Abstract
The light chain of botulinum neurotoxin A (BoNT/A-LC) is a zinc-metalloprotease that requires two extended exosites for optimal substrate binding and recognition of its intracellular target SNAP25. CFP and YFP connected through SNAP25 peptide (141–206) containing both exosites (CsY) has been used in a FRET-based assay for BoNT/A. To further improve the FRET efficiency in this BoNT/A substrate for in vitro high-throughput assays, we explored the feasibility of enhancing the capture of CFP emission by doubling the number of YFP acceptors. In comparison to CsY, the tandem fluorescence substrates CsYY and YsCsY enhanced the ratiometric fluorescence signal between YFP and CFP. YsCsY, containing two substrate sites, offered the greatest fluorometric change upon toxin-catalyzed cleavage. In addition to known approaches for enhancing fluorescence yield through various mutations, this alternative tandem substrate approach can boost the FRET signal and is particularly useful for substrates requiring extensive exosite recognition for specificity.
Keywords: High-throughput screening, inhibitor, FRET-based assay, tandem fluorescent proteins, exosites, extended peptide substrate recognition
INTRODUCTION
Clostridial botulinum neurotoxins (BoNT), serotypes A through G, which target peripheral cholinergic neurons, are considered the most potent protein toxins for humans1. Death occurs by respiratory failure caused by neuromuscular paralysis. The intravenous lethal dose of BoNT serotype A (BoNT/A), which selectively cleaves synaptosome-associated protein of 25 kDa (SNAP25) and prevents neurotransmitter release, is estimated to be 1–10 ng/kg (Arnon et al., 2001). BoNT/A is considered by the Centers for Disease Control and Prevention to be a scategory A-select agent” biosecurity risk due to its potential use as a bioweapon (Arnon et al., 2001). Yet, it is also the active ingredient of BOTOX®, which is gaining extensive use as a valuable therapeutic agent for various neuronal disorders (Montecucco and Molgo, 2005) and as a cosmetic for anti-wrinkle applications (Carruthers and Carruthers, 2001).
There is no effective antidote available to counter botulism arising from deliberate or accidental ingestion of contaminated food or misuse of BoNT-containing pharmaceuticals. Currently, the only available treatment for botulism is a combination of antitoxin immunoglobulin therapy and long-term respiratory care (Arnon et al., 2001), which in the event of mass exposure to BoNT would overwhelm current medical infrastructure. Thus, there is a critical need for development of effective post-exposure therapies to combat botulism and speed recovery as well as for rapid and specific detection. The in vitro and in vivo disconnect found in recent high-throughput screening (HTS) of small molecule libraries for BoNT/A inhibitors (Eubanks et al., 2007) brought to light the urgent need for a quick and reliable in vitro assay for identifying physiologically relevant inhibitors.
The catalytic light chains of BoNTs (BoNT-LC) are zinc-dependent proteases that recognize extended regions of their substrates for cleavage. Recognition between BoNT/A and SNAP25 involves two extended exosites for optimal substrate binding and recognition (Breidenbach and Brunger, 2004). The minimal size of SNAP25 known to retain full activity as a BoNT/A substrate is the C-terminal 66-mer peptide (residues 141–206) with both exosites (Washbourne et al., 1997). Short peptides containing the cleavage site of the substrate can be converted into active site-based FRET substrates for inhibitor screening, but this approach would miss inhibitors specifically targeting the exosites, which are the unique features of BoNT/A necessary for distinguishing BoNT/A from other metalloproteases in cells. Dong et al. (2004) first reported the feasibility of using the CFP-YFP pair with full-length SNAP25 as a FRET-based substrate for BoNT/A in a cell-based assay or with the 66-mer peptide as a FRET substrate in an in vitro assay. However, the length of the 66-mer peptide does not allow for high FRET efficiency under optimal enzymatic cleavage conditions. We found that the simple CFP-SNAP25(141–206)-YFP substrate (CsY) provides only a small change in FRET signal after BoNT/A-LC treatment, and that this FRET signal is also highly dependent on the concentration of substrate and reaction conditions.
One obvious approach to improve the FRET in a substrate such as CsY is to shorten the linker peptide between the two fluorophores since the efficiency of FRET (EFRET) is dependent on the distance (r) between the donor and the acceptor as described in the Förster Equation: EFRET = 1/[1 + (r/Ro)6], where Ro is the Förster distance at which 50% of FRET would be detected. When two fluorophores are attached to the opposite ends of the 66-mer peptide as in CsY, they are separated by more than twice the Förster distance of 48Å for the CFP-YFP pair, presuming the peptide is folded into an alpha-helix such as that found in a SNARE complex (Sutton et al., 1998). Although shorter SNAP25-derived peptides have been reported to be substrates of BoNT/A (Schmidt and Bostian, 1997) and synthetic peptides with proper acceptor/donor pair of fluorescent dyes, such as the commercially available SNAPtide™, have been used to monitor the activity of BoNT/A, we found that CFP-YFP tethered through a 17-mer peptide was resistant to cleavage by BoNT/A-LC (data not shown). Other strategies for optimization of FRET efficiency include use of fluorescent protein variants (Nguyen and Daugherty, 2005) or use of circularly permuted mutants of fluorescent proteins (Nagai et al., 2004).
Here, we present an alternative approach to enhance the efficiency of FRET signal by capturing the CFP emission through doubling of the YFP acceptor. As depicted in Figure 1, when the two fluorescent proteins are far apart as in CsY, only a small fraction of the maximal FRET can be materialized due to the required length of the connecting SNAP25 peptide. However, if two YFPs are in tandem as in CsYY, then the two acceptor proteins would be at similar distances to the donor CFP, and the amount of FRET in CsYY could be nearly double that of CsY. Alternatively, as depicted in the model for YsCsY, two YFPs could be equidistantly connected to CFP through two SNAP25 peptides, likewise doubling the FRET signal, but requiring two cleavage events to lose the FRET. Our results show that joining two YFPs in tandem (CsYY) or connecting two YFPs to CFP through two separate SNAP peptides (YsCsY) nearly doubled the fluorometric changes as well as enhanced the ratiometric changes in the BoNT/A cleavage assay.
Figure 1.
Design of tandem FRET substrates for BoNT/A-LC assays. Shown is an illustration depicting BoNT/A-mediated cleavage of tandem FRET substrates. Spheres represent CFP (cyan) or YFP (yellow) connected to SNAP substrate peptides (curved lines). Dotted lines indicate intramolecular FRET that is lost upon enzymatic cleavage.
MATERIALS AND METHODS
DNA plasmid construction
The plasmids pGFP(uv), pECFP, and pEYFP were obtained from Clonetech. pGST-SNAP25(141–206) was derived from pGST-SNAP25, which was a kind gift from Joseph Barbieri (Baldwin et al., 2004). The pGE vector was constructed from pTRC-His (Invitrogen) after removing a portion of lacIq and inserting a T7 promoter and the GFP coding sequence after the His6-Tag. pGFP-SNAP25(141–206) was constructed from pGE by inserting PCR fragment encoding SNAP25(141–206) and a linker sequence between GFP and SNAP25(141–206). pCs was constructed from pGFP-SNAP25(141–206) through fragment exchange of the GFP coding region with CFP. pCsY was constructed from pCs by inserting YFP plus a linker sequence after the SNAP coding region. pCsYY was constructed from pCsY by inserting YFP plus a linker sequence after the YFP coding region. For construction of pYsCsY, YFP was inserted into the KpnI-SmaI sites of pCsY to generate pYsY, and Cs was inserted into the NotI site of pYsY to generate pYsCsY. pBoNT/A-LC was derived from the pET-33b(+) vector by insertion of the coding region for residues 1–437 of BoNT/A with one extra Gly after the starting Met, and followed by cMyc, HA and His6 tags.
BoNT/A-LC protein expression and purification
Recombinant BoNT/A-LC (residues 1–437) was expressed as a C-terminal His6-tagged protein in Rosetta cells (Novagen) at 30°C in 4L Luria broth (8 × 0.5 L cultures) containing 50 μg/mL kanamacyn. Following 4 hrs of induction with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), cells were harvested by centrifugation, and cell pellets were resuspended in 100 mL of Lysis buffer [PBS, containing 0.1% IGEPAL, 2.5% glycerol, 0.05 Kunitz-unit DNAse I, 20 μg/mL RNAse I, 0.3 mg/mL lysozyme, 1 mg/mL benzamidine, 0.3 mg/mL phenylmethylsulfonyl fluoride and 100 μL protease inhibitor cocktail (Sigma)]. Cells were disrupted by sonication and centrifuged at 20,000 × g for 2 hrs at 4°C. The His6-tagged proteins were purified by Ni2+-chelation chromatography using a Ni2+-NTA column (Qiagen), followed by anion exchange chromatograpy using a HiTrap ANX column (GE Healthcare). Purified proteins were desalted in 50 mM Tris-HCl, pH 7.5, containing 2.5% glycerol using a PD-10 column (GE Healthcare), quantified, and stored at −80°C.
GFP-SNAP25(141–206), CsY, CsYY, and YsCsY protein expression and purification
GFP and FRET substrate proteins were expressed as N-terminal His6-tagged proteins in Rosetta cells (Novagen) at 30°C in 4L Luria broth (8 × 0.5 L cultures) containing 100 μg/mL ampicillin. Following 3 hrs of induction with 1 mM IPTG, cells were harvested by centrifugation, and cell pellets were lysed, and recombinant proteins were purified by Ni2+-NTA and HiTrap ANX chromatography as described above, then further subjected to gel filtration using a Sephacryl S200 column (GE Healthcare), followed again by HiTrap ANX column. Purified proteins were desalted, quantified, and stored at −80°C.
GST-SNAP25(141–206) protein expression and purification
SNAP25(141–206) was also expressed as a GST-tagged protein in E. coli BL21 strain at 30°C in 4L Luria broth (8 × 0.5 L cultures) containing 100 μg/ml ampicillin. Upon reaching OD600nm of 0.6, protein expression was induced with 0.5 mM IPTG and grown overnight at 30°C. Cells were then harvested by centrifugation, and cell pellets were lysed as above and purified by affinity chromatography using a gluthathione-agarose column (Pharmacia). Purified proteins were desalted in 20 mM Hepes buffer, pH 7.5, using a PD10 column, quantified, and stored at −80°C.
Gel-shift cleavage assay
Reactions (60 μL total volume) were performed at 30°C in reaction buffer (20 mM Hepes, pH 7.5, 2 mM DTT, 0.2% Tween 20), containing the indicated concentrations of ZnCl2 and the indicated concentrations of GST-SNAP25(141–206) or GFP-SNAP(141–206). The reaction was initiated by addition of BoNT/A-LC at various concentrations. At the indicated time intervals, aliquots were removed, and the reaction was stopped by addition of SDS-PAGE sample buffer. The proteins were separated by SDS-PAGE (an example gel showing substrates and cleavage products is shown in Supplementary Fig. 1), and the amount of remaining substrate and cleaved product was determined by densitometry. Kinetic constants, determined using the endpoint rates of the earliest possible time points, were obtained from the Lineweaver-Burk reciprocal plots by fitting the data to the equation:
FRET-based cleavage assay with CsY, CsYY or YsCsY
Reactions (100 μL total volume) were performed in 96-well black plates in reaction buffer (20 mM Hepes, pH 7.5, 1.25 mM DTT, 0.2% Tween 20 and 0.1 mg/mL BSA), containing the indicated concentrations of ZnCl2 and the indicated concentrations of FRET substrate. After pre-incubation at 30°C or 37°C for about 40 minutes to stabilize the signal, BoNT/A-LC was added at the indicated concentrations to initiate the reaction. Fluorescence was measured using a microplate fluorescence reader (Bio-Tek Instruments) in filter mode with excitation at 400/30 nm and emissions at 485/20nm and 528/20 nm or monochromatic mode with excitation at 420 nm and emission at 475 nm and 530 nm. Kinetic constants, determined using initial rates obtained from the change in YFP fluorescence reading (emission at 528/20 nm), were obtained from Lineweaver-Burk reciprocal plots as described above. The kinetic parameters of YsCsY are shown as apparent values, treating the two cleavage events as a single turnover.
FRET-based cleavage assay with SNAPtide™
Reactions (100 μL total volume) were conducted in 96-well black plates in reaction buffer (20 mM Hepes, pH 7.5, 1.25 mM DTT, 0.2% Tween 20 and 0.1 mg/mL BSA), containing the indicated concentrations of BoNT/A-LC and ZnCl2. After 10 minutes pre-incubation at 30°C or 37°C, the reaction was initiated by the addition of 5 μM of SNAPtide™ (oAbz/Dnp) (List Biological Laboratories). Fluorescence was measured using a microplate fluorescence reader (Bio-Tek Instruments) with excitation at 320/20 nm and emission at 420/50 nm.
RESULTS AND DISCUSSION
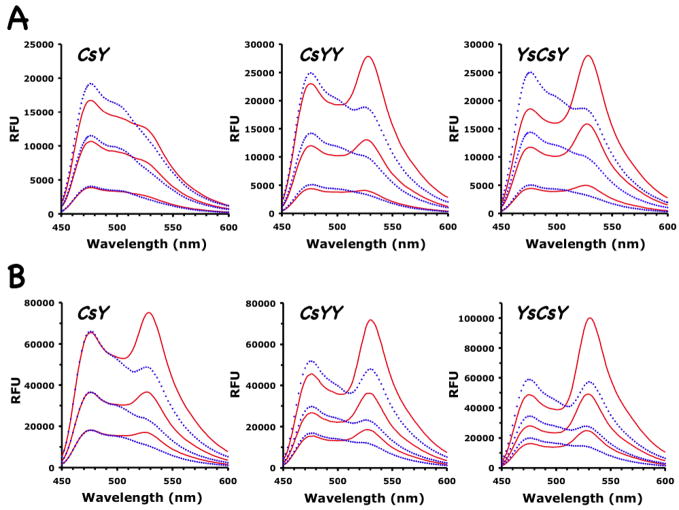
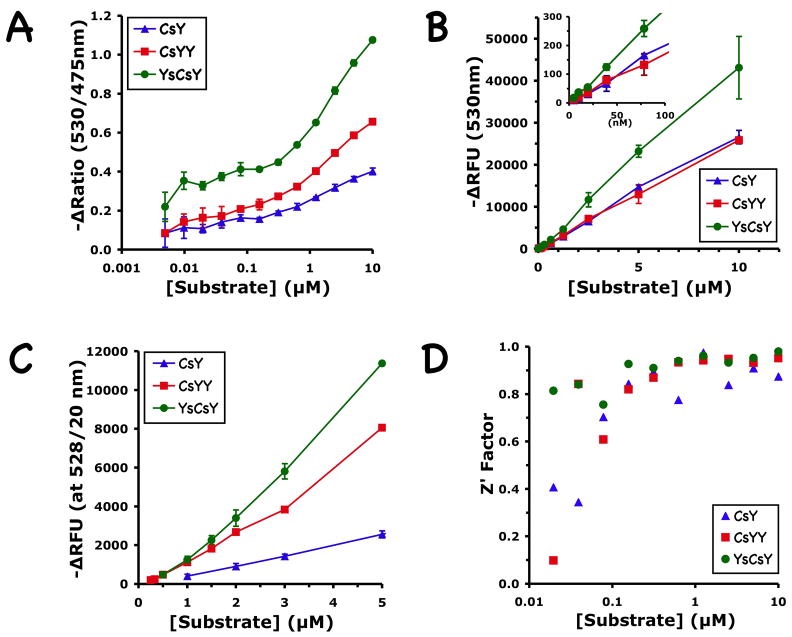
Under optimal reaction conditions with 10 μM Zn2+, the emission spectra of the two FRET substrates CsYY and YsCsY at concentrations of 1, 3, or 5 μM indeed showed much enhanced emission at 530 nm over that of the CsY substrate (Fig. 2A). The change of emission intensity (ΔRFU) at 530 nm after cleavage by BoNT/A-LC was proportional to the substrate concentrations used (Fig. 2 and Fig. 3A). Both CsYY and YsCsY gave ΔRFU that were greater than that for CsY. YsCsY showed the best ratiometric change of 1.6 versus 0.7 at 5 μM, compared to 1.2 versus 0.7 for CsYY and 0.7 versus 0.5 for CsY. The greater ratiometric change for YsCsY was detectable at substrate concentrations as low as 10 nM (Fig. 3A).
Figure 2.
Emission scans of three FRET substrates, CsY, CsYY and YsCsY. Spectra before (solid red lines) and after (dotted blue lines) treatment with BoNT/A-LC were recorded at 30°C using excitation at 420 nm with (A) 10 μM Zn2+ and increasing substrate concentrations of 1, 3 or 5 μM, or (B) 300 μM Zn2+ and increasing substrate concentrations of 2.5, 5 and 10 μM.
Figure 3.
Concentration dependence of ratiometric and fluorometric changes in CsY, CsYY and YsCsY. (A) Change of fluorescence ratio at 530 nm versus 475 nm calculated from full monochromatic scans with excitation at 420 nm, recorded at 30°C in reaction buffer containing 300 μM of Zn2+ before and after BoNT/A-LC-catalyzed cleavage. Shown are the averages of three repeats for each data point. (B) Change of fluorescence at 530 nm calculated from the same scans as in (A). Inset shows the magnification of the same graph in the low substrate concentration region. (C) Change of fluorescence at 530 nm recorded in filter mode (528/20 nm) at 30°C and 10 μM of Zn2+. Shown are the averages of 3 repeats performed in triplicate, except for CsYY, where the two highest points were from a single triplicate measurement. (D) Z′ Factor calculated for the ratiometric assay in (B), using the equation: as defined by Zhang et al. (1999), μe, μc, σe,and σc are the means (μ) and standard deviations(σ)of experimental (e) and control (c) measurements. CsY, blue triangles; CsYY, red squares; YsCsY, green circles.
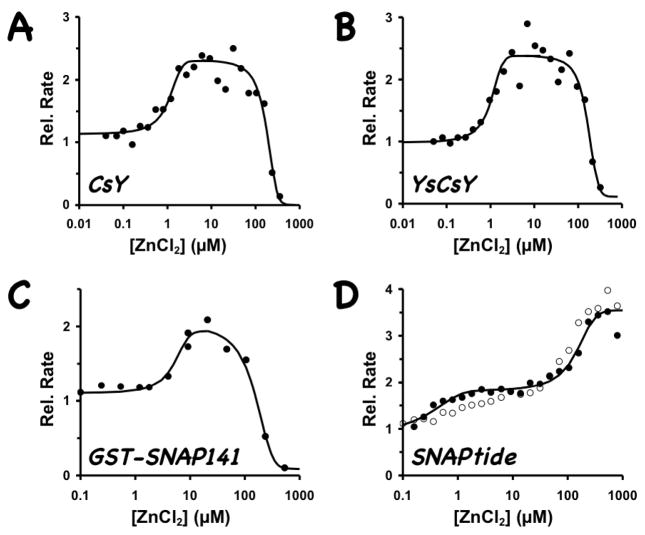
Our FRET substrates behave differently from SNAPtide™ in their Zn2+ dependence. As shown in Figure 4, the rate of cleavage of the CsY and YsCsY substrates sharply decreased at Zn2+ concentrations higher than 100 μM. The optimal Zn2+ concentrations for reaction with our SNAP substrates at 30°C were 5–50 μM (Fig. 4A–B), which is similar to that observed for the nonfluorescent substrate GST-SNAP(141–206) (Fig. 4C), while the optimal Zn2+ concentration for SNAPtide™ substrate was at 300–500 μM (Fig. 4D). Although at 300 μM of Zn2+ concentration the ΔRFU at 530 nm for CsY was nearly doubled that observed at 10 μM of Zn2, this high Zn2+ concentration was not optimal for toxin-catalyzed cleavage of SNAP proteins. In addition, at 300 μM Zn2+ the CFP signal at 475 nm for CsY did not increase after cleavage as was expected (Fig. 2B), suggesting the possibility of zinc-induced structural changes or a zinc-altered energy transfer mechanism. In contrast, the ΔRFUs for CsYY and YsCsY were less sensitive to changes in Zn2+ concentration (Fig. 2 and Fig. 3B–C). In addition to the obvious difference that the protein substrates contain the entire C-terminal SNAP25(141–206) sequence with both recognition exosites, this divergent zinc-dependent phenomenon may contribute to the disconnect observed for the inhibitor potencies found through SNAPtide™-based HTS and those found through cell-based or in vivo assays.
Figure 4.
Zn2+-dependence of BoNT/A-LC activity. CsY (3 μM) or YsCsY (1.5 μM) was incubated with 2 nM BoNT/A-LC; GST-SNAP25(141–-206) (20 μM) with 0.5 nM BoNT/A-LC; and SNAPtide™ (5 μM) with 10 nM BoNT/A-LC. Reactions were performed in the presence of varying concentrations of Zn2+ at 30°C (solid circles) or 37°C (open circles). Relative rates for the FRET substrates, compared to that of the enzyme alone without addition of ZnCl2, were determined from the initial rates and for the non-fluorescent substrate from gel-shift assay after 2 min of reaction, as described in Methods section.
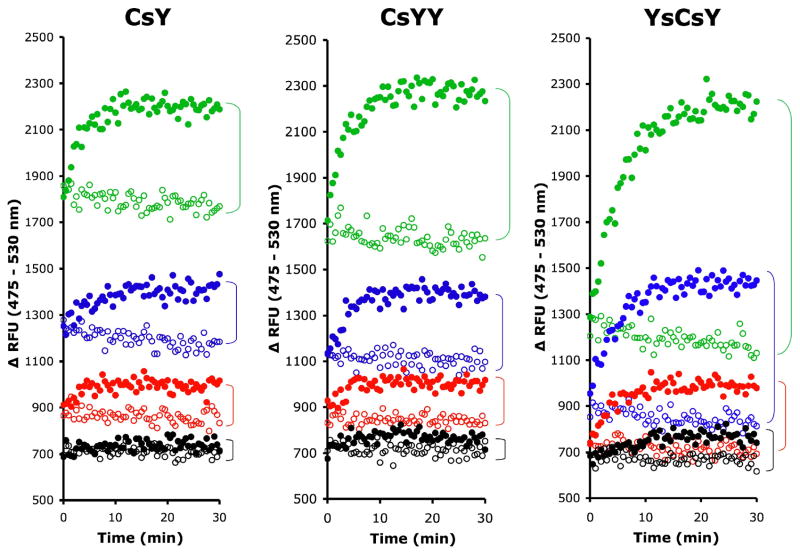
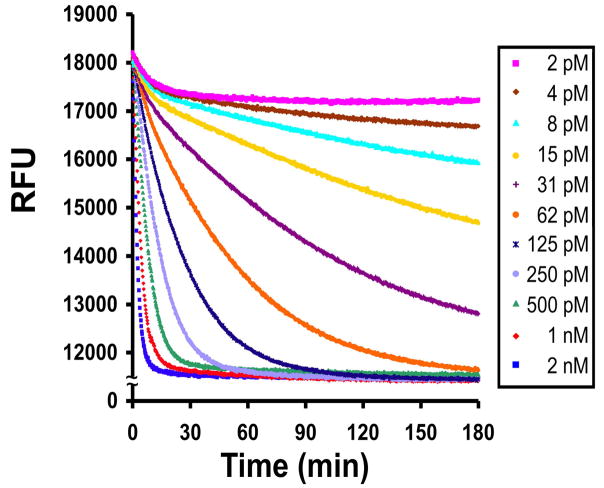
As shown in Figure 3B, the changes in fluorescence observed for the YsCsY substrate were nearly twice those observed for CsY at a wide range of substrate concentrations, including concentrations as high as 10 μM and as low as 10 nM, suggesting that there were no significant inner filter effects over the concentrations used. Control experiments mixing varying concentrations of CFP and YFP in ratios of 1:1 or 1:2 at 10 μM Zn2+ (data not shown) further confirmed that the enhanced fluorescence at 530 nm in YsCsY or CsYY over CsY (Fig. 3C) was not due to intermolecular FRET. Comparative analysis of the minimal substrate concentrations for each of the FRET substrates demonstrated the enhanced sensitivity of the tandem substrates over CsY (Fig. 5), with discernable differences in fluorescence as low as 7.8 nM for YsCsY. This enhanced ability to monitor changes in fluorescence allowed for reliable detection of toxin-catalyzed cleavage within 30 min at concentrations as low as 2 pM (Fig. 6).
Figure 5.
Minimal substrate concentrations for FRET. FRET substrates, CsY, CsYY and YsCsY at a final concentrations of 62.5 nM (green), 31.2 nM (blue), 15.6 nM (red) or 7.8 nM (black) in reaction buffer with 300 μM ZnCl2 were incubated at 30°C for 30 min before addition of BoNT/A-LC pre-incubated at 30°C in reaction buffer at a final concentration of 10 nM (closed circle) or reaction buffer alone (open circle). The fluorescence was recorded in monochromatic mode with excitation at 420 nm and emission at 475 nm and 530 nm. Data shown are the combined time-dependent changes in fluorescence at both emission wavelengths, i.e.ΔRFU = RFU at 475 nm – RFU at 530 nm.
Figure 6.
Continuous monitoring of BoNT/A-LC-catalyzed cleavage of YsCsY. BoNT/A-LC was 2-fold serially diluted at concentrations from 2 nM to 2 pM in reaction buffer containing 10 μM of Zn2+ and incubated in a fluorescence microplate reader at 30°C for 30 min before addition of a solution of 3 μM of YsCsY. The data were recorded in filter mode with excitation at 400/30 nm and emission at 528/20 nm.
The optimal reaction rate for SNAPtide™ was similar at both 30°C and 37°C (Fig. 4D). Whereas the reaction rate for YsCsY under optimal initial rate conditions at 37°C was similar to that for SNAPtide™, it was three times faster at 30°C (Fig. 7). This suggests that the SNAP25 peptide in the YsCsY substrate assumes a more favorable conformation for recognition by the enzyme at the lower temperature, possibly due to better interaction with the exosites. Kukreja and Singh (2005) reported that the optimal cleavage temperature for the full-length SNAP25 substrate using a gel-shift assay is 35–40°C. Although we are not using the full-length protein, Washbourne et al. (1997) had previously shown that the truncated SNAP25 peptide encompassing residues 141–206 is the minimal SNAP25 substrate retaining full catalytic activity at 37°C. It is possible that the difference in optimal reaction temperature observed between our truncated substrates and the full-length substrate could be due to the greater propensity of full-length SNAP25 to form a helical bundle that might require a higher temperature to partially unfold for binding to the BoNT/A-LC, whereas the presence of the fluorescence proteins in our truncated FRET-substrates may contribute to somewhat greater unfolding near the cleavage site and hence the lower temperature requirement.
Figure 7.
Temperature dependence of BoNT/A-LC catalyzed cleavage of YsCsY. The initial rates were determined from ΔRFU recorded in filter mode (528/20 nm) for reactions with 5 μM of YsCsY, 10 μM of Zn2+ and 1, 2, 5, or 10 nM of BoNT/A-LC at 30°C (squares) or 37°C (circles). In comparison, the initial rates were determined from ΔRFU recorded in filter mode (420/50 nm) for reactions with 5 μM of SNAPtide™ (oAbz/Dnp), 300 μM of Zn2+, and 1, 2, 5, or 10 nM of BoNT/A-LC at 37°C (triangles).
Kinetic analysis under initial rate conditions based on loss of YFP signal (ΔRFU at 530 nm) showed that our three FRET substrates had good recognition for the enzyme, with Km values < 1 μM at 10 μM Zn2+ (Table 1). Higher Zn2+ concentrations increased the Kmvalue for CsY up to ten-fold, but only 2- and 3-fold for CsYY and YsCsY, respectively. The more sensitive, double-site substrate YsCsY showed the same catalytic efficiency (kcat/Km) as the single-site substrate CsY. The non-fluorescent substrate GST-SNAP25(141–206) was a competitive inhibitor in the fluorometric assay for CsY and YsCsY with Ki values of 7.6 and 4.5 μM, respectively (Table 1 and Supplementary Fig. 2). These Ki values are similar to the Km values determined by gel-shift assay and are in agreement with published Km values (Chen and Barbieri, 2006). In contrast, the known small molecule active-site inhibitor, 2,4-dichlorocinnamic hydroxamate (DCCH) (Eubanks et al., 2007), gave IC50 values of 81 and 59 μM for CsY and YsCsY, respectively (Table 1 and Supplementary Fig. 4). The BoNT/A-LC cleavage product GST-SNAP25(141–197) failed to inhibit the reaction with CsY at concentrations up to 30 μM (data not shown). A synthetic peptide corresponding to the cleavage site SNAP25(187–206) likewise failed to show inhibition against CsY at concentrations up to 300 μM (data not shown). Taken together, these results are consistent with the need to have both exosites for optimal substrate recognition.
Table 1.
Kinetic parameters of BoNT/A-LC on three SNAP substrates and inhibitory properties of GST-SNAP25(141–206) (GST-SNAP) and DCCH. Data shown are the average of three repeats.
|
ZnCl2 (10 μM) |
|||
|---|---|---|---|
| Km (μM) | kcat (sec−1) | kcat/Km | |
| CsY | 0.84 ± 0.35 | 19 ± 4 | 22.6 |
| CsYY | 0.49 ± 0.16 | 7 ± 2 | 14.3 |
| YsCsY | 0.46 ± 0.03 | 11 ± 3 | 23.9 |
|
| |||
|
ZnCl2 (300 μM) |
|||
| Km (μM) | kcat (sec−1) | kcat/Km | |
| CsY | 8.1 ± 2.2 | 20 ± 4 | 2.47 |
| CsYY | 0.83 ± 0.02 | 4.0 ± 0.2 | 4.82 |
| YsCsY | 1.5 ± 0.3 | 3.9 ± 0.4 | 2.6 |
|
| |||
|
ZnCl2 (10 μM) |
|||
| GST-SNAP | DCCH | ||
|
|
|||
| Ki (μM) | IC50(μM) | ||
| CsY | 7.6 ± 2.5 | 81 ± 5 | |
| YsCsY | 4.5 ± 0.3 | 59 ± 7 | |
We have shown that longer SNAP peptide substrates containing both recognition exosites more closely mimic the natural substrate than shorter peptides and that increasing the number of acceptor fluorophores (YFP) in our tandem FRET substrates indeed enhances the efficiency of capturing the CFP fluorescence emission. We have also demonstrated that our tandem FRET substrates are highly sensitive reagents for detection (Fig. 6 and 7) and characterization of BoNT/A activity (Table 1). Overall, YsCsY was a better FRET substrate with the greater ΔRFU at 530 nm for use in kinetic characterization studies, where initial rates and continuous monitoring of reaction progress is necessary.
YsCsY also had the largest ratiometric change at 530 nm versus 475 nm. The estimated Z′ factors (Zhang et al., 1999) for ratiometric assay were >0.9 for YsCsY concentrations of 0.3 to 10 μM (Fig. 3C–D), compared to ~0.7 for CsY, making YsCsY a more suitable substrate for use in HTS aimed at identifying physiologically relevant inhibitor candidates against BoNT/A. This enhanced dynamic range is critical for HTS, particularly in identifying lead compounds for inhibitors that only provide partial inhibition of enzyme activity. Importantly, the strategy employed to generate our tandem FRET substrates for BoNT/A can readily be adapted for use in the design of enhanced FRET substrates for other BoNT serotypes as well as for other protease substrates requiring extended recognition exosites for improved specificity, such as thrombin and other blood coagulation proteases (Bock et al., 2007), matrix metalloproteases (Overall, 2002), and pregnancy-associated plasma protein-A (Mikkelsen et al., 2008).
Supplementary Material
Supplementary Material includes additional figures showing results from expanded and more detailed experimental data that supports and enhances the condensed summary provided in the main text.
Acknowledgments
This research was supported through funds (to BAW) from the Great Lakes RCE (NIH/NIAID award 1-U54-AI-057153). We thank Joseph Barbieri for helpful discussions and critical reading of the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, et al. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–70. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Baldwin MR, Bradshaw M, Johnson EA, Barbieri JT. The C-terminus of botulinum neurotoxin A light chain contributes to solubility, catalysis and stability. Protein Expr Purif. 2004;37:187–195. doi: 10.1016/j.pep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bock PE, Panizzi P, Verhamme IM. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;(Suppl 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- Carruthers A, Carruthers J. Botulinum toxin type A: History and current cosmetic use in the upper face. Sem Cutaneous Medicine Surgery. 2001;20:71–84. doi: 10.1053/sder.2001.25138. [DOI] [PubMed] [Google Scholar]
- Chen S, Barbieri JT. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J Biol Chem. 2006;281:10906–10911. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- Dong M, Tepp WH, Johnson EA, Chapman ER. Using fluorescent sensors to detect botulinum neurotoxin activity in vitro and in living cells. Proc Natl Acad Sci USA. 2004;101:14701–14706. doi: 10.1073/pnas.0404107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubanks LM, Hixon MS, Jin W, Hong S, Clancy CM, Tepp WH, Baldwin MR, Malizio CJ, Goodnough MC, Barbieri JT, Johnson EA, Boger DL, Dickerson TJ, Janda KD. An in vitro and in vivo disconnect uncovered through high-throughput identification of botulinum neurotoxin A antagonists. Proc Natl Acad Sci USA. 2007;104:2602–2607. doi: 10.1073/pnas.0611213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukreja R, Singh BR. Biologically active novel conformational state of botulinum, the most poisonous poison. J Biol Chem. 2005;280:39346–39352. doi: 10.1074/jbc.M508463200. [DOI] [PubMed] [Google Scholar]
- Mikkelsen JH, Gyrup C, Kristensen P, Overgaard MT, Paulsen CB, Laursen LS, Oxvig C. Inhibition of the proteolytic activity of pregnancy-associated plasma protein-A by targeting substrate exosite binding. J Biol Chem. 2008;283:16772–16780. doi: 10.1074/jbc.M802429200. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Molgo J. Botulinal neurotoxins: revival of an old killer. Curr Opinion Pharmacol. 2005;5:274–279. doi: 10.1016/j.coph.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nature Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- Schmidt JJ, Bostian KA. Endoproteinase activity of type A botulinum neurotoxin: substrate requirements and activation by serum albumin. J Protein Chem. 1997;16:19–26. doi: 10.1023/a:1026386710428. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Washbourne P, Pellizzari R, Baldini G, Wilson MC, Montecucco C. Botulinum neurotoxin types A and E require the SNARE motif in SNAP-25 for proteolysis. FEBS Letters. 1997;418:1–5. doi: 10.1016/s0014-5793(97)01328-8. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material includes additional figures showing results from expanded and more detailed experimental data that supports and enhances the condensed summary provided in the main text.