Abstract
Exercise increases the intracellular T2 (T2,i) of contracting muscles. The mechanism(s) for the T2,i increase have not been fully described, and may include increased intracellular free water and acidification. These changes may alter chemical exchange processes between intracellular free water and proteins. In this study, the hypotheses were tested that 1) pH changes T2,i by affecting the rate of magnetization transfer (MT) between free intracellular water and intracellular proteins and 2) the magnitude of the T2,i effect depends on acquisition mode (localized or non-localized) and echo spacing. Frog gastrocnemius muscles were excised and their intracellular pH was either kept at physiological pH (7.0) or modified to model exercising muscle (pH 6.5). The intracellular transverse relaxation rate (R2,i =1/T2,i) always decreased in the acidic muscles, but the changes were greater when measured using more rapid refocusing rates. The MT rate from the macromolecular proton pool to the free water proton pool, its reverse rate, and the spin-lattice relaxation rate of water decreased in acidic muscles. It is concluded that intracellular acidification alters the R2,i of muscle water in a refocusing rate-dependent manner and that the R2,i changes are correlated with changes in the MT rate between macromolecules and free intracellular water.
Keywords: intracellular pH, T2, magnetization transfer, muscle functional MRI
INTRODUCTION
The proton T2 of water increases in exercising muscles by up to 30% (1,2). This robust change in T2 in response to a normal physiological perturbation has allowed exercising muscle and models of exercising muscle to be outstanding experimental paradigms for studying the biophysical basis of transverse relaxation. Moreover, measuring T2 or T2-weighted image signal intensity may provide information concerning the metabolic and hemodynamic responses of muscle to exercise (3,4).
Several hypotheses regarding the mechanism of the T2 increase have been proposed. Several researchers have shown that increases in intracellular muscle water content may be the most important determinant of exercise-induced T2 changes (5–7). During exercise, intracellular water accumulation may result from osmotically (7,8) and/or hydrostatically driven fluid shifts. For a system containing free intracellular water and macromolecular protons exchanging rapidly by way of a third pool of interfacial water, such fluid shifts will increase the apparent T2 of the free intracellular water by changing the relative populations and/or exchange rates between the pools (9,10). Support for the hypothesis that osmotically induced intracellular water accumulation increases T2 during exercise comes from 1) greater levels of metabolite accumulation, volume change, and T2 change in predominantly glycolytic muscles than in predominantly oxidative muscles (6); 2) greater levels of T2 and relative metabolite accumulation (and, presumably, volume change) in exercised fresh water than marine invertebrates (8); and 3) the direct relationship between T2 and intracellular volume in frog sartorius muscles during osmotic shock experiments (7).
In addition, intense exercise decreases the intracellular pH (pHi) in healthy subjects. Support for a pH effect on T2 comes from the inverse relationship between pHi and T2 following exercise (11) and during direct manipulations of pH in isolated muscles (7,12); we proposed that pH affects the T2 by modulating chemical exchange pathways between macromolecules and free water (7). However, during incremental arm ergometer exercise, pHi changes lag T2 changes and following exercise, pHi recovers more rapidly than T2 (13). Also, Meyer et al. have shown that there is no additional effect of pHi on the in vivo T2 changes measured during imaging experiments (8). These discrepancies concerning the role of pHi changes in the T2 change of exercise may be due to differences in the T2 measurement techniques employed. Because the sensitivity of T2 to chemical exchange effects depends on the refocusing rate (14), the rapid refocusing pulses used the Damon et al. study may have been sensitive to a pHi effect on T2, while the slower refocusing rates using the imaging studies of Meyer et al. may not have been.
Therefore, in order to understand better the mechanism of the increased T2 in exercising muscle and more specifically the effect of pHi on muscle T2, the T2 of ex vivo frog muscle was measured at two different pHi values using non-localized and localized Carr-Purcell Meiboom-Gill (CPMG) pulse sequences with different echo spacing. A second reason for the study was to relate pHi-induced T2 changes with changes in the magnetization transfer (MT) rate between the immobile macromolecular proton pool and the free water proton pool. pHi values of 7.0 and 6.5 were chosen to model resting and exercise conditions, respectively. Frog muscle was used because it is a valid model for many aspects of mammalian muscle physiology and because it is more robust ex vivo than mammalian tissue. The results of the study show that intracellular acidification increases the intracellular T2 (T2,i) without changes in muscle density, and that the magnitude of the effect depends on echo spacing. These T2,i changes are accompanied by changes in the MT rate between the macromolecular and free intracellular water proton pools and in the longitudinal relaxation rate (R1) of water.
METHODS
Tissue Preparation
These procedures were approved by the Institutional Animal Care and Use Committee at Vanderbilt University Medical Center. Rana pipiens were purchased from Carolina Biological Supply (Burlington, NC), anesthetized in a solution of Fenquil 1 g/L for 15–20 min, and then decapitated and doubly pithed. Pairs of gastrocnemius muscles (<0.5 cm (diameter, d) × ~4.5 cm (length, l) were removed and the tendons of origin and insertion were tied with nylon sutures. The muscles were bathed in Ringer’s solution and oxygenated in an ice bath for 1 hour and 45 minutes. The Ringer’s solution contained (in mM) 115 NaCl, 2.0 KCl, 2.5 CaCl2, 2.15 Na2HPO4, and 0.85 NaH2PO4 (pH 7.0 at 20 °C). A modified Ringer’s solution was used to acidify the muscles (15) and it contained (in mM) 75 NaCl, 40 NH4Cl, 2.0 KCl, 2.5 CaCl2, 2.15 Na2HPO4, and 0.85 NaH2PO4 (pH 7.0 at 20 °C). The muscles were refrigerated in either the Ringer’s solution or modified Ringer’s solution at 4 °C for ~12 hours. The muscle stored in modified Ringer’s solution was rinsed several times in Ringer’s solution for at least 3 hours prior to experiment, acidifying the muscle (15). The muscle stored in Ringer’s solution was maintained in its preceding condition during the experiment.
The muscle was placed inside a 12 mm (d) × 7 cm (l) NMR tube (Wilmad, Buena, NJ) with open ends. The tendon sutures were held in place by the tube’s end caps, ensuring that the muscle remained securely in place. This inner tube was placed in an outer tubular holder (15 mm (d) × 8 cm (l)), also containing Ringer’s solution. Holes in the end caps of the inner tube and in the top portion of the outer tube allowed air bubbles to escape, improving B0 homogeneity. The muscle tube assembly was placed in a Chemagnetics (Fort Collins, CO) 40 mm inner diameter millipede coil, parallel to the long axis of the RF coil, and the entire assembly was parallel to B0.
Experiments were performed at room temperature. Temperature measurements were made by placing a non-magnetic thermocouple probe in the sample holder adjacent to the muscle and were used to verify the thermal stability of the sample and to determine the negative log of the equilibrium acid constant of carnosine (pKa,c) for the calculation of pHi, as described below.
MR Data Acquisition
General
All MR experiments were made over a three-hour period using a 120 mm, horizontal bore 4.7 T superconducting magnet (Magnex Scientific, Oxfordshire, UK) with a Varian Inova console (Varian, Inc., Palo Alto, CA). Prior to all studies, global shimming was done using a single pulse-acquire sequence until the water linewidth was less than 20 Hz. For measurements requiring slice selection, localized shimming was performed using a PRESS sequence on 10 × 10 × 30 mm voxel until the water linewidth was less than 9 Hz.
Total Muscle Volume
Muscle volume was measured by using a single spin-echo multi-slice (SEMS) sequence. The experimental parameters included a 4 ms Gaussian pulse, echo time (TE)/repetition time (TR) = 30/2000 ms, 128 × 128 matrix size, 46 – 50 slices, 1 slice-interleaved acquisition with thickness = 1 mm and no gap, number of excitations (NEX) = 1, and field of view (FOV) = 18 mm × 18 mm.
1H MR Spectroscopy
At the middle and conclusion of the MR experiments, 1H water suppressed spectra were obtained using a PRESS sequence modified with MEGA (16) water saturation. The experimental parameters included a 2 ms sinc excitation pulse with 8 ms Gaussian saturation pulse, TE/TR = 28/3000 ms, NEX = 64 – 128 and voxel size = 7 mm × 7 mm × 20 mm.
Localized T2
Transverse relaxation decay data were obtained using a single-slice, multi-echo CPMG sequence. The experimental parameters included a 1 ms sinc excitation pulse followed by 300 μs duration, 90°-180°-90° composite refocusing pulses. For all studies, TR = 3s; for the 8 ms echo spacing, the number of echoes (NE) = 50, and for the 30 ms echo spacing, NE = 30. The FOV was 16 mm × 16 mm with a 2 mm slice thickness, matrix = 64 × 64 matrix, and NEX = 4. The power was calibrated for a 300 μs, 90°-180°-90° composite pulse with maximum signal intensity profile attained with a 4 mm slice and FOV of 10 mm × 10 mm that included as much of the muscle sample and as little of the Ringer’s solution as possible.
Non-localized T2
Transverse relaxation data were obtained by using a CPMG sequence having a 37.5 – 40 μs hard excitation pulse and refocusing pulses of 75 – 80 μs. For the echo time spacing of 2 ms, NE = 4096 echoes and for the echo time spacing of 8 ms, NE = 2048. The TR was 5 s with NEX = 16.
Quantitative MT (qMT)
The MT imaging data were acquired with a selective inversion recovery preparation (a 1 ms hard inversion (180°) pulse) followed by a variable inversion time (TI) and then a fast spin echo readout (a 500 μs Gaussian (90°) pulse with a train of eight 180° Gaussian refocusing pulses separated by 10 ms) (17). Twenty-five TI values were used, with 21 points logarithmically spaced between 3.5 ms and 150 ms with additional times of 300 ms, 1 s, 2 s and 8 s. Other parameters included a predelay (PD) = 3 s, NEX = 4, 64 × 64 matrix, FOV of 18 mm × 18 mm, and 1 mm thick slice.
Data Analysis
Muscle mass, volume, and density
Intracellular acidification brought about by transient exposure of muscle cells to NH4Cl activates Na+-H+ exchange as the principal pHi regulatory mechanism (18), which in turn may lead to a regulatory volume increase. Because the density of muscle under control conditions is greater than that of water, intracellular water accumulation would be reflected in a decrease in muscle density. Therefore, the mass of the muscles (mm) was measured at the end of NMR experiments and the muscle volume (Vm) was calculated from the SEMS data by defining a region of interest (ROI) around the muscle in each slice. The volume of each ROI was computed from the FOV, slice thickness, and matrix size and used to calculate Vm. Muscle density was calculated as ρm = mm/Vm.
Intracellular pH
Time domain data were processed in Matlab v. 7.0 (The Mathworks, Inc. Natick MA) using 0.2 Hz exponential line broadening, baseline correction, and Fourier transformation. Chemical shifts were referenced to creatine –R–NH2 at 3.02 ppm (19). The C-2 carnosine proton peak was fitted to a Lorentzian function and the chemical shift of the maximum peak height (δc) was used to calculate pHi, using
| [1] |
where δa is the acidic limiting chemical shift (8.58 ppm) and δb is the basic limiting chemical shift (7.66 ppm) (19). The pKa,c was calculated from the information provided in reference (19) and the temperature.
T2
A multi-component T2 model and a non-negative least squares (NNLS) algorithm (20) were used. The value for the regularizer term, μ, was ~10/SNR (μ = 0.01 and 0.05 for the nonlocalized and localized data, respectively). The non-localized transverse relaxation decay data were fitted to 128 T2 values logarithmically spaced between the first TE (2 or 8 ms) and 5 s. The localized transverse relaxation decay data were obtained by specifying an ROI around the muscle borders (ROI size range 460 – 745 pixels). These data were fitted to 250 T2 values logarithmically spaced between 8 or 30 ms and 1 s. To characterize the information in the T2 spectra simply, regions of short and long biological T2 values, assumed to represent intracellular and extracellular water, were defined. The intracellular T2 (T2,i) regions were defined as: 15 – 52 ms (non-localized, TE spacing = 2 ms); 15 – 70 ms (non-localized, TE spacing = 8 ms); 15 – 57 ms (control, localized, TE spacing = 8 ms); 15 – 45 ms (acidic, localized, TE spacing = 8 ms); 30 – 45 ms (localized, TE spacing = 30 ms). The extracellular T2 (T2,e) regions were defined as the remaining values less than 400 ms. The rationale for the thresholds distinguishing T2,i and T2,e is illustrated in the Results section. The 400 ms upper value for T2,e was set on the basis of T2 spectra from phantom studies in which a vial containing a short T2 species (~75 ms) placed within Ringer’s solution (T2 ~2.1 s). The T2 spectra contained an extra peak at ~617 ms when T2 was measured using non-localized acquisitions, but not with localized acquisitions (results not shown). This 400 ms threshold is approximately two times greater than the largest value of T2,e that has been reported in the literature. Within each region, the average T2 value (weighted by relative signal intensity) was calculated and the transverse relaxation rates were calculated as R2 ≡ 1/T2. All echoes were used in the T2 analysis since pilot studies revealed similar results regardless of whether all echoes, even echoes-only or odd echoes-only were used.
Intracellular volume fraction
The intracellular and extracellular water contents were assumed to relate directly to the areas under the curve in the T2,i and T2,e spectral regions. The intracellular water fraction, Fi, was calculated as 100% × the ratio of the area of intracellular region to the total area under the curve.
qMT Analysis
The analysis described in reference (17) was applied to the selective inversion recovery data. This analysis assumes that the sample can be described as containing a macromolecular pool with population fraction pm and a free water pool with population fraction pf. The data were fit on a pixel-by-pixel basis using Matlab’s built-in Nelder-Mead Simplex search algorithm to a bi-exponential function of TI as
| [2] |
where Mf(TI) is the longitudinal magnetization at time TI, Mf∞ is the equilibrium magnetization, and are the slow and fast recovery rates, respectively, and and are the corresponding amplitudes (17). The MT rate from the macromolecular pool to the free water pool (kmf) and the pool size ratio (pm/pf) were determined by using Eqs. 4 and 10 of reference (17); the reverse rate (kfm) was calculated as kfm=(pm/pf)·kmf.
Statistics
Descriptive statistics include the mean and standard deviation (SD). Paired, two-tailed Student’s t-tests were performed to compare the mean values of pHi, ρM, R2, kmf, and in the control and acidic muscles. A one-way analysis of variance was used to compare the mean values of the R2,i difference between control and acidic muscle (ΔR2,i) and for the different echo time spacings and acquisition type. The hypotheses that kmf is linearly related to [H+] and pHi were tested by calculating linear correlations between pHi and kmf and between [H+] and kmf, separately for the acidic muscles, control muscles, and all muscles. Because kfm is calculated from pm/pf and kmf and is therefore not an independent measurement, statistics for this variable were not calculated. A p value <0.05 was considered statistically significant.
Results
Intracellular pH
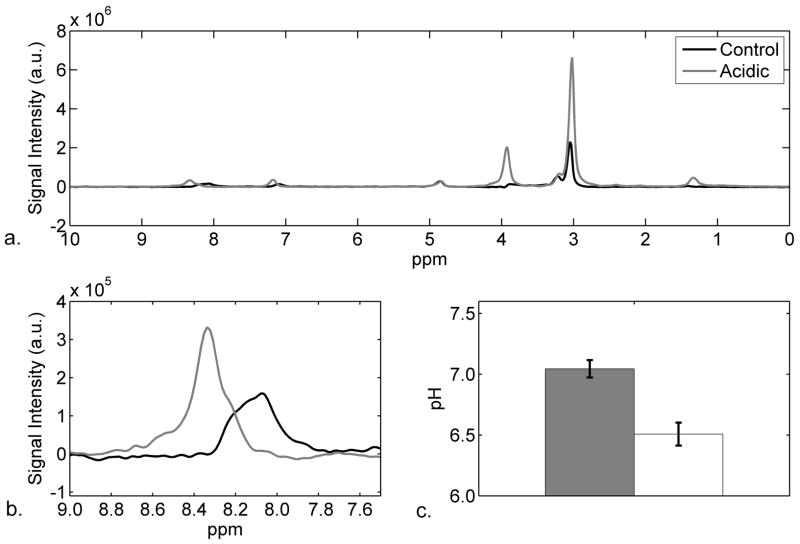
Figure 1a shows typical 1H NMR water-suppressed spectra for the control (black line) and acidic (gray line) muscles. Figure 1b shows the carnosine C-2 proton resonance in a zoomed region (7.5 – 9 ppm) of the spectra in Figure 1a. The chemical shift of the C-2 proton is 8.10 ppm for the control muscle (pHi 7.03) and shifts to 8.30 ppm for the acidic muscle (pHi 6.58). Figure 1c shows the mean and SD for the pHi of all muscles. The control muscles had a mean pHi of 7.04 ± 0.07 whereas the acidic muscles had a mean pHi of 6.51 ± 0.09 (P < 0.001). During the course of the MR experiments, the pHi did not vary more than 0.05 pH units.
Figure 1.
a) Representative 1H NMR water suppressed spectra of control (black line) and acidic (gray line) muscles at pHi 7.03 and 6.58, respectively. b) Zoomed-in region of Panel a, showing the shift of the C-2 carnosine resonance between the control (black line) and acidic (gray line) muscles. For panels a and b, the differences in peak heights reflect receiver gain differences only. c) Bar graph of pHi of muscles (n=7) showing mean and standard deviation of control (pH 7.04 ± 0.07) and acidic (pH 6.51 ± 0.09) muscles (P < 0.001).
Mass, Volume, Density, Temperature
The masses of the control and acidic muscles were 1.63 ± 0.20 and 1.68 ± 0.23 g, respectively (P = 0.019). The average volumes for the control and acidic muscles were 1.32 ± 0.15 cm3 and 1.34 ± 0.16 cm3, respectively (P = 0.033). The density for the control muscles was 1.23 ± 0.02 g/cm3 and for the acidic muscles was 1.25 ± 0.03 g/cm3 (P = 0.22). The muscle temperature did not vary more than 0.5° within an individual experiment and for all experiments ranged between 20 – 23 °C.
Transverse Relaxation Rates and Derived Measures
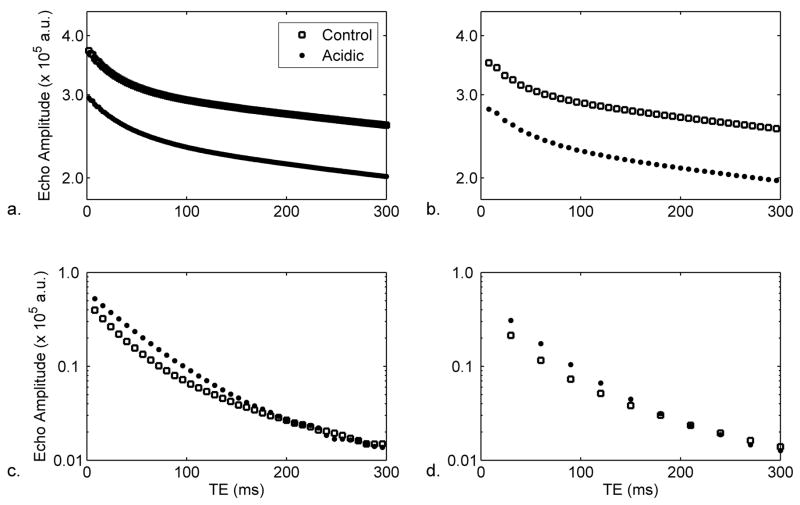
Semi-log plots illustrating transverse magnetization decay data for TE<300 ms are shown for non-localized experiments with echo spacing times of 2 ms and 8 ms in Figures 2a and 2b, respectively. Representative T2 decay data for localized experiments with echo spacings of 8 ms and 30 ms are shown in Figures 2c and 2d, respectively. In all four plots, the decay is non-monoexponential.
Figure 2.
Representative semi-log plots of transverse magnetization decay data for a) 2 ms echo spacing of non-localized experiments; b) 8 ms echo spacing of non-localized experiments; c) 8 ms echo spacing of localized experiments; and d) 30 ms echo spacing of localized experiments. As indicated in the legend to Panel a, the data are shown for control and acidic conditions. For all plots, the TE region from 0–300 ms is shown, in order to highlight the transverse relaxation of the muscle water; note that non-localized acquisitions include contributions from Ringer’s solution. Also, note that the differences in the Y intercepts result from differences in receiver gain settings and do not imply proton density differences between the conditions.
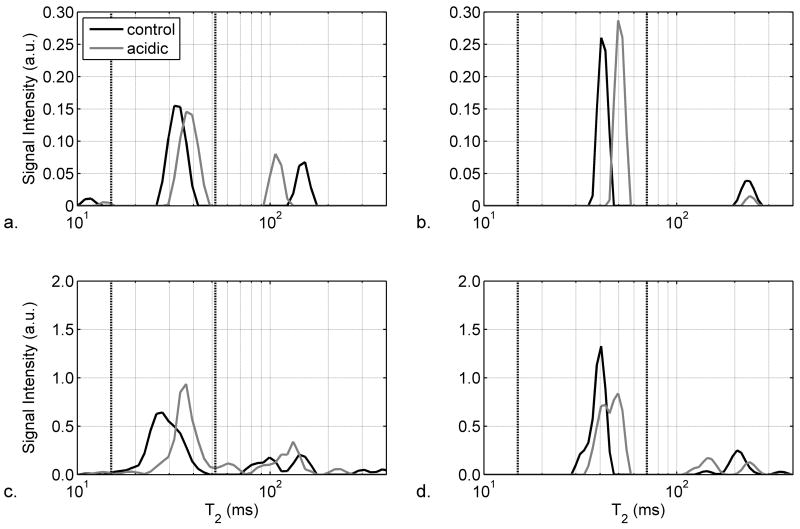
Representative non-localized T2 spectra from a control muscle (black line) and the paired acidic muscle (gray line) are shown as a semilog plot in Figures 3a and 3b for echo spacing times of 2 ms and 8 ms, respectively. There is a peak representing T2,i at ~30 ms and a peak representing T2,e at ~125 ms. Other peaks that appeared but are not shown include a third apparent T2 component between 400 ms and 1 s and a large peak with a T2 of ~2 s that resulted from the Ringer’s solution. The summed T2 spectra are shown in Figures 3c and 3d for the experiments performed at 2 ms and 8 ms, respectively. The vertical lines indicate the boundaries of the T2,i region.
Figure 3.
Semi-log plots of T2 spectra from representative (Panels a and b) and summed (Panels c and d; n=7) non-localized acquisitions with echo spacings of 2 ms (Panels a and c) and 8 ms (Panels b and d). As indicated in the legend to Panel a, the data are shown for control muscles (black lines) and acidic muscles (gray lines). In order to highlight the muscle region of the T2 spectrum, the region from 10–400 ms is shown. The vertical dashed lines indicate the boundaries of the intracellular T2 component, as discussed in the text.
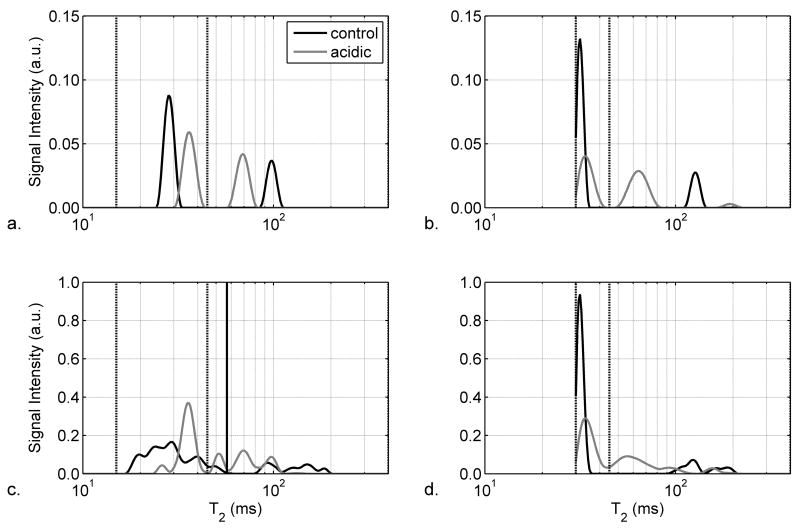
Representative T2 spectra from a control muscle (black line) and the paired acidic muscle (gray line) are shown in Figures 4a and 4b for localized acquisitions using echo time spacings of 8 ms and 30 ms, respectively. The two peaks represent the T2,i component at ~30 ms and the T2,e component at 80 to 150 ms. The summed T2 spectra measured with echo spacing times of 8 and 30 ms are shown in Figures 4c and 4d, respectively. The vertical lines indicate the T2,i region. For both echo spacings, the summed T2 spectrum of the acidic muscles show that the T2,i increases and the T2,e decreases as compared to the control muscles.
Figure 4.
Semi-log plots of T2 spectra from representative (Panels a and b) and summed (Panels c and d; n=7) non-localized acquisitions with echo spacings of 2 ms (Panels a and c) and 8 ms (Panels b and d). As indicated in the legend to Panel a, the data are shown for control muscles (black lines) and acidic muscles (gray lines). In order to highlight the muscle region of the T2 spectrum, the region from 10–400 ms is shown. The vertical dashed lines indicate the boundaries of the intracellular T2 component, as discussed in the text.
The mean and SD of the R2,i values are reported in Table 1. Regardless of echo spacing or whether the experiments were localized or non-localized, R2,i decreased as pHi decreased. In non-localized experiments, the difference in R2,i between the acidic and control muscles (ΔR2,i) was 8.1 ± 1.5 s−1 for the 2 ms TE spacing and 3.4 ± 1.0 s−1 for the 8 ms TE spacing (P = 0.002 and 0.013, respectively). For localized experiments, the ΔR2,i values were 4.5 ± 1.9 s−1 and 3.6 ± 0.8 s−1 for the TE = 8 ms and 30 ms spacings, respectively. The ΔR2,i values are shown in Figure 5 and reveal that ΔR2,i was significantly greater for the non-localized, TE = 2 ms spacing than for the non-localized, TE = 8 ms spacing (P = 0.020) and for the localized, 30 ms TE spacing (P = 0.023); a similar trend was noted for the 8 ms TE, localized experiment (P = 0.066).
Table 1.
Influence of pHi on R2,i, the latter assessed using non-localized and localized acquisitions and with different TE spacings. The first two rows of data report the mean and standard deviation (N = 7).
| Non-localized R2,i (s−1) | Localized R2,i (s−1) | ||||
|---|---|---|---|---|---|
| 2 ms | 8 ms | 8 ms | 30 ms | ||
| Means at pHi ~ | 7.0 | 35.2 ± 3.1 | 25.8 ± 2.2 | 34.0 ± 2.7 | 31.5 ± 0.3 |
| 6.5 | 27.0a ± 3.0 | 22.4b ± 2.6 | 29.5c ± 3.8 | 27.9b ± 1.9 | |
p < 0.05;
p < 0.01;
p = 0.052
Figure 5.
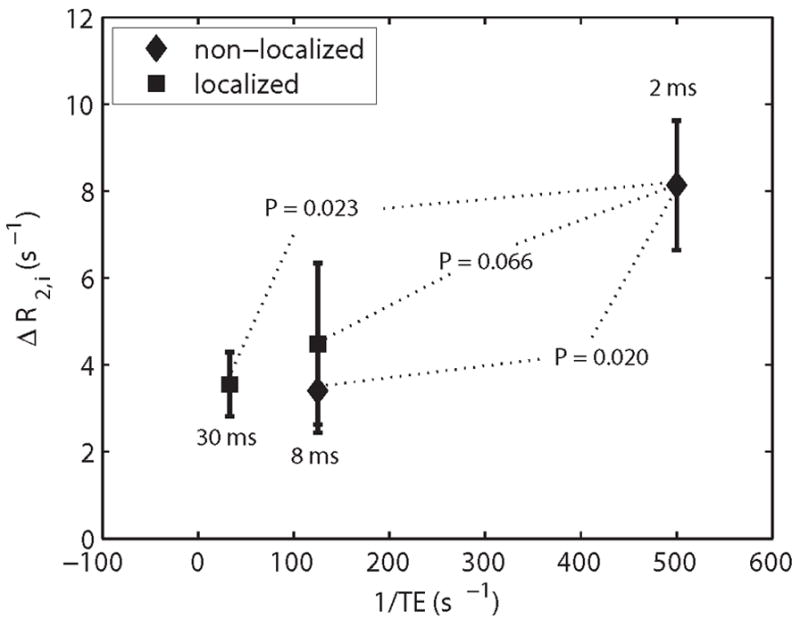
The differences in R2,i (ΔR2,i) between acidic and control muscles are plotted as a function of the refocusing rate for the non-localized and localized experiments. The error bars represent the standard error of the mean. In both cases, localized and non-localized, as the echo spacing time is decreased, ΔR2,i becomes larger. The P values reported are relative to the shortest TE of 2 ms.
In general, R2,e did not change significantly with pHi. The only exception was the localized experiment with an echo spacing of 8 ms, in which the R2,e for the control muscle was 8.3 ± 2.2 s−1 and for the acidic muscle was 13.7 ± 3.3 s−1 (P < 0.001). Irrespective of the type of measurement, Fi was ~80% in the control condition. In the non-localized measurements the Fi values did not differ significantly between the control and acidic muscles. However, the Fi values derived from the localized R2,i measurements differed significantly between the control and acidic muscles, decreasing to 60% in the acidic condition.
Magnetization Transfer
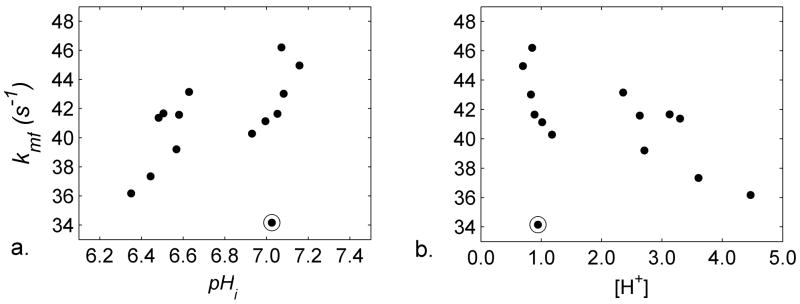
Sample selective inversion recovery data are shown for the acidic and control conditions in Figure 6. Figure 7 presents the raw data illustrating the relationships among kmf, pHi, and [H+] and Table 2 reports the MT parameters under control and acidic conditions. A data point in the control group (the circled point in Figure 7) was identified as an outlier with respect to kmf, and so the data from it and its paired, acidic muscle were removed from the following statistical comparisons. kmf and R1− decreased in acidic muscles; pm/pf did not change. Table 2 also reports the correlations between kmf and pHi and between kmf and [H+]. Significant or nearly-significant linear correlations existed between each variable pair, regardless of whether the acidic muscles only, control muscles only, or all muscles were considered.
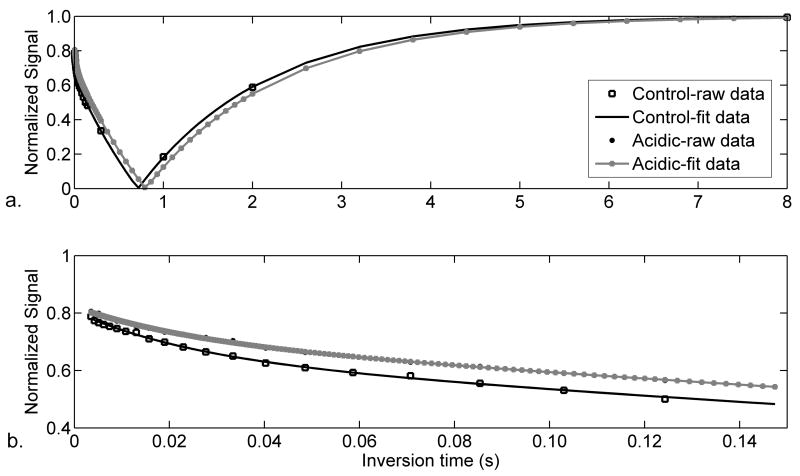
Figure 6.
Representative MT-inversion recovery data for control (squares and black lines) and acidic (gray points and lines) muscles. A) Full inversion-recovery curve. b) The early portion of the recovery (0 – 200 ms) in Figure 7a, revealing the bilinear recovery resulting from MT.
Figure 7.
Scatterplots illustrating relationships among pH and kmf parameters. a) Plot of kmf as a function of pHi. The circled point was identified as an outlier and so the data from it and its acidic muscle pair were removed from all descriptive and inferential statistical tests involving the MT parameters. b) Plot of kmf as a function of [H+].
Table 2.
Effect of pHi on the MT rates, kmf and kfm, from the solid pool (with population fraction pm) to the free water pool (with population fraction pf) and in the reverse direction, respectively. The third and fourth rows report the correlations between pHi, [H+], and the MT parameter estimates.. Pool size ratio (pm/pf) and slow recovery rate ( ) are also reported. Statistical tests reflect removal of outlying point and its pair (see Text). Significant findings are indicated with superscripts; the same notation as in Table 1 has been continued here. For means, significance reflects differences from control condition. Note that because kfm is not an independent measurement, statistics for this variable are not presented.
| Kmf (s−1) | kfm (s−1) | pm/pf | (s−1) | ||
|---|---|---|---|---|---|
| Mean±SD at pHi ~ | 7.0 | 42.88 ± 2.31 | 3.56 ± 0.42 | 0.08 ± 0.01 | 0.68 ± 0.02 |
| 6.5 | 39.82 a ± 2.71 | 3.16 ± 0.18 | 0.08 ± 0.00 | 0.65b ± 0.02 | |
| Correlations with pHi | 0.85a (acidic) | ||||
| 0.78c (control) | |||||
| 0.76b (all) | |||||
| Correlations with [H+] | −0.85a (acidic) | ||||
| −0.78c (control) | |||||
| −0.73b (all) | |||||
p <0.05;
p <0.01;
p=0.066.
DISCUSSION
An important finding of this study is that decreases in pHi decrease R2,i in isolated muscle preparations. This finding is consistent with that of Fung and Puon’s study of permeabilized muscle strips in solutions with pH values of 5, 7 and 9, in which they showed that the R2 is directly related to pHi (12); with the observations of Bertram et al, who demonstrated that the T2 of extracted myofibril preparations also increases at low pH (21); and with those of our previous work at 7.0T, in which R2,i was directly related to pHi throughout the physiological range (6.5 –7.4) (7). In addition, this study shows that the influence of pHi on R2,i exists for all acquisition conditions studied, including localized and non-localized data acquisitions and for refocusing intervals ranging from 2 – 30 ms. The magnitude of R2,i varied with the refocusing period, however, with the pHi effect on R2,i being larger at 2 ms intervals than with longer intervals. Additionally, the pHi induced variations in R2,i correspond to changes in the rates of MT between macromolecular protons and free intracellular water and of longitudinal relaxation for free intracellular water. Collectively, these data provide new insights into the specific biophysical phenomena that contribute to the R2,i change of exercise, the exchange processes among the various tissue proton pools, and the biophysical influences on relaxation in general.
Multi-exponential Transverse Relaxation in Muscle
A common finding in transverse relaxometry studies of ex vivo muscle (7,9,22,23), and in certain studies of in vivo muscle (5,24), is that this tissue exhibits multi-exponential relaxation. In the present study, the 1H transverse relaxation of muscle water was non-monoexponential in all of the measurements performed, as reflected in the non-linear semi-log plots in Figure 2 and the complex T2 spectral patterns in Figures 3 and 4. We conducted phantom studies that demonstrated that T2 components larger than 400 ms, but unassociated with the Ringer’s solution (T2 ~2 s), were artifactual in origin. In order to characterize the information in the biological portion of the spectrum simply and in a manner that would permit statistical comparisons, we interpreted the shorter T2 portions as reflecting water in a single compartment of the intracellular space of muscle and the longer T2 portion of the spectrum as representing water in a single compartment of the interstitial space (9,22,23). In addition, a T2 component <10 ms has been inconsistently reported for muscle, which has been attributed to hydration shell of macromolecules (9). The long TE spacings used for most of the T2 measurements in the present study precluded the observation of this component.
pHi and Intracellular Water Content
Despite the potential for activation of Na+-H+ exchange and regulatory volume changes in the cells, we argue for several reasons that the MT and R2,i changes observed in this study are a direct and exclusive effect of intracellular acidification. Most importantly, the densities of the control and acidic muscles did not differ significantly and were uncorrelated with the R2 and MT findings. Second, intracellular water accumulation would tend to increase Fi. However, Fi either did not change (non-localized R2 measurements) or decreased (localized R2 measurements). We consider these two possibilities further below; but for the present argument, the salient point that neither behavior is consistent with the requirement of a higher Fi under this alternative hypothesis. Finally, pm/pf should decrease with intracellular water accumulation; but this variable did not differ between the two conditions. For these reasons, we conclude that the decrease in pHi was the most significant, and probably the only, effecter of the R2,i decrease in these experiments.
pHi and R2,i
A consistent finding in this work is that the pHi is directly related to R2,i. Moreover, the magnitude of this effect varies with TE spacing, with larger effects being observed with shorter spacings. This finding resolves the discrepancy between our previous findings with short TE spacings (7), which indicated a direct effect of pHi on R2,i, and those of Meyer et al. with long TE spacings (8), which suggested that there was no additional effect of exercise-induced pHi changes on the whole-muscle T2 beyond that already exerted by intracellular water accumulation. Because the present data demonstrate that the dynamic range of the pHi effect on T2 at long TE spacings is small, it may have been that in the Meyer et al. study, the effect of pHi on T2 was below the limit of detectability. Moreover, the ability to resolve distinct intracellular and interstitial T2 components in the present study may have increased the sensitivity of our measurement as well. The dependence of R2,i on pHi under all acquisitions studied indicates that the possibility for systematic, group-wise variations in the pHi response to exercise, such as might occur in metabolic conditions such as myophosphorylase deficiency and phosphofructokinase deficiency (25) or with peripheral vascular disease (26), must be taken into account when interpreting R2 or T2 data from exercising muscles.
While the analyses performed resulted consistently in the identification of a pronounced effect of pHi on R2,i, there was ambiguity with regard to the effects of pHi on R2,e and Fi. In the non-localized experiments, neither R2,e nor Fi changed significantly, while in the localized experiments, R2,e decreased and Fi increased. This discrepancy may have been due to differences in SNR, difficulty in fitting the non-localized data due to the large signal contribution from the buffer, and/or B1 inhomogeneity in the refocusing pulses used in the non-localized acquisitions. Without a standard measurement such as a sucrose or inulin space determination, it is not possible to resolve this discrepancy. For this reason, and because several studies using different methods have demonstrated that the T2 change of exercise is primarily an intracellular phenomenon, we focus the remaining portions of this discussion dealing with transverse relaxation on R2,i.
pHi and MT
Gochberg and Gore (17) previously reported values for kmf and pm/pf of 46 s−1 and 0.108, respectively, in the skeletal muscle of a single ferret maintained at 37°C. The mean value for kmf that we report here under control conditions, 42.88 s−1, is slightly lower than the value reported by Gochberg and Gore and probably relates to the temperature difference between the experimental preparations. We found also that the R1 of water, kmf, and kfm all decreased with pHi (that is, with increasing [H+]). A likely explanation for this effect lies in hydrogen exchange between water and amide protons, as the reaction is base-catalyzed for pH values greater than ~5 (27–29) and a recent preliminary report has indicated the presence of a pH-sensitive peak at the amide proton frequency in the CEST spectrum of similarly treated amphibian skeletal muscles (30). Also similar to the present observations, Gochberg et al. have observed decreases in R1+ (≈kmf) in acrylamine and methacrylamine polymer gels with decreasing pH (31). While simple acid-base chemistry would predict that the reaction rate would depend inversely and linearly on [H+] and exponentially with pH (27–29,32), we observed linear dependences of kmf with both pH and [H+], regardless of experimental condition (Table 2 and Figure 7). The linear dependence of kmf on pH may exist if the proteins function as polyprotic acids, whose multiple pKa’s would make for a complex dependence of reaction rates on pH that could appear linear within the 0.8 pH-unit range that we studied. In addition, there are well recognized roles for acidic and basic side chains of proteins in determining protein structure, as their pH-dependent ionization states affect the number of opportunities for hydrogen bonding (33). In structural motifs such as the leucine zipper, there are periodic alterations in amide exchange rates that depend on the local tertiary structure, with the amide proton exchange rate corresponding to hydrogen bond length (34). Because the secondary and tertiary structures of muscle proteins including sperm whale apomyoglobin (35) and the myofibrillar proteins (21) are altered by pH, alternative and/or additional mechanisms for the non-exponential dependences of the MT rates on pHi also exist. These include secondary or tertiary structural changes in the proteins that remove exchange opportunities between functional groups on the proteins and water or that alter the amount of interfacial (hydration) water.
pHi, R2,i, and MT
As noted above, the dependence of R2,i on TE spacing, the correspondence between the MT and R2,i changes, and the field strength dependence of exercise-induced R2,i changes previously reported (7) are consistent with a chemical exchange mechanism of transverse relaxation (14). However, it should be noted that alternative explanations exist, at least in principle. Diffusion through magnetic field gradients such as those generated by heme-containing molecules would create a refocusing rate dependence of R2. However, this mechanism cannot explain the present findings, as Carr and Purcell (36) showed that the effect of diffusion on R2 should increase with the square of the echo spacing time; conversely, we observed larger R2,i changes with lower echo spacing times. Also, while we did not measure the transmembrane water exchange rate explicitly, we consider that any such changes are unlikely, as there was no coherent pattern to the effect of pHi on R2,e and the permeability of the aquaporin-4 water channel is not affected by acidic deviations from neutrality (37). In light of these considerations, and taking into account also the correspondence of the R2,i changes to the MT rate changes, we conclude that a pHi-mediated effect on chemical exchange was the predominant mechanism of the transverse relaxation rate change.
The measured muscle MT and relaxation rates dependences on pHi, with kmf, kfm, , and R2,i all showing small decreases with pHi, are similar to those seen previously in BIS gel dosimeters with varying chemical side groups (31) and in 1-palmitoyl-2-oleoylphosphatidyl-choline linked with either cholesterol or galactocerebroside (38). This connection between kmf and R2 is consistent with a third small pool of interfacial protons providing a connection between the free water and macromolecular proton pools and effecting both MT and relaxation. However, a simple three pool model would predict that (ΔR2)/(Δkmf) would be roughly equal to the pool size ratio (31). Our results for (ΔR2,i)/(Δkmf) depend on the echo spacing, but are always at least an order of magnitude greater than the pool size ratio. These results preclude using a simple three pool model for explaining the relaxation and MT results. Instead, they are consistent with the possible existence of polyprotic acids and pH-induced alterations in protein structure and interfacial water content discussed above.
Conclusions
We have shown that pHi affects the intracellular muscle R2,i without significant changes in either the muscle density or the ratio of macromolecular to free protons. Further, the R2,i change occurs regardless of TE spacing and with both non-localized and localized CPMG pulse sequences; however, shorter TE spacings are more sensitive to pHi changes than long TE spacings. There are corresponding changes in the MT properties of the muscle, with decreases in kmf and kfm with decreasing pHi. These changes suggest a role for amide proton exchange in the MT and transverse relaxation processes in muscle, and that these processes are modulated by based-catalyzed exchange and/or changes in protein structure. Comparison of the MT and transverse relaxation data reveals that transverse relaxation in muscle cannot be explained by a simple three-pool model.
Acknowledgments
We thank Nathan A. Oyler for use of his Matlab code for processing NMR spectroscopy data and David Damon of Pfizer Global Research and Development (Groton, CT) for helpful discussions. Funding was provided by NIH/NIAMS R01 AR050101 (BMD), NIH/NIBIB R01 EB001744 (MDD), NIH/NIBIB R01 EB001452 (DFG), and NIH/NIBIB T32 EB001628.
References
- 1.Bratton CB, Hopkins AL, Weinberg JW. Nuclear magnetic resonance studies of living muscle. Science. 1965;147:738–739. doi: 10.1126/science.147.3659.738. [DOI] [PubMed] [Google Scholar]
- 2.Fleckenstein JL, Canby RC, Parkey RW, Peshock RM. Acute effects of exercise on MR imaging of skeletal muscle in normal volunteers. AJR Am J Roentgenol. 1988;151(2):231–237. doi: 10.2214/ajr.151.2.231. [DOI] [PubMed] [Google Scholar]
- 3.Damon BM, Gore JC. Physiological basis of muscle functional MRI: predictions using a computer model. J Appl Physiol. 2005;98(1):264–273. doi: 10.1152/japplphysiol.00369.2004. [DOI] [PubMed] [Google Scholar]
- 4.Meyer RA, Prior BM. Functional magnetic resonance imaging of muscle. Exerc Sport Sci Rev. 2000;28(2):89–92. [PubMed] [Google Scholar]
- 5.Saab G, Thompson RT, Marsh GD. Effects of exercise on muscle transverse relaxation determined by MR imaging and in vivo relaxometry. J Appl Physiol. 2000;88(1):226–233. doi: 10.1152/jappl.2000.88.1.226. [DOI] [PubMed] [Google Scholar]
- 6.Prior BM, Ploutz-Snyder LL, Cooper TG, Meyer RA. Fiber type and metabolic dependence of T2 increases in stimulated rat muscles. J Appl Physiol. 2001;90(2):615–623. doi: 10.1152/jappl.2001.90.2.615. [DOI] [PubMed] [Google Scholar]
- 7.Damon BM, Gregory CD, Hall KL, Stark HJ, Gulani V, Dawson MJ. Intracellular acidification and volume increases explain R2 decreases in exercising muscle. Magn Reson Med. 2002;47(1):14–23. doi: 10.1002/mrm.10043. [DOI] [PubMed] [Google Scholar]
- 8.Meyer RA, Prior BM, Siles RI, Wiseman RW. Contraction increases the T2 of muscle in fresh water but not in marine invertebrates. Nmr in Biomedicine. 2001;14(3):199–203. doi: 10.1002/nbm.702. [DOI] [PubMed] [Google Scholar]
- 9.Hazlewood CF, Chang DC, Nichols BL, Woessner DE. Nuclear magnetic resonance transverse relaxation times of water protons in skeletal muscle. Biophys J. 1974;14(8):583–606. doi: 10.1016/S0006-3495(74)85937-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woessner DE. Brownian motion and its effects in NMR chemical exchange and relaxation in liquids. Concepts in Magnetic Resonance. 1996;8(6):397–421. [Google Scholar]
- 11.Weidman ER, Charles HC, Negro-Vilar R, Sullivan MJ, MacFall JR. Muscle activity localization with 31P spectroscopy and calculated T2-weighted 1H images. Invest Radiol. 1991;26(4):309–316. doi: 10.1097/00004424-199104000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Fung BM, Puon PS. Nuclear magnetic resonance transverse relaxation in muscle water. Biophys J. 1981;33(1):27–37. doi: 10.1016/S0006-3495(81)84870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng HA, Robergs RA, Letellier JP, Caprihan A, Icenogle MV, Haseler LJ. Changes in muscle proton transverse relaxation times and acidosis during exercise and recovery. J Appl Physiol. 1995;79(4):1370–1378. doi: 10.1152/jappl.1995.79.4.1370. [DOI] [PubMed] [Google Scholar]
- 14.Meiboom S, Luz Z. Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution - order of the reaction with respect to solvent. J Chem Phys. 1963;39(2):366–370. [Google Scholar]
- 15.Roos A, Boron WF. Intracellular pH. Physiol Rev. 1981;61(2):296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 16.Mescher M, Tannus A, Johnson MO, Garwood M. Solvent suppression using selective echo dephasing. J Magn Reson A. 1996;123(2):226–229. [Google Scholar]
- 17.Gochberg DF, Gore JC. Quantitative magnetization transfer imaging via selective inversion recovery with short repetition times. Magn Reson Med. 2007;57(2):437–441. doi: 10.1002/mrm.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marjanovic M, Elliott AC, Dawson MJ. The temperature dependence of intracellular pH in isolated frog skeletal muscle: lessons concerning the Na+-H+ exchanger. J Membr Biol. 1998;161(3):215–225. doi: 10.1007/s002329900328. [DOI] [PubMed] [Google Scholar]
- 19.Damon BM, Hsu AC, Stark HJ, Dawson MJ. The carnosine C-2 proton’s chemical shift reports intracellular pH in oxidative and glycolytic muscle fibers. Magnetic Resonance in Medicine. 2003;49(2):233–240. doi: 10.1002/mrm.10384. [DOI] [PubMed] [Google Scholar]
- 20.Whittall KP, Mackay AL. Quantitative interpretation of NMR relaxation data. J Magn Reson. 1989;84(1):134–152. [Google Scholar]
- 21.Bertram HC, Kristensen M, Andersen HJ. Functionality of myofibrillar proteins as affected by pH, ionic strength and heat treatment - a low-field NMR study. Meat Science. 2004;68(2):249–256. doi: 10.1016/j.meatsci.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Belton PS, Jackson RR, Packer KJ. Pulsed NMR studies of water in striated muscle. I Transverse nuclear spin relaxation times and freezing effects. Biochim Biophys Acta. 1972;286(1):16–25. doi: 10.1016/0304-4165(72)90084-0. [DOI] [PubMed] [Google Scholar]
- 23.Cole WC, LeBlanc AD, Jhingran SG. The origin of biexponential T2 relaxation in muscle water. Magn Reson Med. 1993;29(1):19–24. doi: 10.1002/mrm.1910290106. [DOI] [PubMed] [Google Scholar]
- 24.Saab G, Thompson RT, Marsh GD. Multicomponent T2 relaxation of in vivo skeletal muscle. Magn Reson Med. 1999;42(1):150–157. doi: 10.1002/(sici)1522-2594(199907)42:1<150::aid-mrm20>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Fleckenstein JL, Haller RG, Lewis SF, Archer BT, Barker BR, Payne J, Parkey RW, Peshock RM. Absence of exercise-induced MRI enhancement of skeletal muscle in McArdle’s disease. J Appl Physiol. 1991;71(3):961–969. doi: 10.1152/jappl.1991.71.3.961. [DOI] [PubMed] [Google Scholar]
- 26.Yoshioka H, Anno I, Kuramoto K, Matsumoto K, Jikuya T, Itai Y. Acute effects of exercise on muscle MRI in peripheral arterial occlusive disease. Magn Reson Imaging. 1995;13(5):651–659. doi: 10.1016/0730-725x(95)00018-c. [DOI] [PubMed] [Google Scholar]
- 27.Englander SW, Downer NW, Teitelbaum H. Hydrogen exchange. Ann Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- 28.Liepinsh E, Otting G. Proton exchange rates from amino acid side chains - implications for image contrast. Magnetic Resonance in Medicine. 1996;35(1):30–42. doi: 10.1002/mrm.1910350106. [DOI] [PubMed] [Google Scholar]
- 29.Mori S, van Zijl PCM, Shortle D. Measurement of water-amide proton exchange rates in the denatured state of staphylococcal nuclease by a magnetization transfer technique. Proteins: Structure, Function, and Genetics. 1997;28(3):325–332. [PubMed] [Google Scholar]
- 30.Louie EA, Does MD, Gochberg DF, Damon BM. Effect of pH on CEST in muscle. Toronto, ON: 2008. p. 2587. [Google Scholar]
- 31.Gochberg DF, Kennan RP, Maryanski MJ, Gore JC. The role of specific side groups and pH in magnetization transfer in polymers. J Magn Reson. 1998;131(2):191–198. doi: 10.1006/jmre.1998.1371. [DOI] [PubMed] [Google Scholar]
- 32.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JWM, van Zijl PCM. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-L-lysine and a starburst dendrimer. Magnetic Resonance in Medicine. 2006;55(4):836–847. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perutz MF. Electrostatic effects in proteins. Science. 1978;201(4362):1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 34.Goodman EM, Kim PS. Periodicity of amide proton exchange rates in a coiled-coil leucine zipper peptide. Biochemistry. 1991;30(50):11615–11620. doi: 10.1021/bi00114a002. [DOI] [PubMed] [Google Scholar]
- 35.Yang A-S, Honig B. Structural origins of pH and ionic strength effects on protein stability: acid denaturation of sperm whale apomyoglobin. J Molec Biol. 1994;237(5):602–614. doi: 10.1006/jmbi.1994.1258. [DOI] [PubMed] [Google Scholar]
- 36.Carr H, Purcell E. Effects of diffusion on free precession in NMR experiments. Phys Rev. 1954;94:630–638. [Google Scholar]
- 37.Nemeth-Cahalan KL, Kalman K, Hall JE. Molecular basis of pH and Ca2+ regulation of aquaporin water permeability. J Gen Physiol. 2004;123(5):573–580. doi: 10.1085/jgp.200308990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucharczyk W, Macdonald PM, Stanisz GJ, Henkelman RM. Relaxivity and magnetization transfer of white matter lipids at MR imaging: importance of cerebrosides and pH. Radiology. 1994;192:521–529. doi: 10.1148/radiology.192.2.8029426. [DOI] [PubMed] [Google Scholar]