1Δ::hisG/hog1Δ::hisG; RPS1/rps1Δ::pCHS2plac | This study |
| NGY323 | CNC15 | hog1Δ::hisG/hog1Δ::hisG; RPS1/rps1Δ::pCHS3plac | This study |
| NGY324 | CNC15 | hog1Δ::hisG/hog1Δ::hisG; RPS1/rps1Δ::pCHS8plac | This study |
All strains apart from CAF2-1 are also ura3Δ::λimm434/ura3Δ::λimm434.
Fig. 1.
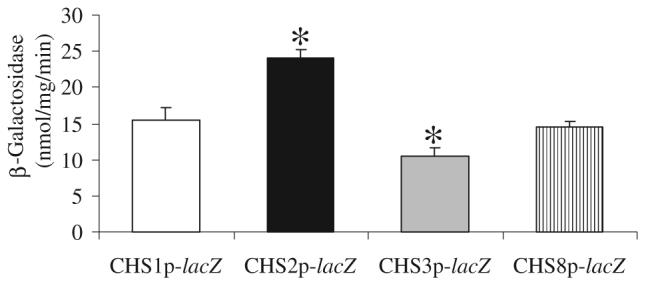
Native CHS promoter activities. The β-galactosidase activity of cell-free protein extracts of parental C. albicans strain CAI-4 transformed with CHS1p-lacZ, CHS2p-lacZ, CHS3p-lacZ or CHS8p-lacZ, grown on YPD at 30°C. Data are from three independent experiments (average ± SD n = 9). Asterisks indicate significant differences from CHS1p-lacZ where P ≤ 0.05.
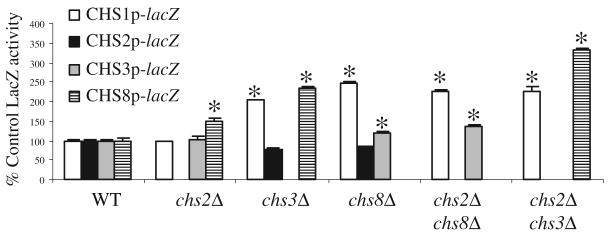
The pCHSplac plasmids were transformed into isogenic mutant strains derived from CAI-4 with single or double CHS gene disruptions (Table 1) to test whether deletion of CHS genes results in a compensatory upregulation of other members of the CHS family. The CHS1 promoter activity was significantly increased in the single mutants chs3Δ (strain Myco3) and chs8Δ (NGY138) and the double mutants chs2Δ chs8Δ (NGY128) and chs2Δ chs3Δ (C157) (Fig. 2). Chs1p may contribute to the maintenance of lateral wall integrity (Munro et al., 2001) and play a compensatory role when CHS3 and CHS8 gene functions are lost. Expression from the CHS8 promoter was stimulated when either CHS2 or CHS3 were deleted and was increased further in the chs2Δ chs3Δ double null mutant. CHS3 expression was slightly elevated in chs8Δ and chs2Δ chs8Δ mutants while the CHS2 promoter did not show any significant changes in any of the mutants tested (Fig. 2). Therefore, the deletion of single CHS genes resulted in activation of the expression of others.
Fig. 2.
Compensatory activation of CHS promoters in response to mutation in CHS genes. β-galactosidase activities are from cell-free protein extracts of C. albicans strain CAI-4 and chitin synthase mutants transformed with CHSp-lacZ constructs. Assays were performed on three independent YPD cultures grown at 30°C (average ± SD n = 9). Asterisks indicate differences where P ≤ 0.05 compared with the wild-type parent strain transformed with the same promoter-lacZ fusion.
CHS promoter activity responds to wall perturbing agents
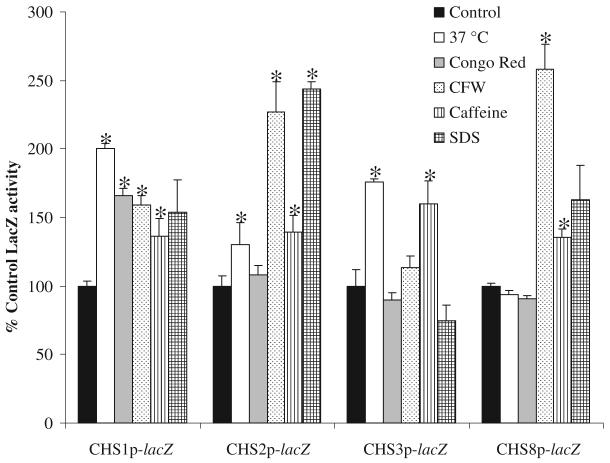
Transcriptional regulation of the four CHS genes was determined in response to various environmental changes and perturbations (Fig. 3). Growth at 37°C stimulated CHS1, CHS2 and CHS3 promoters compared with growth at 25°C (not shown) and 30°C (control conditions). The addition of SDS, which perturbs membrane integrity, or CFW that interferes with cell wall assembly, induced expression from three of the four promoters (Fig. 3). CR stimulated only CHS1 expression. Caffeine, an inhibitor of cAMP phosphodiesterase, stimulates dual phosphorylation of ScSlt2, the MAP kinase component of the PKC cell wall integrity signal transduction pathway (Martin et al., 2000). The addition of 12 mM caffeine to the growth medium resulted in significantly elevated expression from all four CHS promoters.
Fig. 3.
CHS promoters are activated by cell wall and membrane perturbing agents. β-galactosidase activities of cell-free protein extracts of C. albicans strain CAI-4 transformed with the CHSp-lacZ constructs (strains NGY210 – NGY213, Table 2). Cells were grown on YPD supplemented with the compounds at 30°C or at 37°C and harvested at OD600 0.8. Assays were performed in triplicate on three independent cultures (average ± SD n = 9). Asterisks denote significant differences to controls that were untreated strains, NGY210 – NGY213, grown on YPD at 30°C (P ≤ 0.05).
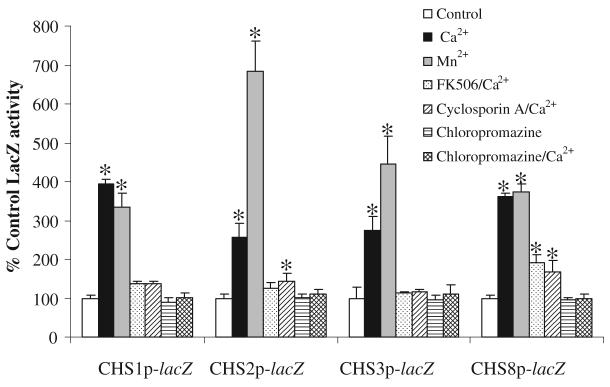
The CHS transcriptional response to cations and salts – 200 mM Ca2+, Mn2+, K2+, Li2+, Mg2+ or 800 mM NaCl was tested. Addition of K2+, Li2+, Mg2+ or Na2+ had no detectable effects (data not shown), however, exogenous Ca2+ and Mn2+ activated all four CHS promoters (Fig. 4). Some response was observed even with 5 mM Ca2+ (data not shown). Exogenously applied Ca2+ leads to activation of the calcineurin pathway, which induces dephosphorylation of the Crz1p transcription factor (Cyert, 2003). The calcineurin specific inhibitors FK506 and Cyclosporin A inhibited Ca2+ activation of CHS transcription (Fig. 4). Activation of CHS expression with Mn2+ was also reduced, but not totally blocked, by simultaneous addition of FK506 (data not shown) suggesting the Mn2+-specific activation occurred in part through the calcineurin signalling pathway but also involved a calcineurin-independent mechanism. These results suggest that Ca2+ and Mn2+ activated CHS expression via both calcineurin/Crz1-dependent and independent mechanisms. The calmodulin inhibitor chloropromazine had no effect on CHS transcriptional activity but chloropromazine with 200 mM Ca2+ completely inhibited the Ca2+-activation response. The calcium ionophore A23187 also inhibited the Ca2+-activation response (data not shown). Inhibition by chloropromazine suggested that the observed Ca2+ stimulation involved the classical Ca2+ signalling pathway acting through calmodulin and calcineurin.
Fig. 4.
CHS promoters response to Ca2+ and Mn2+. β-galactosidase activities were measured in cell-free protein extracts of C. albicans strain CAI-4 transformed with the CHSp-lacZ constructs (strains NGY210 – NGY213, Table 2). Cells were grown at 30°C on YPD supplemented with Ca2+ and Mn2+ ± FK506, cyclosporin A or chloropromazine. Assays were performed in triplicate on three independent cultures (average ± SD n = 9). Asterisks indicate that there are significant differences to untreated control samples prepared in the same experiment from strains NGY210 – NGY213 (P ≤ 0.05).
To corroborate these findings the four CHS-reporter constructs were transformed into null mutant strains lacking genes involved in the calcineurin pathway. The Crz1p transcription factor is dephosphorylated when the phosphatase calcineurin is activated by Ca2+/calmodulin. It then enters the nucleus and induces expression of a number of genes, many of which encode proteins with cell wall-related functions (Yoshimoto et al., 2002; Lagorce et al., 2003; Garcia et al., 2004; Karababa et al., 2006; Pardini et al., 2006). Putative calcium-dependent response element (CDRE) motifs that are recognized by the Crz1p transcription factor were found in the promoter region of the CHS genes in C. albicans (Table 2). In the cna1Δ strain, which is mutated in one of the calcineurin subunits, the CHS2 and CHS8 promoters still responded to Ca2+, but the response of the CHS2 promoter was reduced (Fig. 5). The Ca2+ responses of the CHS1 and CHS3 promoters were not significantly different to the untreated cna1Δ control. Stimulation with Ca2+ was abrogated by simultaneous addition of FK506. In the crz1Δ mutant background, expression from the CHS1 promoter was elevated when cells were grown in YPD, and addition of Ca2+ did not further stimulate CHS1 expression (Fig. 5). The response of the CHS2 and CHS3 promoters to exogenous Ca2+was significantly different in the crz1Δ mutant compared with wild-type cells. Therefore, CHS2 and CHS3 were activated in part via the Ca2+/calcineurin/Crz1 pathway but Crz1p repressed the expression of CHS1. Transformation of the CHSp-lacZ constructs into the double calcium channel mutant mid1Δ cch1Δ had no significant effect on Ca2+-stimulated activation of expression (data not shown).
Table 2.
Putative transcription factor binding sites.
| Gene | Positiona/strand | CDREb | Positiona/strand | Sko1 binding sitec |
|---|---|---|---|---|
| CHS1 | −351/+ | GGGCTTC | −121/+ | TACGT |
| −380/− | TGGCTTG | −851/− | TACGT | |
| −744/+ | AGGCTCC | |||
| −810/+ | TGGCTCT | |||
| CHS2 | −539/− | TGGCTTT | ||
| −879/+ | GGGCGTG | |||
| −918/+ | AGGCTGA | |||
| CHS3 | −498/− | AGGCGGG | −130/+ | TACGT |
| −904/− | AGGCTCA | −665/− | TACGT | |
| CHS8 | −847/− | TGGCTTC | −782/+ | TACGT |
| −893/+ | AGGCTTA |
Start codon A taken as position 1.
Crz1p consensus NGGC(G/T)CA.
Sko1p binding site consensus TACGT.
Fig. 5.
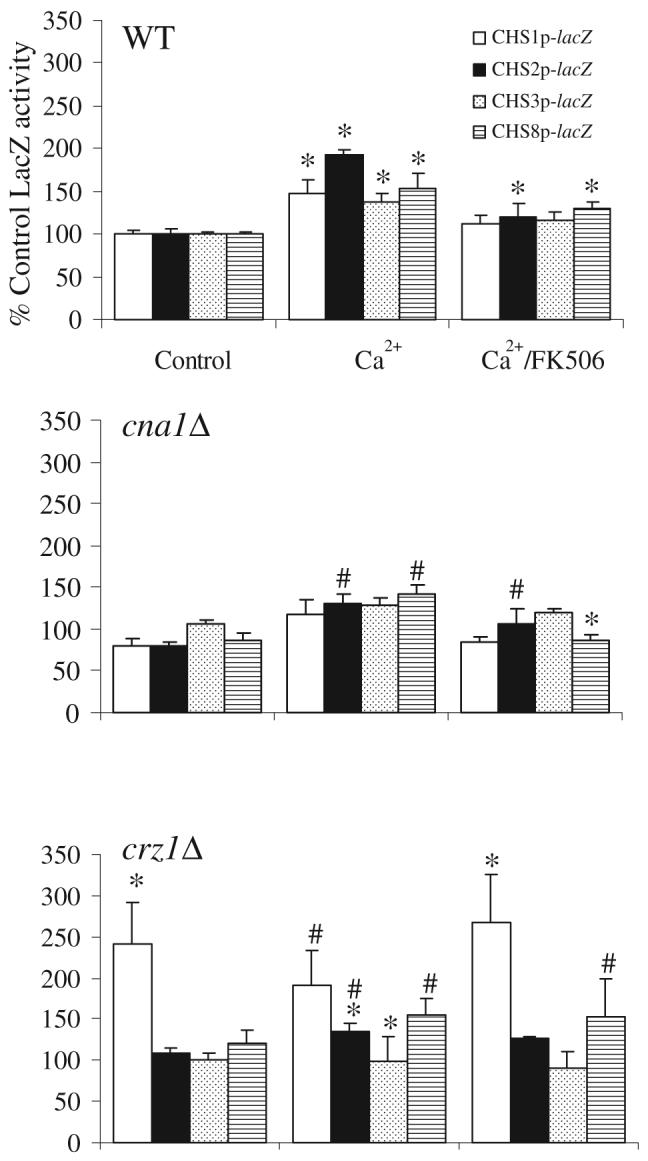
Stimulation of the CHS promoters by Ca2+ is dependent upon calcineurin and Crz1p. β-galactosidase activities of protein extracts of CAI-4 (control), cna1Δ and crz1Δ transformed with the CHSp-lacZ constructs were measured. Cells were grown at 30°C on YPD, YPD supplemented with Ca2+ ± FK506. Triplicate assays were performed on three independent cultures (average ± SD, n = 9). In WT asterisks indicate significantly different to untreated control of that promoter under the same conditions (P ≤ 0.05). For mutants, an asterisk indicates a significant difference relative to wild type under the same conditions, a number sign indicates a significant difference to the untreated control in the same mutant background (P ≤ 0.05).
Synergistic stimulation of CHS promoters by combined Ca2+ and CFW treatment
Addition of Ca2+ or CFW stimulated the CHS promoters – therefore we tested the effects of combinations of Ca2+ and CFW treatments. All four promoters were hyper-stimulated by combined treatment with Ca2+ and CFW (Table 3). The pCHSplac plasmids were transformed into mutant strains lacking MAP kinase genes of the PKC and HOG signal transduction pathways (Table 1). The first mutant tested had a disrupted MKC1 gene that encodes the MAP kinase of the PKC pathway (Navarro-Garcia et al., 1998). In the mkc1Δ strain background the CHS8 promoter, and to a lesser extent, the CHS2 promoter had reduced activity. All four CHS promoters were still stimulated by Ca2+ in the mkc1Δ mutant (Table 3), therefore the Ca2+-induced upregulation of CHS promoters can be independent of the PKC pathway. However, the ability of the CHS2 and CHS8 promoters to respond to CFW was impaired in the mkc1Δ mutant. All four promoters were stimulated by the combined Ca2+/CFW treatment but the level of stimulation of CHS2 and CHS8 promoters was significantly less in mkc1Δ cells compared with wild-type cells. The response of the CHS promoters to CFW and combined Ca2+/CFW was also examined in the crz1Δ mutant (Table 3). Again the response of CHS2 and CHS8 promoters to CFW was significantly reduced in the crz1Δ mutant and the Ca2+/CFW induction of all four promoters was dramatically reduced (three- to five-fold).
Table 3.
The CHS promoters are hyper-stimulated by combined Ca2+/CFW treatment.
| Strain | Treatment | Mean % /acZ activity |
Fold changea | P-value | Fold changeb |
P-value | |
|---|---|---|---|---|---|---|---|
| CHS1p-lacZ | |||||||
| wt | Control | 100 ± 7 | |||||
| +Ca2+ | 174 ± 38 | 1.74 | 1.60E-05 | ||||
| +CFW | 190 ± 34 | 1.90 | 3.71E-02 | ||||
| +Ca/CFW | 888 ± 59 | 8.88 | 1.30E-06 | ||||
| mkc1Δ | Control | 91 ± 9 | 0.91 | 7.06E-01 | |||
| +Ca2+ | 189 ± 25 | 2.09 | 4.00E-04 | 1.20 | 1.00E+00 | ||
| +CFW | 141 ± 41 | 1.55 | 2.27E-01 | 0.82 | 7.08E-01 | ||
| +Ca2+/CFW | 735 ± 40 | 8.11 | 4.40E-10 | 0.91 | 3.08E-01 | ||
| crz1Δ | Control | 209 ± 11 | 2.09 | 3.10E-10 | |||
| +Ca2+ | 202 ± 22 | 0.97 | 1.00E+00 | 0.55 | 7.81E-01 | ||
| +CFW | 154 ± 19 | 0.74 | 3.70E-04 | 0.39 | 7.91E-01 | ||
| +Ca2+/CFW | 411 ± 174 | 1.96 | 2.78E-01 | 0.22 | 1.43E-03 | ||
| hog1Δ | Control | 39 ± 12 | 0.39 | 2.00E-06 | |||
| +Ca2+ | 48 ± 9 | 1.22 | 9.94E-01 | 0.70 | 1.44E-09 | ||
| +CFW | 47 ± 13 | 1.18 | 1.00E+00 | 0.62 | 2.80E-03 | ||
| +Ca2+/CFW | 592 ± 219 | 15.00 | 8.79E-03 | 1.69 | 3.85E-01 | ||
| CHS2p-lacZ | |||||||
| wt | Control | 100 ± 3 | |||||
| +Ca2+ | 226 ± 43 | 2.26 | 1.52E-07 | ||||
| +CFW | 221 ± 23 | 2.21 | 7.67E-08 | ||||
| +Ca2+/CFW | 889 ± 121 | 8.89 | 3.05E-06 | ||||
| mkc1Δ | Control | 68 ± 3 | 0.68 | 9.98E-11 | |||
| +Ca2+ | 196 ± 13 | 2.88 | 1.77E-08 | 1.27 | 6.02E-01 | ||
| +CFW | 76 ± 3 | 1.12 | 7.00E-03 | 0.51 | 1.08E-08 | ||
| +Ca2+/CFW | 379 ± 18 | 5.56 | 5.96E-10 | 0.63 | 5.20E-11 | ||
| crz1Δ | Control | 142 ± 10 | 1.42 | 3.80E-05 | |||
| +Ca2+ | 153 ± 12 | 1.08 | 8.91E-01 | 0.48 | 2.13E-04 | ||
| +CFW | 147 ± 6 | 1.04 | 1.00E+00 | 0.47 | 9.56E-06 | ||
| +Ca2+/CFW | 402 ± 43 | 2.84 | 2.13E-06 | 0.32 | 1.06E-05 | ||
| hog1Δ | Control | 43 ± 3 | 0.43 | 2.30E-14 | |||
| +Ca2+ | 54 ± 2 | 1.24 | 8.95E-06 | 0.55 | 1.58E-09 | ||
| +CFW | 38 ± 1 | 0.88 | 8.63E-03 | 0.40 | 1.40E-09 | ||
| +Ca2+/CFW | 354 ± 113 | 8.15 | 2.00E-03 | 0.92 | 2.67E-01 | ||
| CHS3p-lacZ | |||||||
| wt | Control | 100 ± 7 | |||||
| +Ca2+ | 154 ± 26 | 1.54 | 1.69E-04 | ||||
| +CFW | 118 ± 10 | 1.18 | 1.36E-02 | ||||
| +Ca2+/CFW | 693 ± 33 | 6.93 | 2.94E-10 | ||||
| mkc1Δ | Control | 101 ± 8 | 1.01 | 1.00E+00 | |||
| +Ca2+ | 184 ± 23 | 1.82 | 1.20E-04 | 1.18 | 4.27E-01 | ||
| +CFW | 104 ± 25 | 1.03 | 1.00E+00 | 0.87 | 9.94E-01 | ||
| +Ca2+/CFW | 732 ± 93 | 7.23 | 2.08E-06 | 1.04 | 1.00E+00 | ||
| crz1Δ | Control | 110 ± 8 | 1.10 | 4.45E-01 | |||
| +Ca2+ | 89 ± 6 | 0.81 | 8.87E-02 | 0.52 | 1.20E-04 | ||
| +CFW | 91 ± 4 | 0.83 | 2.32E-01 | 0.70 | 3.00E-01 | ||
| +Ca2+/CFW | 288 ± 41 | 2.61 | 1.66E-04 | 0.38 | 6.10E-10 | ||
| hog1Δ | Control | 156 ± 14 | 1.56 | 1.29E-05 | |||
| +Ca2+ | 251 ± 21 | 1.61 | 2.06E-07 | 1.04 | 6.24E-07 | ||
| +CFW | 135 ± 9 | 0.86 | 2.12E-05 | 0.73 | 5.15E-02 | ||
| +Ca2+/CFW | 988 ± 237 | 6.32 | 2.16E-04 | 0.91 | 2.12E-01 | ||
| CHS8p-lacZ | |||||||
| wt | Control | 100 ± 9 | |||||
| +Ca2+ | 158 ± 16 | 1.58 | 2.28E-11 | ||||
| +CFW | 263 ± 14 | 2.63 | 8.27E-07 | ||||
| +Ca2+/CFW | 1237 ± 421 | 12.37 | 9.01E-09 | ||||
| mkc1Δ | Control | 27 ± 5 | 0.27 | 1.75E-09 | |||
| +Ca2+ | 76 ± 17 | 2.86 | 7.52E-04 | 1.81 | 1.61E-01 | ||
| +CFW | 16 ± 2 | 0.59 | 6.41E-03 | 0.22 | 4.38E-08 | ||
| +Ca2+/CFW | 378 ± 47 | 14.12 | 8.50E-07 | 1.14 | 3.81E-06 | ||
| crz1Δ | Control | 141 ± 8 | 1.41 | 1.99E-06 | |||
| +Ca2+ | 160 ± 12 | 1.13 | 1.13E-01 | 0.72 | 3.95E-07 | ||
| +CFW | 233 ± 16 | 1.65 | 4.08E-07 | 0.63 | 4.13E-09 | ||
| +Ca2+/CFW | 529 ± 94 | 3.74 | 9.74E-05 | 0.30 | 4.42E-05 | ||
| hog1Δ | Control | 43 ± 8 | 0.43 | 2.03E-08 | |||
| +Ca2+ | 48 ± 5 | 1.11 | 9.99E-01 | 0.70 | 1.09E-07 | ||
| +CFW | 34 ± 6 | 0.79 | 6.00E-01 | 0.30 | 3.70E-09 | ||
| +Ca2+/CFW | 315 ± 69 | 7.29 | 1.23E-04 | 0.59 | 7.10E-04 | ||
Fold change with respect to the control in the same genetic background.
Ratio of fold change in the mutant compared with fold change in wild-type cells under the same conditions.
Statistically significant changes are highlighted in bold.
Three of the four CHS promoter sequences contained ATF/CREB elements – potential binding sites for the Sko1p transcription factor that is regulated by Hog1p (Table 2) (Proft et al., 2005). Expression from the CHS1, CHS2 and CHS8 promoters was reduced in the hog1Δ mutant compared with wild-type cells in YPD and in the presence of Ca2+ or CFW (Table 3). In contrast, expression from the CHS3 promoter was increased in the hog1Δ mutant, suggesting Hog1p normally repressed CHS3 transcription. In the hog1Δ mutant the CHS3 promoter still responded to exogenous Ca2+, but not to CFW and combinations of Ca2+ and CFW stimulated the CHS3 promoter in both the hog1Δ and wild-type backgrounds. The Ca2+/CFW-induced stimulation of the CHS1 and CHS2 promoters was not significantly altered in the hog1Δ strain but the level of expression from the CHS8 promoters was significantly less than in wild-type cells. Together these data suggest that Mkc1p, Crz1p and Hog1p play significant roles in the Ca2+/CFW hyper-stimulation of CHS promoters.
Exogenous Ca2+ stimulates chitin synthase activity
We tested whether the Ca2+-activated CHS gene expression translated into measurably higher chitin synthase enzyme activity. C. albicans yeast cells were cultured in YPD or YPD plus 100 mM Ca2+ for 5 h and membrane fractions of wild type (CAI-4), chsΔ and signalling mutant strains were prepared and assayed for chitin synthase activity. The specific chitin synthase activity of wild-type mixed membrane fractions (MMF) increased slightly when exogenous Ca2+ was added to the growth medium (Fig. 6). In the mkc1Δ mutant, Chs activity was comparable to the control strain and did not increase upon addition of Ca2+. The hog1Δ mutant had markedly elevated Chs activity compared with the control and addition of Ca2+ did not further stimulate chitin synthase activity. Chitin synthase activity of the crz1Δ mutant was comparable to wild type and decreased in Ca2+-treated cells. As shown previously (Munro et al., 2003), the Chs activity of the chs2Δ chs8Δ double mutant was only around 5% of wild-type levels and no further stimulation was observed when cells were grown in the presence of Ca2+. The chs3Δ mutant had reduced Chs activity but this was elevated in response to Ca2+. Therefore, Chs2p and Chs8p are mainly responsible for the elevated Chs activity in response to Ca2+ and this was mediated via Crz1p, Mkc1p and Hog1p. Attempts were made to measure chitin synthase activity from membranes prepared from cells grown in the presence of CFW and Ca2+/CFW. No detectable Chs activity was found (not shown). CFW inhibition of in vitro chitin synthase activity has been reported previously (Roncero and Duran, 1985) and was shown to be dependent upon pH of the growth medium (Roncero et al., 1988).
Fig. 6.
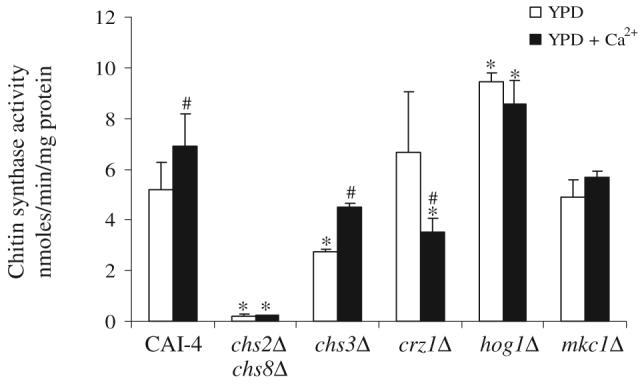
Exogenous Ca2+ elevates in vitro chitin synthase activity of yeasts cells of C. albicans strains. Activities are from mixed membrane fractions after trypsin-treatment isolated from mid-exponential cells grown at 30°C in YPD (open bars) or YPD + 100 mM Ca2+ (black bars). Triplicate assays were performed (average ± SD, n = 3). Asterisks indicate significant differences from CAI-4 under the same conditions, a number sign indicates significant differences to untreated samples in the same strain background (P ≤ 0.05).
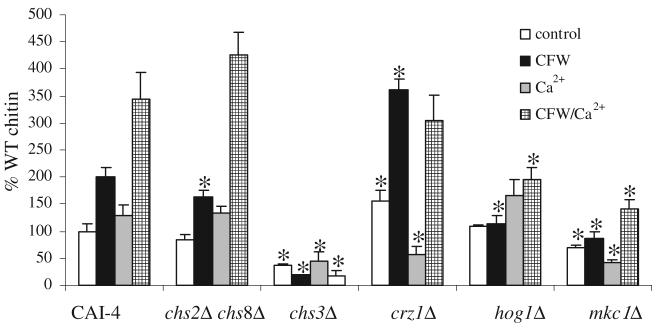
Treatment with Ca2+ and CFW elevates cell wall chitin levels
The chitin content of cells was measured under conditions where Ca2+ stimulated CHS gene expression and in vitro Chs activity. Addition of Ca2+, CFW and Ca2+/CFW resulted in elevated cell wall chitin levels in wild-type CAI-4 cells (Fig. 7) with the combination of Ca2+/CFW giving the greatest stimulation. Chs3p is responsible for the synthesis of the majority of the chitin in the C. albicans cell wall (Bulawa et al., 1995; Mio et al., 1996). In the chs3Δ mutant, chitin levels were dramatically reduced and Ca2+-treatment had only a slight effect on chitin content. The chs2Δ chs8Δ mutant behaved similarly to wild type. In the mkc1Δ mutant, chitin levels were lower than in parental controls, but were elevated after combined Ca2+/CFW treatment. However, addition of Ca2+ or CFW alone had little effect on chitin content in the mkc1Δ mutant. Chitin levels of untreated crz1Δ cells were significantly higher than wild-type cells again suggesting that under some conditions Crz1p represses chitin synthesis. Chitin levels of crz1Δ were elevated by CFW or Ca2+/CFW treatments but did not respond, or were repressed, when treated with Ca2+ alone. Untreated hog1Δ cells had wild-type chitin levels that were increased marginally when cells were grown with Ca2+ but the activation with CFW or Ca2+ plus CFW was significantly reduced compared with wild-type cells. These results suggest that elevated cell wall chitin content in response to combined treatments with Ca2+ and CFW is due mainly to Chs3p and that the PKC and HOG and to a lesser extent the Ca2+/Crz1 pathways are involved in this Chs3p-dependent stimulation of chitin synthesis.
Fig. 7.
Hyper-stimulation of cell wall chitin in response to Ca2+/CFW. C. albicans strains were grown at 30°C on YPD and YPD supplemented with Ca2+, CFW or both Ca2+ and CFW. Cell wall chitin assays were performed five times on three biologically independent samples (average ± SD n = 15). Asterisks indicate significant differences from CAI-4 cells grown under the same conditions.
Ca2+- and CFW-dependent phosphorylation of Mkc1p and Cek1p
The phosphorylation status of the Mkc1p and Cek1p MAP kinases was examined in order to assess the status of the PKC and SVG (STE vegetative growth) pathways in the strains and treatments described above (Navarro-Garcia et al., 2005; Eisman et al., 2006). Cek1p is the C. albicans orthologue of ScKss1p, which is the S. cerevisiae MAP kinase component of pathways that regulate filamentous growth, the pheromone response and promote vegetative growth (the latter via the SVG pathway) (Lee and Elion, 1999; Eisman et al., 2006). The SVG pathway is constitutively activated in the och1Δ N-glycosylation mutants of S. cerevisiae and C. albicans (Lee and Elion, 1999; Bates et al., 2006) and in the hog1Δ mutant and has been implicated in the response to cell wall perturbing agents (Eisman et al., 2006). Phospho-Mkc1p and phospho-Cek1p were identified by western analysis as 59 kDa and 48 kDa bands, respectively, using phospho-specific antibodies (Fig. 8). Phosphorylation status was assessed 10, 30, 60 and 120 min after treatment addition, however, only 10 and 120 min time points are presented here. In non-stressed conditions, no phospho-Mkc1p was detected in wild type or hog1Δ yeast cells however, Mkc1p was phosphorylated in the crz1Δ mutant. Activation of Mkc1p was observed in the crz1Δ strain after treatment with Ca2+. CFW stimulated strong activation of Mkc1p in the wild type, weaker activation in hog1Δ and activation comparable to untreated controls in crz1Δ. CFW-stimulated phosphorylation of Mkc1p was observed at 2 h in wild type and hog1Δ and after 1 h in crz1Δ. Combined treatment with Ca2+ and CFW had a synergistic effect on activation of Mkc1p in the crz1Δ strain where phospho-Mkc1p was detected after 10 min. In wild-type cells, Ca2+/CFW did not give as strong a response in terms of Mkc1p phosphorylation as CFW alone. In agreement with Navarro-Garcia et al. (2005); Roman et al. (2005) and Eisman et al. (2006), phospho-Mkc1p was only detected in extracts prepared from hog1Δ cells when cells were treated with CFW. Under these conditions Cek1p appeared to be constitutively activated in the hog1Δ mutant. Treatment of the hog1Δ strain with Ca2+, CFW and Ca2+/CFW increased the level of phospho-Cek1p significantly. We conclude that the PKC pathway is activated when cells are treated with CFW and Ca2+/CFW but Ca2+ alone could not stimulate phosphorylation of Mkc1p in a wild-type background.
Fig. 8.
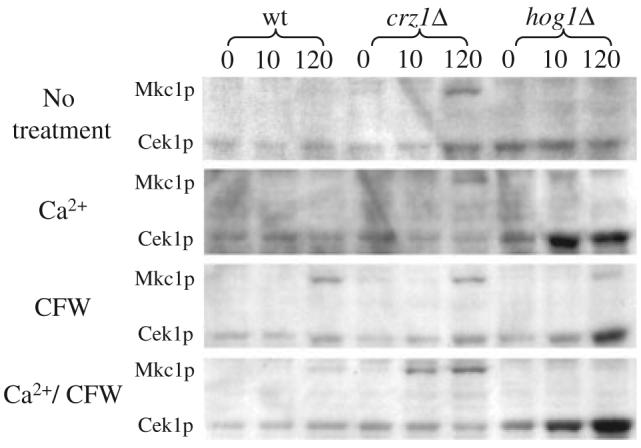
Western blot analysis of Mkc1 phosphorylation status. C. albicans strains were grown at 30°C on YPD and YPD supplemented with Ca2+, CFW or both Ca2+ and CFW. Total protein was extracted and phosphorylated Mkc1 and Cek1 detected using phospho-specific anti-p44/42 MAP kinase antibody.
Discussion
This study has shown that at least three signalling systems are involved in chitin synthesis regulation: (i) Ca2+/calcineurin/Crz1p, (ii) PKC-Mkc1p and (iii) HOG pathways. At the transcriptional level CHS expression was monitored using a lacZ reporter gene fused to each of the four C. albicans CHS promoters. Each promoter was regulated differentially – the CHS2 promoter was the most active under control conditions (YPD at 30°C) and the CHS3 promoter was the least active. Real-time quantitative PCR confirmed these observations (data not shown). The CHS promoters responded to deletion of other CHS genes with a twofold increase in expression levels from CHS1, CHS3 and CHS8 promoters in several chsΔ mutants. In Wangiella dermatitidis, a melanized fungal pathogen of humans, a compensatory increase in WdCHS expression has also been described in response to chitin synthase gene disruptions (Wang et al., 2002). Although there is no evidence of true functional redundancy within the chitin synthases examined to date, fungi appear to upregulate certain CHS in compensation for loss of others perhaps to maintain a robust cell wall.
The C. albicans CHS promoters were found to respond to a number of environmental stimuli notably when cells were treated with cell wall perturbing drugs and when growth medium was supplemented with Ca2+. Addition of exogenous Ca2+, stimulated CHS gene expression; stimulated in vitro chitin synthase activity, and resulted in increased cell wall chitin mediated through Chs3p. In addition, simultaneous treatment of cells with CFW and Ca2+ resulted in synergistically enhanced expression from all four CHS promoters and a threefold increase in the amount of chitin in the cell wall.
The CHS promoters were activated by exogenous Ca2+ and Mn2+ but not by equivalent concentrations of Mg2+ or Na+. In S. cerevisiae, Ca2+ activates calcineurin via calmodulin, which induces gene expression by regulating the Crz1p/Tcn1p transcription factor. This plays a role in regulating cell wall structure including the induction of ScCHS1 in response to Ca2+ (Yoshimoto et al., 2002) and tolerance of fungi to a wide range of antifungal agents (Edlind et al., 2002; Sanglard et al., 2003; Onyewu et al., 2004; Karababa et al., 2006). Ca2+-activation of CaCHS expression was blocked by inhibitors of both calmodulin and calcineurin confirming the role of this pathway in the regulation of CHS genes. In addition, transcription from all CHS promoters was reduced in the cna1Δ mutant in response to exogenous Ca2+. In the crz1Δ mutant, basal activity of the CHS2, CHS3 and CHS8 promoters was not altered but the CHS1 promoter was de-repressed. In addition, crz1Δ cells were attenuated, but not completely blocked, in their ability to activate CHS expression in response to exogenous Ca2+ and the hyper-stimulation of CHS expression caused by cross-activation with Ca2+ and CFW was reduced dramatically in the crz1Δ mutant background. In silico analysis of the CHS promoter sequences also identified potential CDREs, motifs recognized by Crz1p. Therefore, activation of the CHS promoters due to Ca2+ was mainly regulated by the classical Cna1/Crz1 pathway; however, some of the Ca2+ stimulation was Cna1p and/or Crz1p-independent indicating that calcineurin and Crz1p may have roles that are distinct from their role in this signalling pathway. Our results suggest that in C. albicans the Ca2+ signalling pathway plays a major role in regulating chitin synthesis. This pathway may be vital to the co-ordination of responses to a variety of conditions that compromise cell wall integrity because it also regulates the expression of genes encoding cell wall proteins Utr2p and Crh11p (Pardini et al., 2006) and the glucan synthase catalytic subunit Fks1p/Gsc1p (Sanglard et al., 2003).
Many of the conditions that stimulated the CHS promoters including growth at 37°C, treatment with cell wall perturbing agents CFW, CR and SDS and the cAMP-phosphodiesterase inhibitor caffeine lead to hyper-phosphorylation of Slt2p/Mkc1p (De Nobel et al., 2000; Martin et al., 2000; Navarro-Garcia et al., 2005). We used the mkc1Δ MAP kinase null mutant to test the role of the PKC pathway in chitin synthesis regulation. In the mkc1Δ mutant background the CHS2 and CHS8 promoters were less responsive to CFW but were still stimulated by Ca2+ and compared with wild-type cells only the CHS2 promoter had a significantly reduced response to Ca2+/CFW. However, there was a dramatic decrease in chitin levels in the mkc1Δ mutant under all conditions tested suggesting post-transcriptional regulation of Chs3p occurs via the PKC pathway. This pathway has been shown to regulate Chs3p in S. cerevisiae (Valdivia and Schekman, 2003).
The HOG signalling pathway was the third pathway implicated in CHS transcriptional regulation. Promoter sequences recognized by the HOG-regulated transcription factor Sko1p were identified in the CHS1, CHS3 and CHS8 promoters. Loss of the HOG pathway in non-stressed conditions resulted in reduced expression of CHS1, CHS2 and CHS8 but increased expression of CHS3. Therefore, as with Crz1p, blocking a particular signalling pathway had both positive and negative effects on CHS expression. Although the CHS1, CHS2 and CHS8 promoters had attenuated responses to either CFW or Ca2+ in the hog1Δ mutant, only the CHS8 promoter had markedly reduced activity in response to the combined Ca2+/CFW treatment compared with wild-type cells. Despite the apparently low basal level of expression of CHS2 and CHS8 in the hog1Δ mutant, the levels of chitin synthase enzyme activity were greater than in wild-type cells. This suggests the presence of a compensatory mechanism that is activated in response to loss of Hog1p that acts post-transcriptionally and results in enhanced Chs enzyme activity. Nevertheless, the amount of chitin in the wall of the hog1Δ mutant synthesized in response to co-stimulation with Ca2+ and CFW was significantly (40%) lower than wild type implying that the HOG pathway is involved in activation of chitin synthesis via Chs3p.
The Ca2+/calcineurin, PKC and HOG pathways contribute to the hyper-stimulation of chitin synthesis in response to Ca2+/CFW treatment. The role of the Ca2+-signalling pathway appears to be mainly in regulating CHS transcription, whereas the PKC and HOG pathways also contribute to regulation of chitin synthase enzyme activity and total cell wall chitin content. The Mkc1 pathway positively regulates CHS expression, chitin synthase activity and chitin levels in the cell wall while the Ca2+/Crz1 and HOG pathways have both positive and negative regulatory effects on different CHS genes and Chs isoenzymes (Fig. 9).
Fig. 9.
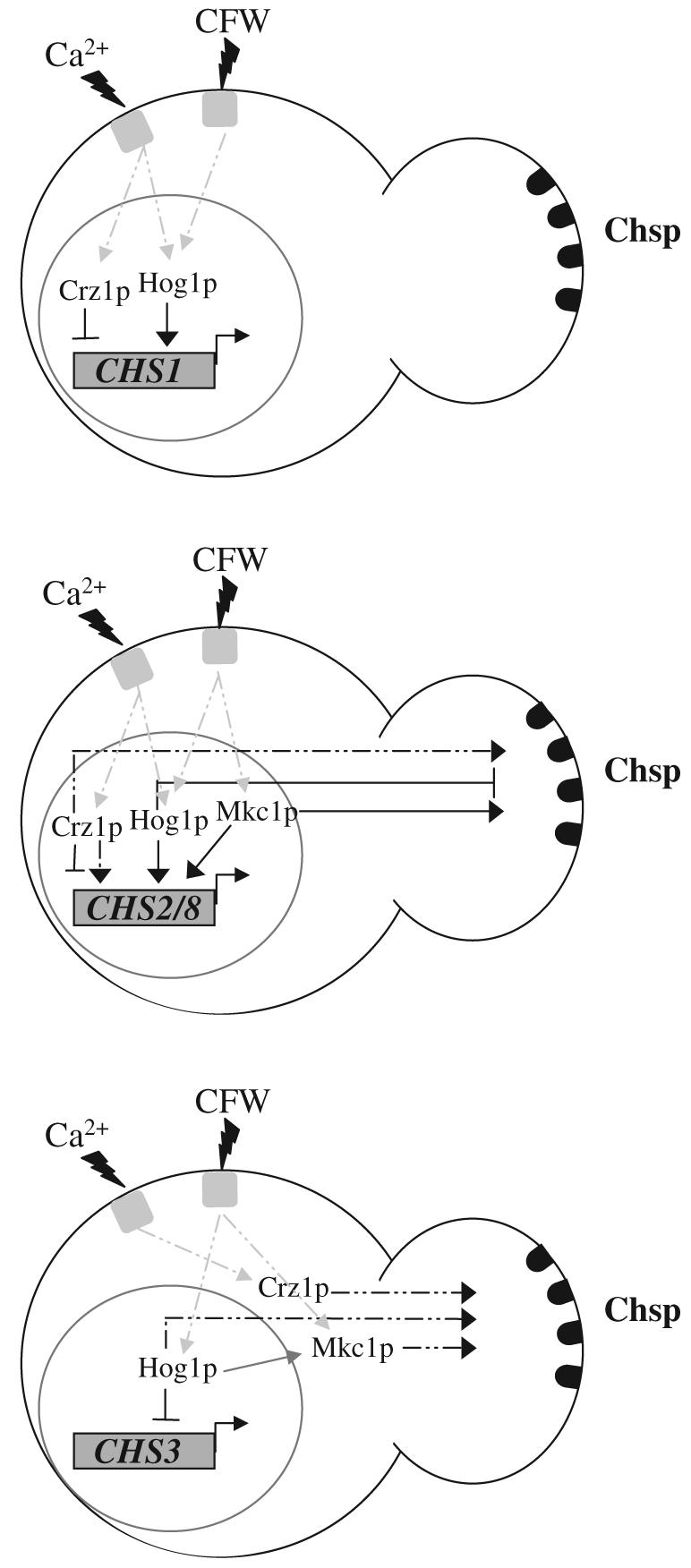
Summary model of the co-ordinated regulation of chitin synthesis. Solid lines indicate regulation under non-stress conditions and dotted lines indicate regulation in response to various stresses. Under normal growth conditions Hog1 is required for basal levels of CHS1 transcription and Crz1p acts as a repressor. CaCl2 or CFW-stimulated CHS1 expression is dependent upon Crz1p and Hog1p, while the PKC pathway does not play a major role. The contribution of these three pathways to post-transcriptional regulation of Chs1p cannot be determined by measurement of in vitro chitin synthase activity and chitin content because Chs1p is only a minor contributor to these. CHS2 and CHS8 are regulated by the Crz1p, Hog1p and Mkc1p pathways under normal growth conditions. Although Crz1p represses CHS2 expression it is required for CaCl2 stimulation. Hog1p and Mkc1p are both required for normal control levels of CHS2 and CHS8 mRNA and CFW-activated transcription is dependent upon both pathways. In addition, Hog1p is required for CaCl2 activation of CHS2 and CHS8 expression. In Chs activity assays, Hog1p negatively regulates Chs2p and Chs8p and Crz1p is responsible for their activation due to the presence of exogenous CaCl2. Hog1p had a significant negative contribution to CHS3 promoter activity but all three pathways were involved in elevation of chitin levels due to treatment with CaCl2 and CFW – a measure of Chs3p post-transcriptional regulation.
Interpretation of experiments studying CHS expression is complicated by cross-talk between signalling pathways and by compensatory mechanisms that are triggered in mutants defective in single signalling pathways. For example, mutants blocked in the HOG pathway have a constitutively active Cek1 MAP kinase, which contributes to a CR resistance phenotype (Roman et al., 2005; Eisman et al., 2006). We examined the phosphorylation status of Mkc1p and Cek1p in cells treated with Ca2+, CFW and Ca2+/CFW. We confirmed phosphorylation of Cek1p in the hog1Δ mutant and enhanced phosphorylation of Cek1p when hog1Δ was treated with Ca2+, CFW and Ca2+/CFW. Despite activation of Cek1p, chitin levels are reduced in the hog1Δ mutant suggesting Cek1p may not make a major contribution to chitin regulation. Our findings also corroborate the observations of Navarro-Garcia et al. (2005) that Hog1p was required for phosphorylation of Mkc1p under a variety of conditions, but not with CFW treatment. In addition, our Western analyses suggested that Mkc1p was phosphorylated in the crz1Δ mutant. These data suggest that these pathways do not operate in isolation and that mutations in one pathway results in activation of others as in the case of Cek1p activation in the hog1Δ mutant and Mkc1p phosphorylation in the crz1Δmutant. Hence, the activation of Mkc1p is not solely responsible for the elevated chitin synthesis under the conditions tested. Instead, the PKC, HOG and Ca2+ signalling pathways all contribute to the regulation of chitin synthesis.
In conclusion, the Ca2+/Crz1p, PKC-Mkc1p and HOG signalling pathways co-ordinate the regulation of chitin synthesis in C. albicans. The use of multiple pathways may enable the fungus to fine-tune the co-ordinated assembly of cell wall chitin to exogenous stresses by modulating chitin synthesis. CHS gene expression responded to a wide range of environmental conditions and individual CHS genes and Chs enzymes responded differently to these stresses. This regulation is vital for the maintenance of a robust cell wall during growth and morphogenesis but also under conditions where the integrity of the cell wall is compromised by treatments with antifungal drugs that target fungal cell wall synthesis.
Experimental procedures
Strains, media and growth conditions
Candida albicans strains used in this study are listed in Table 1. C. albicans cultures were maintained on solid YPD medium comprising 1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose, 2% (w/v) agar. Yeast cells of C. albicans were grown at 30°C in YPD with shaking at 200 r.p.m.
Transformation of C. albicans
Ura−C. albicans strains were cultured in 10 ml of YPD supplemented with 25 μg ml−1 uridine at 30°C for 36–72 h. After centrifugation, the cell pellets from 200 μl of cells were resuspended in 100 μl of OSB (200 mM LiAc pH 7.5, 100 mM DTT, 50% v/w PEG 6000, 10 mg ml−1 Clontech herring testis carrier DNA) and then transforming DNA was added. Samples were incubated at 43.5°C for 60 min and spread over SD agar plates (2% (w/v) d-glucose, 0.67% (w/v) yeast nitrogen base (YNB) (Bio 101, Carlsbad), 1.5% (w/v) purified agar, Oxoid) and incubated at 30°C. Single colonies were picked and grown in 5 ml of SD medium, and genomic DNA was extracted for Southern analysis.
Construction of plasmids and C. albicans strains
The placpoly-6 vector was used for the promoter-fusion reporter system and was based on a plasmid previously described by Uhl and Johnson (2001). This contains the CaURA3 and the CaRPS1 genes and was used to create fusion between the promoter of each CaCHS gene and the S. thermophilus lacZ ORF. A 1 kb upstream region from the ATG start codon of each CHS1, CHS2, CHS3 and CHS8 ORF was cloned into the PstI–XhoI sites of placpoly-6 generating pCHS1plac, pCHS2plac, pCHS3plac, pCHS8plac respectively. Ura−C. albicans cells were transformed with these plasmids previously cut within the RPS1 gene with StuI to target homologous recombination at the neutral chromosomal RPS1 locus and the URA3 gene was the selectable marker (Murad et al., 2000). Southern analysis was used to screen transformants and only those with single integrations of each pCHSplac plasmid were selected. Genomic DNA from each transformant was digested with XhoI/BamHI and hybridized to 693 bp RPS1 specific probe.
Measurement of β-galactosidase activity
The expression of each CHS gene in lacZ promoter fusions was measured using a modified version of the assays described previously (Rose and Botstein, 1983). C. albicans cells were grown with shaking at 200 r.p.m. at the chosen growth condition and harvested at OD600 < 1. Yeast cells were centrifuged at 3000 g for 5 min at 4°C and the pellet was resuspended in 0.5 ml of ice–cold water and transferred to microcentrifuge tubes. The cells were then centrifuged at 13 000 g for 5 min and resuspended in 0.5 ml of breaking buffer [100 mM TRIS-HCl pH 7.5, 0.01% (w/v) SDS, 1 mM dithiothreitol (DTT), 10% (v/v) glycerol, pepstatin 4 μg ml−1, 1 × proteinase cocktail tablets EDTA-free (Roche)]. Approximately equal volumes of glass beads (Sigma, Poole, UK G9268) and cell pellet were used and the cells were disrupted using a Fastprep cell breakage machine (Thermo Savant, Middlesex, UK) using six cycles of 30 s with chilling on ice for 1 min in between each cycle. The extract was centrifuged at 13 000 r.p.m. for 10 min and the protein concentration of the supernatant was measured using Coomassie® Protein Assay Reagent Kit (Pierce Biotechnology, Perbio, Rockford, UK). Varying quantities of protein extract, 30–300 μl, were added to Z-buffer (60 mM NaH2PO4, 40 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4), in a total volume of 800 μl. The reaction was initiated by adding 200 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) stock solution (4 mg ml−1 in phosphate buffer) and incubated until the yellow o-nitrophenol product was produced. The reaction was stopped by addition of 400 μl of 1 M Na2CO3. The specific β-galactosidase activity was measured in terms of the yield of product o-nitrophenol at the absorbance of 420 nm.
Measurement of chitin synthase activity
Mixed membrane fractions were prepared from exponential phase yeast cells and their chitin synthase activities measured as described previously (Munro et al., 1998). MMF proteins were activated by limited incubation with 100 ng trypsin μl−1 MMF at 30°C and the reactions were stopped by addition of 150 ng soybean trypsin inhibitor μl−1 MMF. Briefly, standard reactions for measuring chitin synthase activity were carried out in a 50 μl volume and were composed of; 50 μg MMF protein, 25 mM N-acetylglucosamine, 1 mM UDP-N-acetylglucosamine which included 25 nCi UDP-[U-14C] N-acetylglucosamine, 50 mM Tris-HCl pH 7.5 and 10 mM MgCl2. Incubations were carried out at 30°C for 30 min and the reaction was stopped by addition of 1 ml of 66% (v/v) ethanol. The reaction mixture was then filtered through GF/C filter discs (Whatman), which had been presoaked in 10% (v/v) trichloroacetic acid. The reactions tubes were rinsed out with 2 × 1 ml of 1% (v/v) Triton X-100 and each filter was then washed with 4 × 2 ml of 66% (v/v) ethanol. The radiolabelled chitin synthesized in the reaction was trapped on the filters and unincorporated substrate was removed by washing. Filters were dried at 80°C and their radioactivity counted in a scintillation counter.
Measurement of cell wall chitin content
Cell walls were prepared from 10 ml of C. albicans stationary phase yeast cultures grown in YPD and the chitin content was measured as described previously (Munro et al., 2003). Cells were disrupted with glass beads (Sigma, G9268) using a Fastprep cell breakage machine (Thermo Savant, Middlesex, UK) until at least 95% of cells were disrupted. They were then washed five times with 1 M NaCl and extracted in SDS-MerOH buffer (50 mM Tris, 2% sodium dodecyl sulphate (SDS), 0.3 M β-mercaptoethanol, 1 mM EDTA; pH 8.0) at 100°C for 10 min, then washed in dH2O. Cell wall pellets were resuspended in sterile dH2O, freeze dried, and the dry weight of recovered cell walls was measured. Chitin contents were determined by measuring the glucosamine released by acid hydrolysis of purified cell walls (Kapteyn et al., 2000).
Culture conditions
Cells were grown in normal laboratory media and under conditions of various environmental stresses. Cells were grown overnight in YPD then transferred to YPD supplemented with different agents: 1 M sorbitol, 0.8 M NaCl, 0.2 M CaCl2, 100 μg ml−1 CFW, 200 μg ml−1 CR, 0.05% SDS, 12 mM caffeine, 25 mM DTT, 23 mM glucosamine, 50 μg ml−1 cyclosporin A, 1 μg ml−1 FK506, 1 mM chloropromazine, 4 μM A23187. Cells were harvested at OD600 0.8.
Western analysis
Western analysis was performed using the method of Millar et al. (1995) with some modifications. Overnight cultures of the wild type, crz1Δ and hog1Δ strains were diluted 1:50 into 25 ml of YPD supplemented with uridine and incubated shaking for 4 h at 30°C. The mid-log phase cultures were then treated with a final concentration of 100 mM CaCl2, 100 μg ml−1 CFW, or both for 0, 10, 30, 60 or 120 min. No-treatment controls were also performed. After treatment, cells were harvested by centrifugation (1500 g, 2 min, 4°C) and washed in 1 ml of cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% NP40, 2 μg ml−1 Leupeptin, 2 μg ml−1 Pepstatin, 1 mM PMSF, 2 mM Na3VO4, 50 mM NaF). Cells were collected by centrifugation (800 g, 5 min, 4°C) and resuspended in 250 μl of cold lysis buffer. Cells were broken using a FastPrep machine in the presence of acid-washed glass beads (4 × 15 s bursts at speed 6.5 with 1 min on ice between bursts). The extracts were clarified by centrifugation (16 000 g, 5 min, 4°C). Protein concentration in the cleared lysate was estimated using the method described by Bradford (1976) with BSA as a standard.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using the XCell SureLock™ Mini-Cell system (Invitrogen) with NuPAGE®Novex Bis-Tris 4–12% precast gels (Invitrogen) in NuPAGE® MOPS-SDS Running Buffer (Invitrogen) containing NuPAGE® Antioxidant (Invitrogen) as per the manufacturer's instructions. Approximately 15 μg of protein was loaded in each lane. The proteins were transferred to Invitrolon™ PVDF Membranes (Invitrogen) in NuPAGE® Transfer Buffer containing methanol using the XCell II™ Blot Module (Invitrogen) following the manufacturer's instructions.
Following transfer, the membranes were rinsed in PBS and blocked in PBS-T + 10% BSA (PBS, 0.1% Tween-20, 10% (w/v) BSA, 50 mM (NaF) for 30 min at room temperature. The membranes were then incubated overnight at 4°C in PBS-T + 5% BSA (PBS, 0.1% Tween-20, 5% (w/v) BSA, 50 mM (NaF) containing a 1:1000 dilution of Phospho-p44/42 Map Kinase (Thr202/Tyr204) Antibody (Cell Signaling Technology). The membranes were washed five times for 5 min in PBS-T (PBS, 0.1% Tween-20) and then incubated for 1 h at room temperature in PBS-T + 5% BSA containing a 1:2000 dilution of Anti-rabbit IgG, HRP-linked Antibody (Cell Signaling Technology). The membranes were washed three times for 5 min in PBS-T and the signal was detected using LumiGLO™ Reagent and Peroxide (Cell Signaling Technology) as per the manufacturer's instructions.
Statistical analyses
Statistical significant differences in the assay results were determined with SPSS software using ANOVA and Post Hoc Dunnett's T-test, P < 0.05. When the results displayed unequal variance the Kruskal–Wallis non-parametric test or Dunnett's T3 test were applied.
Acknowledgements
We thank D. Sanglard and J. Pla for provision of mutant strains and acknowledge financial support from the BBSRC (BB/C510191/1 and studentship to SS) the EC (Eurocellwall, Fungwall and SignalPath consortia), the Wellcome Trust (063204, 080088) and the MRC for a New Investigator Award to CAM. We are indebted to the referees for their valuable critiques.
References
- Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Hughes HB, Munro CA, Thomas WP, MacCallum DM, Bertram G, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Boorsma A, de Nobel H, ter Riet B, Bargmann B, Brul S, Hellingwerf KJ, Klis FM. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast. 2004;21:413–427. doi: 10.1002/yea.1109. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bulawa CE, Miller DW, Henry LK, Becker JM. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. The synthesis and degradation of chitin. Adv Enzymol. 1987;59:59–101. doi: 10.1002/9780470123058.ch2. [DOI] [PubMed] [Google Scholar]
- Carotti C, Ferrario L, Roncero C, Valdivieso MH, Duran A, Popolo L. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast. 2002;19:1113–1124. doi: 10.1002/yea.905. [DOI] [PubMed] [Google Scholar]
- Chattaway FW, Holmes MR, Barlow AJE. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J Gen Microbiol. 1968;51:367–376. doi: 10.1099/00221287-51-3-367. [DOI] [PubMed] [Google Scholar]
- Chen-Wu JL, Zwicker J, Bowen AR, Robbins PW. Expression of chitin synthase genes during yeast and hyphal growth phases of Candida albicans. Mol Microbiol. 1992;6:497–502. doi: 10.1111/j.1365-2958.1992.tb01494.x. [DOI] [PubMed] [Google Scholar]
- Cyert MS. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun. 2003;311:1143–1150. doi: 10.1016/s0006-291x(03)01552-3. [DOI] [PubMed] [Google Scholar]
- De Nobel H, Ruiz C, Martin H, Morris W, Brul S, Molina M, Klis FM. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology. 2000;146:2121–2132. doi: 10.1099/00221287-146-9-2121. [DOI] [PubMed] [Google Scholar]
- Demarini DJ, Adams AEM, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- Deshpande MV, O'Donnell R, Gooday GW. Regulation of chitin synthase activity in the dimorphic fungus Benjaminiella poitrasii by external osmotic pressure. FEMS Microbiol Lett. 1997;152:327–332. doi: 10.1111/j.1574-6968.1997.tb10447.x. [DOI] [PubMed] [Google Scholar]
- Edlind T, Smith L, Henry K, Katiyar S, Nickels J. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol Microbiol. 2002;46:257–268. doi: 10.1046/j.1365-2958.2002.03165.x. [DOI] [PubMed] [Google Scholar]
- Eisman B, Alonso-Monge R, Roman E, Arana D, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, et al. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Duran A, Roncero C. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J Bacteriol. 2000;182:2428–2437. doi: 10.1128/jb.182.9.2428-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday GW. The dynamics of hyphal growth. Mycol Res. 1995;99:385–394. [Google Scholar]
- Gow NAR, Robbins PW, Lester JW, Brown AJP, Fonzi WA, Chapman T, Kinsman O. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, Verkleij AJ, et al. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol. 2000;35:601–611. doi: 10.1046/j.1365-2958.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- Kapteyn JC, ter Riet B, Vink E, Blad S, de Nobel H, Van den Ende H, Klis FM. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol Microbiol. 2001;39:469–480. doi: 10.1046/j.1365-2958.2001.02242.x. [DOI] [PubMed] [Google Scholar]
- Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Klis FM, Mol P, Hellingwerf K, Brul S. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2002;26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Lagorce A, Hauser CN, Labourdette D, Martin-Yken H, Arroyo J, Hoheisel JD, François J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J Biol Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci USA. 1999;96:12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanism for modulation of signalling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Mio T, Yabe T, Sudoh M, Satoh Y, Nakajima T, Arisawa M, Yamada-Okabe H. Role of three chitin synthase genes in the growth of Candida albicans. J Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro CA, Gow NAR. Chitin biosynthesis as a target for antifungals. In: Dixon GK, Copping LG, Hollomon DW, editors. Antifungal Agents: Discovery and Mode of Action. Bios Scientific Publishers; Oxford: 1995. pp. 161–171. [Google Scholar]
- Munro CA, Gow NAR. Chitin synthesis in human pathogenic fungi. Med Mycol. 2001;39S:41–53. [PubMed] [Google Scholar]
- Munro CA, Schofield DA, Gooday GW, Gow NAR. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology. 1998;144:391–401. doi: 10.1099/00221287-144-2-391. [DOI] [PubMed] [Google Scholar]
- Munro CA, Winter K, Buchan A, Henry K, Becker JM, Brown AJ, et al. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol Microbiol. 2001;39:1414–1426. doi: 10.1046/j.1365-2958.2001.02347.x. [DOI] [PubMed] [Google Scholar]
- Munro CA, Whitton RK, Hughes B, Rella M, Selvaggini S, Gow NAR. CHS8-a fourth chitin synthase gene in Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fung Genet Biol. 2003;40:146–158. doi: 10.1016/s1087-1845(03)00083-5. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJP. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F, Alonso-Monge R, Rico H, Pla J, Sentandreu R, Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144:411–424. doi: 10.1099/00221287-144-2-411. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F, Eisman B, Fiuza SM, Nombela C, Pla J. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology. 2005;151:2737–2749. doi: 10.1099/mic.0.28038-0. [DOI] [PubMed] [Google Scholar]
- Odds FC, Brown AJP, Gow NAR. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11:272–279. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- Onyewu C, Wormley FL, Perfect JR, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect Immun. 2004;72:7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardini G, De Groot PW, Coste AT, Karababa M, Klis FM, De Koster CG, Sanglard D. The CRH family coding for cell wall GPI proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J Biol Chem. 2006;281:40399–40411. doi: 10.1074/jbc.M606361200. M606361200 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Popolo L, Gualtieri T, Ragni E. The yeast cell wall salvage pathway. Med Mycol. 2001;39S:111–121. [PubMed] [Google Scholar]
- Proft M, Gibbons FD, Copeland M, Roth FP, Struhl K. Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1343–1352. doi: 10.1128/EC.4.8.1343-1352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman E, Nombela C, Pla J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol Biol Cell. 2005;25:10611–10627. doi: 10.1128/MCB.25.23.10611-10627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- Roncero C, Duran A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Effect of Calcofluor white on chitin synthases from Saccharomyces cerevisiae. J Bacteriol. 1988;170:1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Botstein D. Construction and use of gene fusions to LACZ (Beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J, San-Blas G. Chitin synthesis as target for antifungal drugs. Curr Drug Targets Infect Disord. 2003;3:77–91. doi: 10.2174/1568005033342064. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J, Elorza MV, Valentin E, Sentandreu R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006;6:14–29. doi: 10.1111/j.1567-1364.2005.00017.x. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Duran A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AJ, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Durán A, Cabib E. The function of chitin synthase 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sietsma JH, Wessels JGH. Apical wall biogenesis. In: Wessels JGH, Meinhardt H, editors. The Mycota I. Springer-Verlag; Berlin: 1994. pp. 125–141. [Google Scholar]
- Sullivan PA, Yin CY, Molloy C, Templeton MD, Shepherd MG. An analysis of the metabolism and cell wall composition of Candida albicans during germ tube formation. Can J Microbiol. 1983;29:1514–1525. doi: 10.1139/m83-233. [DOI] [PubMed] [Google Scholar]
- Trilla JA, Cos T, Duran A, Roncero C. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast. 1997;13:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Trilla JA, Duran A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl MA, Johnson AD. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology. 2001;14:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc Natl Acad Sci USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso MH, Ferrario L, Vai M, Duran A, Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu H, Szaniszlo PJ. Compensatory expression of five chitin synthase genes, a response to stress stimuli, in Wangiella (Exophiala) dermatitidis, a melanized fungal pathogen of humans. Microbiology. 2002;148:2811–2817. doi: 10.1099/00221287-148-9-2811. [DOI] [PubMed] [Google Scholar]
- Wessels JGH. Role of cell wall architecture in fungal tip growth generation. In: Heath IB, editor. Tip Growth in Plant and Fungal Cells. Academic Press; San Diego, CA: 1990. pp. 1–28. [Google Scholar]
- Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- Ziman M, Tsung M, Schekman RW. Role of CHS6/CSD3 in the membrane trafficking of Chs3p. A Saccharomyces cerevisiae chitin synthase whose localization is temporally and spatially regulated. Mol Biol Cell. 1996;7:1893. [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]