Abstract
Indirect immunofluorescence (IF) microscopy is one of the most frequently employed methods to determine intracellular protein localization in yeast. It is especially useful for low abundance proteins, eg., the GATA-factors (Gln3, Gat1) which activate NCR-sensitive transcription. Limiting the nitrogen supply or treating cells with the Tor pathway inhibitor, rapamycin, elicits nuclear GATA-factor localization and increased NCR-sensitive transcription, whereas excess nitrogen restricts these proteins to the cytoplasm and decreases transcription. The initial step of the IF procedure is formalin-fixation that quenches cellular activity and fixes protein locations via cross-linking. Indeed, it is on the success of this immobilization step that the assay depends. We have found that under some conditions, formalin itself can influence GATA-factor localization. With low concentrations of formalin (1.6%), Gat1-Myc13 became more nuclear, and with higher concentrations (5.6%), it became more cytoplasmic. Gln3-Myc13 localization, on the other hand, did not respond to low formalin, but became more cytoplasmic at the higher concentration. Interestingly, the high concentration of formalin had no demonstrable effect when the GATA-factors were completely nuclear, i.e., after rapamycin-(Gat1-Myc13) or Msx-(Gln3-Myc13) treatment. Our data indicate that these effects are most likely elicited by methylene and polyoxymethylene glycols, which account for more than 99% of the formaldehyde in formalin. These compounds greatly increased the osmolarity of the medium (0.5–2) and leads us to suggest that varying degrees of osmotic stress, to which both Gln3 and Gat1 are known to respond, and protein movement in response to it can occur after the beginning of fixation but before proteins become immobilized. Therefore, precautions are required to minimize the problem if possible or to account for it during interpretation of IF or other experimental data derived from cells treated with formalin when it cannot be avoided.
INTRODUCTION
Indirect immunofluorescence (IF) microscopy is one of the most often used approaches to investigate protein localization in Saccharomyces cerevisiae. In fact, much of our understanding of intracellular protein localization as a principle mode of transcriptional regulation derives from our ability to track tagged proteins (Kaiser et al., 1994; Mondoux et al., 2007; Sbia et al., 2008). Chromatin immunoprecipitation (ChIP) and Chip-ChIP analyses of transcription factor binding to DNA are second and third generation extensions of the basic method (Aparicio et al., 2004; Dasgupta & Chellappan, 2007; Southall & Brand, 2007; Collas & Dahl, 2008; Gregory et al., 2008,). The IF approach rests on the assumption that the initial step in the procedure, i.e., formalin fixation, quenches cellular activity and freezes the proteome in place by chemical cross-linking. If an environmental perturbation altered a protein’s localization before it was immobilized and the effect went unnoticed, erroneous interpretation of the results could ensue.
Our interest in indirect immunofluorescence derives from its application to the study of nitrogen catabolite repression (NCR) sensitive transcription factors and participation of the Tor signal transduction pathway in their regulation. When a cell encounters a rich nitrogen supply, transcription of NCR-sensitive genes is repressed (Hofman-Bang, 1999; Cooper, 2002; Magasanik & Kaiser, 2002). At the other end of the spectrum, limiting or starving environmental conditions elicit increased transcription of NCR-sensitive genes in preparation for the cell to identify and exploit alternative poorer nitrogen sources (Hofman-Bang, 1999; Cooper, 2002; Magasanik & Kaiser, 2002). This finely tuned, rapid response to the environment is regulated in part by controlling the intracellular localization of the GATA-factors that activate NCR-sensitive transcription, Gln3 and Gat1/Nil1 (Beck & Hall, 1999; Cardenas et al., 1999; Hardwick et al., 1999; Bertram et al., 2000; Cox et al., 2000; Cunningham et al., 2000; Carvalho et al., 2001; Cox et al., 2002; Crespo et al., 2002; Cox et al., 2004; Cox et al., 2004b; Tate et al. 2006; Tate et al., 2005; Kulkarni et al., 2006; Tate & Cooper, 2007; Georis et al., 2008;). The GATA-factors are restricted to the cytoplasm in excess nitrogen and move to the nucleus as the nitrogen supply dwindles or cells are treated with inhibitors of Tor (rapamycin) or in the case of Gln3, with the glutamine synthetase inhibitor methionine sulfoximine, Msx (Crespo et al., 2002; Tate et al., 2005; Kulkarni et al, 2006).
Tor regulation of Gln3 and Gat1 bifurcates at the level of their localization (Kulkarni et al. 2006; Georis et al. 2008). Gln3-Myc13 sequestration in the cytoplasm requires the negative NCR-sensitive transcriptional regulator, Ure2 (Blinder et al., 1996; Beck & Hall, 1999; Hardwick et al., 1999; Bertram et al., 2000; Shamji et al., 2000), whereas nuclear Gln3-Myc13 localization in some but not all strains provided with a poor nitrogen source requires the Tor pathway type 2A-related phosphatase, Sit4 (Beck & Hall, 1999; Bertram et al., 2000; Georis et al., 2008). In contrast, the requirements of Sit4 and Ure2 for Gat1 localization are modest to minimal (Georis et al., 2008).
During the above studies, we noticed that the variability of measurements for Gat1-Myc13 was occasionally somewhat greater than with Gln3-Myc13 (Georis et al., 2008). This prompted us to carefully track the variation, even though it was not compromising interpretation of our experiments. As data in this work will show, this limited variability led to the demonstration that formalin is able, in some conditions, to markedly alter the intracellular location of a transcription factor before it is immobilized.
MATERIALS AND METHODS
Strains and Culture Conditions
Saccharomyces cerevisiae strains used in this work were TB123 (MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX]), JK9-3da (MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa), FV063 (MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3]), and FV034 (MATα, GAT1-MYC13[HIS3], ura3, his3, trp1). FV034 is in the Σ1278b genetic background, while the others are in the TB123 background. Plasmids pRS416-GLN3-GFP and pRS416-GAT1-GFP in vector pRS416-GFP were those reported earlier by Butow and colleagues (Liu et al., 2003; Giannattasio et al., 2005). These plasmids were used to transform JK9-3da using standard methods. Cultures (50 ml) were grown to mid-log phase (A600 nm = 0.5) in YNB minimal medium containing 0.1% glutamine, ammonia or proline as sole nitrogen source and appropriate supplements (120 μg/ml leucine, 20 μg/ml uracil, 20 μg/ml histidine, tryptophan and arginine). Where indicated, cells were treated with 200 ngm/ml rapamycin for 20 min or 2 mM methionine sulfoximine (Msx) for 30 min as described earlier (Tate et al., 2006). Transferring cells from one medium to another was performed as described (Tate et al., 2005; Tate et al., 2006).
Indirect Immunofluorescence Microscopy
Cell collection and immunofluorescent staining were performed as previously described by (Cox et al., 2002; Tate et al., 2006; Tate et al., 2006b; Georis et al., 2008). Unless indicated otherwise, cells were fixed by addition of 1/10 volume (0.5 ml) of 1 M phosphate buffer (pH 6.5) and 1/10 volume (0.5 ml) 37% formalin to a 5 ml aliquot of the culture to be assayed. This was followed by incubation for 60–70 min at 30° C. This range of fixation times was used in the past (until late in the present work) rather than a precisely timed period because of: (i) literature indicating that 60 min fixation times were adequate (Kaiser et al., 1994), and (ii) our finding when we first began using this assay that Gln3-Myc13 localization was the same following fixation for one vs. two hr (J.J. Tate and T.G. Cooper, unpublished observations). An exception to this fixation time was required when cells were treated with rapamycin or Msx. Due to the time associated with processing samples, untreated control samples were permitted to fix 20 or 30 min longer than those treated with rapamycin and methionine sulfoximine, respectively.
Formalin concentrations shown in present figures are calculated concentrations taking into account the volumes of the formalin and buffer additions. The approximate final formalin concentrations following addition of 1/10, 1/5, 1/20, and 1/40 of the culture vol of formalin (Fisher, 37% w/w) were 3.1%, 5.6% 1.6%, and 0.8%, respectively. These values as well as the formaldehyde concentrations and osmolarities calculated below are limited by uncertainties in: (i) the amount of methanol preservative added to commercial formalin solutions (10–15%; whether concentration was indicated in terms of w/w or v/v was not indicated), (ii) the degree of methylene glycol polymerization and (iii) age-dependent changes in formalin component concentrations due to losses into the atmosphere. Para-formaldehyde additions were made from a 16% (w/v), methanol-free solution (Electron Microscopy Sciences) and were present in the cultures at the final concentrations shown in the figures.
Formalin fixed and untreated (when pGln3-GFP and pGat1-GFP transformants were used) cells were imaged using a Zeiss Axioplan 2 imaging microscope with a 100x Plan-Apochromat 1.40 oil objective at room temperature. Images were acquired (at room temperature) using a Zeiss Axio camera and AxioVision 3.0 (Zeiss) software, and processed with Adobe Photoshop and Illustrator programs. Gamma settings were altered where necessary to avoid any change or loss in cellular detail relative to what was observed in the microscope; changes were applied uniformly to the image presented.
Determination Of Intracellular Gln3-Myc13 and Gat1-Myc13 Localization
To provide more representative and complete descriptions of the Gln3-Myc13 and Gat1-Myc13 behaviors we observed than is possible from an isolated image, we manually scored Gln3-Myc13 or Gat1-Myc13 localization in 200 or more cells in multiple, randomly chosen microscopic fields from which each image presented was taken. Cells were classified into one of three categories: cytoplasmic (cytoplasmic fluorescence only), nuclear-cytoplasmic (fluorescence appeared in the cytoplasm as well as co-localizing with DAPI-positive material), and nuclear (co-localizing only with DAPI-positive material). The reproducibility of cell scoring and limitations recommended when interpreting data obtained from it have been described in great detail earlier (Tate et al., 2006; Tate et al., 2007; Georis et al., 2008). Only relatively minor differences were observed between the TB123 and Σ1278b genetic backgrounds. Data from both backgrounds have been included in different portions of the present work to illustrate this fact.
GFP fluorescence was used to visualize Gln3 and Gat1 in some of the experiments. Although we parsed Gat1-GFP localization into three categories, data presented in the histograms are qualitatively less meaningful than those obtained by indirect immunofluorescence with Myc13 labelling due to the high background fluorescence that occurs unless Gat1-GFP is highly nuclear. In the case of Gln3-GFP, the fluorescence signal was sufficiently weak and background fluorescence sufficiently intense that scoring had to be confined to cytoplasmic vs. nuclear-cytoplasmic localization.
RESULTS
Formalin influence on the intracellular distribution of GATA-transcription factors
During investigation of the type 2A-related phosphatase (Sit4) and Ure2 requirements for intracellular Gat1-Myc13 localization (Georis et al., 2008), we encountered a somewhat greater variation than we had seen in previous studies with Gln3-Myc13. This variation occurred most often when cells were grown in YNB-glutamine medium where Gat1-Myc13 appears in both the cytoplasmic and nuclear-cytoplasmic categories. In contrast, little variation was observed in cells treated with rapamycin where Gat1-Myc13 is nuclear in essentially all cells. As data accumulated, we began to suspect the variation might not be random but possibly derived from an unknown biological response. Careful regulation of growth conditions eliminated this variable and suggested that some component encountered during preparation of the cells for indirect immunofluorescence microscopy was perturbing transcription factor localization. The simplicity of the immunofluorescence assay and reagents involved made it difficult, with one exception, to envision what the perturbant might be. The possible exception was formalin, the reagent used to quench cellular processes and fix the morphology of the cell.
Formalin is a 37% aqueous solution of formaldehyde (w/w) in which greater than 99.9% of the formaldehyde is hydrated (reaction 1) (Chau & Mok, 1992; Schecker & Schulz, 1969; Winkelman et al., 2002), i.e., it is methylene glycol, or low molecular weight polyoxymethylene glycol polymers (n = ~2–10, reaction 2) (Winkelman et al., 2002). Over time and repeated exposure to the atmosphere, the formaldehyde in formalin can also escape as a gas (reaction 3) (Bell & Evans, 1996) or further polymerize to form high molecular weight insoluble polymers in which n can be as high as 100.
| (1) |
| (2) |
| (3) |
Loss of formaldehyde, the active component of the fixative, by either or both of the above processes would decrease its concentration during fixation. Also, any evaporation of the medium during overnight growth of small cultures (5 ml) that are broadly used in protein localization experiments would increase the final formaldehyde concentration. Formaldehyde has not been previously thought to exert any influence on protein localization. However, the importance of unaccounted consequences if it did prompted us to investigate varying formalin concentrations on the intracellular localization of Gat1-Myc13 and Gln3-Myc13.
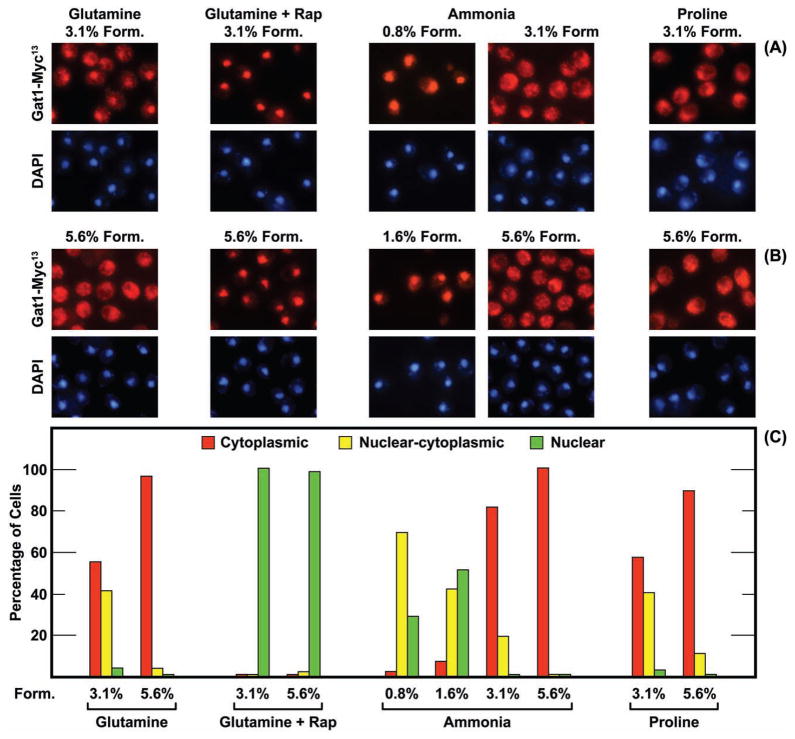
Much to our surprise, variation in formalin concentration could alter the intracellular Gat1-Myc13 localization (Fig. 1). Gat1-Myc13 was nuclear-cytoplasmic in many more glutamine- or proline-grown cells treated with 3.1% formalin than with 5.6% (the actual formalin concentrations when 1/10 or 1/5 vol of formalin is added along with buffer to a 5 ml aliquot of a culture). A more modest effect occurred when ammonia was provided as nitrogen source (Fig. 1), perhaps because Gat1-Myc13 was already cytoplasmic in most ammonia-grown cells treated with 3.1% formalin. In contrast, Gat1-Myc13 was more nuclear and nuclear-cytoplasmic in cells treated with low concentrations (0.8% and 1.6%) of formalin (Fig. 1, ammonia). Parallel behavior was observed in glutamine-grown cells (data not shown).
FIG. 1.
Intracellular Gat1-Myc13 localization is influenced by formalin treatment. Wild-type strain FV063 was grown in YNB-medium containing the nitrogen source indicated (final concentration 0.1%). Cells treated with rapamycin (for 20 min prior to cell harvest) are indicated as +Rap, and the formalin concentration appears above the images or below the histograms as percentages (Materials and Methods). Histograms and corresponding images for each condition were derived from the same slide. Images appear in pairs with Gat1-Myc13 (red fluorescence) shown in the upper panel and images of DNA positive material (DAPI) in the lower one.
The influence of formalin on Gat1-Myc13 localization described above was not, on the other hand, observed when cells were treated with rapamycin (Fig. 1). Here, Gat1-Myc13 was nuclear in nearly all cells whether treated with 3.1% or 5.6% formalin. Together these data argued that formalin treatment could affect Gat1-Myc13 localization under some conditions but not others. The least effects occurred when Gat1-Myc13 was most completely localized to either the cytoplasm or nucleus, i.e, in ammonia-grown or rapamycin-treated glutamine-grown cells (Fig. 1).
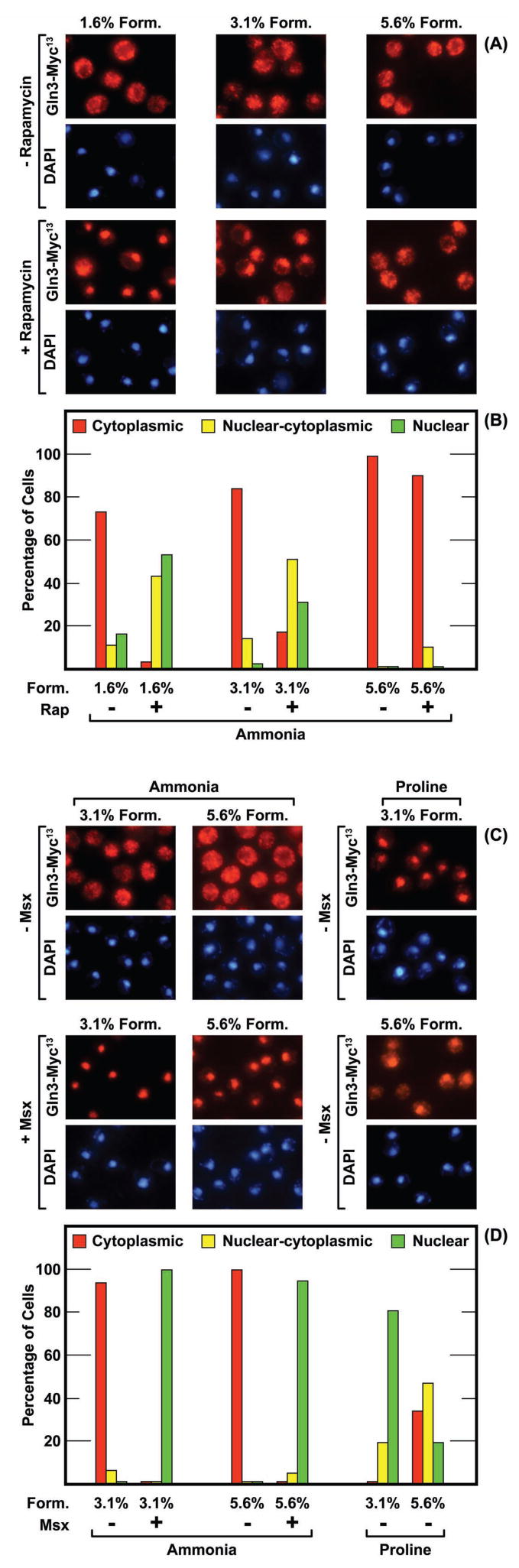
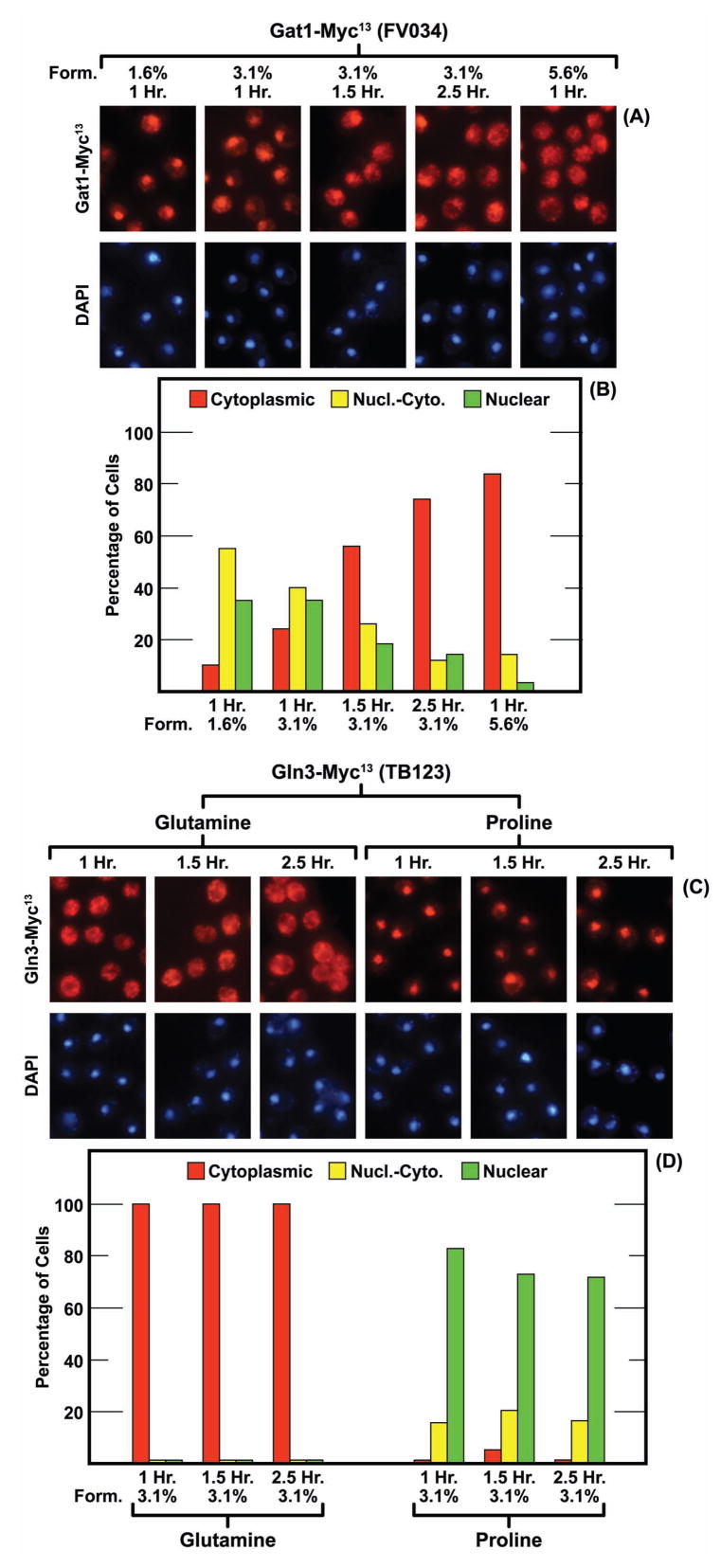
The results with Gat1-Myc13 prompted us to query whether Gln3-Myc13 localization was similarly influenced by formalin. There was no great difference in Gln3-Myc13 localization in ammonia-grown cells treated with 1.6%, 3.1% or 5.6% formalin (Fig. 2A and 2B, −Rap). At all three formalin concentrations, Gln3-Myc13 was cytoplasmic in most cells. The intracellular distributions of Gln3-Myc13 in glutamine-grown cells treated with 1.6%, 3.1% or 5.6% formalin were indistinguishable from those obtained with ammonia-grown cells treated with 5.6% formalin, i.e., Gln3-Myc13 was cytoplasmic in nearly all cells (data not shown). In contrast, a strong shift in Gln3-Myc13 localization was observed in rapamycin-treated cells (Fig. 2A and 2B, +Rap). Gln3-Myc13 was predominantly nuclear and nuclear cytoplasmic when cells were fixed with 1.6% formalin. There was a slight increase in the number of cells scored as cytoplasmic when they were fixed with 3.1% formalin and a drastic increase when 5.6% was used. A similar shift of Gln3-Myc13 away from nuclear localization was also observed when proline-grown cells were treated with 5.6% formalin (Fig. 2C and 2D). This cytoplasmic shift was not observed, however, in cells where nuclear Gln3-Myc13 localization was induced by treating cells with Msx (Fig. 2C and 2D). Gln3-Myc13 was exclusively nuclear irrespective of the formalin concentration as was observed with Gat1-Myc13 in rapamycin-treated cells (Fig. 1). In both cases, the inhibitor (rapamycin or Msx) elicited a very strong response, i.e., complete nuclear localization of the GATA-factor.
FIG. 2.
Gln3-Myc13 localization is influenced by only very high concentrations (5.6%) of formalin. The experimental format and data presentation were as in Fig. 1. The strain was TB123. The nitrogen source provided is indicated below the histograms. Ammonia-grown cells were treated with rapamycin for 20 min (Rap +) or 30 min with Msx (+Msx) as indicated in the figure. Note that proline-grown cells in panels C and D were not treated with rapamycin or Msx.
These data indicated that: (i) Gat1-Myc13 localization responded to both low and high concentrations of formalin, (ii) Gln3-Myc13 localization responded only to high formalin, and (iii) both Gat1-Myc13 and Gln3-Myc13 localizations were unaffected by high concentrations of formalin when the their localizations otherwise exhibited a strong physiological response.
Gln3-Myc13 and Gat1-Myc13 localization before formalin-treatment
Interpretation of the above experiments rested on an important but unsubstantiated assumption, i.e., that the GATA-factors’ localizations following addition of 3.1% formalin were in fact their actual localizations in untreated cells. This was not a serious caveat for the Gln3-Myc13 data in untreated glutamine or ammonia-grown cells because its localization was not significantly influenced by low or high concentrations of formalin. However, it was a caveat for Gat1-Myc13 data because it could be legitimately argued that Gat1-Myc13 was nuclear in most untreated cells, but became increasingly cytoplasmic as the concentration of formalin increased from zero. By this reasoning, Gat1-Myc13 localization at 3.1% formalin represented an intermediate position on a formalin concentration response curve.
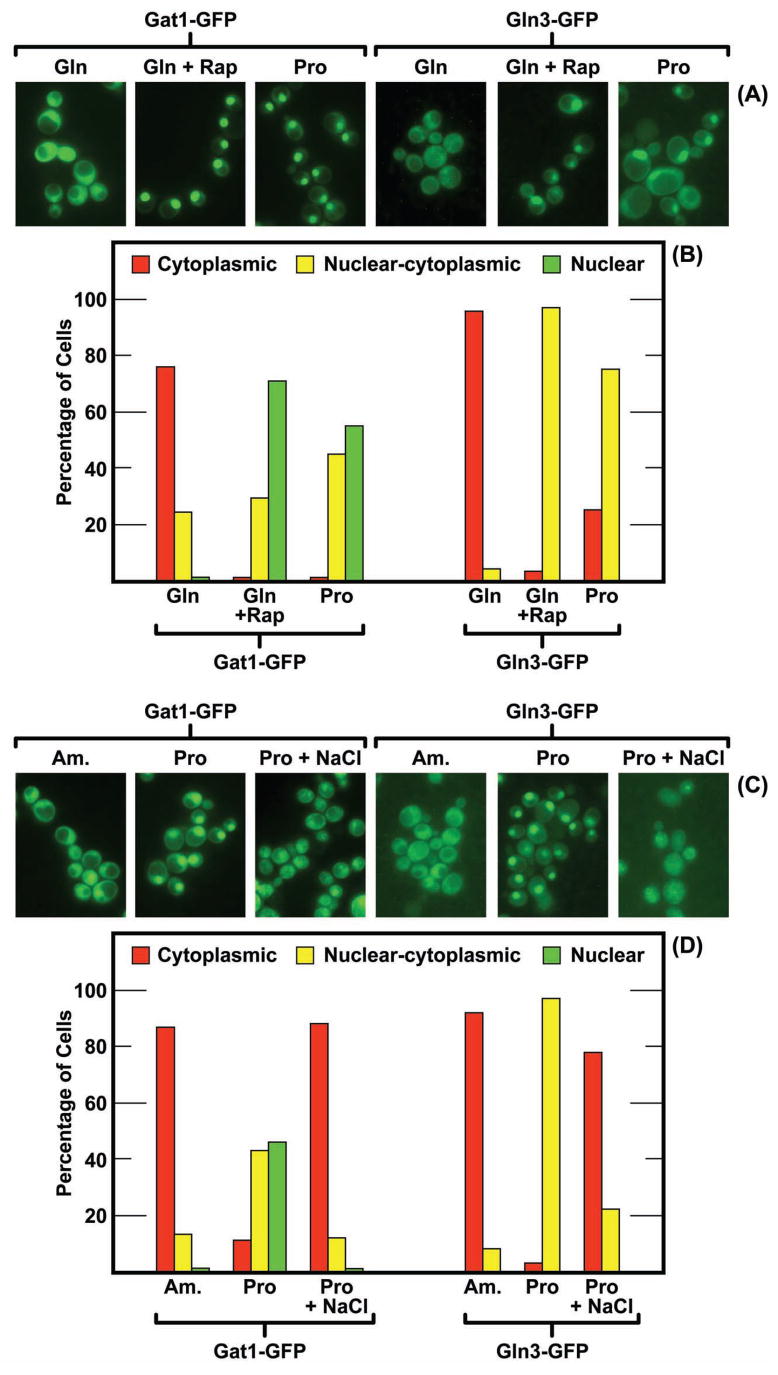
Resolution of the above problem required identification of the intracellular Gat1-Myc13 and Gln3-Myc13 localizations prior to formalin treatment. This was accomplished by visualizing the GATA-factors using GFP fluorescence from plasmids pRS416-GLN3-GFP and pRS416-GAT1-GFP [constructed and reported earlier by Butow and colleagues (Liu et al., 2003; Giannattasio et al., 2005)] transformed into wild-type strain JK9-3da. As shown in Fig. 3, Gat1-GFP and Gln3-GFP both localized to the cytoplasm of most glutamine- (Fig. 3A and 3B) and ammonia- (Fig. 3C and 3D) grown cells. Both Gln3-GFP and Gat1-GFP became more nuclear in most cells following treatment with rapamycin (Fig. 3A and 3B) or growth with proline as sole nitrogen source (Fig. 3C and 3D). These data suggested that Gln3 and Gat1 were predominantly cytoplasmic in living ammonia- and glutamine-grown cells and nuclear or nuclear-cytoplasmic in proline-grown cells. Thus, within the resolution possible using GFP labeled proteins, the intracellular distributions of Gln3-Myc13 and Gat1-Myc13 observed in cells fixed with 3.1% formalin were similar to those of unfixed cells. If there was a difference, it was that Gat1-Myc13 was slightly more nuclear in glutamine-grown, 3.1% formalin fixed cells and somewhat more cytoplasmic in proline-grown cells than occurred with Gat1-GFP. Note also that, as pointed out in Materials and Methods, the quality of Gln3-GFP fluorescence was too low to permit three category scoring.
FIG. 3.
(Panels A and B) Influence of rapmycin treatment and nitrogen source on the intracellular distribution of Gat1-GFP and Gln3-GFP. The nitrogen sources provided were glutamine (Gln) or proline (Pro) as indicated. Two of the cultures were treated with rapamycin (Gln+Rap) as described in Materials and Methods. (Panels C and D) NaCl elicits relocalization of Gat1-GFP and Gln3-GFP. The nitrogen sources provided were ammonia (Am.) or proline (Pro). NaCl was added to a final concentration of 1 M for 20 min prior to cell harvest where indicated (Pro + NaCl). The strain used in all four panels was JK9-3da (TB123 background).
Formaldehyde rather than methanol affects GATA-factor localization
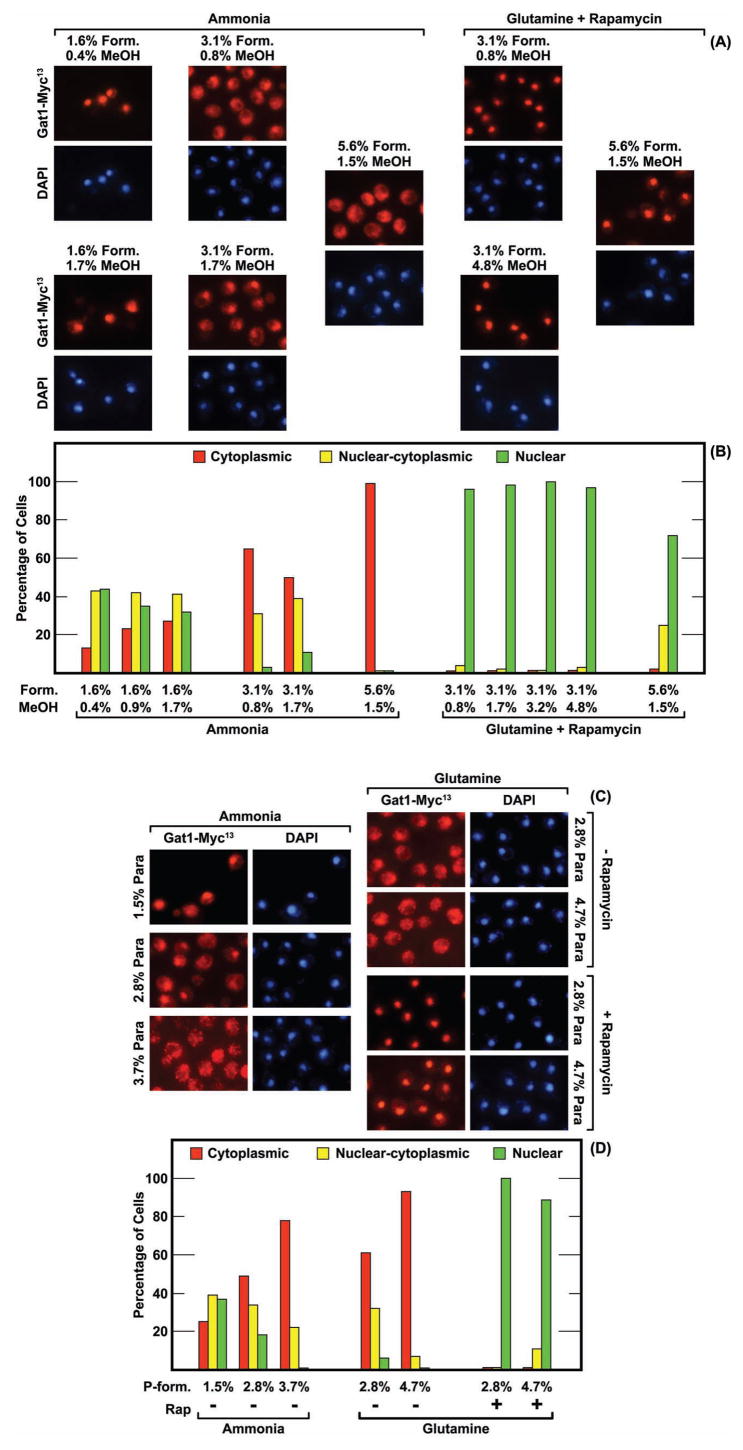
While the above data demonstrated that formalin could influence GATA-factor localization, they failed to identify the causative agent(s). Commercially prepared formalin contains three components: ~37% formaldehyde most of which is polyoxymethylene glycol and ~10–15% methanol. Methanol is added to slow the formation of increasingly high molecular weight polyoxymethylene gycols that eventually precipitate from solution. We identified the causative agent by varying the concentrations of formalin’s individual components. A 1.6% formalin solution contains ~0.4% methanol. At this formalin concentration, Gat1-Myc13 was nuclear or nuclear-cytoplasmic in most cells (Fig. 4A and Fig. 4B). Increasing the methanol concentration to 0.9% or 1.7% (the approximate concentrations of methanol in 3.1% and 5.6% formalin solutions) while holding the formalin concentration constant at 1.6% had no strong effect on Gat1-Myc13 localization (Fig. 4A and 4B). A similar lack of effect was observed when rapamycin-treated, glutamine-grown cells were treated with increasing amounts of methanol while holding the formalin concentration constant at 3.1% (Fig. 4A and Fig. 4B). In contrast, Gat1-Myc13 shifted towards a cytoplasmic localization when formalin concentrations were increased to 3.1% or 5.6% (Fig. 4A and 4B). These data pointed, by exclusion, to formaldehyde or polyoxymethylene glycol rather than methanol as the causative agent.
FIG. 4.
(Panels A and B)The methanol component of commercial formalin preparations has no demonstrable effect on intracellular Gat1-Myc13 localization. The final concentrations (percentage) of formalin (Form.) and methanol (MeOH) used in the fixation procedure are as indicated. Varying methanol concentrations were achieved by adding additional methanol to the formalin solution. The nitrogen source and presence or absence of rapamycin treatment are as indicated. Images are as indicated in Fig. 1. (Panels C and D) The concentration of Para-formaldehyde present during fixation influences intracellular Gat1-Myc13 localization. Para-formaldehyde is indicated as P-form. in histograms and as Para in images. See Materials and Methods for para-formaldehyde preparation. The strain used in all four panels was FV063.
To test the effects of formaldehyde on Gat1-Myc13 localization more directly, we used a 16% stock solution of formaldehyde prepared by dissolving solid para-formaldehyde in water that did not contain methanol preservative (Electron Microscopy Sciences). Gat1-Myc13 was nuclear or nuclear cytoplasmic in most cells treated with 1.5% para-formaldehyde (1.5% Para), but became increasingly localized to the cytoplasm as the para-formaldehyde concentration was raised to 2.8% and then 3.7% (Fig. 4C and 4D). At 3.7%, Gat1-Myc13 was cytoplasmic in most cells. However, increased para-formaldehyde did not affect Gat1-Myc13 localization in glutamine-grown, rapamycin-treated cells (Fig. 4C and 4D). Together these experiments demonstrated that formaldehyde, and/or methylene glycol and polyoxymethylene glycol were responsible for altering intracellular GATA-factor localization.
High sodium chloride elicits the same effects as treating cells with high concentrations of formalin
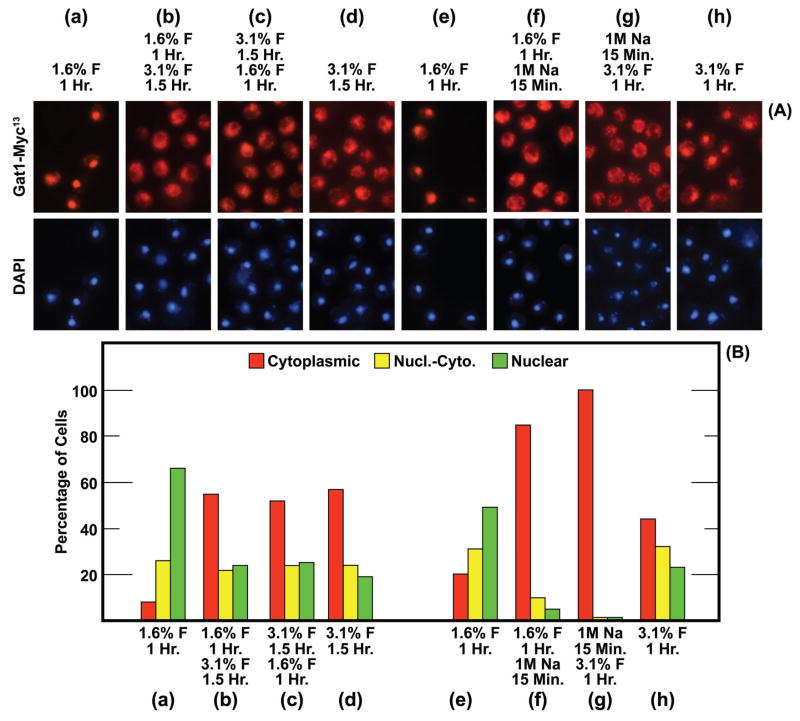
The effects of formalin on GATA-factor localization left two important questions unanswered: (i) Does treating cells with low concentrations of formalin effectively immobilize Gat1-Myc13? (ii) What causes Gat1-Myc13 re-localization following formalin treatment, formaldehyde or methylene/polyoxymethylene glycol? Answering the first question was facilitated by the fact that low and high concentrations of formalin caused Gat1-Myc13 to localize to different cellular compartments. We reasoned that if Gat1-Myc13 was absolutely immobilized in the nuclei of cells treated with a low concentration of formalin, then subsequently increasing the formalin concentration would not relocate it from the nucleus to the cytoplasm. To test this hypothesis, we treated ammonia-grown cells with 1.6% formalin for 1 hr As shown in Fig. 5A and 5B, panel (a), Gat1-Myc13 was nuclear or nuclear-cytoplasmic in nearly all cells. A second sample of the initial culture was treated for 1 hr with 1.6% formalin and then the formalin concentration was increased to 3.1% for another 1.5 hr This second formalin-treatment shifted Gat1-Myc13 localization away from the nucleus and towards the cytoplasm (Fig. 5A and 5B panel (b)); the localization profile was the same as observed if the initial culture had been treated for 1.5 hr with 3.1% formalin (Fig. 5A and 5B, panel (d)). When the experimental format was reversed, i.e., cells were treated with 3.1% formalin for 1.5 hr, harvested and transferred to the same medium containing 1.6% formalin for 1 hr, the second formalin treatment had no effect (Fig. 5A and 5B, panel (c)). Note that the distribution of Gat1-Myc13 was the same in panels (b), (c), and (d). These data argued that Gat1-Myc13 was not completely immobilized in cells treated with 1.6% formalin for 1 hr and hence could be relocated in some of the cells by treating them with high concentrations of formalin. The reverse, however, did not occur. Gat1-Myc13 in cells treated with 3.1% formalin could not be relocated to the nucleus by subsequently lowering the formalin concentration to 1.6%.
FIG. 5.
Gat1-Myc13 is not completely immobilized even after 1 hr of fixation with 1.6% formalin. The experimental format is indicated above each pair of images (a)-(h), and below each histogram (a)-(h). The concentration of formalin is indicated as % F with the time of the initial treatment immediately below it. Below that is the final concentration of formalin that existed during the second round of fixation and below that value, the time of that second fixation. For example, the format in (b) was fixation for 1 Hr with 1.6% formalin (final concentration) followed by an additional 1.5 Hr fixation with 3.1% formalin (final concentration. In (f) cells were treated with 1 M NaCl (1 M Na, final concentration) for 15 min in place of the second round of formalin fixation. In (g) 1 M NaCl (final concentration) was substituted for the first round of formalin fixation. The strain used in these experiments was FV063.
The second question was more difficult to answer because the equilibrium constant of the formaldehyde hydration-dehydration reaction (reaction 1) precluded treating cells with pure compounds. Therefore, we drew on experience from earlier work with Gln3-Myc13. We previously showed that Gln3-Myc13 localization is strongly influenced by a variety of environmental insults, including temperature, osmotic and oxidative stress (Tate & Cooper, 2007). In each case, nuclear Gln3-Myc13 was relocated from the nucleus to the cytoplasm. Since Gln3-Myc13 responded similarly to high concentrations of formalin, we at first suspected that formalin treatment might be subjecting cells to oxidative stress. This was not difficult to envision because formaldehyde is both an oxidant and a reductant. However, our suspicion was greatly diminished by the size of the formaldehyde hydration-dehydration reaction equilibrium constant (4–5 × 10−4). Using this equilibrium constant and a measured density of the commercial formalin solution of 1.07 gm/ml, the formaldehyde concentrations in 1.6%, 3.1% and 5.6% formalin solutions are approximately 5.2, 10, and 18 μM, respectively. This is on the order of 10 to 50 times less than the 300 μM hydrogen peroxide concentration we had used to generate oxidative stress in our earlier experiments with Gln3-Myc13 (Tate & Cooper, 2007) and over two orders of magnitude lower than the 3 mM hydrogen peroxide used in other reports to elicit an oxidative stress response (Park et al., 2005). Given that the hydrogen peroxide concentrations we used in our earlier experiments elicited less of a response on Gln3-Myc13 localization than the other environmental stresses we tested (Tate & Cooper, 2007), we thought it rather unlikely that the low concentrations of formaldehyde used here would generate the observed strong effect on GATA-factor localization.
In addition to formaldehyde being an oxidant and reductant, the components of a formalin solution (methylene glycol, polyoxymethylene gycol and methanol) markedly change the osmolarity of the solution to which it’s added. Assuming an average polymerization of n = 5, the osmolarities of the phosphate buffered 1.6%, 3.1% and 5.6% formalin solutions used above would be approximately 0.5 M, 0.7 M, and 1.0 M, respectively. The corresponding values, if methylene glycol polymerization is assumed to be minimal, would be approximately 0.9 M, 1.6 M, and 2.6 M, respectively. These values are for the most part lower but in the same order of magnitude as the osmolarities (0.8 M and 2 M) generated by the 0.4 M and 1 M NaCl additions used by Crespo et al. (2001) and Tate et al. (2007) to determine the effects of osmotic stress on Gln3/Gat1-mediated ENA1-lacZ expression and Gln3-Myc13 localization, respectively. Therefore, we considered osmotic stress was more likely than oxidative stress to have occurred following addition of formalin to the cultures. This line of reasoning assumes that the osmotic effects of the NaCl addition are greater than those caused by the Na+ ions themselves.
From this reasoning, we hypothesized that it should be possible to mimic the effects of formalin treatment in the experiment depicted in Fig. 5, panels (a) and (b), by substituting sodium chloride for formalin. This, indeed was observed when we added 1 M sodium chloride in place of the second formalin addition (compare Figs. 5A and 5B, panels (a) and (b) with panels (e) and (f)).
We next attempted to use Gln3- and Gat1-GFP fusion proteins to further test the conclusion that osmotic stress was affecting GATA-factor localization. Unfortunately, formalin treatment severely quenched Gat1-GFP and Gln3-GFP fluorescence to the point of being unusable. However, both Gln3-GFP and Gat1-GFP relocated from a nuclear or nuclear-cytoplasmic distribution in untreated proline-grown cells to the cytoplasm of most cells following addition of sodium chloride to a final concentration of 1 M (Figs. 3C and 3D). These data were consistent with the suggestion that relocalization of Gln3-Myc13 and Gat1-Myc13 after treating cells with a high concentration of formalin (5.6%), likely resulted from the osmotic stress it placed upon them. However, some reservation about this interpretation must remain because we could not convincingly mimic the results observed after treating cells with low concentrations of formalin (1.6%), by the addition of small amounts of sodium chloride. We cannot determine whether this is a legitimate negative result or derives from the high background fluorescence that occurs with the GFP-labelled GATA factors, especially Gln3-GFP.
Formalin elicited relocalization of Gat1-Myc13 is time dependent
Experimental protocols for formalin fixation of S. cerevisiae cells recommend treating cells “for at least 1 hr” (Chau & Mok, 2008) with others in a range of 1–2 hr In the histological literature formalin has long been known to be a rapidly penetrating, but slowly acting fixative (Burnett, 1982). The former has been suggested to derive from the low molecular weight of formaldehyde (M.W. 30), and the latter by the fact that the rate limiting step in the cross-linking reaction is the dehydration of methylene glycol (Chau & Mok, 1992).
| (4) |
| (5) |
Formaldehyde reacts only with nitrogen atoms that possess an unshared pair of electrons. Positively charged (fully protonated) nitrogen atoms will not react with formaldehyde. As a result, formaldehyde reacts at neutral pH (6.5–7.0) with the acetamide nitrogen atoms of asparagine and glutamine, the imino nitrogens of peptide bonds, and the imidazole nitrogen of histidine (pKa2 = 6.0). Cross-linking of the unprotonated ε-amino nitrogen atom of lysine residues (pKa3 = 10.5) occurs only slowly at neutral pH due to their very low concentration.
These considerations led us to suspect that Gat1-Myc13 localization might be influenced not only by the formalin concentration but also by the duration of treatment. To test this, we incubated glutamine-grown wild-type cells containing Gat1-Myc13 or Gln3-Myc13 with several concentrations of formalin for varying lengths of time. As shown in Fig. 6A and 6B, Gat1-Myc13 increasingly relocalized from the nucleus to the cytoplasm as the time of treatment (3.1% formalin) increased from 1 to 2.5 hr The same results were achieved by treating cells for 1 hr with 5.6% formalin. On the other hand, Gln3-Myc13 in both glutamine- and proline-grown cells yielded the same localization profiles after 2.5 hrs as 1 hr of formalin treatment.
FIG. 6.
The intracellular localization of Gat1-Myc13 but not that of Gln3-Myc13 is influenced by the formalin concentration and duration of fixation. The times of fixation are indicated in Hr and the formalin concentrations (Form.) in percentages. The nitrogen source in panels A and B was glutamine. Those in panels C and D are indicated above the images and below the histograms. Strain FV034 was used in Panels A and B and TB123 in Panels C and D.
DISCUSSION
Five important results derive from the experiments described in this work: (i) Not all transcription factors respond similarly to formalin fixation. For some, such as Gln3-Myc13, the effects of formalin are minimal to non-existent until abnormally high concentrations are employed. For others, such as Gat1-Myc13, the effects are potentially substantial and can result in movement of the protein into or out of the nucleus depending on the formalin concentration. (ii) Formalin treatment does not instantly quench all cellular activity. Transcription factors can potentially move during the course of formalin treatment. Again some factors, eg., Gat1-Myc13, are more susceptible to movement following the onset of formalin treatment than others, eg., Gln3-Myc13. (iii) Formalin-induced movements were more likely when the transcription factor was localized to more than a single cellular compartment, i.e., when transcription factor distribution was not tilted too far towards the extremes of either nuclear or cytoplasmic localization. Perturbations that elicit strong regulatory responses, such as rapamycin treatment of glutamine-grown cells for Gat1-Myc13 or Msx treatment of ammonia-grown cells for Gln3-Myc13, were more immune to these effects. (iv) Two potentially different cellular processes may be involved in formalin-induced protein movement, one occurring at low formalin concentrations where Gat1-Myc13 localization became more nuclear and another, with opposite effects, at high concentrations where both Gat1-Myc13 and Gln3-Myc13 localization became cytoplasmic. Gat1-Myc13 localization responded to both processes, whereas Gln3-Myc13 responded only to the one occurring at high formalin concentrations. (v) The influence of formalin on transcription factor localization most likely derives from osmotic stress caused by the buffer and high concentrations of methylene/polyoxymethylene gycol used to prepare cells for protein localization experiments employing indirect immunofluorescence methods. Although we cannot eliminate the possibility that formalin-treatment is generating some oxidative stress, because we compared the effects generated by various concentrations of hydrogen peroxide to those with formalin, the great difference between the concentrations of formaldehyde (micromolar) and polyoxymethylene glycol (molar), argues in favor of the latter compound being responsible for the bulk of the effects observed.
If the conditions of formalin fixation are capable of altering the intracellular distribution of GATA and potentially other transcription factors, two questions are pertinent. Have the effects of formalin treatment adversely compromised our previously reported protein localization data, and if so, how? What are the most advisable conditions of fixation?
Formalin fixation has had little if any influence on our past measurements of intracellular Gln3-Myc13 localization. As shown in Fig. 2A and 2B, the Gln3-Myc13 distribution profiles observed following 1.6% and 3.1% formalin treatment were quite similar. Gln3-Myc13 was slightly more cytoplasmic in rapamycin-treated cells fixed with 3.1% formalin than seen at 1.6%. However, given the qualitative nature of scoring GATA-factor intracellular distribution, this difference is unlikely to be significant and would not have influenced any of the conclusions reported. It is not until the formalin concentration is increased to 5.6%, nearly twice the concentration normally used in our assays, that it reversed nuclear Gln3-Myc13 localization induced by rapamycin treatment or growth in proline medium. Gln3-Myc13 behaved as expected from past reports showing that its localization responds to high osmotic stress. Formalin had no effect on Msx-treated, ammonia-grown cells even at the high concentration (5.6%) (Figs. 2C and 2D). This result is consistent with the fact that increased osmolarity generated by formalin treatment did not reach levels previously achieved with 1 M NaCl. With 1 M NaCl, Gln3-Myc13 became cytoplasmic in both rapamycin- and Msx-treated cells (Tate & Cooper 2007). As best we could measure it, given the low fluorescence yields of Gln3-GFP, the behavior of Gln3-Myc13 and Gln3-GFP were indistinguishable (compare Figs. 2 and 3). Finally, the duration of fixation did not affect Gln3-Myc13 distribution, the same profiles were observed whether the cells were fixed for 1, 1.5, or 2.5 hrs (Fig. 6C and 6D). The most likely situation in which formalin, if its concentration was a little high, might influence Gln3-Myc13 localization is when it exhibits both nuclear and nuclear-cytoplasmic localization. Under these conditions, Gln3-Myc13 might be shifted slightly more towards cytoplasmic localization.
The above data show that Gat1-Myc13 localization, on the other hand, is more sensitive to the effects of formalin than that of Gln3-Myc13. At low formalin concentrations, Gat1-Myc13 localization was nuclear or nuclear-cytoplasmic in the majority of cells. As the formalin concentration or time of fixation was increased, Gat1-Myc13 became more cytoplasmic. Fortunately, and serendipitously, the combination of conditions we used in previously reported Gat1-Myc13 distribution measurements, 3.1% formalin for 60–70 min, struck a reasonable compromise between the two effects (short vs. longer fixation times and formalin concentrations). It was a compromise that was retrospectively found to generate Gat1-Myc13 distributions that were not overly biased towards nuclear or cytoplasmic localizations and were nearest to those observed with Gat1-GFP. Therefore, we suggest that the influence of formalin on previously reported measurements of Gat1-Myc13 distribution were minor. The most likely situations for formalin-generated bias would be those in which Gat1-Myc13 and experiment to experiment variation was greatest, i.e., in untreated, glutamine-grown wild-type cells. Such variation is a likely hallmark of a proteins’ sensitivity to the affects of formalin treatment. Retrospective evaluation of our previously reported Gat1-Myc13 localization data in light of this caveat did not alter the conclusions we had reached.
Whether other proteins respond to formalin fixation in manners similar to those seen with the GATA-transcription factors cannot be reliably predicted in the absence of experimental data. However, it would be quite surprising if such influences are not more broadly observed upon more detailed analyses. Proteins whose localization is stress-responsive will be the most likely candidates for these effects. Even in those instances where formalin fixation might have influenced cellular distributions, it would be difficult to detect it a priori in the existing literature because most past distribution data have been reported qualitatively in the form of isolated images containing a limited number of cells.
In light of the above results, one might reasonably query whether indirect immunefluorescence should be abandoned as a method for determining intracellular protein localization in favor of real time GFP measurements in unfixed cells. In our view, the limitations of GFP-based measurements are different, but potentially just as great as those with indirect immunofluorescence. The most significant limitations derive from the low fluorescence yield of GFP rendering the method unusable when low abundance proteins are to be assayed. Solving the yield problem via overproduction of the protein being investigated generates its own compromises by running the obvious risk of saturating regulatory factors. Further, the low signal to background fluorescence ratio that occurs with low abundance proteins makes even qualitative scoring questionable unless the protein is localized to a single cellular compartment. We suggest that both methods can be advantageously and prudently used when their limitations are recognized and taken into account. The above experiments emphasize, however, the necessity of evaluating whether a protein whose distribution is being investigated is sensitive, like Gat1-Myc13, or insensitive, like Gln3-Myc13, to the effects of normal formalin fixation. Such information can then form the basis for selecting the most objective and non-biasing fixation conditions or defining the limitations most prudently placed on the interpretation of localization measurements when such complications cannot be avoided.
Finally, we suggest that the effects of formalin treatment we observed in this work potentially derive from physiological responses to various degrees of environmental stress. Strongest support for this contention is the fact that we can duplicate some of the effects with sodium chloride, the a commonly used agent for eliciting osmotic stress and further that the response to formalin can be surmounted if a counter-acting perturbant, eg., rapamycin or Msx treatment, is simultaneously imposed. This behavior would unlikely be the case if we were dealing with a non-physiological response. We also speculate that two distinguishable cellular processes are likely involved: the low concentration effect, correlating with low level stress, that triggers movement of Gat1-Myc13 but not Gln3-Myc13 into the nucleus, and the high concentration effect, correlating with more drastic stress to which both GATA-factors respond by leaving the nucleus.
Acknowledgments
We thank Drs. Evelyne Dubois, Isabelle Georis, Ronald Butow, and Zhengchang Liu for strains and plasmids. We also appreciated discussions and suggestions from the University of Tennessee Yeast Group.
This work was supported by National Institutes of Health grant GM-35642.
References
- Aparicio O, Geisberg JV, Struhl K. Curr Protoc Cell Biol. Unit 177. Chapter 17. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Bell RP, Evans PG. Kinetics of the dehydration of methylene glycol in aqueous solution. Proc Roy Soc Lon. 1966;291:297–323. [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Blinder D, Coschigano PW, Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett MG. The mechanism of the formaldehyde clock reaction. Methylene glycol dehydration. J Chem Educ. 1982;59:160–162. [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J, Bertram PG, Wente SR, Zheng XF. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J Biol Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- Chau FT, Mok KW. A computer-enhanced pH study of the formaldehyde-sulphite clock reaction. J Automatic Chem. 1992;14:79–83. doi: 10.1155/S146392469200018X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J Biol Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Actin cytoskeleton is required for nuclear accumulation of Gln3 in response to nitrogen limitation but not rapamycin treatment in Saccharomyces cerevisiae. J Biol Chem. 2004;279:19294–19301. doi: 10.1074/jbc.M309240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Daicho K, Ushimaru T, Hall MN. The GATA transcription factors GLN3 & GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J Biol Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J Biol Chem. 2000;275:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Chellappan SP. Chromatin immunoprecipitation assays: molecular analysis of chromatin modification and gene regulation. Methods Mol Biol. 2007;383:135–152. doi: 10.1007/978-1-59745-335-6_9. [DOI] [PubMed] [Google Scholar]
- Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J Biol Chem. 2008;283:8919–8929. doi: 10.1074/jbc.M708811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio S, Liu Z, Thornton J, Butow RA. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem. 2005;280:42528–42535. doi: 10.1074/jbc.M509187200. [DOI] [PubMed] [Google Scholar]
- Gregory BD, Yazaki J, Ecker JR. Utilizing tiling microarrays for whole-genome analysis in plants. Plant J. 2008;53:636–644. doi: 10.1111/j.1365-313X.2007.03320.x. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- Idicula AM, Blatch GL, Cooper TG, Dorrington RA. Binding and activation by the zinc cluster transcription factors of Saccharomyces cerevisiae. Redefining the UASGABA and its interaction with Uga3p. J Biol Chem. 2002;277:45977–45983. doi: 10.1074/jbc.M201789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; 1994. pp. 115–117. [Google Scholar]
- Kulkarni A, Abul-Hamd AT, Rai R, El Berry H, Cooper TG. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J Biol Chem. 2001;276:32136–32144. doi: 10.1074/jbc.M104580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Buford TD, Rai R, Cooper TG. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:218–229. doi: 10.1111/j.1567-1364.2006.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Spírek M, Thornton J, Butow RA. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell. 2003;12:401–411. doi: 10.1016/s1097-2765(03)00285-5. [DOI] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Mondoux MA, Scaife JG, Zakian VA. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics. 2007;177:2019–2029. doi: 10.1534/genetics.107.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Collinson EJ, Grant CM, Dawes IW. Rom2p, the Rho1 GTP/GDP exchange factor of Saccharomyces cerevisiae, can mediate stress responses via the Ras-cAMP pathway. J Biol Chem. 2005;280:2529–35. doi: 10.1074/jbc.M407900200. [DOI] [PubMed] [Google Scholar]
- Sbia M, Parnell EJ, Yu Y, Olsen AE, Kretschmann KL, Voth WP, Stillman DJ. Regulation of the Yeast Ace2 Transcription Factor during the Cell Cycle. J Biol Chem. 2008;283:11135–11145. doi: 10.1074/jbc.M800196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schecker HG, Schulz G. Investigation of the hydration kinetics of formaldehyde in water solution (untersuchungen zur hydratationskinetik von formaldehyd in wiassriger liosungen) Zeit Phys Chem (Frankfurt) 1969;65:221–224. [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Southall TD, Brand AH. Chromatin profiling in model organisms. Brief Funct Genomic Proteomic. 2007;6:133–140. doi: 10.1093/bfgp/elm013. [DOI] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Tor1/2 regulation of retrograde gene expression in Saccharomyces cerevisiae derives indirectly as a consequence of alterations in ammonia metabolism. J Biol Chem. 2003;278:36924–36933. doi: 10.1074/jbc.M301829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Stress-responsive Gln3 localization in Saccharomyces cerevisiae is separable from and can overwhelm nitrogen source regulation. J Biol Chem. 2007;282:18467–18480. doi: 10.1074/jbc.M609550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Cox KH, Rai R, Cooper TG. Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J Biol Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Feller A, Dubois E, Cooper TG. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J Biol Chem. 2006;281:37980–37992. doi: 10.1074/jbc.M606973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J Biol Chem. 2005;280:27195–27204. doi: 10.1074/jbc.M504052200. Erratum in: J. Biol. Chem. 282: 13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Ammonia-specific regulation of Gln3 localization in Saccharomyces cerevisiae by protein kinase Npr1. J Biol Chem. 2006;281:28460–28469. doi: 10.1074/jbc.M604171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis ER. The protein –formaldehyde reaction. J Biol Chem. 1944;154:87–97. [Google Scholar]
- Theis ER. The protein-formaldehyde reaction. 1945;157:7–14. [Google Scholar]
- Theis ER, Lams MM. The protein-formaldehyde reaction. J Biol Chem. 1944;154:99–103. [Google Scholar]
- Winkelman JGM, Voorwinde OK, Ottens M, Beenackers AACM, Janssen LPBM. Kinetics and chemical equilibrium of the hydration of formaldehyde. Chem Engineering Sci. 2002;57:4067–4076. [Google Scholar]