Abstract

Cytochrome P450 3A4 is a major human drug-metabolizing enzyme, and displays pharmacologically-relevant allosteric kinetics caused by multiple substrate and/or effector binding. Here, in the first single-molecule fluorescence studies of CYPs, we use total internal reflection fluorescence microscopy to measure residence times of the fluorescent dye Nile Red in CYP3A4 incorporated in surface-immobilized lipid Nanodiscs, with and without the effector α-naphthoflavone. We find direct evidence that CYP3A4 effectors can decrease substrate off-rates, providing a possible mechanism for effector-mediated enhancement of substrate metabolism. These interesting results highlight the potential of SM methods in studies of CYP allosteric mechanisms.
Cytochrome P450 (CYP) isoform 3A4 (CYP3A4) is the predominant human drug-metabolizing enzyme, responsible for metabolizing ∼50% of commercial pharmaceuticals.1 CYP3A4 displays a wide variety of allosteric (non-hyperbolic) kinetic behavior that may cause atypical CYP3A4-mediated pharmacokinetics. CYP allosterism results from the simultaneous binding of multiple substrate and/or effector molecules.2-9 Here, in the first single-molecule (SM) fluorescence studies of CYPs, we measure residence times of the fluorescent dye Nile Red (NR) bound to CYP3A4 incorporated in surface-immobilized lipid Nanodiscs,10, 11 with and without the effector α-naphthoflavone (ANF). We find good agreement with ensemble kinetic measurements, as well as direct evidence that CYP3A4 effectors can modulate substrate off-rates. These results highlight the utility of SM methods in studies of CYP allosteric mechanisms.
In recent work, we described NR as a reporter substrate of allosteric behavior in CYP3A4:12, 13 NR forms spectrally distinct singly- and doubly-occupied complexes with CYP3A4 (KDs of 0.3 μM and 2.2 μM respectively). The second binding induces a ∼8-fold greater heme spin shift than the first. NR fluorescence also responds to changes in the active-site environment induced by ANF bound to a separate site.8 ANF raises the kcat for NR by ∼3-fold (C. Fernandez and J. Lampe, unpublished data).
Here, we extend these studies to CYP3A4 in Nanodiscs. Nanodiscs consist of a lipid bilayer ∼10 nm in diameter stabilized by a helical protein coat, and provide a biologically relevant membrane environment for CYP3A4. SM kinetic studies of CYP3A4-Nanodiscs require a detailed thermodynamic understanding of the multiple possible binding modes. The Nanodisc bilayer may compete with CYP3A4 for NR binding; moreover, ligands can bind to CYP3A4-Nanodiscs with significantly different affinity than to recombinant protein in solution.14 To extend the previous equilibrium binding analysis to NR and aid in interpretation of SM measurements, we combined steady-state fluorescence spectroscopy and UV-Vis spectroscopy of NR binding Nanodiscs and CYP3A4-Nanodiscs.
NR displays significant fluorescence enhancement relative to buffer upon binding either CYP3A4 or the Nanodisc membrane. NR fluorescence at increasing Nanodisc concentrations showed a KD of 0.97 ± 0.07 μM for a single NR binding (Fig. 1a). Complementary titrations of NR at three Nanodisc concentrations showed an increase and red-shift in fluorescence, followed by self-quenching. Data were globally fit (Fig. 1b) to numerical simulations15 yielding a model with five NR binding modes in the bilayer with a sequential cooperativity factor of 2.3 ± 0.04. In other words, each NR binding alters the bilayer properties so as to disfavor subsequent binding by ΔΔG ≈ 0.5 kcal/mol. The apparent negative cooperativity resembles other amphiphile/membrane interactions.14 The resulting membrane partition coefficient is 1.8×104, compared to literature values16 of 7.7×103.
Figure 1.
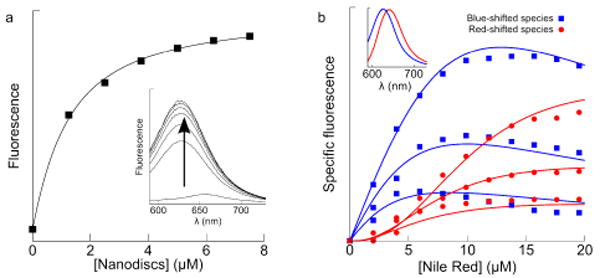
(a) NR (0.5 μM; λex = 550 nm) fluorescence increasing with [Nanodiscs] yields a KD of 1 μM for a singly-occupied disc. (b) NR fluorescence increases and red-shifts with increasing [NR], at three different Nanodisc concentrations, 0.5, 1.0, and 2.0 μM. Solid lines are global fits with 5 sequential NR binding modes and a negative cooperativity factor of 2.3. (Inset) Basis spectra used in singular value decomposition to deconvolute the spectral shift.
Titrations of NR into a fixed concentration of CYP3A4-Nanodiscs displayed decreased fluorescence intensity at low [NR] coupled with a lack of self-quenching at high [NR], relative to an equal concentration of Nanodiscs (Fig. 2a). This is suggestive of high-affinity binding to a site with a quantum yield less than that of membrane-bound NR (albeit higher than NR in buffer) coupled with lower-affinity binding to a site with higher quantum yield, mirroring the pattern of binding with CYP3A4 in solution.12, 13 CYP3A4-Nanodiscs displayed a ligand-induced shift to the high-spin state with a KD of 10 ± 2.5 μM (Fig. 2b), in good agreement with the putative lower-affinity site observed in the fluorescence data. Fig. 3 contains the scheme used to globally fit all of the equilibrium binding data. These results with NR confirm the relevance of multiple binding to CYP3A4-Nanodiscs described by Sligar and co-workers6, 17, 18 for testosterone, and establish the need for methods that selectively probe defined occupancy states to better understand CYP allosteric mechanisms.
Figure 2.
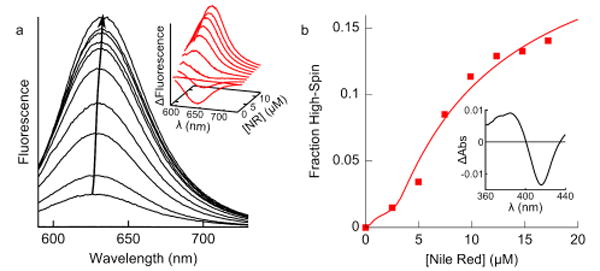
(a) NR binding CYP3A4-Nanodiscs (0.8 μM) also shows a fluorescence increase and red-shift. (Inset) Difference in fluorescence between NR binding CYP3A4-Nanodiscs and Nanodiscs shows decreased intensity at low [NR] and an increase at high [NR], suggesting multiple binding to CYP3A4. (b) UV-Vis spectroscopy of the NR-induced spin shift indicates the KD of the lower-affinity site is ∼10 μM. (Inset) Difference spectrum of the spin-shift in CYP3A4-Nanodiscs. CYP3A4 was expressed, purified and incorporated into Nanodiscs as previously described.14
Figure 3.
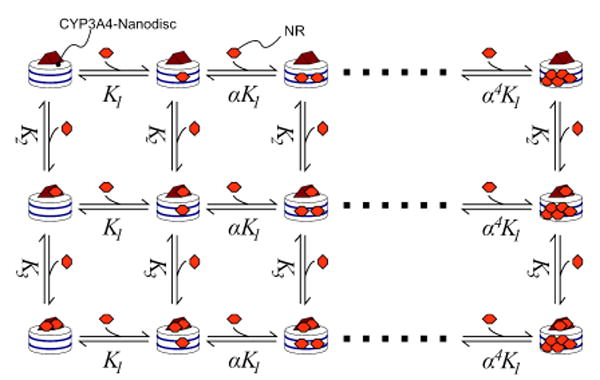
Binding model for NR/CYP3A4-Nanodiscs; global fits to binding curves gave K1 = 0.97 μM, α=2.3, K2 ≈ 0.3 μM and K3 ≈10 μM
We used total internal reflection fluorescence microscopy (TIRFM)19 to measure heterotropic allosteric effects on NR residence times in the low-occupancy complexes on the SM level. The evanescent excitation generated in TIRFM decays exponentially resulting in emission from NR bound to surface-attached Nanodiscs or CYP3A4-Nanodiscs with insignificant contributions from free NR. For surface attachment, Nanodiscs and CYP3A4-Nanodiscs were prepared with 5% Biotinyl-cap-DPPE (Avanti Polar Lipids) and added to avidin-treated glass slides. Low concentrations (30 nM) were used, allowing for detection of and data-collection from individual complexes. Then 20-50 nM NR was added, so that only singly-occupied complexes were observed.
Dwell-time histograms (Fig. 4) were fit to exponential decays to yield dissociation constants. NR has a koff of 33±2.2 s-1 from plain Nanodiscs. Given a KD of 1 μM, this implies a kon of 3×107 M-1s-1, close to on-rates observed by stopped-flow for NR and similar amphiphiles binding Nanodiscs (data not shown). The observed dwell-times are comparable to NR in DMPC vesicles.20 NR in CYP3A4-Nanodiscs has a biphasic dwell-time distribution, with a fast phase (74%) corresponding, speculatively, to koff from the bilayer, and a slow phase (26%) which reflects the off-rate of Nile Red from CYP3A4 (1.5±0.09 s-1). Assuming NR accesses CYP3A4 via the bilayer, this implies that the on-rate from the singly-occupied bilayer to the active site is ∼5 s-1. The CYP3A4 inhibitor miconazole (at 10 μM, a saturating concentration) almost totally eliminates the slow phase (Fig. S3B) by competing with NR for active-site binding.
Figure 4.

Dwell-time histograms for NR bound to Nanodiscs, with koff = 30 s-1; CYP3A4-Nanodiscs, with koff = 1.5 s-1; and CYP3A4-Nanodiscs + 5 μM ANF, with koff = 0.32 s-1, show the dramatic effect of ANF on NR dissociation kinetics. (Inset) Typical photon trajectory of CYP3A4-Nanodiscs in the presence of NR only. Dwell-times were binned manually, and histograms were fit to single- or double-exponentials to yield off-rates.
In contrast, the effector ANF (at 5 μM) slows the dissociation of NR from CYP3A4-Nanodiscs by ∼5-fold to 0.32±0.05 s-1, although with negligible effect on off-rate from plain Nanodiscs (Fig. S3A). This is direct evidence that CYP3A4 effectors can modulate substrate off-rates. Such an increase in substrate residence time may contribute to the decrease in CYP3A4 catalytic uncoupling observed with other effectors.6 Our group7, 8 and others3-6 have proposed that many effectors enhance substrate binding at equilibrium, but the SM results provide direct kinetic corroboration. It has also been proposed that ANF acts by altering the oligomeric distribution of CYP3A4.9 Because we observe a clear kinetic effect with monomerized CYP3A4-Nanodiscs, we suggest that ANF alters monomeric CYP3A4 structure/dynamics in addition to its proposed effect on oligomerization state.
SM methods provide superior sensitivity and complementary detail to ensemble techniques, enabling novel insights into CYP3A4 allosteric mechanisms. Indeed, allosteric effects on ligand off rates have not been accessible for membrane-bound CYPs by ensemble measurements. SM spectroscopy may enable a wide variety of kinetic studies into other fundamental aspects of CYP catalysis and allosterism.
Supplementary Material
Experimental methods, fitting details, dwell-time histograms of NR in CYP3A4-Nanodiscs with miconazole, and of NR in Nanodiscs with ANF, and TIRF images of NR binding Nanodiscs and CYP3A4-Nanodiscs. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by NIH grant GM-32165. We thank Professor Sligar for the membrane scaffold protein used to generate Nanodiscs.
References
- 1.Thummel KE, Wilkinson GR. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 2.Atkins WM. Expert Opin Drug Metab Toxicol. 2006;2(4):573–9. doi: 10.1517/17425255.2.4.573. [DOI] [PubMed] [Google Scholar]
- 3.Khan KK, He YQ, Domanski TL, Halpert JR. Mol Pharmacol. 2002;61(3):495–506. doi: 10.1124/mol.61.3.495. [DOI] [PubMed] [Google Scholar]
- 4.Fernando H, Halpert JR, Davydov DR. Biochemistry. 2006;45(13):4199–209. doi: 10.1021/bi052491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isin EM, Guengerich FP. J Biol Chem. 2006;281(14):9127–36. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 6.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J Biol Chem. 2007;282(10):7066–76. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 7.Lampe JN, Atkins WM. Biochemistry. 2006;45(40):12204–15. doi: 10.1021/bi060083h. [DOI] [PubMed] [Google Scholar]
- 8.Roberts AG, Atkins WM. Arch Biochem Biophys. 2007;463(1):89–101. doi: 10.1016/j.abb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Biochemistry. 2005;44(42):13902–13. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Grinkova YV, Sligar SG. Nano Lett. 2002;2(8):853–856. [Google Scholar]
- 11.Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46(8):2059–69. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 12.Lampe JN, Fernandez C, Nath A, Atkins WM. Biochemistry. 2008;47(2):509–16. doi: 10.1021/bi7013807. [DOI] [PubMed] [Google Scholar]
- 13.Nath A, Fernandez C, Lampe JN, Atkins WM. Arch Biochem Biophys. 2008 doi: 10.1016/j.abb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nath A, Grinkova YV, Sligar SG, Atkins WM. J Biol Chem. 2007;282(39):28309–20. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- 15.Mendes P. Trends Biochem Sci. 1997;22(9):361–3. doi: 10.1016/s0968-0004(97)01103-1. [DOI] [PubMed] [Google Scholar]
- 16.Hernando V, Rieutord A, Pansu R, Brion F, Prognon P. J Chromatogr A. 2005;1064(1):75–84. doi: 10.1016/j.chroma.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Baas BJ, Denisov IG, Sligar SG. Arch Biochem Biophys. 2004;430(2):218–28. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Denisov IG, Grinkova YV, McLean MA, Sligar SG. J Biol Chem. 2007;282(37):26865–73. doi: 10.1074/jbc.M704747200. [DOI] [PubMed] [Google Scholar]
- 19.Axelrod D. J Cell Biol. 1981;89(1):141–5. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F, Mei E, Lim M, Hochstrasser RM. J Am Chem Soc. 2006;128(14):4814–22. doi: 10.1021/ja058098a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental methods, fitting details, dwell-time histograms of NR in CYP3A4-Nanodiscs with miconazole, and of NR in Nanodiscs with ANF, and TIRF images of NR binding Nanodiscs and CYP3A4-Nanodiscs. This material is available free of charge via the Internet at http://pubs.acs.org.


