Abstract
Background:
Morphological abnormalities in hippocampus have been implicated in neuropsychiatric disorders, including depression, schizophrenia and dementia. Vascular endothelial growth factor (VEGF) has been demonstrated to have neurogenic effects in the hippocampus in rats. However, influence of VEGF variation on hippocampus morphology in humans has yet to be shown. Here, an integrated genetic and neuroimaging approach was used to investigate whether VEGF variation influences hippocampus morphology in humans.
Methods:
High-resolution magnetic resonance imaging and voxel-based morphometry were used to identify the influence of genetic variation of VEGFA [rs833068 (SNP-1), rs833070 (SNP-2), rs2146323 (SNP-3) and rs3025020 (SNP-4)] on brain morphology in forty-seven healthy individuals.
Results:
Variation in VEGFA SNP-2 and SNP-3 showed significant effects on hippocampus concentration.
Conclusions:
The findings suggest that effects of VEGF in hippocampus found in rats extend to humans; further understanding of effects of VEGFA variation may have important implications in identifying individuals more vulnerable to hippocampus pathology, as well as those neuropsychiatric populations most likely to benefit from VEGF-mediated interventions.
Keywords: Vascular Endothelial Growth Factor, hippocampus, magnetic resonance imaging, voxel-based morphology, single-nucleotide polymorphisms, neurogenesis
Abnormalities in hippocampus morphology are implicated in neuropsychiatric disorders including depression, schizophrenia and dementia. Recent preclinical study of rats by our group suggests that beyond its role in angiogenesis, vascular endothelial growth factor (VEGF) has neurogenic effects in the hippocampus, that are associated with antidepressant effects and memory improvement (1). In humans, the genetic locus for VEGFA has been identified on chromosome 6. However, there is no previous report demonstrating influence of VEGFA variation on hippocampus morphology in humans in vivo. In this study, an integrated genetic and neuroimaging approach was used to investigate whether VEGFA variation [identified via a set of 4 single-nucleotide polymorphisms (SNPs) that span the locus: rs833068 (SNP-1), rs833070 (SNP-2), rs2146323 (SNP-3) and rs3025020 (SNP-4)] influences hippocampus morphology in healthy humans.
Methods
The study group included 47 participants [mean age 29.74±SD 10.8 years, 26 females (55%)] who were recruited from the community and were without personal history of a DSM-IV Axis I Disorder confirmed by the Structured Clinical Interview for DSM-IV Axis I Disorders Version 2.0 (SCID) (2). No subject was taking medications with potential central nervous system effects or had a history of psychiatric or neurological disorder, head trauma with loss of consciousness over five minutes, or major medical disorder. Sixty-two percent of the participants were European-American (EA), 17% were African-American (AA), 13% were Asian, 4% were Hispanic and 4% were biracial. After a complete description of the study, written informed consent was obtained from all participants in accordance with the human investigation committees of the Yale School of Medicine and Department of Veterans Affairs. Subjects were divided into two subgroups for each SNP: those homozygous for the more frequent allele and “carriers” of the less frequent allele who were heterozygous and homozygous for that allele. For SNP-1, subjects were divided into those homozygous for G allele and “A carriers” with AA or AG genotypes; for SNP-2, divided into those homozygous for C and “T carriers” with TT or TC genotypes; for SNP-3, divided into those homozygous for C and “A carriers” with AA or AC genotypes; and for SNP-4, divided into those homozygous for C and “T carriers” with TT or TC genotypes. Sample characteristics by genotype are shown in Table 1.
Table 1.
Sample characteristics by genotype
| rs833068:SNP-1 | rs833070:SNP-2 | rs2146323:SNP-3 | rs3025020:SNP-4* | |||||
|---|---|---|---|---|---|---|---|---|
| Genotype | AA or AG | GG | TT or TC | CC | AA or AC | CC | TT or TC | CC |
| Number of subjects:male,total** | 13,27 (8) | 8,20 | 15,28 (8) | 6,19 | 11,23 (6) | 10,24 | 8,19 (4) | 12,27 |
| ethnicity*** (EA,AA,A,Others) | 19,5,2,1 | 10,3,4,3 | 19,3,3,3 | 10,5,3,1 | 15,2,3,3 | 14,6,3,1 | 15,1,1,2 | 13,7,5,2 |
| Age (mean+/−SD) | 31.37/10.41 | 27.55/11.27 | 27.86/10.89 | 32.53/10.42 | 27.61/11.26 | 31.79/10.22 | 30.68/11.98 | 28.07/8.73 |
the genotype could not be obtained from one subject
subjects homozygous for the less frequent allele in the parentheses
EA: European-American; AA: Africa-American; A: Asian
High resolution structural magnetic resonance imaging scans were performed on a 3T Trio MR scanner (Siemens, Erlangen, Germany). A three-dimensional Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) T1-weighted sequence was used to acquire sagittal images with parameters TR=1500ms, TE=2.83ms, FOV=256×256mm, matrix=256×256, slice thickness=1.0 mm without gap, 160 slices, 2 averages. Images were processed and analyzed with Statistical Parametric Mapping 5 (SPM5) (http://www.fil.ion.ucl.ac.uk/spm) consistent with our previously published methods (3). Four SNPs, spanning the 16,278 basepair VEGFA locus (average spacing is 2194 basepairs), with good coverage of the major haplotype block, were chosen using ABI SNPbrowser 3.5 software. Genotyping was done in duplicate with complete concordance.
Voxel-based analysis of covariance (ANCOVA) was used to detect effects for each of the 4 SNPs on gray matter volume (modulated images) and gray matter concentration (unmodulated images) (4). ANCOVA was performed to explore the group differences in hippocampus morphology across ethnic groups (EAs, AAs, Asian and other) in order to confirm if there were differences among the populations. Age and sex were included as covariate in these analyses. For a priori hypothesis testing, findings were considered significant at P<0.005 uncorrected for multiple comparisons and spatial extent of 25 voxels. Data were also inspected for survival for conservative Bonferroni correction for study of four SNPs which required a P-value of 0.00125. Whole brain analyses were performed to explore for potential effects in gray matter regions that we did not hypothesize a priori, with significance assumed at P<0.0125, corrected for multiple comparisons with false discovery rate, and extent threshold of 25 voxels.
Results
For SNP-2 we found that, compared to subjects homozygous for the C allele, T carriers had significantly reduced concentration in the left hippocampus (Talairach coordinates for the maximal point of difference: x=−22mm, y=−33mm, z=−4mm, T=3.28, P=0.001 uncorrected, 35 voxels) (Figure 1a). For SNP-3, compared to the subjects homozygous for the C allele, A carriers had significantly reduced concentration in the left hippocampus (Talairach coordinates for the maximal point of difference: x=−20mm, y=−31mm, z=−5mm, T=3.74, P<0.001 uncorrected, 67 voxels) (Figure 1b). Findings remained significant for these two SNPs with Bonferroni correction. Analyses were also performed with small volume correction (SVC) for multiple comparisons in the hippocampus that were survived for SNP-3 (P<0.05, corrected), however, not for SNP-2.
Figure 1.

The left and right sagittal images display the regions of reduced concentration in the left hippocampus in T allele carriers of SNP-2 and A allele carriers of SNP-3 for the vascular endothelial growth factor gene, respectively (P<0.005, a cluster > 25 voxels). The findings are displayed on a tissue probability map of gray matter. The color bar represents the range of statistical T values (referring in this case to the T statistic, not a “T” allele). The left and right graphs plot the corresponding values for each subject extracted from the voxel of peak difference between genotype groups for SNP-2 and for SNP-3, respectively. The horizontal bars display the genotype-specific means that differed significantly between genetic subgroups (P<0.005).
Analyses of larger samples using Haploview (5), that expanded the samples of this study to a total of 245 EAs (minor allele frequencies for SNP-1: 0.30; SNP-2: 0.50; SNP-3: 0.36; SNP-4: 0.30) and 37 AAs (minor allele frequencies for SNP-1: 0.50; SNP-2:0.26; SNP-3: 0.16; SNP-4: 0.08), provided further evidence that the two SNPs have a high degree of linkage disequilibrium (LD) (Figure 2). ANCOVA did not detect significant differences in hippocampus volumes among the populations studied (p>0.6); since there are no differences in the phenotype in the different populations studied, population stratification is unlikely to be the cause of the observed differences. Whole brain gray matter analyses did not reveal any other regions with significant difference between the genotypes. There were no significant findings associated with the other two (flanking) SNPs (SNP-1 and SNP-4).
Figure 2.
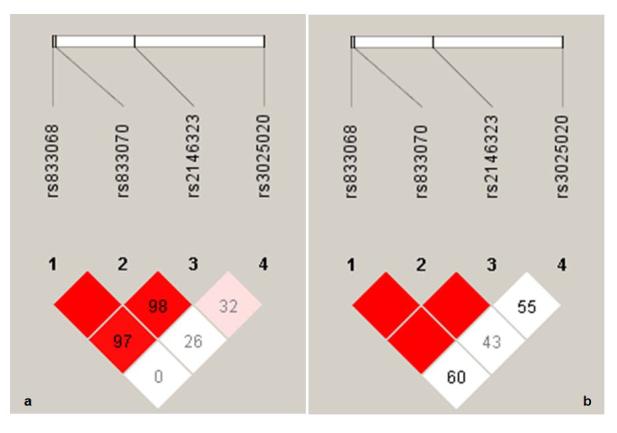
The Haploview (http://www.broad.mit.edu/mpg/haploview/index.php) figures show linkage disequilibrium (LD) relationships between all marker pairs, separately for European Americans (EAs) and African Americans (AAs), using an extended sample. The upper bar in each figure represents the relative location of each marker. SNP-2 (rs833070) and SNP-3 (rs2146323) for the vascular endothelial growth factor gene have a high degree of LD. They reside in the same LD block in both EAs (a) and AAs (b) (a block that also includes SNP-1: rs833068). Red color represents high LD, decreasing to pink then white as LD reduces.
Discussion
In summary, we found T carriers of SNP-2 and A carriers for SNP-3 had reduced hippocampus concentration compared to individuals homozygous for the C allele of these variants. VEGF is an angiogenic protein demonstrated in rats to induce neurogenesis in the subventricular and subgranular zones of the hippocampus dentate gyrus (6), and to protect the hippocampus from neurotoxicity such as through glutamate excesses (7). Rat models further support VEGF-induced neurogenesis as a key mediator of beneficial effects of environmental enrichment and exercise on hippocampus-dependent memory and learning (8, 9). The current findings suggest that effects of VEGFA in hippocampus found in rats extend to humans; specific variation in VEGFA may contribute to individual differences in hippocampus structure and potentially function.
The SNPs are intronic with unclear function. Evidence suggests that some VEGFA introns contain transcription factor binding sites and these areas may be important in the regulation of VEGF production and/or influence splicing (10). Therefore, it seems possible that the SNP-2 and SNP-3 (or other variants in LD with them) may alter VEGF expression in the hippocampus, which then influences hippocampal development, and/or plasticity in adulthood. Of note, the specific cytoarchitectural contributions, such as differences in neuronal size or packing density, to the morphological changes cannot be concluded from these neuroimaging data (11). Although VEGF has direct effects on neurogenesis, it also has effects on astroglia and endothelial cells (6) and thus changes in these cell populations may also contribute to the morphological changes.
The ethnic heterogeneity within the subject sample is an important potential limitation. Although we did not detect differences in hippocampus morphology between ethnic groups and effects of the VEGFA genotypes were similar in the groups, it is still possible that population stratification influenced our findings, and replication in larger, more homogeneous samples will be important. Additionally, in order to have comparable group sizes, we grouped minor allele heterozygotes and homozygotes; this is consistent with a dominant model but would also have reasonable power if the alleles were codominant (the most likely case where the markers studied are in LD with an ungenotyped functional variant). If the correct mode of effect is recessive, then there is the potential for Type II error in analyses for SNP-1 and SNP-4. These results should be considered preliminary, especially owing to the modest sample size, and replication studies are warranted.
The findings support the importance of further study of VEGFA variation as a potentially substantial factor in influencing human variation in hippocampus-related functions, including mnemonic functioning, learning and resilience to stress. Moreover, as VEGF has been demonstrated to mediate therapeutic effects of antidepressant treatments and recovery from neurological injury in rodents (1, 12-14), further understanding of effects of VEGFA variation may have important implications in identifying individuals more vulnerable to hippocampus pathology, as well as those neuropsychiatric populations most likely to benefit from VEGF-mediated interventions.
Acknowledgments
We thank Ms. Kathleen Colonese for her dedicated work with the study participants, Ms. Ann Marie Lacobelle for her genotyping assistance, Hedy Sarofin RTRMR, Karen Martin, RTRMR and Terry Hickey RTRMRN for their technical expertise, the nurses of the Biostudies Unit Ms, Angelina M. Genovese RNC, MBA, Elizabeth O'Donnell RN, Brenda Breault RN, BSN, and the research subjects for their participation. The authors were supported by research grants from the Department of Veterans Affairs Research Career Development (HPB), Merit Review (HPB and JG) and Research Enhancement Award Program (REAP) (HPB, RSD and JG) Awards, the National Institute of Mental Health R01MH69747 (HPB), R01MH070902 (HPB), K24 15105 (JG), T32MH14276 (LGC, JHK, RSD), the National Alliance for Research on Schizophrenia and Depression (Great Neck, NY) (HPB), The Attias Family Foundation (HPB), Marcia Simon Kaplan (JHK), The Ethel F. Donaghue Women's Investigator Program at Yale (New Haven, CT) (HPB) and the Klingenstein Foundation (JHK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
Dr. Blumberg: Consultant: Pfizer, Inc. Dr. Gelernter has received financial support or compensation for the following: related to consultation for Columbia University, the Thailand Center for Excellence for Life Sciences (TCELS), the University of CT Health Center, NIH, related to grant reviews for the National Institutes of Health; and related to academic lectures and editorial functions in various scientific venues. Drs. Wang, Chepenik, Kalmar, Duman and Ms. Edmiston reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Warner-Schmidt JL, Duman RS. VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc Natl Acad Sci U S A. 2007;104:4647–52. doi: 10.1073/pnas.0610282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I & II Disorders (Version 2.0) Biometrics Research, New York State Psychiatric Institute; New York: 1995. [Google Scholar]
- 3.Tang Y, Wang F, Xie G, Liu J, Li L, Su L, et al. Reduced ventral anterior cingulate and amygdala volumes in medication-naive females with major depressive disorder: A voxel-based morphometric magnetic resonance imaging study. Psychiatry Res. 2007;156:83–86. doi: 10.1016/j.pscychresns.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 5.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 6.Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2002;99:11946–50. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzaki H, Tamatani M, Yamaguchi A, Namikawa K, Kiyama H, Vitek MP, et al. Vascular endothelial growth factor rescues hippocampal neurons from glutamate-induced toxicity: signal transduction cascades. Faseb J. 2001;15:1218–20. [PubMed] [Google Scholar]
- 8.Cao L, Jiao X, Zuzga DS, Liu Y, Fong DM, Young D, et al. VEGF links hippocampal activity with neurogenesis, learning and memory. Nat Genet. 2004;36:827–35. doi: 10.1038/ng1395. [DOI] [PubMed] [Google Scholar]
- 9.Fabel K, Fabel K, Tam B, Kaufer D, Baiker A, Simmons N, et al. VEGF is necessary for exercise-induced adult hippocampal neurogenesis. Eur J Neurosci. 2003;18:2803–12. doi: 10.1111/j.1460-9568.2003.03041.x. [DOI] [PubMed] [Google Scholar]
- 10.Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, et al. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem. 1991;266:11947–54. [PubMed] [Google Scholar]
- 11.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 12.Jin KL, Mao XO, Greenberg DA. Vascular endothelial growth factor: direct neuroprotective effect in in vitro ischemia. Proc Natl Acad Sci U S A. 2000;97:10242–7. doi: 10.1073/pnas.97.18.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, et al. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet. 2001;28:131–8. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- 14.McCloskey DP, Croll SD, Scharfman HE. Depression of synaptic transmission by vascular endothelial growth factor in adult rat hippocampus and evidence for increased efficacy after chronic seizures. J Neurosci. 2005;25:8889–97. doi: 10.1523/JNEUROSCI.2577-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


