Abstract
A cDNA for a second mouse mitochondrial carbonic anhydrase (CA) called CA VB was identified by homology to the previously characterized murine CA V, now called CA VA. The full-length cDNA encodes a 317-aa precursor that contains a 33-aa classical mitochondrial leader sequence. Comparison of products expressed from cDNAs for murine CA VB and CA VA in COS cells revealed that both expressed active CAs that localized in mitochondria, and showed comparable activities in crude extracts and in mitochondria isolated from transfected COS cells. Northern blot analyses of total RNAs from mouse tissues and Western blot analyses of mouse tissue homogenates showed differences in tissue-specific expression between CA VB and CA VA. CA VB was readily detected in most tissues, while CA VA expression was limited to liver, skeletal muscle, and kidney. The human orthologue of murine CA VB was recently reported also. Comparison of the CA domain sequence of human CA VB with that reported here shows that the CA domains of CA VB are much more highly conserved between mouse and human (95% identity) than the CA domains of mouse and human CA VAs (78% identity). Analysis of phylogenetic relationships between these and other available human and mouse CA isozyme sequences revealed that mammalian CA VB evolved much more slowly than CA VA, accepting amino acid substitutions at least 4.5 times more slowly since each evolved from its respective human–mouse ancestral gene around 90 million years ago. Both the differences in tissue distribution and the much greater evolutionary constraints on CA VB sequences suggest that CA VB and CA VA have evolved to assume different physiological roles.
The carbonic anhydrases (CAs) are a family of zinc metalloenzymes that catalyze the reversible hydration of CO2 in the reaction CO2 + H2O ⇄ HCO3− + H+ (1, 2). Ten enzymatically active isozymes have been discovered, including CAs I-VII, IX, XII, and XIV (1–7). They differ in tissue-specific expression, subcellular localization, kinetic properties, and susceptibility to various inhibitors. The CAs have been implicated in a variety of physiological processes, including respiration, renal acidification, bone resorption, formation of aqueous humor and cerebral fluid, and gastric acid secretion. A role for a CA in mitochondria was suggested when it was realized that mitochondria are impermeant to HCO3− ions and that HCO3− is required in mitochondria by pyruvate carboxylase for gluconeogenesis and by carbamoyl phosphate synthetase I for detoxification of NH3 in liver (reviewed in refs. 8 and 9). Inhibition of both processes by acetazolamide provided pharmacological evidence that mitochondria need a CA for these and other metabolic processes (10–13).
A mitochondrial CA was first isolated from guinea pig liver and named CA V (11). Subsequently, mouse, rat, and human cDNAs for CA V were cloned (14–16). The CA V transcript was identified by Northern blots only in mRNA from mouse liver (14). Western blot data (15) and immunohistochemical results (17) suggested a much wider distribution in rat tissues. A possible solution to the apparent discrepancies in tissue distribution in different studies was suggested by GenBank expressed sequence tag (EST) data, indicating that there are at least two mitochondrial CAs with differing tissue distributions. We discovered an EST from mouse kidney (AA123271) that encodes a CA that has greatest sequence identity to CA V and also contains a mitochondrial leader sequence. EST databases showed identical ESTs to be present in libraries from kidney and male mammary gland. While characterizing this second mouse mitochondrial CA, we became aware that its human orthologue had been identified independently and mapped to human chromosome Xp22.1 (6). Northern blots showed this mitochondrial CA to have a wider tissue distribution than that originally described for human CA VA. Both groups agreed through the nomenclature committee to refer to the new mitochondrial CA V as CA VB and to the original CA V as CA VA (6). The genes should be referred to as Car5A/Car5B (mouse) and CA5A/CA5B (human).
In this report, we describe the sequence and the properties of the protein encoded by the murine cDNA for CA VB. We also compare the tissue-specific expression of murine CA VB and CA VA and present a phylogenetic analysis in an attempt to illustrate and explain the striking sequence conservation of CA VB between human and mouse that was revealed by these studies.
Materials and Methods
Identification of Mouse Kidney CA VB cDNA.
A cDNA clone was recognized in the mouse EST database on the basis of nucleotide homology with mouse CA V (AA123271). Careful analysis of the 447-bp nucleotide sequence reported revealed that deletion of two cytosine residues at positions 16 and 17 from the initiation codon would create an ORF encoding a protein containing a complete CA domain. This cDNA clone was obtained from the Image Consortium and the complete sequence was determined by using T7 and internal primers. Comparison of the sequence we obtained with that published for the EST showed an additional adenosine following the cytosine at position 16. The presence of this additional nucleotide restored the ORF and the protein encoded showed 67.2% similarity to mouse CA VA. The complete sequence of the novel murine CA VB was entered in GenBank (AF192978).
Expression of CA VB cDNA in COS-7 Cells.
The 1201-bp fragment produced by EcoRI/NotI digestion of the full-length clone was introduced into the eukaryotic expression vector pCXN at the XhoI cloning site (18) by blunt end ligation. COS-7 cells were transfected by using the DEAE-dextran procedure (19). After 72 hr of transfection, cells were harvested and homogenized in 25 mM Tris⋅SO4 (pH 7.5) with 1 mM each of benzamidine, PMSF, and phenanthroline. CA activity was measured in homogenates of transfected COS cells and in isolated mitochondrial suspensions according to Maren (20) as described (21). The protein concentration was determined according to modified microLowry's procedure (22).
Isolation of Mitochondria from COS-7 Cells.
COS-7 cells transfected with CA VB cDNA were Dounce homogenized in 0.33 M sucrose, 1.33 mM Tris⋅EDTA (pH 7.4) containing 1 mM PMSF, 1 mM benzamidine, and 5 mM iodoacetate. The mitochondria were isolated by using a discontinuous sucrose density gradient as described (23). The mitochondrial fraction at the junction of 1 M and 1.5 M sucrose was pooled for CA VB characterization.
Immunochemical Methods.
Antibodies against the carboxyl-terminal 17-aa peptide were produced by injecting the C-terminal peptide of mouse CA VB conjugated to thyroglobulin into rabbits in complete Freund's adjuvant (23). The titer and specificity of the antiserum were checked by Western blot. Affinity-pure anti-CA VB C-tail-specific IgG was purified by using CA VB C-tail peptide-Affigel 10 column. Antibodies against mouse CA VA C-tail peptides were the same as described (15). Affinity-pure antibodies against mouse CA VA C-tail were purified by using a peptide containing the last 8 amino acid C-tail peptide-Affigel 10 column.
Western Blot Analysis.
SDS/PAGE was performed under reducing conditions according to Laemmli (25). After electrophoretic transfer of the polypeptides from the gel to the Immobilon-P membranes, the membranes were incubated with first antibodies followed by second antibodies as described (21). The polypeptide was visualized by using chemiluminescence substrate.
RNA Blot Analysis.
Total cellular RNA was isolated from the sources indicated in the figures utilizing a guanidinium/phenol solution (RNA-Stat60; Tel-Test, Friendswood, TX). RNA concentration and purity were determined by absorbance at 260 Å and 280 Å. A total of 15 μg from each source was denatured in formaldehyde-containing buffer and electrophoresed in 1% agarose, 2.2 M formaldehyde gels. The size of mRNA transcripts was estimated by using RNA standards (Promega). The RNA was transferred to Nytran membranes (Schleicher & Schuell) and immobilized by UV crosslinking. The blots were prehybridized in 50% formamide, 5× standard saline phosphate/EDTA [SSPE: 0.18 M NaCl/1 mM phosphate (pH 7.4)/1 mM EDTA], 50 mM Denhardt's, 50 mM sodium phosphate (pH 6.5), 200 μg/ml salmon sperm DNA, 1 mM EDTA, and 0.1% SDS. Hybridization was performed by using antisense RNA probes (Riboprobe; Promega) for mouse CA VA or CA VB labeled with [α-32P]CTP (ICN). The template for the CA VA probe consisted of nucleotides 95–1050 at the full-length murine CA VA cDNA (16). The template for the CA VB probe consisted of nucleotides 122-1086 of the full-length murine CA VB cDNA (AF192978). The blots were hybridized with the probes overnight at 65°C in the same solution used for prehybridization and washed in 2× SSPE at room temperature for 20 min, 0.2× SSPE at room temperature for 20 min, and 0.2× SSPE/0.1% SDS at 68°C for 20 min twice.
Results
Identification and Expression of Mouse Kidney CA V cDNA Clone.
The complete sequence of the full-length CA VB cDNA clone has been deposited in GenBank (AF192978). It contains a 147-bp 5′ untranslated sequence, a 951-bp ORF, and a 103-bp untranslated region with an AATAATA polyadenylation signal 20 bp upstream of the poly(A) tail. To determine whether it encoded a functional CA, we compared its expression in COS cells with that seen with the previously described murine CA VA cDNA. Table 1 shows that the cell homogenates and isolated mitochondria from COS cells transfected with mouse CA VB and CA VA had comparable increases in CA activity over that seen in cells transfected with vector only. In both cases, the CA activities were comparably enriched in isolated mitochondria (Table 1) and inhibited by acetazolamide (data not shown).
Table 1.
Carbonic anhydrase activity in cell homogenate and mitochondria of COS-7 cells transfected with CA VB
| Enzyme | CA activity, enzyme units/mg protein*
|
|
|---|---|---|
| Cell homogenate | Mitochondria | |
| Vector only | 0.09 | 0.01 |
| mCA VA | 0.48 | 3.65 |
| mCA VB no. 19 | 0.46 | 3.80 |
| mCA VB no. 21 | 0.43 | 3.80 |
mCA VB cDNAs from two independent clones nos. 19 and 21 were used; both expressed CA activity.
*Average of two measurements.
Expression of Immunoreactive CA VB and CA VA in Transfected COS Cells and in Mouse Tissues.
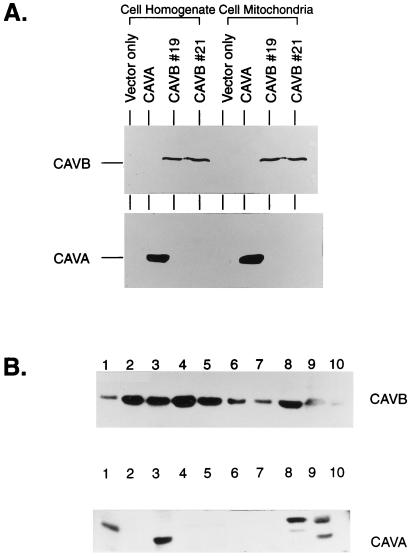
The CA VB and CA VA polypeptides expressed in transfected COS cells were also identified by antibodies raised to the different C-terminal peptides predicted from the respective cDNA sequences (Fig. 1). In each case, the antibody identified a 33-kDa protein not present in homogenates or isolated mitochondria from COS cells transfected with vector only (Fig. 2A).
Figure 1.
Comparison of amino acid sequences of mouse and human CA VBs and CA VAs. Fourteen amino acids conserved in mammalian CAs and thought to be near the active site are boxed. The proteolytic cleavage sites where the mitochondrial leader sequences are known or proposed to be cleaved are indicated by the triangles. The arginine at −2 positions from the cleavage sites are circled. The potential zinc-binding histidines equivalent to H94, H96, and H119 of CA I (2) are indicated by asterisks. The C-terminal peptides used to raise antibodies to mouse CA VB and CA VA are underlined.
Figure 2.
Expression of CA VB in COS-7 cells and different mouse tissues. (A) The cell homogenate or cell mitochondria from COS-7 cells transfected with vector only, CA VA and CAVB cDNAs clone nos. 19 and 21 were analyzed by SDS/PAGE followed by immunoblotting. The polypeptides for CA VA and CA VB are marked (Fig. 1). (B) Homogenates of different tissues containing 50 μg of protein were analyzed by SDS/PAGE followed by immunoblotting by using anti-mouse CA VB C-tail or CA VA C-tail antisera. Lanes: 1, brain; 2, heart; 3, liver; 4, lung; 5, kidney; 6, spleen; 7, intestine; 8, testis; 9, muscle; 10, pancreas. The polypeptides of 31-kDa mature CA VB were seen in all tissues used here. The polypeptides of 31-kDa mature CA VA and proteolytically nicked 28-kDa polypeptides were seen in few tissues.
Western blot data from tissue homogenates probed with anti-CA VB C-tail antibody showed a strong band at around 31 kDa, approximately the size predicted for the mature mitochondrial CA VB, in heart, liver, lung, kidney, testes, and muscle, with a weaker band of that size in most other tissues (Fig. 2B). The CA VA antibody gave a strong band at the 29-kDa molecular weight predicted for the mature CA VA only in liver and skeletal muscle. However, it also reacted with a higher molecular weight band in brain, testis, and muscle that is attributed to an unidentified crossreacting protein.
Analysis of CA VB and CA VA Transcripts by Northern Blot Analysis.
Fig. 3 shows the Northern blot data produced when total mRNA from various mouse tissues was analyzed by using probes for CA VB and CA VA coding sequences. The CA VB transcripts were found to be more widely distributed, with the strongest signal seen in RNA from kidney. Multiple CA VB transcripts were seen. The predominant bands were approximately 3.7 kb and 2.9 kb. A third transcript of 2.3 kb was particularly prominent in liver, skeletal muscle, and kidney. The CA VA transcript was more limited in distribution. It was most intense in liver, moderately intense in skeletal muscle, and present in lower amounts in kidney.
Figure 3.
Northern blot analysis of CA VA and CA VB mRNA tissue distribution. Total cellular RNA from the tissue sources indicated in the figure was electrophoresed, blotted, and hybridized with 32P-labeled probes transcribed from the CA VB (Top) and CA VA (Bottom) cDNA coding sequence. The blots were washed under high stringency conditions and autoradiography performed. Positions of the 28S and 18S ribosomal bands and the RNA size markers are indicated.
Thus, the Western blot data and the Northern blot data both demonstrate wider tissue distribution of CA VB expression than that reported for CA VA.
CA Vs in EST Databases.
Direct analysis of expression of CA VB and CA VA in human and mouse can be augmented by examining the partial cDNAs deposited in the dbEST database at GenBank. In mouse, both the original CA VA cDNA (14) and all of the CA VA ESTs (19 sequences from 13 clones) are from liver. In rat and human, the original CA VA cDNAs were also isolated from liver. While there are no CA VA ESTs from rat, the five human CA VA ESTs are either from fetal liver/spleen or B cell-enriched tonsil cDNA libraries.
For CA VB, the ESTs come from many more tissues. The only two mouse clones are from kidney and male mammary gland. The eight rat clones are from lung, kidney (3), embryo (2), and eye (2). The 19 human CA VB ESTs can be divided into three groups: (i) 5 that show no evidence of aberrant mRNA splicing—fetal heart (1), germ cell tumor (1), and pooled tissue (3); (ii) 12 that show evidence of aberrant splicing, e.g., skipping of exon 3, retention of introns—brain (4), tonsil (3), pooled tissue (2), fetus (1), squamous cell carcinoma (1), and breast (1); and (iii) 2 brain ESTs that are very short exonic sequences from which no conclusions about splicing can be drawn.
In summary, all of the liver cDNA ESTs are CA VA. On the other hand, ESTs containing CA VB sequences come from many tissues. In addition, although the numbers are small, aberrant mRNA splicing of CA VB transcripts appears common in human tissues and may explain the additional bands on Northern blots from human tissues reported by Fujikawa-Adachi et al. (6).
Sequence Analysis and Sequence Comparisons.
Fig. 1 shows the 317-aa sequence predicted for the murine CA VB aligned with the sequence of the recently reported human orthologue and with the previously reported sequences from mouse and human CA VAs (14–16). The N-terminal 33 amino acids correspond to a typical mitochondrial leader sequence (27). The boxed residues demonstrate conservation of the 14 invariant residues in mammalian CAs, including the three histidines that form the zinc-binding site (indicated by asterisks) (2). The predicted molecular mass of the murine CA VB precursor is 36,596 Da. The triangles indicate the predicted cleavage sites of CA VA and CA VB, where each would be processed in the mitochondrial matrix. Assuming this cleavage site, the molecular mass predicted for mature CA VB is 32,668 Da.
Comparison of the human and mouse sequences for CA VB and CA VA reveals a striking difference in sequence conservation. The mouse and human CA VB sequences show identities of 54.5% over the 33-residue mitochondrial leader sequence, 95% over the next 263 residues that comprise the CA domain, and 67% over the 21-residue C-terminal segment that follows the CA domain. By contrast, the mouse and human CA VA sequences show only four conserved residues over their 30- and 38-residue leader sequences, 79% identity over the 260-residue CA domain (1–260), and only 22% identity in the C-terminal nine residues that follow the CA domain. This unexpectedly large difference in the degree of conservation of sequences led us to analyze the phylogenetic relationships of these CAs to the four cytosolic CAs.
Evolution of CA VA and CA VB.
From a phylogeny of the entire α-CA family, it appears that Drosophila CAH1 (see refs. 3 and 35; GenBank accession no. AC001659) represents the closest outgroup. CAHI (BG:DS00941.1) is encoded by 3 exons: AC001659 bp. 78748–78715; 73868–73710; and 73642–73026. It was used in this analysis to provide a precise root for the tree presented in Fig. 4.
Figure 4.
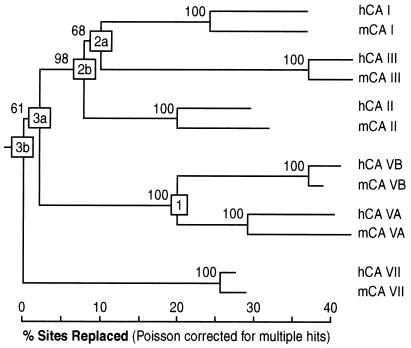
Phylogenetic tree of the six mitochondrial and cytosolic carbonic anhydrase isozymes of human and mouse. Sequences are as in Hewett-Emmett and Tashian (3), except human CA VB (16), mouse CA VB (this study), and mouse CA VII (32). The five gene duplications (1, 2a, 2b, 3a, 3b) are boxed. Branches are drawn to scale, their lengths representing percent of sites in the protein alignment replaced after Poisson correction for undetected multiple hits (cf. Kumar et al., ref. 28). Numbers beside each node represent percent of 1,000 bootstrap replicates supporting that particular grouping, e.g., 98% of the bootstrap resamples place the human and mouse CA I, CA II, and CA III sequences in a single group. Duplications 2a and 2b, and 3a and 3b are separated by short branches and low bootstraps (68% and 61%), and there is no statistical support for the order of the duplications and for the duplications occurring sequentially rather than at the same time.
The five gene duplications are labeled 1, 2a, 2b, 3a, and 3b; the last (3b) is also the root of the tree. Initially, a pair of gene duplications (boxes 3a and 3b) led to the separate CA I/II/III, CA VA/VB, and CA VII lineages. These two duplications appear to have occurred at close to the same time, and the branching order found has only weak support (61% bootstrap). Later, before the divergence of the amniotes (over 300 million years ago), another pair of closely spaced gene duplications (boxes 2a and 2b) occurred in a CA I/II/III lineage that generated distinct genes (encoding CA I, II, and III) that remain linked on human chromosome 8q22 and mouse chromosome 3. Once again, the low bootstrap value (68%) supporting the clustering of CA I and CA III (duplication 2a) is not significant. Both of these pairs of duplication events should be recorded as ill-resolved trifurcations rather than closely spaced pairs of bifurcations.
More recently, the CA VA/VB lineage duplication occurred (box 1) leading to distinct genes encoding CA VB and CA VA. This probably took place early in the lineage leading to the monotremes, marsupials, and placental mammals (i.e., around 200–300 million years ago). These genes are no longer linked; CA5B is located on human chromosome Xp22.1 (6), while CA5A is on human 16q24 (16).
An analysis of the evolution of the CA VA and CA VB lineages shows several very interesting features. From the time immediately after the gene duplication (box 1 in Fig. 4) up until the radiation of placental mammals (around 90 million years ago), the CA VB lineage accepted amino acid substitutions at almost twice the rate of CA VA (17.8% vs. 9.6%; see Fig. 5). Since the human–mouse ancestor (HMA), however, the pattern is reversed, with CA VB evolving more than 4.5 times more slowly than CA VA (2.6% vs. 11.6% average per mammalian lineage; see Fig. 5). The same pattern is also observed in the CA I/CA III (or CA II/CA III) comparisons (Fig. 5; columns 3 and 4). The very slow evolution of CA VB, CA III, and CA VII in the mammals (2.6%, 4.9%, and 2.4%, respectively; Fig. 5) contrasts sharply with that of the isozymes whose function is best characterized, CA VA, CA I, and CA II (11.8%, 12.3%, and 10.4%, respectively; Fig. 5). Worth noting is that since the origin of the cytosolic/organelle CAs (duplication 3b), CA II (30.1%) and CA VII (28.5%) have been evolving most conservatively overall. CA VA (40.7%) and CA VB (41.8%) have almost the same number of changes since duplication 3b overall, but, as noted above, their pattern of evolution is quite different.
Figure 5.
Rates of evolution of the six mitochondrial and cytosolic carbonic anhydrase isozymes of human and mouse. Rates are from the phylogenetic tree in Fig. 5. Column 1 shows the average of the human and mouse branches since the human and mouse ancestor (HMA) for each of the six isozymes. Columns 2–6 show percent sites replaced (after Poisson correction for undetected multiple hits) on internal branches of the tree from each duplication (1, 2a, 2b, 3a, 3b) until the human–mouse ancestor for the relevant isozymes. Column 6 shows the corrected percent sites replaced (average of human and mouse) from the earliest duplication (3b) until present for each of the six isozymes.
Discussion
In this study, we cloned and characterized the cDNA for the murine CA VB, the second known mitochondrial CA. When expressed in COS cells, it produces a catalytically active enzyme that resembled CA VA in its catalytic properties and its localization in mitochondria. Studies of mouse tissues showed that CA VB and CA VA have different tissue distributions. CA VA is most abundant in liver and skeletal muscle, where it has been proposed that a mitochondrial CA is required to provide HCO3− for pyruvate carboxylase, the first step in gluconeogenesis and for carbamyl phosphate synthase I, the first step in ureagenesis (8–11). The biological significance of the much more widespread expression of CA VB and the limited expression of CA VA are not yet understood.
While this work was in progress, Fujikawa-Adachi et al. (6) characterized the human orthologue of mouse CA VB and noted differences in tissue-specific distribution similar to those reported here, with one exception. They reported that CA VA is the only isoform expressed in liver. Although we found that CA VA is highly expressed in liver, we also see a transcript for CA VB in liver and a signal for CA VB on Western blots of mouse liver. It is not yet clear whether the two isoforms expressed in mouse liver are expressed in the same cell types or have the same lobular distribution.
The availability of the human CA VB sequence for comparison led to the interesting discovery of a surprising difference in sequence conservation between human and mouse CA VB and CA VA sequences. Phylogenetic comparisons indicated that CA VB and CA VA have different evolutionary histories. The data suggest that CA VB evolved even more rapidly than CA VA early in their separate histories, but underwent a far slower rate of evolution following divergence from the human–mouse ancestral genes for CA VA and CA VB.
This pattern of rapid evolution of the new gene duplicate followed by a slowdown after a new function is developed has also been observed in other cases. Within the α-CA gene family, the CA-RP VIII and CA-RP XI lineages display similar patterns (3, 30) and the α and β globins were shown by Goodman (31) and Czelusniak et al. (32) to follow this pattern. Under Ohno's hypothesis (33), following a gene duplication, one copy is released from the constraints of purifying (negative) selection and begins to evolve more rapidly than the member of the pair continuing to carry out the ancestral function. In many cases, the rapidly evolving gene will become a pseudogene and eventually any trace of it will disappear from the genome, as may be happening to the CA VA pseudogene (CA5P) (16). Occasionally, however, a new function or a new pattern of expression may emerge, whereupon the gene may again be subject to (purifying) selection, which may be more intense than that imposed on the sister gene that has retained the original function. This appears to be the case for CA VB.
An alternative view of this process has been put forward more recently by Hughes (34). He argues that it is unlikely that such diverse functions in gene family members could have arisen in this way. His view is that ancestral genes tend to have multiple functions and/or tissue expression (gene sharing) and that the process of gene duplication allows rapid separation of these functions and/or tissue-specific expression and regulation. Under this model, it is less clear why one member of a gene pair should evolve rapidly and then slow down as we see with CA VB and other members of the α-CA gene family.
Hewett-Emmett and Tashian (3) estimated the conservation of the α-CAs between pairs of mammalian sequences. The most conserved group are the CA-RP VIII, CA VII, CA III, and the extracellular CA-RP domain of two transmembrane phosphatases, to which can now be added CA-RP XI (30) and, from this study, CA VB. Interestingly, these represent the isozymes and inactive forms (CA-RPs) about whose function little is known. By contrast, the least conserved group were CA IV, CA VI, CA VA, CA I, and CA II, about whose function most is known. It would appear that these more rapidly evolving isozymes can tolerate amino acid replacements distant from the active-site pocket (providing folding is not altered). The slowly evolving isozymes and inactive isoforms may have extensive surface interactions with other proteins that preclude tolerance of most surface amino acid replacements. If this speculation is true, one would predict that CA VB has a wider range of important protein–protein interactions than CA VA, perhaps related to different metabolic roles for these two isozymes in liver (CA VA) and nonliver (CA VB) mitochondria. Because of these differences in tissue-specific expression and striking differences in sequence conservation between CA VB and CA VA, it will be of great interest to compare the phenotypes of mice selectively targeted for disruptions of each of the individual CA V genes. If they indeed have different, nonredundant functions, one might expect gene disruption of CA VB and CA VA to produce different phenotypes. From such knockout mice, we may be able to infer why these two isozymes have different tissue distributions and why CA VB sequences away from the active site are so much more highly conserved than those of CA VA.
Abbreviations
- CA
carbonic anhydrase
- EST
expressed sequence tag
Footnotes
References
- 1.Tashian R E. Adv Genet. 1992;30:321–356. doi: 10.1016/s0065-2660(08)60323-5. [DOI] [PubMed] [Google Scholar]
- 2.Sly W S, Hu P Y. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 3.Hewett-Emmett D, Tashian R E. Mol Phylogenet Evol. 1996;50:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 4.Türeci O, Sahin U, Vollmar E, Siemer S, Göttert E, Seitz G, Parkkila A-K, Shah G N, Grubb J H, Pfreundschuh M, Sly W S. Proc Natl Acad Sci USA. 1998;95:7608–7613. doi: 10.1073/pnas.95.13.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastorek J, Pastorekova S, Callebaut I, Mornon J P, Zelnik V, Opavsky R, Zat'ovicova M, Liao S, Portetelle D, Stanbridge E J, et al. Oncogene. 1994;9:2877–2888. [PubMed] [Google Scholar]
- 6.Fujikawa-Adachi K, Nishimori I, Taguchi T, Onishi S. J Biol Chem. 1999;274:21228–21233. doi: 10.1074/jbc.274.30.21228. [DOI] [PubMed] [Google Scholar]
- 7.Mori K, Ogawa Y, Ebihara K, Tamura N, Tashiro K, Kuwahara T, Mukoyama M, Sugawara A, Ozaki S, Tanaka I, Nakao K. J Biol Chem. 1999;274:15701–15705. doi: 10.1074/jbc.274.22.15701. [DOI] [PubMed] [Google Scholar]
- 8.Forster R E, 2nd, Dodgson S J, Storey B T, Lin L. Ann N Y Acad Sci. 1984;429:415–429. doi: 10.1111/j.1749-6632.1984.tb12368.x. [DOI] [PubMed] [Google Scholar]
- 9.Dodgson S J, Forster R E, 2nd, Storey B T. Ann N Y Acad Sci. 1984;429:516–524. doi: 10.1111/j.1749-6632.1984.tb12380.x. [DOI] [PubMed] [Google Scholar]
- 10.Dodgson S J, Forster R E., 2nd Arch Biochem Biophys. 1986;251:198–204. doi: 10.1016/0003-9861(86)90066-4. [DOI] [PubMed] [Google Scholar]
- 11.Dodgson S J. J Appl Physiol. 1987;63:2134–2141. doi: 10.1152/jappl.1987.63.5.2134. [DOI] [PubMed] [Google Scholar]
- 12.Hazen S A, Waheed A, Sly W S, LaNoue K F, Lynch C J. FASEB J. 1996;10:481–490. doi: 10.1096/fasebj.10.4.8647347. [DOI] [PubMed] [Google Scholar]
- 13.Parkkila A-K, Scarim A L, Parkkila S, Waheed A, Corbett J A, Sly W S. J Biol Chem. 1998;273:24620–24623. doi: 10.1074/jbc.273.38.24620. [DOI] [PubMed] [Google Scholar]
- 14.Amor-Gueret M, Levi-Strauss M. Nucleic Acids Res. 1990;18:1646. doi: 10.1093/nar/18.6.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagao Y, Srinivasan M, Platero J S, Svendrovski M, Waheed A, Sly W S. Proc Natl Acad Sci USA. 1994;91:10330–10334. doi: 10.1073/pnas.91.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagao Y, Batanian J R, Clements M F, Sly W S. Genomics. 1995;28:477–484. doi: 10.1006/geno.1995.1177. [DOI] [PubMed] [Google Scholar]
- 17.Väänänen H K, Carter N D, Dodgson S J. J Histochem Cytochem. 1991;39:451–459. doi: 10.1177/39.4.1900871. [DOI] [PubMed] [Google Scholar]
- 18.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 19.Lopata M A, Cleveland D W, Sollner-Webb B. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maren T H. J Pharmacol Exp Ther. 1960;130:26–29. [PubMed] [Google Scholar]
- 21.Sundaram V, Rumbolo P, Grubb J, Strisciuglio P, Sly W S. Am J Hum Genet. 1986;38:125–136. [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson G L. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 23.Tapper D P, Van Etten R A, Clayton D A. Methods Enzymol. 1983;97:426–423. doi: 10.1016/0076-6879(83)97153-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhu X L, Sly W S. J Biol Chem. 1990;265:8795–8801. [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Waheed A, Zhu X L, Sly W S. J Biol Chem. 1992;267:3308–3311. [PubMed] [Google Scholar]
- 27.Hendrick J P, Hodges P E, Rosenberg L E. Proc Natl Acad Sci USA. 1989;86:4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earnhardt J N, Qian M, Tu C, Lakkis M M, Bergenhem N C H, Laipis P J, Tashian R E, Silverman D N. Biochemistry. 1998;37:10837–10845. doi: 10.1021/bi980046t. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetic Analysis. University Park, PA: Pennsylvania State Univ.; 1993. , Version 1.0. [Google Scholar]
- 30.Lovejoy D A, Hewett-Emmett D, Porter C A, Cepoi D, Sheffield A, Vale W W, Tashian R E. Genomics. 1998;54:484–493. doi: 10.1006/geno.1998.5585. [DOI] [PubMed] [Google Scholar]
- 31.Goodman M. Prog Biophys Mol Biol. 1981;37:105–164. doi: 10.1016/0079-6107(81)90012-2. [DOI] [PubMed] [Google Scholar]
- 32.Czelusniak J, Goodman M, Hewett-Emmett D, Weiss M L, Venta P J, Tashian R E. Nature (London) 1982;298:297–300. doi: 10.1038/298297a0. [DOI] [PubMed] [Google Scholar]
- 33.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970. [Google Scholar]
- 34.Hughes A L. Proc R Soc London Ser B. 1994;256:119–124. [Google Scholar]
- 35.Ashburner M, Misra S, Roote J, Lewis S E, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]