Abstract
Atrial fibrillation (AF) is a frequent chronic dysrythmia with an incidence that increases with age (>40). Because of its medical and socio-economic impacts it is expected to become an increasing burden on most health care systems. AF is a multi-factorial disease for which the identification of subtypes is warranted. Novel approaches based on the broad concepts of systems biology may overcome the blurred notion of normal and pathological phenotype, which is inherent to high throughput molecular arrays analysis. Here we apply an internal contrast algorithm on AF patient data with an analytical focus on potential entry pathways into the disease. We used a RMA (Robust Multichip Average) normalized Affymetrix micro-array data set from 10 AF patients (geo_accession #GSE2240). Four series of probes were selected based on physiopathogenic links with AF entryways: apoptosis (remodeling), MAP kinase (cell remodeling), OXPHOS (ability to sustain hemodynamic workload) and glycolysis (ischemia). Annotated probe lists were polled with Bioconductor packages in R (version 2.7.1). Genetic profile contrasts were analysed with hierarchical clustering and principal component analysis. The analysis revealed distinct patient groups for all probe sets. A substantial part (54% till 67%) of the variance is explained in the first 2 principal components. Genes in PC1/2 with high discriminatory value were selected and analyzed in detail. We aim for reliable molecular stratification of AF. We show that stratification is possible based on physiologically relevant gene sets. Genes with high contrast value are likely to give pathophysiological insight into permanent AF subtypes.
Keywords: atrial fibrillation, gene expression, Robust Multichip Average, genetic profile
Background
Atrial fibrillation (AF) is a frequent chronic dysrythmia with an incidence that increases with age (>40) [1]. Due to its medical and socio-economic impacts and, of the aging European/ Western /Japanese populations, it is projected to become an increasing burden on most health care systems [2]. Besides the obvious medical emergencies, the costs include the chronic corrective therapies (pacemakers, surgical ablations, etc.), the severe complications (thromboembolisms, strokes e.a., [3]) and various other hospitalisation costs. Atrial fibrillation is a multi-factorial disease for which the identification of subtypes - molecular categories - will be both useful and needed to provide better and cost-effective therapies [4] and to improve the quality of future clinical studies.
With the advent of high-throughput molecular analyses, the classical paradigm of a failing gene corresponding to an altered/pathological phenotype is being lost. It is widely accepted that an altered genotype can lead to a healthy phenotype via numerous compensation mechanisms, but it is becoming also apparent - and much less known - that even healthy genotypes could lead to defective/pathological phenotypes. Indeed a given cell phenotype could be considered as a state more akin to the orbit of a chaotic strange attractor than a definitive frozen combination of gene expression [5]. There seem to exist many ways to dissociate genotypes from phenotypes [6], hence the conceptual impossibility of a bona fide control group. In this context of a blur between the notions of normal and pathological phenotypes, novel analytical approaches are warranted based on the broad concepts of system biology [7].
To provide evidence of robust/reliable stratification based on molecular profiles is one of the biggest challenges in molecular medicine. Exploratory tools from multivariate statistics and data mining offer solutions, but often miss a biological and physiological focus, and neglect the dynamic reality of the biological system. As part of a series of feasibility studies, we are using an internal contrast analysis - i.e without comparison with a control population - of an existing micro-array data-set from chronic AF patients [8] in which up-regulation of metabolism was found to be a key marker. Prior to their chronic stage where tissue remodelling acts as a positive feedback for the phenotype, AF patients enter this pathology either via ischemia-related, work-load and hypertension, and/or degenerative problems, each of which can be potentially traced at a molecular level.
Methodology
Datasets
We used the RMA (Robust Multichip Average, [9]) normalized gene expression micro-array data set for 4 series of probes linked to either apoptosis, glycolysis, OXPHOS and MAP kinase pathway. Those probe sets were selected because of their potential physiopathogenic links with the entryways toward AF: apoptosis (remodeling), MAP kinase (cell and tissular differentiation), OXPHOS (ability to sustain workload including high tensions), glycolysis (ischemic or pre-ischemic situations). The data were obtained from 10 patient samples in an Affymetrix -set (geo_accession #GSE2240).
Gene selection
Bioconductor, version 2.2 [10] and R, version 2.7.1 (packages: hgu133b, KEGG, annotate, cluster) provided the tools to select functional category-related genes that are prevalent in the KEGG database (function hgu133bPATH2PROBE).
Contrast analysis
Overall similarity in genetic profiles between patients was apparent from the dendrograms after hierarchical clustering (HC) [11] based on the “dissimilarity” correlation matrix. To select genes with high contrast value between all patients a principal component analysis (PCA) [12] was done. Gene probes were selected based on the loadings vector of the first and second principal component. Selected genes were further classified according to absolute univariate expression levels. We found contrast mostly interpretable for genes with (i.) low variance in between-probe (ii.) high difference between patient groups and (iii.) high deviation from the over-all median patient value for a given gene.
Discussion
Micro-array analysis is a high-throughput technique, which allows probing the expression levels of thousands of genes simultaneously. Though at first sight arrays provide a powerful tool, conclusions and robustness of the technique are still heavily debated and solutions are presented to address standardization of experimental conditions, calculation of proper sample size, pre-processing of data and statistical inference of gene signals, to name a few. Systems biology provides a useful framework, mainly addressing the “dynamics” and the concepts of normality, and of modularity in biological networks. In supra-ventricular arrhythmia, time-dependent changes for functional groups of genes were recently investigated in canine models [13].
Barth and colleagues found that atrial tissue acquires ventricular characteristics, up-regulation of metabolic genes was also apparent in chronic AF patients. The multi-factorial pathophysiological patterns may come more apparent from a patient-centric viewpoint. Therefore we applied contrast analysis within the AF patient group, aiming at finding the genes for which divergence were maximal, and which contributed the most to the variations in the expression signals.
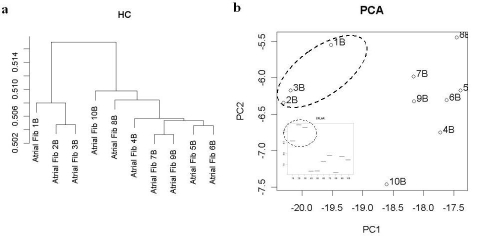
In Figure 1, HC and PCA of AF patients based on the apoptotic gene set is illustrated. Based on gene expression profiles, multivariate analysis (HC and PCA) of patients revealed distinct clustering patterns of the patients in all 4 datasets. The most striking observation is the consistent grouping of 3 patients (patient id: 1, 2, 3) for the 4 datasets, clearly separated from the rest, and the isolation of one patient (patient id: 10), bearing little resemblance to all of the other AF patients. The analysis also revealed high similarity of another group (id: 5, 6, 9) in all datasets.
Figure 1.
Analysis of apoptosis gene set. HC (a) of AF patients shows 2 distinct clusters of patients. PCA analysis (b) corroborates clear differentiation of patient AF1-2-3 (dashed group). Inset: example of univariate expression levels within patients of CFLAR (CASP8 and FADD-like apoptosis regulator), a gene with highest loadings on PC1.
For all datasets a substantial proportion of the variance (54% till 67%) in the dataset is captured in the first 2 principal components (cumulative proportion for apoptosis: 0.53 0.67, oxphos: 0.51 0.63, glycolysis: 0.31 0.54, MAPK: 0.42 0.67). Gene probes with high discriminatory value were selected based on high loadings in PC1/2, either positive (+) or negative (-) (see Table 1 under supplementary material). These probes are likely to be the most relevant in terms of the between-patient contrast. It should be noted that genes are often mapped to multiple probesets. Affymetrix arrays such as the hgu133b are designed to target the 600 bp at the 3' end of the transcript, not the start of the transcript. Therefore many-to-one mappings between probesets and genes are often due to alternative splicing, use of alternative poly (A) sites, or annotation errors [14,15]. This could add to the variations, a point we will discuss later.
We are especially interested in genes for which the expression range for all probes (i.e. with identical annotation), clearly differs among patients, and thus provide unambiguous discrimination between those patients. To identify such genes, we also explored the univariate expression levels of all probes. Doing so, we could identify 3 types of genes. The genes for which all probe levels were markedly different between patients and displayed nearly no variance within a single patient, those were defined as Class I, and are reliable signals. The genes, for which the intra-patient variance of the expression levels between their various probes was large, were identified as Class II. For those, the high contrast was more likely due to the spread response instead of bona fide differences in expression levels; hence these are not correct signals. Then we also identified genes for which both a marked difference in levels between different patients could be recorded, and at the same time a large intra-patient variance for the response of gene expression for the various probes, those were termed Class III and could be considered acceptable signals. Hence we evidence that we could identify and classify 36 different genes, which are reliable markers and could be used to categorize the patients. The split per pathological entryway was: for MAPK (8 class I, 0 class II, 3 class III), for glycolysis (7, 0, 4), for OXPHOS (7,1,1) and for apoptosis (4, 2, 2).
The Class II – i.e. the non-reliable and/or irrelevant markers – were GAPDH (glucose phosphate dehydrogenase), Acetyl-coA synthase 1 (whereas Acetyl-coA synthase 2 fell into class I), ADHFE (alcohol dehydrogenase), ENOβ3 (muscular isoform of β˜enolase), PPP3R (protein phosphatase 3 regulatory subunit α), CYC (cytochrome C). Surprisingly these are all metabolic markers.
A closer look at Class I markers reveals interesting facts (Figure 1, PCA lower inset for an example). For instance, in 3 patients (id:1,2,3), MAP3K16 levels - an apoptotic effector involved in the re-organization of actin filaments [16] - are up-regulated and, this occurs in parallel with the marked up-regulation of the SOS1 gene - a well known guanine nucleotide exchange protein which links mitogen- activated kinase and cytoskeletal elements like actin. Hence these patients differ from all the others in that they are re-modelling their heart tissue. In those 3 patients, PPP3CA (the catalytic subunit of protein phosphatase A), (NDUFFS8) NADH-coQ reductase, and ubiquinol-cytochrome C reductase core protein II (UQCRC2) are also up-regulated. Those would reflect an enhanced electron transport chain activity. It is worth noticing that dehydrolipoamide dehydrogenase (DLD), a flavoprotein linked to electron transport and the Krebs cycle is also up-regulated. It could thus be concluded that these 3 patients are markedly different from a metabolic standpoint. The CFLAR (caspase 8 FADD-like apoptosis regulator and CASP8 (caspase 8 itself) are respectively up- and down regulated. CFLAR is transducing external triggering of apoptosis; this is coupled with the fact that effector caspases are not up-regulated and the fact that XIAP - a post-mitochondrial inhibitor of apoptosis - is normal. In contrast within 3 other patients (id: 5,6,8), XIAP is down-regulated, those latter patients are hence more apoptotic and less remodelling than the 3 former ones. The patients (id: 4, 5, 6) had marked increases in FG18 - a fibroblast growth factor isoform, important for cell proliferation and inflammation, and whose overexpression can lead to abnormalities - whereas patients (id: 1,2,3) had decreases. Once more, this is consistent with those 2 groups of patients being functionally very different. Another and potentially important marker distinguishing those two groups is the AKT3 gene, a protein kinase B isoform with some oncogenic activity but selectively expressed in the heart [17]. It promotes cell growth and is known to have some cardioprotective effect during re-modelling, but a continuous overexpression would lead to dysfunctional maladaptive hypertrophy. Patients (id: 1, 2, 3) are up-regulated whereas patients (id:4,5,6) are down-regulated. The first group is hence cardio-protected vs. the second, but could also become more dysfunctional on the long term. At any rate AKT3 is likely to be an important follow-up marker. Two isoforms of aldolase (ALDOA and ALDOB, a glycolytic enzyme) showed an interesting pattern: all patients who were up for ALDOA were down for ALDOB and conversely, with the exception of 2 of the 10 patients (id: 1, 5). Noteworthy patient 10 was markedly different from all the other for most of the 36 markers we studied.
Conclusion
We aim for a reliable patient stratification based on molecular profiles without the use of a “normal” group. We showed that by contrast analysis; we could identify patient clusters and individual markers. By using objective criteria, those markers could be further appraised for their biological reliability. Even with a small cohort (10 AF patients in total), we could identify at least 2 markedly different sub-populations, which differed based on potential entryway into the disease. One group (id: 1,2,3) differs metabolically and functionally from another. While both groups have some apoptotic markers, their molecular context is highly different as the first had more MAP-kinase activation (hypertrophy) and effector caspases were less controlled in the second. We could also identify an interesting prognostic follow-up marker. However we have to highlight several caveats. There is a high variance between probes for one gene and hence quality control is warranted. This variance can arise from actual differences (alternate splices,…), problems on the DNA CHIP or in annotation. In our feasibility study, we face clear limitations, e.g. the number of patients is small and not all apoptosis genes are included (e.g. no VDAC's). However we showed that stratification is possible based on physiologically relevant gene sets. Genes with high contrast value in PCA are likely to give pathophysiological insight into “permanent” AF subtypes. It is our interest to extend this exploratory analysis with classification approaches using independent training and validation data sets. In a future programme we also like to contrast dynamic changes in expression patterns with a focus on AF entry point-related molecular signatures in a reductionist animal model.
Supplementary material
References
- 1.Chen LY, Shen WK. Heart Rhythm. 2007;4:S1. doi: 10.1016/j.hrthm.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 2.McBride D, et al. Value Health. 2008 [Google Scholar]
- 3.Ali S, et al. Cardiovasc Hematol Disord Drug Targets. 2006;6:233. doi: 10.2174/187152906779010728. [DOI] [PubMed] [Google Scholar]
- 4.El Sherif N, Baroudi G. J Cardiovasc Electrophysiol. 2004;15:224. doi: 10.1046/j.1540-8167.2004.03618.x. [DOI] [PubMed] [Google Scholar]
- 5. http://home.comcast.net/~reillyjones/order.html
- 6.Suel GM, et al. Nature. 2006;440:545. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 7.Noble D. Science. 2002;295:1678. doi: 10.1126/science.1069881. [DOI] [PubMed] [Google Scholar]
- 8.Barth AS, et al. Circ Res. 2005;96:1022. doi: 10.1161/01.RES.0000165480.82737.33. [DOI] [PubMed] [Google Scholar]
- 9.Bolstad BM, et al. Statistics for Biology and Health. Springer; 2005. p. 13. [Google Scholar]
- 10.Gentleman RC, et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. http://sekhon.berkeley.edu/stats/html/hclust.html
- 12. http://sekhon.berkeley.edu/stats/html/princomp.html
- 13.Cardin S, et al. Circ Res. 2007;100:425. doi: 10.1161/01.RES.0000258428.09589.1a. [DOI] [PubMed] [Google Scholar]
- 14. https://stat.ethz.ch/pipermail/bioconductor/2008-July/023305.html
- 15.Stalteri MA, Harrison AP. BMC Bioinformatics. 2007;8:13. doi: 10.1186/1471-2105-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zihni C, et al. Journal of Biological Chemistry. 2006;281:7317. doi: 10.1074/jbc.M513769200. [DOI] [PubMed] [Google Scholar]
- 17.Taniyama Y, et al. J Mol Cell Cardiol. 2005;38:375. doi: 10.1016/j.yjmcc.2004.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.