Abstract
The evolution of complex animals such as insects and mammals is achieved with surprisingly few additions in protein coding genes. MicroRNAs (miRNAs), a class of noncoding RNAs, have emerged as important regulators of organogenesis in insects, fish and mammals. The microRNA repertoire of animals has expanded significantly during evolution especially in vertebrates, insects and nematodes, accompanying the appearance of complex body plans. MicroRNAs therefore have gained enormous interest in recent years. They are now regarded as key modulators of gene expression in many tissues during embryogenesis, in adult organisms and in disease processes. Therefore, these small RNA molecules have entered the center stage of molecular biology and are promising candidates not only for the regulation of key biological processes such as proliferation and apoptosis, but also for therapy of human diseases.
Key Words: microRNA, Dicer, canalization, embryogenesis, cancer
Introduction
The field of RNA research has experienced a renaissance with the discovery of RNA interference (RNAi). Recently, studies in this area have intensified with the identification of other small RNAs and the realization that microRNAs (miRNAs) are natural RNAi agents. This renewed interest has been further fueled by the choice of the Nobel prize committee to honor Andrew Fire and Craig Mello for their work on RNAi just eight years after their important publication.1 With these findings, there are great expectations for a better understanding of human disease along with hopes of novel therapeutic approaches. This review is intended to give an overview of current ideas of how microRNAs are generated, function and influence development and human disease.
MicroRNA Biogenesis and Function
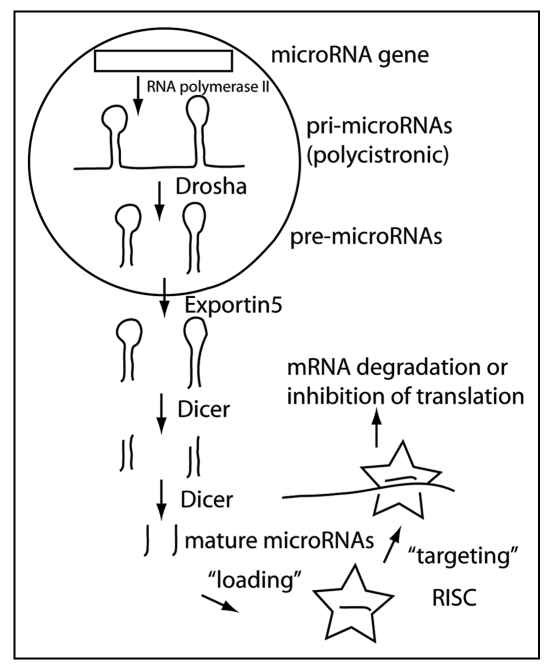
miRNAs are indeed small RNAs with the mature active form being approximately 22 nucleotides long. MiRNAs are derived from much larger transcripts (known as pri-miRNA) that are likely generated by RNA polymerase II. A hallmark of these transcripts is their ability to form hairpin structures containing sections of double stranded RNA. miRNAs can be located within introns of protein coding or noncoding genes, and in exons between genes (Miranda database, release 1.0 2005: about 62% intergenic, 34% in introns and 4% in exons). miRNAs can also occur in clusters, resulting in polycistronic transcripts. The double-stranded RNA structures in the primary transcript are substrates for a protein complex that contains RNase activity.2 The responsible RNase, Drosha, is active in the nucleus. Drosha works in conjunction with DGCR8 (DiGeorge Syndrome critical region gene 8). DGCR8 directs Drosha to the cleavage sites of the pri-microRNA (about 11 nucleotides away from the beginning of the stem structure3). The product of Drosha-mediated RNA cleavage is termed the pre-miRNA. Pri-miRNA cleavage appears to be tightly regulated in early embryogenesis in mammals, though probably not in C. elegans.4 Many mammalian mature miRNAs are not expressed in early embryogenesis even though their primary transcripts are present. In fact, the regulation of miRNA biogenesis can likely occur at all processing steps (transcription, Drosha-mediated processing, exportation to cytoplasm, Dicer-mediated cleavage). Pre-miRNAs are actively transported out of the nucleus into the cytoplasm by exportin-5, where they become the target of another protein complex, containing the RNaseIII enzyme Dicer. Dicer produces the final product, the mature miRNA, which is loaded into the RNA-induced silencing complex (RISC5). MiRNA-loaded RISCs target mRNAs that contain at least partially complementary sequences to the miRNA (Fig. 1). Several methods have been applied to calculate the number of miRNA targets in different species. Target prediction algorithms integrate sequence complementary data, free energy values of the miRNA-mRNA complex, conservation of the target site between different species and the importance of the 5′ seed sequence (nucleotides 2 to 7 of the mature miRNA) for binding the target mRNA. However, these different approaches predict somewhat different sets of target genes. Most estimates suggest that about one third of all human genes could be targets for microRNA inhibition6 and that each microRNA family may target on average approximately 200 transcripts.7 The reliability of these predictions is still controversial.8 The latest astonishing and probably the most elaborate predictions go far beyond previous estimates, suggesting that there may be more than 25000 miRNAs in humans targeting more than 20000 mRNAs.9
Figure 1.
MicroRNA biogenesis. MiRNAs are mainly derived from RNA polymerase II transcribed genes. The primary transcript (pri-microRNA) is often polycistronic, containing several miRNas. In the nucleus, Drosha in conjunction with DRCG8 processes the pri-microRNAs into pre-microRNAs that are exported to the cytoplasm by Exportin5. In the cytoplasm, the mature miRNA is formed by the enzymatic activity of Dicer. The mature miRNAs is incorporated or “loaded” into the RISC (star shaped icon) that mediates miRNA-directed targeting of specific mRNAs.
The fate of the mRNA targeted by microRNAs remains less clear. Generally, these RISC-mRNA complexes end up in cellular substructures called P-bodies10 where the mRNA is either stored for later release under certain conditions, i.e., stress, or degraded. miRNAs exert post-transcriptional repression via two main mechanisms: degradation of the targeted mRNA or its storage in P-bodies. Deadenylation of the targeted mRNA is associated with its appearance in P-bodies. In any case, the mRNA is prevented from undergoing translation.
MicroRNAs and the Evolution of the 3′UTR in Eumetazoa
MicroRNAs have existed in animals for probably 600 million years and are present in almost all eumetazoa with the exception of the phylum porifera, which seems to be devoid of this class of small RNAs. The core set of eumetazoan microRNAs includes miR-10 and miR-100 (for nomenclature of miRNAs see Griffiths-Jones et al., ref. 11), which can be found in a variety of organisms from cnidaria to humans. There are another 18 ancient microRNAs that are shared by all deuterostomes and protostomes. From this point, there appears to have been a rapid enlargement of the microRNA pool during the evolution of several lineages, most prominently in vertebrates and again in eutherian mammals. There are even sets of microRNAs that appear to be entirely specific to humans.
The analysis of microRNAs can therefore be a powerful tool in phylogenetics. In this vein, miRNAs may also be helpful in clarifying the classification of certain animal groups. For example, planaria appear to be protostomes rather than deuterostomes (the two major subgroups of bilaterian animals) when analyzed by their microRNA repertoire. Many of the protostome-specific microRNA families, but not deuterostome-specific ones, have been found in planaria.12,13
There appears to be a strong correlation between the evolutionary appearance of new body plans and microRNA evolution. Because the target sequences and the sequences of the mature microRNAs are so small, new microRNAs may form de novo very easily and potentially contribute to the regulation of the expression patterns of a large set of genes. Consistent with this, there are indications that microRNAs have had a substantial impact on the evolution of the 3′UTR region of many genes. Large sets of genes seem either lack large 3′UTRs and/or microRNA target sites in their 3′UTR. These genes have been called anti-targets. However, for mRNAs that have target sites in their 3′UTR, these sites are frequently conserved between species indicating their biological significance. During evolution 3′UTRs have been surprisingly well conserved,14 particularly for transcription factor genes in vertebrates. This supports the observation made in Drosophila that microRNAs can drive the evolution of 3′UTRs of target genes.15
Interestingly, the average size of 3′UTRs has increased during evolution,16 concomitant with the expansion of the microRNA repertoire. In contrast, 5′UTR lengths is similar in fungi, plants, invertebrates and mammals.
microRNA Function: Conferring Robustness to Developmental Programs?
Intriguingly, miRNAs and their targets are often not coexpressed, but rather have inverse expression patterns, with microRNAs being expressed in tissue domains adjacent to cells that express target mRNAs. Analysis of microRNA and target gene expression in Drosophila supports the idea that microRNAs have evolved to confer robustness to the gene expression pattern established by transcription factors. In this view microRNAs function as suppressors of leaky unwanted transcripts and as inhibitors of severe undesirable fluctuations of gene expression. Leaky and unwanted transcripts may produce ambiguous signals, potentially perturbing execution of the developmental program. The more complex the developmental program becomes, the greater the potential contribution of microRNAs to its fidelity. However, Farh et al.17 have shown that microRNAs can also function during development by dampening the expression of target genes that are coexpressed with the microRNA (see also miR-278 in fat bodies of Drosophila, ref. 18). This mechanism may be important during differentiation and other processes requiring broad changes in expression that have to be accomplished quickly and with precision. Once we have a better picture of the total number of microRNAs in different phyla and their functions, we will be better able to address the question of whether and how microRNAs have contributed to the evolution of the different animal phyla, the diversification in certain animal groups (such as vertebrates and mammals) and whether the broadening of the influence of microRNAs on gene expression can influence the evolution of new body plans.
Erecting a Limes Against Unwanted mRNA Transcripts: microRNA Legions Securing the Outcome of Developmental Programs
The hypothesis that microRNAs may play an essential role in the process called canalization may help to explain one of the roles of microRNAs in animals. Canalization refers to a biological process conferring robustness and precision to gene expression patterns, a mechanism to “even out” deviations from the desired phenotype (reviewed by Hornstein and Shomron, ref. 19). MicroRNAs seem excellent tools for the execution of canalization. This hypothesis is based on several observations. One is that the phenotype of loss of microRNA function is far less dramatic than expected. For example, zebrafish development without the crucial mature microRNA-generating enzyme Dicer, and therefore without microRNA control over potentially thousands of genes, is surprisingly normal.20 All the major tissues, organs and cell lineages seem to form in Dicer mutant fish. However, tissue morphogenesis proceeds abnormally, leading to a deformed embryo. The authors conclude from their analysis that patterning of fish embryos in the absence of Dicer occurs properly but is followed by irregular morphogenesis in a variety of tissues. This is also in concordance with microRNA expression patterns in zebrafish.21 Most mature microRNAs are hardly detectable in early development and are strongly expressed at the end of organogenesis. This appears also to be true for mouse embryogenesis.22
This finding is in striking contrast to the restrictive model of messenger RNA gene expression during embryogenesis.23 Messenger RNA expression patterns become less complex during development, and this restriction is believed to go along or be responsible for the restricted potential of specialized, differentiated somatic cells and tissues. In contrast, microRNA expression patterns become more complex during mouse, human and zebrafish development, and each tissue seems to acquire a very distinct microRNA expression signature. This in turn may actually enhance the already restrictive mRNA expression pattern during development by further limiting mRNA expression domains. This can be interpreted as supporting evidence for a crucial function of microRNAs in the process of canalization. In our own studies, we have observed a phenomenon that represents the loss of canalization: lack of Dicer expression leads to the disruption of the regular scale pattern on the tail the Dicer mutant mice (see Fig. 3).
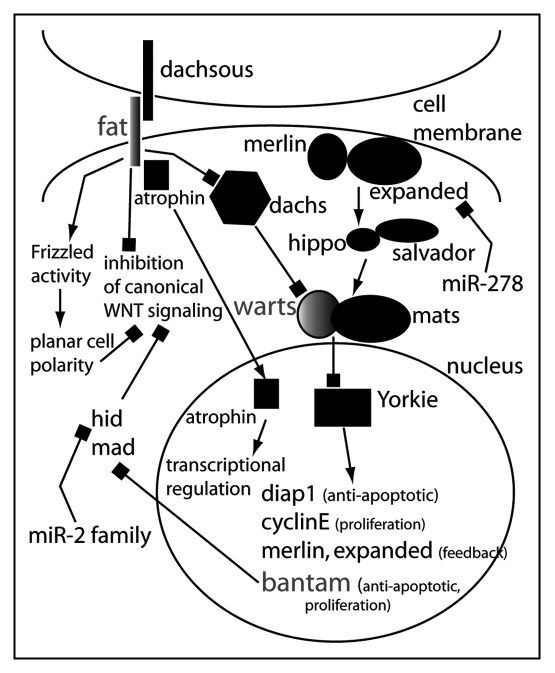
Figure 3.
Model for the role of microRNAs in the regulation of ‘tumor suppressor’ pathways in Drosophila. The miRNA bantam is a key target of the transcriptional regulator Yorkie. Yorkie is under tight control of the Hippo pathway that consists of merlin and expanded at the membrane integrating signals from an unknown transmembrane receptor. In certain cell types in Drosophila, expanded mRNA is regulated by miR-278. Warts and Mats transduce the signal from Merlin/Expanded and Hippo/Salvador. Warts and Mats are also elements of Fat/Dachsous signaling (another Drosophila tumor suppressor pathway) through Dachs. Fat also can interfere with canonical Wnt signaling and activate noncanonical Wnt signaling. This could theoretically occur via the inactivation of Yorkie and Bantam: one of Bantam's target mRNAs is Hid, which has been shown to inhibit canonical Wnt signaling.
Examples of microRNA Function: miR-1 in Heart and Muscle. One of the few miRNAs that has been well studied in vertebrates and flies, miR-1, has a very specific mesodermal and, consequently, muscle- and heart-specific expression pattern (reviewed by Nguyen and Frasch, ref. 24). As such, miR-1 may be a good candidate for mediating the muscle and heart phenotypes in zebrafish Dicer mutants. miR-1 gene expression can be induced by muscle-associated transcription factors SRF, MyoD, and Myogenin25 (Twist and Mef226 in myogenesis of Drosophila), and miR-1 can target the 3′UTR of HDAC4, a potent transcriptional repressor of muscle gene expression.27 This initiates or maintains the transition from myoblast to myotubes. Therefore, miR-1 plays an important role in myoblast differentiation.
Loss of function mutations of miR-1 have been studied in Drosophila; however, the results are somewhat ambiguous. Two groups have generated miR-1 mutant flies and both show that the majority of the mutants die at a similar stage.26,28 However, Kwon et al.28 describe a much more severe phenotype in a substantial subset of embryos. These embryos show an expansion of cardiac progenitor cell populations and the loss of differentiation into cardiac and muscle cells. miR-1 may have several functions in mesodermal cells during muscle differentiation: Suppression of unwanted transcripts, regulation of Notch/Delta signaling in early stages of cardiac and muscle cell differentiation, and regulation of muscle growth during larval stages. Altogether the results of Kwon et al.28 complement previous studies using antisense-mediated depletion of miR-1 in Drosophila.
Loss of Function Analyses of microRNAs in Drosophila Reveal Broad Roles for this Class of Regulators
In addition to miR-1, four other miRNA loss-of-function mutants in Drosophila have been analyzed. The Bantam miRNA controls apoptosis and proliferation in Drosophila, indicating that miRNAs can influence the development in animals, especially the determination of organ and body size. Bantam targets mRNAs of apoptosis-inducing genes and negative regulators of proliferation. One direct target appears to be hid, a pro-apoptotic gene.29 Bantam expression itself is regulated by the transcriptional coactivator Yorkie (Yki), a Drosophila homologue of the vertebrate yes-associated protein, Yap.30,31 Bantam is thus an integral component of the Hippo tumor suppressor pathway and may also contribute to other tumor suppressor pathways.32,33 The main target of the Hippo pathway appears to be Yki. Activation of the pathway leads to inactivation of Yki and consequently the suppression of proliferation and activation of apoptosis. Yap has been shown to have oncogenic activity, at least in mammary cells.34 Yap also appears a key element of the 11q22 amplicon found in several types of tumors.35 However, there has been no connection made between Yap and microRNAs in vertebrates so far. Moreover, Bantam, a target of Yki in Drosophila, can only be found in protostomes. Although the Hippo pathway is well established in Drosophila, its existence and regulation by microRNAs in deuterostomes is uncertain (for an overview of the Drosophila Hippo pathway and its interactions with miRNAs see Fig. 2).
Figure 2.
Disruption of regular scale pattern on tails of mice with epidermal specific loss of Dicer. Arrows indicate the regular lines separating scales on the tail of a control mouse (upper tail with hair). However, in mice with loss of Dicer function in the epidermis this regular pattern is destroyed (lower, darker and hairless tail).
Similar to bantam, the loss of function of miR-14 in Drosophila also seems to involve the disturbance of apoptosis control resulting in reduced viability of the organism.36 However, the exact molecular mechanisms by which miR-14 exerts its function remain unclear. Interestingly, several apoptotic genes contain miR-14 target sites and one of them, Drice, shows elevated expression in miR-14 mutant flies.
MiR-278 is highly expressed in the fat body of flies. The loss of miR-278 in Drosophila results in lean flies while gain-of-function of this microRNA causes tissue overgrowth.18 The loss of miR-278 is accompanied by elevated insulin signaling, insulin resistance of the fat body, and aberrant expression of the gene expanded in the fat body. Interestingly, expanded is a member of the ezrin/radixin/moesin family and is part of the Hippo signaling pathway. miR-278 and bantam therefore seem to be involved in the regulation of the same pathway. While bantam is activated by Yorkie, miR-278 seems to be able to regulate an upstream component of the pathway, i.e., expanded. However, it seems unlikely that bantam and miR-278 are involved in Hippo signaling in the same cell since no reports mention overlapping expression patterns.
An example that demonstrates how the differential expression of a miRNA in neighboring cells directs the fate of the cells comes from miR-9a mutant flies. Li et al37 generated miR-9a mutant and overexpressing flies. MiR-9a is associated with the development of the nervous system. The authors show that miR-9a is important in the peripheral nervous system as well in establishing the correct pattern of sensory organ precursors (SOPs). Without miR-9a, ectopic SOPs form and overexpression of miR-9a leads to the loss of SOPs. MiR-9a achieves this mainly by regulating the transcription factor senseless. High levels of senseless activate proneural genes and SOP fate while low levels suppress proneural genes. Differential expression of miR-9a in SOPs and adjacent cells is one of the supportive factors that help to define accurately which cell becomes a SOP. MiR-9a therefore can be regarded as an example of the “classical” function of miRNAs: fine-tuning gene expression and conferring robustness to the gene regulatory network (reviewed by Cohen et al., ref. 38).
Dicer as a Tool to Study microRNA Function on a Larger Scale
Loss of Dicer is an alternative and important tool in studying microRNA function. There is most likely only one Dicer gene in zebrafish, mice and humans; thus, targeting this gene has been a productive way to elucidate microRNA function in vertebrate development. Although the loss of Dicer in zebrafish correlates with the idea that microRNAs mediate canalization, the findings that mouse embryos without Dicer do not survive beyond embryonic day E7.5 is harder to explain.39 One possibility is that microRNA function is different in mice and fish during early embryogenesis and that microRNAs in mice have essential functions in stem cell maintenance. An alternative explanation is that Dicer has functions in addition to generating mature microRNAs: In several organisms, including those lacking microRNAs, Dicer is required for heterochromatin formation and centromeric silencing using siRNAs. In mouse embryonic stem cells (ES cells) the same function of Dicer has been demonstrated.40 Furthermore, Dicer mutant mouse ES cells cannot differentiate in vitro into embryonic bodies that show differentiation into the three germ layers. However, both papers describing ES cells with conditional alleles of Dicer differ on an important point: viability of Dicer mutant ES cells appears to be severely compromised in one study41 while the second paper by Kanellopoulou et al.40 does not report this phenomenon. This striking difference remains to be explained.
The availability of conditional alleles of the Dicer gene in mice has allowed for analysis of Dicer and microRNA function in specific organs. Dicer has been targeted in T cells,42,43 the limb mesenchyme,44 the lung,45 and the epidermis.46,47 In most of these experiments loss of Dicer was accompanied by strong induction of apoptosis. The effects of loss of Dicer were relatively mild, similar to observations made in Dicer mutant zebrafish, while complete and timely elimination of Dicer and hence of mature microRNAs may not have been achieved in all of these experiments. Nevertheless, these experiments show that Dicer and consequently microRNAs are important for the proper formation and function of many mammalian tissues (see Table 1).
Table 1.
Phenotypes of mice with a tissue-specific loss of Dicer
| Tissue | Cre | Phenotype | Literature |
| Lung | Shh-cre (lung epithelium) | Branching defect, Fgf10 dysregulation, apoptosis induction. | Harris et al.45 |
| Limb | Prx1-cre (limb bud mesenchyme) | Apoptosis | Harfe et al.44 |
| Skin | K14-cre (epidermis and other squamous epithelia and their adnexa) | Apoptosis, loss of hair follicle stem cells, disturbance of epithelial-mesenchymal interactions | Andl et al.47 |
| Skin | K14-cre (epidermis and other squamous epithelia and their adnexa) | Apoptosis, disturbance of epithelial-mesenchymal interactions | Yi et al.46 |
| Immune system | CD4-cre (T cells) | CD8+ T cells development blocked; CD4+ T cells show increased apoptosis upon stimulation | Muljo et al.42 |
| Immune system | Lck-cre (early stages of T cell development) | thymocyte cell number reduced, few peripheral T cells | Cobb et al.43 |
From the phylogenetic microRNA expression data and data of Dicer mutant mice and fish, a general concept of microRNA function has emerged. MicroRNAs do not seem to play a major role in early development with regard to cell fate specification or patterning of the embryo, at least in zebrafish. Rather, microRNAs are developmental perfectionists, meaning that one of their main functions seems to be to make sure that genetic programs are executed with precision, and they enforce the correct expression pattern of a broad set of genes. Therefore, one current hypothesis of a major function of microRNAs is that they make sure development proceeds smoothly resulting in a near utopian result.
microRNAs and Cancer: Another Magic Bullet?
On the other hand, there is always potential trouble in paradise. microRNAs themselves can contribute to pathological disturbances, and this has already been well documented in tumorigenesis. A major theme that emerges from analysis of Dicer mutant mice (especially the tissue-specific ones) and flies is a role for microRNAs in apoptosis and differentiation. These are critical processes that can inhibit uncontrolled growth of cells. The process of apoptosis is highly conserved in animals and is used to eliminate unwanted cells in a highly controlled fashion. However, malfunction of apoptosis has been implicated in many human diseases including cancer, auto-immune diseases, and neurodegenerative diseases. microRNAs have been identified as regulators of apoptosis, and therefore they have become potential players in human diseases associated with defective apoptotic regulation.
Evidence for an anti-apoptotic function of microRNAs in cancer comes from analysis of B-cell lymphomas. The chromosomal region 13q31 has been implicated in tumorigenesis, including the tumorigenesis of B-cells. The 13q31 chromosomal region frequently shows amplifications in several tumor types, and one gene, c13orf25, was eventually identified as the potential culprit.50 However, this gene actually contains a cluster of microRNAs, the mir17-92 cluster (mir-17, mir-18, mir-19a, mir-20, mir-19b and mir-92). The expression of these microRNAs is indeed up-regulated in B-cell lymphomas.51 In an animal model of B-cell lymphoma, over-expression of microRNAs of the mir17-92 cluster dramatically enhanced c-myc induced lymphomagenesis. The onset of disease was accelerated and accompanied by a substantial suppression of apoptosis in the lymphomas.51 In this tumor model, c-myc expression alone leads to the formation of tumors that are plagued by high levels of apoptosis. Expression of high levels of microRNAs of the mir17-92 cluster suppress this apoptosis. O'Donnell et al52 showed that c-Myc itself appears to be a key regulator of c13orf25 and therefore of the expression of the mir17-92 cluster. It was also shown by this group that one of the target genes of two of the microRNAs of the cluster was the cell cycle regulator E2F1. Mis-expression of E2F1 can lead to enhanced apoptosis and therefore suppressing E2F1 via microRNAs could lead to a more favorable growth versus apoptosis ratio in c-myc induced lymphomas. There may be additional crucial target genes of the mir17-92 cluster since loss of E2f1 does not enhance c-myc induced lymphomagenesis and does not abrogate excessive apoptosis in these tumors. Rather, loss of E2f1 leads to a slowdown in proliferation via enhanced expression of the CDK-inhibitor p27kip1.53 Perhaps the regulatory circuit between c-Myc, E2F1 and the mir17-92 cluster has to be controlled very delicately in order to achieve a balanced result, and in B-cell lymphoma over-expression of c-myc and the mir-17-92 cluster may tip the balance towards growth without triggering extensive apoptosis. Interestingly, in Drosophila, dacapo is a major target of microRNA regulation during proliferation of stem cells. Dacapo is the major CDK inhibitor and G1/S regulator in Drosophila (similar to p21 and p27 in mammals). Loss of Dicer-1 in Drosophila leads to a defect in G1/S transition and decreased proliferation due to increased levels of Dacapo.54 Not surprisingly, potential targets of the mir17-92 cluster include cancer-associated genes, especially ones that are involved in the regulation of G1/S transition of the cell cycle, such as cyclin D1, cyclin D2, cdk6, p21, p57, E2f1 and Rb family members.55
One recent paper goes even further in defining the role of microRNAs in cancer development. The mir17-92 cluster was investigated, this time in a mouse model of colon carcinogenesis, where c-myc was used to trigger tumorigenic growth in k-ras and p53 mutant colonocytes. Without c-myc, tumor growth is slow and lacks strong angiogenic potential. However, c-myc can overcome this lack of neo-vascularization through microRNA-induced suppression of the anti-angiogenic genes Tsp1 and Ctgf.56 Therefore, microRNAs not only can contribute to tumorigenesis via modulation of proliferation and cell death, but also by acting non-cell autonomously and enhancing angiogenesis. These papers, analyzing the relationship between c-Myc and microRNAs, have greatly extended our comprehension of how microRNAs can act as powerful oncogenes and have opened a new door for therapeutic interventions.
Other known oncogenic microRNAs are miR-372 and miR-373.57 They cooperate with oncogenic ras to transform primary fibroblasts. Lats2/Kpm—the mammalian homologue of the tumor suppressor like gene lats/warts (a component of the Drosophila Hippo tumor suppressor pathway)—is directly inhibited by miR-372 and miR-373. Voorhoeve et al.57 also presented evidence that this plays a role in human testicular germ cell tumors.
A New Class of Tumor Suppressor Genes from the Realm of microRNAs?
microRNAs have not only been implicated in tumorigenesis as oncogenes. The chromosomal region 13q14 is commonly deleted in many tumors including B-cell chronic lymphocytic leukemia (CLL). Absence of protein-coding genes associated with this deletion, was explained with the realization that the actual target of this chromosomal loss is a microRNA cluster. Mir-15a and mir-16-1 are contained in the minimal deleted region at 13q14.58 CLLs exhibit a reduction in miR-15a and miR-16-1 gene expression and the level of these microRNAs is inversely correlated with BCL2 protein levels.59 bcl2 could be identified as a target gene and its modulation by miR-15a and miR-16-1 was associated with sensitivity to apoptosis. Another miRNA with potential tumor suppressor activity may be miR-17-5p. This miRNA is able to regulate Ncoa3 (also Aib1 or Src3), an oncogene associated with the chromosome 20q12 amplicon in breast and other carcinomas.60
It must have come as a big surprise that Ras GTPases are also targets of microRNAs. The microRNAs involved in post-transcriptional regulation of Ras expression are let-7 family members. Loss of let-7 leads to failure in terminal differentiation of certain cells in C. elegans, although these cells continue to proliferate.61 This is surprisingly similar to changes in tumor cells. Johnson et al. have shown the C. elegans ras homologue let-60 has functional let-7 target sites in its 3′UTR and let-7 microRNAs can regulate Ras expression in C. elegans and in human cells in vitro. Let-7 also can reduce the tumorigenic potential of lung cancer cells in vitro as measured by a colony-forming assay, and reduced let-7 expression is associated with poor prognosis in lung cancer patients.62 Taken together, these results give strong evidence for a tumor suppressor activity of let-7 family members, and this is at least partially mediated by suppressing Ras activity. This also is further evidence for the notion that certain signaling pathways, in particular ras signaling, may actually be regulated and mediated not so much on the transcriptional level.63,64 Rajasekhar et al.64 found that Ras signaling has only a very modest immediate impact on the overall change in transcription. However, analysis of changes in the polysomal pool of mRNAs (the mRNAs that are actually translated into protein) show much more dramatic results, indicating that ras signaling mediates cellular changes via control of translation of mRNAs.
Mechanisms of microRNA Regulation in Cancer
Although there is good evidence that certain microRNAs function as oncogenes and others as tumor suppressor genes, there are other, more generalized levels of microRNA regulation in tumor cells as well. microRNA expression in tumors shows broad alterations compared to normal tissue: in general, microRNA levels are reduced in tumors.65 One conceptual explanation for this is the idea that microRNAs are associated with a more differentiated status, predicting that each step to a less differentiated cell type should result in a loss of microRNA diversity and expression levels. microRNA expression and activity can be regulated at multiple steps. Recently, it was shown by Thomson et al.4 that processing of microRNAs is negatively affected at the Drosha step not only during early mouse embryogenesis but also in tumors. This adds another layer of complexity to microRNA regulation in cancer in addition to chromosomal abnormalities in microRNA genes,66,67 transcriptional activation (mir-17-92 cluster by c-Myc) and potentially epigenetic silencing.68
Dicer may also contribute to the centromeric silencing via the RNAi pathway. Defective centromeric chromatin structure can result in aneuploidy. It is unclear whether loss of Dicer also can contribute to chromosomal instability and consequently to tumor formation. This, however, would be independent of microRNAs.
It appears that microRNAs have perplexed our somewhat over-confident assumption that we have made considerable progress in understanding cancer. Considering the number of patients dying from cancer each year in the US alone (about 500000), and the limited tools available to fight this disease intelligently, there is a lot of room for improvement. For example, microRNA expression profiles using just 217 microRNAs are superior to large mRNA expression profiles with about 16000 genes in distinguishing normal cells from tumor ones, and especially in characterization of undifferentiated tumors of unknown origin.69
Our rapidly expanding knowledge of miRNA function during tumorigenesis may indeed lead to novel approaches to cure cancer. It is no accident that RNAi has been suggested as a powerful therapeutic tool. However, now miRNAs themselves are implicated in the tumorigenic disease process, providing the opportunity to beat cancer with its own weapons. It remains to be seen, whether we will be able to mold the appropriate tools for this purpose and whether there will be any magic to them.
However, much can be learned in the field of cancer research from how microRNAs and transcription factors coordinate the expression of the genome during embryogenesis. Embryogenesis is like a symphony with many reoccurring themes and melodies, in which miRNAs function as metronomes to guide the orchestra of proteins to execute the score precisely according to the composer's ideas (that is evolution). In contrast, cancer represents a cacophony with misguided instruments directing and distorting the original score.
Surprising New Insights into How Our Adult Life is Managed by microRNAs
microRNAs appear to be important beyond embryogenesis and tumorigenesis. For example, if we consult the expression pattern of miR-1, it becomes obvious that miR-1 is much more highly expressed in the adult muscle and heart compared to embryonic and neonatal tissues.27 Similar observations have been made in zebrafish using microarrays21 and in mammals.22,70 Many microRNAs show strong expression in the adult fish and their expression appears to be tissue-specific for the most part. Several reports show that animals have included microRNAs in many complex processes of the adult organism from the regulation of endocrine function to the regulation of life span. For example, the previously discussed miRNAs miR-14 and mir-278 have clear implications in the regulation of the fat metabolism in the adult fruit fly. In mammals, miR-122, a microRNA highly expressed in the liver, is involved in the regulation of plasma cholesterol levels in mice.71 Furthermore, mir-375 is a pancreatic islet-specific microRNA that is involved in the regulation of insulin secretion probably via inhibition of myotrophin.72 Even the interference of microRNAs with amino acid catabolism has been demonstrated.73 A hint for the essential function of the microRNA machinery in adulthood comes also from a study on the toxicity of high levels of short hairpin RNA in the liver. The toxicity appeared to be due to an overload of the microRNA processing system and resulted in the death of animals within two months.74 The effects of microRNAs on metabolic regulation are more thoroughly reviewed by Krutzfeld and Stoffel.75
Lecellier et al. found that miR-32 acts as an anti-viral microRNA,76 and Drosophila Dicer-2 has been shown to be important for innate immunity against viruses.77,78
Viruses themselves have developed mechanisms to suppress the mircroRNA machinery that targets them.79 Even more intriguing, certain DNA viruses have adapted the microRNA idea and introduced novel microRNAs in their genomes80,81 (for an overview of the interplay between viruses and miRNAs see Sarnow et al., ref. 82).
Even the response to stress on the cellular level involves changes in microRNA mediated post-transcriptional control. Bhattacharyya et al.83 have shown that mRNAs targeted by miR-122 can be redirected from the P-body to polysomes after stress.
Further examples of the importance of miRNA function in human disease come from studies on the fragile X syndrome gene Fmr1 (fragile X mental retardation gene 1). The protein derived from the Fmr1 gene is a RNA binding protein that can inhibit translation of certain mRNAs. FMR1 can recruit RISC components like argonaute proteins, Dicer and microRNAs, and facilitate contact with the target mRNAs. Through FMR1, microRNAs and/or other small RNAs may impact brain neuronal plasticity, learning, and behavior. Furthermore, FMR1 has been implicated in the control of the circadian rhythm, suggesting a possible involvement of microRNAs of the biological clock.84–86 Table 2 gives an overview of the potential involvement of the microRNA machinery in human genetic diseases.
Table 2.
The MicroRNA machinery and its association with human diseases
| Disease | Gene | Mechanism | Literature |
| Fragile X Syndrome | Fmr1 | recruits RISCs to specific mRNAs | Caudy et al.84; Jin et al.86 |
| Spinal muscular atrophy | Gemini3 and 4 | can bind to argonaute and associated with miRNAs | Dostie et al.92 |
| DiGeorge syndrome | Dgcr8 | partner of Drosha | Landthaler et al.93 |
| Tourette's disease | Slitrk1 | changes in miR-189 binding site of Slitrk1 gene | Abelson et al.94 |
microRNAs in Therapy: New Miracles or Just a Mirage?
In the near future miRNAs will probably have been implicated in the control of almost all cellular processes. Their involvement in proliferation, apoptosis and metabolic control have already made them potential targets for the therapy of cancer and diabetes. Exciting results have been obtained in mice using “antagomirs”, cholesterol-conjugated stabilized antisense microRNAs, which can be directed very specifically against a certain microRNA and inhibit its function.71 The microRNA field can also profit from many years of experience from work with antisense technology. Several stabilized forms of oligonucleotides have been tested successfully (LNA, PNA, morpholino). However, it appears that there is still a lot of work to be done to better understand the world of small RNAs and how they can be used to cure human diseases.
Gene Regulation: Beyond Transcription and Translation
With the realization that microRNA-mediated post-transcriptional gene expression regulation is a conserved process in almost all eumetazoa, a shift from the primary focus on transcriptional regulation to a broader more holistic view of how the expression of genes is regulated, appears possible. Additionally, our obsession with protein coding genes may eventually end since recent studies also have shown that perhaps half of the genome is actually transcribed and the vast majority of it seems not to be translated into proteins.87,88 Importantly, it was recently shown that a noncoding RNA gene has evolved dramatically during human evolution and may have had a considerable impact on human development.89 MicroRNAs have contributed substantially to this view of the genome and renewed interest in RNA biology. Our understanding of microRNA function has truly come a long way since their formal discovery in 199390,91 and undoubtedly these small molecules are likely to be at the crux of our future understanding of many human diseases and the development of novel therapeutic strategies.
Acknowledgements
I am very grateful for the support by Dr. Sarah E. Millar and for her helpful comments. Furthermore, I would like to thank Omar Harb, Candace M. Moore, Emily Chu and Peggy Myung for their comments and encouragement.
Footnotes
Previously published online as an Organogenesis E-publication: http://www.landesbioscience.com/journals/organogenesis/abstract.php?id=3670
The limes was the line of demarcation or border line of the Roman Empire in the 2nd century A.D. (see also www.limes-in-deutschland.de/limes_english.html).
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 4.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 9.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Rivas FV, Wohlschlegel J, Yates JR, IIIrd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: A model system for stem cell biology. Rna. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol. 2006 doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 14.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 16.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 17.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 18.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 20.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 21.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 23.Strauss WM, Chen C, Lee CT, Ridzon D. Nonrestrictive developmental regulation of microRNA gene expression. Mamm Genome. 2006;17:833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen HT, Frasch M. MicroRNAs in muscle differentiation: Lessons from Drosophila and beyond. Curr Opin Genet Dev. 2006;16:533–539. doi: 10.1016/j.gde.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 30.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 31.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 33.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006 doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding-a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 40.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 47.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 49.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 50.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 51.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 53.Baudino TA, Maclean KH, Brennan J, Parganas E, Yang C, Aslanian A, Lees JA, Sherr CJ, Roussel MF, Cleveland JL. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11:905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 54.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 55.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 58.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 62.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 63.Lasko P. Ribosomes rule: Translation, not transcription, is the primary target of two major intercellular signaling pathways. Dev Cell. 2003;5:671–672. doi: 10.1016/s1534-5807(03)00333-2. [DOI] [PubMed] [Google Scholar]
- 64.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 65.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 66.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 69.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 70.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 72.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 73.Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum Mol Genet. 2005;14:3371–3377. doi: 10.1093/hmg/ddi368. [DOI] [PubMed] [Google Scholar]
- 74.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 75.Krutzfeldt J, Stoffel M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 76.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 77.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 78.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 81.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 82.Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: Expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- 83.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 84.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 86.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 87.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 88.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 89.Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 90.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 91.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 92.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. Rna. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 94.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, Davis NR, Ercan-Sencicek AG, Guez DH, Spertus JA, Leckman JF, Dure LSt, Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]