Abstract
Despite compelling evidence from twin and family studies indicating a strong genetic involvement in the etiology of autism, the unequivocal detection of autism susceptibility genes remains an elusive goal. The purpose of this review is to evaluate the current state of autism genetics research, with attention focused on new techniques and analytic approaches. We first present a brief overview of evidence for the genetic basis of autism, followed by an appraisal of linkage and candidate gene study findings and consideration of new analytic approaches to the study of complex psychiatric conditions, namely, genome-wide association studies, assessment of structural variation within the genome, and the incorporation of endophenotypes in genetic analysis.
Keywords: Autism, Copy number variation (CNV), Endophenotype, Genetic
INTRODUCTION
First formally documented in 1943 by child psychiatrist Leo Kanner (1), autism is a severe neurodevelopmental disorder defined by profound impairments in language, social-emotional functioning, and restricted, repetitive interests and behaviors (2). Although once considered to be relatively rare, striking new prevalence estimates of 1 in 500 for strict diagnosis and 1 in 150 using broader diagnostic criteria (3) have prompted the Centers for Disease Control and Prevention to declare autism a national public health crisis. Such reports underscore the pressing urgency for determining the etiology of autism, which remains cryptic. Bolstered by historic highs in public awareness, advocacy, and funding, research into the basis of autism is advancing at an accelerated pace. Several large autism consortia now exist and bring to bear increased resources to enable more powerful and rapid pursuit of etiologic clues.
The last decade has witnessed the development of an armamentarium of genetic techniques and tools for studying the genetic basis of disease, such as sequencing of the human genome (4), identification of common genetic variants via the HapMap project (5), and development of cost-efficient high-throughput genotyping and analysis methodologies. Although these tools have led to major breakthroughs in medical genetics, we have not yet witnessed successful disease gene discovery in psychiatric diseases. Autism has proven particularly frustrating to genetic dissection. Despite compelling evidence from twin and family studies indicating a strong genetic involvement, the unequivocal detection of autism susceptibility genes remains an elusive goal. The purpose of this review is to evaluate the current state of autism genetics research critically with focused attention on new techniques and analytic approaches. We first present a brief overview of evidence for the genetic basis of autism, followed by an appraisal of linkage and candidate gene study findings and consideration of new analytic approaches to the study of complex psychiatric conditions, including genome-wide association studies (GWAS), assessment of structural variation within the genome, and the incorporation of endophenotypes in genetic analysis.
EVIDENCE FOR A GENETIC BASIS: FAMILY AND TWIN STUDIES OF AUTISM
Strong but indirect evidence supports the role of genetic factors in the etiology of autism. Monozygotic twins show ~60% concordance in contrast to only 3% to 5% concordance in dizygotic twins with a heritability estimate of ~90% (6-9). Family studies indicate 5% to 8% recurrence rate within families (10); this translates into a 25- to 40-fold increase in risk over current population base rates (3, 11). The detailed clinical characterizations performed in many of these family and twin studies also documented among relatives a phenotype similar in quality to the defining features of autism, but much milder in expression. This constellation of subtle language, cognitive, and personality traits mirrors the symptom domains of autism and occurs more frequently among unaffected relatives of autistic individuals than controls (Table 1). Concordance for this broader phenotype leaps to around 80% in monozygotic versus 10% in dizygotic twins (6-9), thereby supporting the notion that such traits reflect a genetic liability to autism.
TABLE 1.
Summary of Broad Phenotypes Identified in Relatives
| Language Impairment |
| Restricted Interests and Behaviors |
| Neurocognition |
Whereas language abnormalities figure prominently, the literature also reveals multiple reports of social and repetitive features, along with more recent reports of neuro-cognitive abnormalities. Recent evidence suggests that such features segregate independently in relatives and appear more commonly in families with higher genetic loading (12-14), consistent with their relevance to autism susceptibility. As discussed later in this review, such features, measurable in unaffected relatives, could provide an index of genetic effects of salience to the etiology of autism. Inclusion of such phenotypic information in relatives may provide a potentially important, complementary approach for detecting the genes causing autism.
GENOME SCREENS: AN UNBIASED SEARCH FOR GENES
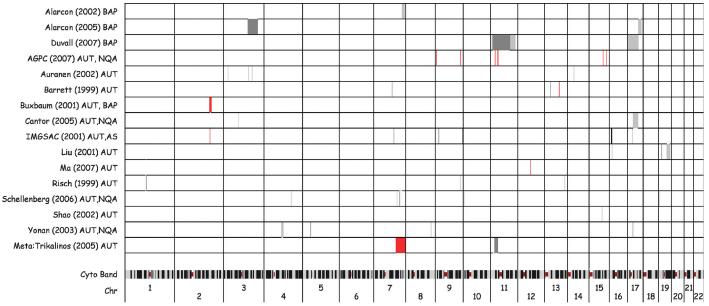
Genome-wide linkage analysis was initially viewed as a valuable approach for guiding the search for autism disease genes because this approach holds the advantage of scanning the genome for disease-associated loci in the absence of a priori hypotheses about the genetic architecture of a disease. Such studies have led to gene discoveries for more than 1,600 Mendelian disorders (15); however, this approach when applied to complex traits and disorders has met with considerably less success. There now exist more than a dozen genome-wide linkage studies of autism (16-31). Because most of these studies have applied different genotyping and analysis tools and include substantially overlapping samples, comparisons across them are complex. In Figure 1, we present results from the primary genome-wide scans of autism, including those analyzing the broader phenotype. These studies comprise between 12 and 1,181 pedigrees that typically are multiplex. Ancestry was generally well controlled within each sample but varied across studies. Genotyping density ranged from 264 to 9,505 genetic markers.
FIGURE 1.
Summary of genome-wide linkage scans in autism with LOD/equivalents plotted. Red indicates LOD ≥3, dark gray indicates LOD ≥2, and light gray indicates LOD ≥1.5. First author and year of publication for each study are listed. AS, Asperger syndrome; BAP, broad autism phenotype; Chr, chromosome; NQA, not quite autism (diagnostic criteria not fully met).
These studies reveal numerous suggestive linkage peaks but with relatively little congruence across them. The most consistent evidence for linkage occurs on 7q, with 7q22-q32 most strongly implicated by meta-analysis (31). In the largest sample analyzed to date, however, this region yielded no evidence for linkage (20). Although this review does not relect a number of fine-mapping and targeted follow-up studies, the picture emerging from those data is of numerous suggestive signals with little compelling evidence for replication. What may underlie these largely inconsistent findings? One problem may lie in the phenotypic and etiologic complexity of the disorder itself, which may be compounded by varying phenotypic definitions used across studies (e.g. strict vs broad). As in other complex human traits and disorders, different genes may contribute to distinct components of the phenotype, thereby giving rise to the full disorder through concerted actions (27, 32, 33). In this case, success in detecting susceptibility loci may rest on our ability to disaggregate such complex clinical phenomena into more basic phenotypes that may be more amenable to genetic dissection. In a subsequent section, we review the growing body of research adopting this more refined phenotypic approach.
Another reason that linkage analyses of autism have generated only inconsistent findings may lie in the limitations inherent in this analytic approach. Although useful for highly penetrant single-gene disorders, linkage analysis seems ill suited for gene detection in oligogenic disorders involving multiple risk alleles of small effects (34). Without minimizing the tremendous effort required to undertake this work, it is important to note that the sample sizes are generally relatively small. Of the genome-wide scans listed in Figure 1, fewer than half had more than 100 pedigrees. Such limited sample sizes are insufficient to delineate true genetic signals from the noise of multiple comparisons and study-specific artifacts.
Genome-wide association studies may provide a more powerful alternative approach. As in association studies of candidate genes, GWAS compares genetic risk factors (in the form of specific genetic markers) in cases and controls; in GWAS, markers are distributed throughout the genome rather than limited to candidate regions, thus providing a more unbiased canvassing of the genome. In a seminal article, Risch and Merikangas (34) demonstrated that association affords significantly greater power over linkage for detecting susceptibility loci that confer weak effects. This indicated that GWAS is a more appropriate approach for genetic studies of complex disorders such as autism. Previously cost and technically prohibitive, GWAS has only recently been applied to psychiatric disorders, and the first high-density GWAS of autism are currently underway (35). By the conclusion of 2008, 3 groups should have published GWAS for autism, and a meta-analysis will soon follow.
CANDIDATE GENE ASSOCIATION: TARGETED INVESTIGATIONS
The first molecular genetic studies of autism took form incandidate gene association studies. Plausible candidates were selected based on known involvement in pathways related to neurodevelopment and/or evidence from pharmacological interventions that implicate specific biomolecular pathways. By and large, these investigations have been forestalled by inadequate sample sizes and sparse genotyping. Indeed, of more than 100 genes having been investigated for involvement in autism, only a few have been supported by replication. We review briefly those that have surfaced as the most plausible candidates: MET; SLC6A4 (the serotonin transporter); RELN (reelin); the tumor suppressor genes PTEN, TSC1, and TSC2; and neuroligins and their binding partners.
MET
The MET gene, which is located in the 7q31 candidate gene region, is implicated in genome-wide linkage studies. MET is also a strong functional candidate for involvement in autism because it encodes a receptor tyrosine kinase involved in neuronal growth and organization, as well as immunological and gastrointestinal functioning; these are systems in which abnormalities have been suggested in autism. Variants in the MET promoter region show strong association with autism. In particular, Campbell et al (36) found significant overtransmission of the common C allele in autism cases in multiple samples. Case-control comparisons found significant overrepresentation of the C allele in autism, with a relative risk of 2.27. In a separate study, significantly decreased MET protein levels were found in autopsied cortical tissue from individuals with autism (37). The C risk allele is believed to be a functional regulator of the MET gene. Campbell et al (36) also found that mouse cells transfected with human MET promoter variants showed a 2-fold decrease in MET promoter activity associated with the C allele.
SLC6A4
Implicated by pharmacological evidence (38) and repeated findings of elevated levels of platelet serotonin (5HT) in approximately 25% to 30% cases of autism (39), the serotonin pathway was one of the initial targets for candidate gene studies of autism. Studies examining the SLC6A4 locus generally support its involvement in autism, but findings have not converged on a specific allele, nor have they consistently reported association with the same polymorphism (40-44). Several reports have focused on SLC6A4 and its promoter region, 5HTTLPR. Whereas biased transmission of 5HTTLPR alleles has been reported in several data sets (45, 46), the findings are mixed in reporting overtransmission of the long or short allele of this polymorphism (47, 48); there are also reports that contradict a role of 5HTTLPR (49).
RELN
Reelin encodes a protein that controls intercellular interactions involved in neuronal migration and positioning in brain development (50). RELN maps to the 7q22 chromosomal region, where suggestive or significant linkage to autism has been reported in several studies (Fig. 1). Both family- and population-based association studies also indicate that variations in RELN may confer risk to autism. In particular, a large polymorphic trinucleotide repeat in the 5′ UTR of the RELN gene has been implicated in autism in several studies (51-53). Preferential transmission of the large repeat polymorphisms to autistic versus unaffected siblings has also been reported (54, 55). A contribution of RELN in autism is further supported by studies of mutant reeler mice, which carry a large deletion in RELN and show atypical cortical organization similar to the cytoarchitectural cerebral abnormalities documented in postmortem studies in autism (56).
Tumor Suppressor Genes—PTEN, TSC1, and TSC2
As detailed below, mutations in these genes cause disorders that have been associated robustly with autism. Because their signaling pathways have been well characterized, their association with autism may offer important clues into etiologic mechanisms of this complex and heterogeneous disorder.
PTEN (phosphatase and tensin homolog) is a tumor suppressor gene involved in the chemical pathway that prevents uncontrolled cell growth and division. Mutations in PTEN cause Cowden syndrome and related disorders involving hamartomas and are often associated with macrocephaly. Building on the observation that autism sometimes occurs with Cowden syndrome and related PTEN disorders, Butler et al initially examined the PTEN gene in individuals with autism and macrocephaly. They sequenced the PTEN gene in 18 of such patients and reported 3 individuals with PTEN mutations (57). Several additional studies have also documented PTEN mutations in cases of autism and macrocephaly (58-60). Moreover, studies of transgenic mice further support a role of PTEN in autism. In particular, mice lacking PTEN in regions of the hippocampus and frontal lobe show arborization of neuronal processes in these brain regions and display some autistic-like behaviors (61).
The tumor suppressor genes TSC1 and TSC2 have also been associated with autism. TSC1 and TSC2 encode the growth suppressor proteins hamartin and tuberin, respectively. Mutations in either gene cause tuberous sclerosis complex (TSC), a neurodevelopmental disorder characterized by benign tumors or lesions in many organs, including characteristic lesions in the brain. Clinically, TSC typically presents with cognitive delays and epilepsy, and autism has been reported in approximately 15% to 60% of cases (62, 63). Because TSC involves easily identifiable cortical lesions, studies have attempted to correlate lesion localization with the presence of autism symptomatology. Whereas several studies have reported lesions in the temporal lobe associated with autism (64, 65), others have reported associations with more diffusely localized lesions (66, 67); in other cases, the presence of autism in TSC was not correlated with lesions in any particular brain regions.
Neuroligins and Neurexins
Neuroligins are cell adhesion molecules that play a prominent role in synaptic maturation and function; this renders them as plausible candidates for involvement in neurodevelopmental disorders such as autism (69). A link between neuroligins and autism was first supported by findings of mutations in the X-linked neuroligins, NLGN3 and NLGN4, in 2 affected sib pairs (70). Subsequently, Laumonier and colleagues detected a 2-bp deletion in the NLGN4 gene in individuals affected with mental retardation within a large French family (71). Although not specific to autism, it was notable that all affected individuals were found to have the same frameshift mutation. Another study detected missense mutations in the NLGN4 gene in 4 of 148 individuals with autism, whereas no mutations were found in healthy or psychiatric controls (72). More recently, Lawson-Yuen et al (73) reported exonic deletions in NLGN4 in a family affected with autism and a range of other learning and psychiatric disorders. Not all studies report significant findings for neuroligins (74-76), yet evidence implicating the neuroligin binding partners neurexins, CNTNAP2, and SHANK3 (reviewed next), bolsters support for the role of neuroligins in autism.
Neurexins encode a highly polymorphic family of neuronal proteins that interact with neuroligins to promote synaptic functioning (77). Evidence for neurexin involvement in autism comes from a number of recent investigations. Feng et al (78) screened 3 neurexin beta genes in 72 individuals with autism and 535 controls, followed by sequencing of exon 1 of NRXN1β in an additional 192 additional cases. Missense mutations were found in 4 individuals with autism and in-frame deletions, and insertions were detected in 9 additional cases. No such mutations were reported in controls. Neurexin mutations were also detected in a recent genome screen conducted by the Autism Genome Project Consortium (20); a hemizygous deletion of coding exons from NRXN1 was found in a pair of affected siblings. Finally, Kim et al (79) recently identified a number of rare coding variants in a scan of NRXN1 coding exons in 57 individuals with autism. These mutations were not observed in controls with Tourette syndrome or obsessive compulsive disorder.
Contactin-associated protein-like 2 (CNTNAP2) is part of the neurexin superfamily that encodes CASPR2, a transmembrane scaffolding protein (80). CNTNAP2 was recently associated with autism in a study of an Old Order Amish community that is densely affected with cortical dysplasia-focal epilepsy; the syndrome was associated with autism in 67% of cases (81). By screening individuals affected with cortical dysplasia-focal epilepsy, the investigators detected a frameshift mutation in CNTNAP2 exon 22 present among all 9 affected individuals. Screening of 105 healthy Old Order Amish controls revealed 4 carriers, but none who were homozygous for the mutation.
Three recent studies further support a role of CNTNAP2 in autism. In a 2-stage study, Arking et al (82) detected significant linkage at 7q35 (which covers the CNTNAP2 locus), and a follow-up association study in 72 multiplex families found significant overtransmission of the T allele in a common polymorphism residing in the intron between exons 2 and 3 of CNTNAP2. Notably, this result was then replicated in an independent sample of 1,295 parent-child trios. In another report, Bakkaloglu et al (83) resequenced CNTNAP2 in a cohort of 635 individuals with autism and 942 controls, finding several rare variants in individuals with autism that were not present in controls. Alarcon et al (84) reported further evidence implicating CNTNAP2 as an autism susceptibility gene and specifically investigated the association of CNTNAP2 with an autism language phenotype. In a 2-stage association study, investigators found significant association between variants in CNTNAP2 and an index of language delay in autistic children. In addition, a microdeletion in CNTNAP2 was identified in 1 proband and his father but was not seen in 1,000 controls. An independent expression study of fetal brain development was then performed, with results indicating preferential expression of CNTNAP2 in the language centers of the brain (i.e. frontal and anterior temporal lobes). Collectively, these studies provide compelling evidence that CNTNAP2 mutations could be associated with autism and perhaps particularly the language endophenotypes of autism. CNTNAP2 is one of the largest genes in the human genome (2.3 million bases or ~1.5% of chromosome 7), and future studies will therefore be important to tease out specific variants that underlie these associations.
SHANK3 is another neuroligin binding partner that has been associated with autism. SHANK3 belongs to a family of neuronal scaffolding proteins that play a critical role in synaptic functioning and also regulate dendritic spine morphology. Durand et al found mutations in SHANK3 in 3 of 226 families of autistic individuals. In 1 family, the child with autism carried a de novo deletion in SHANK3 (85). Two siblings in another family carried a frameshift mutation; and in a third family, the proband carried a deletion in SHANK3 and her affected brother had an additional copy. In another study evaluating sequence and copy number variants in the SHANK3 region, Moessner et al (86) detected 1 de novo mutation and 2 gene deletions in a group of 400 individuals with autism. Although SHANK3 mutations may account for only a minority of cases, when considered together with findings from several other neuroligin binding partners along with findings for SLC6A4 and MET, there seems to be accumulating evidence for the role of synaptic function genes in autism.
STRUCTURAL VARIANTS: AN ADDITIONAL GENETIC MECHANISM
The development of high-resolution platforms with capabilities for characterizing alterations in the DNA copy number with unprecedented resolution has led to a new appreciation of the frequency with which de novo structural variations occur throughout the human genome. Such microdeletions and duplications (or copy number variations [CNVs]) occur in abundance in the general population and appear widespread throughout the genome. For example, Sebat et al (87) found CNVs averaging ~400 kb in length and covering 12% of the genome in the HapMap samples. It is possible that CNVs could cause subsets of cases in complex diseases such as autism (88). It is worth noting that larger-scale genomic changes also have been associated with autism (89). For instance, inherited duplications in the 15q11Yq13 region (which is causal in Prader-Willi and Angelman syndromes) have been reported to occur in ~1% to 3% of autism cases (90). Our focus here, however, is on the previously under-appreciated role of de novo events in autism.
Jacquemont et al (91) detected de novo CNVs in 24% of individuals with autism, and in a genome-wide association screen, 1 pair of affected siblings showed spontaneous CNVs in NRXN1 (see above) (20). Adding to these findings, Weiss and colleagues (35) recently reported a compelling CNV in autism in multiple samples. In the initial stage of a GWAS of autism among 751 multiplex families, the investigators found deletions and duplications at 16p11.2 associated with autism in 1% of cases, which were not apparent in 2 separate psychiatric control groups, and detected in only 0.01% of a large unscreened Icelandic population. The identical 593-kb deletion and a reciprocal microduplication were subsequently found in 2 separate replication samples. Copy number variations in this region have been detected in 2 other studies of autism (92, 93). Although these findings were compelling, they were not necessarily specific to autism because the 16p11.2 deletions and duplications were also observed at elevated rates among individuals with developmental delays. Therefore, additional work will be necessary to clarify the significance of this region to autism susceptibility.
Although still in its infancy, the study of CNVs has already enriched our understanding of autism genetics. In addition to more traditional explanatory models positing multiplicative effects of common variants, it seems that rare, spontaneous, and highly penetrant mutations may explain a portion of autism cases (93). The latter of these mechanisms is most compatible with sporadic cases of autism, whereas multiplicative models may better account for families in which multiple cases of autism and/or broader phenotypes exist among relatives (94). In support of this view, recent work by Sebat and colleagues (93) suggest that de novo CNVs are present much more frequently among pedigrees with only a single case with autism than in multiplex pedigrees. In their study, spontaneous CNVs were present in 10% of affected individuals from single-incidence families (i.e. sporadic cases), contrasting with substantially lower rates observed in controls (1%) and autism cases from multiplex families (3%). Using a similar design in a genome-wide scan for CNVs, Marshall et al (92) found this same pattern—de novo CNVs were detected in 7% of autistic individuals from single-incidence families, 2% of cases from multiplex families. These findings may prove useful for guiding selection of appropriate analytic techniques and specific subgroups of autistic cases in future genetic studies. This method may be complemented by an endophenotypic approach, described below, which may also help to refine more homogenous samples through detailed phenotypic assessment.
NARROWING THE SCOPE: THE STUDY OF AUTISM ENDOPHENOTYPES
Endophenotypes are subclinical markers of disease (e.g. behavioral, physiological, neuropsychological, and others) that are present among both affected and unaffected individuals and which are hypothesized to hold more straightforward ties to underlying neurobiological and genetic etiologies than downstream clinical outcomes (95). Rather than searching for “autism genes,” studies using an endophenotypic approach confront the less daunting task of searching for smaller constellations of genes that contribute to distinct phenotypic features. This approach is supported by family and twin studies that show independent segregation of component features of autism and suggest that although the component features of autism all have strong genetic effects, they seem largely independent in patterns of transmission, with relatively little phenotypic or genetic overlap (13, 96-99). Endophenotypes may, therefore, benefit genetic studies by providing a means for defining more etiologically homogenous subgroups. In addition, endophenotypes are by definition measurable in both affected and unaffected individuals (95), thereby affording analysis of larger sample sizes with greater power.
Table 2 lists the endophenotypic features for which significant linkages or associations with autism have been reported to date. Language phenotypes, such as age at first word or phrase, emerge as the most promising of such endophenotypes because they show significant linkage or association across several independent samples. Of particular interest are the significant linkages observed on chromosome 7 that have been observed in 5 separate investigations. The 7q region has been an intense focus of studies of developmental language disorders (100), and the candidate gene and expression findings discussed above further suggest that this region may harbor loci associated with the autism language phenotype (84). These data underscore the value of delineating more specific and powerful associations with endophenotypes and highlight this region as an important focus for continued focused investigation.
TABLE 2.
Endophenotype Findings
| Phenotype | Chromosome | Locus | Reference |
|---|---|---|---|
| Language, communication | 1p | 1p36.23-p36.13 | (18) |
| 1p21.2 | (28) | ||
| 2q | 2q31.1 | (16) | |
| 2q33.1 | (134) | ||
| 3q | 3q13.31-q26.1 | (18) | |
| 5q | 5q21.3-q23.1 | (18) | |
| 5q12.3 | (28) | ||
| 5p | 5p15.2-p13.1 | (18) | |
| 6q | 6q25.3 | (28) | |
| 7q | 7q21.2-q21.3 | (135) | |
| 7q34-36.2 | (17) | ||
| 7q35 | (18) | ||
| 7q36.1 | (84) | ||
| 7q36.3 | (28) | ||
| 8q | 8q23-q24 | (136) | |
| 9q | 9q34.2 | (28) | |
| 10q | 10q22.1-q23.22 | (18) | |
| 10q22.3 | (18) | ||
| 10q22.3 | (17) | ||
| 10p | 10p14-p15.3 | (18) | |
| 11q | 11q23.3-q24.1 | (17) | |
| 11q23.3-q24.1 | (18) | ||
| 11q25 | (28) | ||
| 13q | 13q22.1 | (135) | |
| 15q | 15q26.3 | (18) | |
| 16q | 16q24.1 | (18) | |
| 16p | 16p12-p13 | (136) | |
| 17q | 17q11.2 | (137) | |
| 17q23.2-q25.1 | (18) | ||
| 18q | 18q23 | (28) | |
| 20p | 20p11.22-q13.32 | (18) | |
| 20p11.22-q13.32 | (17) | ||
| 22q | 22q13.1 | (28) | |
| Social responsiveness | 4q | 4q34.1 | (19) |
| 10q | 10q11.23-q26.3 | (19) | |
| 11p | 11p15.4-q22.1 | (19) | |
| 17p | 17p13.3-q23.2 | (19) | |
| Repetitive behavior/OCD | 1q | 1q42.2 | (137) |
| 7q | 7q31 | (138) | |
| 15q | 15q11-q13 | (139) | |
| 16p | 16p13.3 | (18) | |
| 17q | 17q11.2 | (140) | |
| 17q23.2-q25.1 | (18) | ||
| Savant skills | 15q | 15q11-q13 | (141) |
| Developmental regression | 7q | 7q36.1 | (142) |
| 10p | 10p15.3 | (28) | |
| 14q | 14q32.2 | (28) | |
| 21q | 21q21.1 | (142) | |
| Rapid milestones | 19p | 19p13 | (40) |
| Head circumference | 7p | 7p15.2 | (143) |
SUMMARY
More than 30 years have passed since Folstein and Rutter (7) first reported compelling evidence for a genetic etiology to autism in their landmark twin study. Scores of linkage and candidate gene studies have since attempted to move beyond such promising genetic epidemiologic findings to identify specific DNA sequence variations causing autism. In aggregate, however, these efforts have been fraught with several methodological and analytic challenges. Limited power, varying designs, genotyping and analyses, and imprecise phenotypic definitions are some of the factors that contribute to the scarcity of hard replicated findings to date. Further complicating this picture may be several environmental factors associated with autism that may interact with genetic vulnerabilities in complex ways. We have attempted to highlight those findings that have best withstood rigorous replication standards, with an eye toward recent advancements in the methodological and analytic tools for the study of complex traits and disease, including GWAS, screening for CNVs, and the incorporation of endophenotypes in molecular genetic studies. When implemented into the large-scale collaborative efforts currently underway, such techniques may afford increased power and sensitivity for defining different etiologic pathways and, ultimately, translate into important new knowledge of the pathogenetics of autism.
Acknowledgments
ML was supported by K12RR023248, R03MH079998, and a grant from Autism Speaks.
PFS acknowledges support from Autism Speaks.
REFERENCES
- 1.Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–50. [PubMed] [Google Scholar]
- 2.APA . Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) 4th ed. American Psychiatric Publishing Inc.; Washington DC: 1994. [Google Scholar]
- 3.CDC Autism spectrum disorders common. JAMA. 2007;297:940. doi: 10.1001/jama.297.9.940. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffenburg S, Gillberg C, Hellgren L, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–16. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 7.Folstein S, Rutter M. Infantile autism: A genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 8.Bailey A, Le Couteur A, Gottesman I, et al. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 9.Le Couteur A, Bailey A, Goode S, et al. A broader phenotype of autism: The clinical spectrum in twins. J Child Psychol Psychiatry. 1996;37:785–801. doi: 10.1111/j.1469-7610.1996.tb01475.x. [DOI] [PubMed] [Google Scholar]
- 10.Szatmari P, Jones M, Zwaigenbaum L, Maclean J. Genetics of autism: Overview and new directions. J Autism Dev Dis. 1998;28:351–68. doi: 10.1023/a:1026096203946. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: Confirmation of high prevalence. Am J Psychiatry. 2005;162:1133–41. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- 12.Constantino JN, Lajonchere C, Lutz M, et al. Autistic social impairment in the siblings of children with pervasive developmental disorders. Am J Psychiatry. 2006;163:294–96. doi: 10.1176/appi.ajp.163.2.294. [DOI] [PubMed] [Google Scholar]
- 13.Losh M, Childress D, Lam K, Piven J. Defining key features of the broad autism phenotype: A comparison across multiple- and single-incidence families of autistic individuals. Am J Hum Gen B Neuropsychiatr Genet. 2008;147B:424–33. doi: 10.1002/ajmg.b.30612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szatmari P, MacLean JE, Jones MB, et al. The familial aggregation of the lesser variant in biological and nonbiological relatives of PDD probands: A family history study. J Child Psychol Psychiatry. 2000;41:579–86. doi: 10.1111/1469-7610.00644. [DOI] [PubMed] [Google Scholar]
- 15.McKusick VA. Mendelian inheritance in man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buxbaum JD, Silverman JM, Smith CJ, et al. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet. 2001;68:1514–20. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alarcon M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70:60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarcon M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Mol Psychiatry. 2005;10:747–57. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- 19.Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH. A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am J Psychiatry. 2007;164:656–62. doi: 10.1176/ajp.2007.164.4.656. [DOI] [PubMed] [Google Scholar]
- 20.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–28. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auranen M, Vanhala R, Varilo T, et al. A genomewide screen for autism-spectrum disorders: Evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet. 2002;71:777–90. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett S, Beck JC, Bernier R, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–15. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Cantor RM, Kono N, Duvall JA, et al. Replication of autism linkage: Fine-mapping peak at 17q21. Am J Hum Genet. 2005;76:1050–56. doi: 10.1086/430278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.IMGSAC A genomewide screen for autism: Strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–81. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Nyholt DR, Magnussen P, et al. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69:327–40. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma DQ, Cuccaro ML, Jaworski JM, et al. Dissecting the locus heterogeneity of autism: Significant linkage to chromosome 12q14. Mol Psychiatry. 2007;12:376–84. doi: 10.1038/sj.mp.4001927. [DOI] [PubMed] [Google Scholar]
- 27.Risch N, Spiker D, Lotspeich L, et al. A genomic screen of autism: Evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schellenberg GD, Dawson G, Sung YJ, et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatry. 2006;11:1049–60. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- 29.Shao Y, Wolpert CM, Raiford KL, et al. Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet. 2002;114:99–105. doi: 10.1002/ajmg.10153. [DOI] [PubMed] [Google Scholar]
- 30.Yonan AL, Alarcon M, Cheng R, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73:886–97. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trikalinos TA, Karvouni A, Zintzaras E, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- 32.Pickles A, Bolton P, Macdonald H, et al. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: A twin and family history study of autism. Am J Hum Genet. 1995;57:717–26. [PMC free article] [PubMed] [Google Scholar]
- 33.Szatmari P. Heterogeneity and the genetics of autism. J Psychiatry Neurosci. 1999;24:159–65. [PMC free article] [PubMed] [Google Scholar]
- 34.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–17. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 35.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 36.Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–39. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell DB, D'Oronzio R, Garbett K, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–50. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 38.McDougle CJ, Naylor ST, Cohen DJ, Volkmar FR, Heninger GR, Price LH. A double-blind, placebo-controlled study of fluvoxamine in adults with autistic disorder. Arch Gen Psychiatry. 1996;53:1001–8. doi: 10.1001/archpsyc.1996.01830110037005. [DOI] [PubMed] [Google Scholar]
- 39.Cook EH, Jr, Leventhal BL, Heller W, Metz J, Wainwright M, Freedman DX. Autistic children and their first-degree relatives: Relationships between serotonin and norepinephrine levels and intelligence. J Neuropsychiatry Clin Neurosci. 1990;2:268–74. doi: 10.1176/jnp.2.3.268. [DOI] [PubMed] [Google Scholar]
- 40.McCauley JL, Olson LM, Dowd M, et al. Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet B Neuropsychiatr Genet. 2004;127:104–12. doi: 10.1002/ajmg.b.20151. [DOI] [PubMed] [Google Scholar]
- 41.Sutcliffe JS, Delahanty RJ, Prasad HC, et al. Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet. 2005;77:265–79. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulder EJ, Anderson GM, Kema IP, et al. Serotonin transporter intron 2 polymorphism associated with rigid-compulsive behaviors in Dutch individuals with pervasive developmental disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;133:93–96. doi: 10.1002/ajmg.b.30122. [DOI] [PubMed] [Google Scholar]
- 43.Tordjman S, Gutknecht L, Carlier M, et al. Role of the serotonin transporter gene in the behavioral expression of autism. Mol Psychiatry. 2001;6:434–39. doi: 10.1038/sj.mp.4000873. [DOI] [PubMed] [Google Scholar]
- 44.Wassink TH, Hazlett HC, Epping EA, et al. Cerebral cortical gray matter overgrowth and functional variation of the serotonin transporter gene in autism. Arch Gen Psychiatry. 2007;64:709–17. doi: 10.1001/archpsyc.64.6.709. [DOI] [PubMed] [Google Scholar]
- 45.Kim SJ, Cox N, Courchesne R, et al. Transmission disequilibrium mapping at the serotonin transporter gene (SLC6A4) region in autistic disorder. Mol Psychiatry. 2002;7:278–88. doi: 10.1038/sj.mp.4001033. [DOI] [PubMed] [Google Scholar]
- 46.Cook EH, Jr, Courchesne R, Lord C, et al. Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry. 1997;2:247–50. doi: 10.1038/sj.mp.4000266. [DOI] [PubMed] [Google Scholar]
- 47.Yirmiya N, Pilowsky T, Nemanov L, et al. Evidence for an association with the serotonin transporter promoter region polymorphism and autism. Am J Med Genet. 2001;105:381–86. doi: 10.1002/ajmg.1365. [DOI] [PubMed] [Google Scholar]
- 48.Klauck SM, Poustka F, Benner A, Lesch KP, Poustka A. Serotonin transporter (5-HTT) gene variants associated with autism? Hum Mol Genet. 1997;6:2233–38. doi: 10.1093/hmg/6.13.2233. [DOI] [PubMed] [Google Scholar]
- 49.Maestrini E, Lai C, Marlow A, et al. Serotonin transporter (5-HTT) and gamma-aminobutyric acid receptor subunit beta3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSA families. The International Molecular Genetic Study of Autism Consortium. Am J Med Genet. 1999;88:492–96. doi: 10.1002/(sici)1096-8628(19991015)88:5<492::aid-ajmg11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 50.Rice DS, Nusinowitz S, Azimi AM, Martinez A, Soriano E, Curran T. The reelin pathway modulates the structure and function of retinal synaptic circuitry. Neuron. 2001;31:929–41. doi: 10.1016/s0896-6273(01)00436-6. [DOI] [PubMed] [Google Scholar]
- 51.Ashley-Koch AE, Jaworski J, Ma de Q, et al. Investigation of potential gene-gene interactions between APOE and RELN contributing to autism risk. Psychiatr Genet. 2007;17:221–26. doi: 10.1097/YPG.0b013e32809c2f75. [DOI] [PubMed] [Google Scholar]
- 52.Persico AM, Pascucci T, Puglisi-Allegra S, et al. Serotonin transporter gene promoter variants do not explain the hyperserotoninemia in autistic children. Mol Psychiatry. 2002;7:795–800. doi: 10.1038/sj.mp.4001069. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Liu X, Zhang C, et al. Reelin gene alleles and susceptibility to autism spectrum disorders. Mol Psychiatry. 2002;7:1012–17. doi: 10.1038/sj.mp.4001124. [DOI] [PubMed] [Google Scholar]
- 54.Persico AM, D'Agruma L, Maiorano N, et al. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatry. 2001;6:150–59. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 55.Dutta S, Guhathakurta S, Sinha S, et al. Reelin gene polymorphisms in the Indian population: A possible paternal 5′UTR-CGG-repeat-allele effect on autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144:106–12. doi: 10.1002/ajmg.b.30419. [DOI] [PubMed] [Google Scholar]
- 56.Bailey A, Luthert P, Dean A, et al. A clinicopathological study of autism. Brain. 1998;121(Pt 5):889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- 57.Butler MG, Dasouki MJ, Zhou XP, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–21. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buxbaum JD, Cai G, Chaste P, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–91. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boccone L, Dessi V, Zappu A, et al. Bannayan-Riley-Ruvalcaba syndrome with reactive nodular lymphoid hyperplasia and autism and a PTEN mutation. Am J Med Genet A. 2006;140:1965–69. doi: 10.1002/ajmg.a.31396. [DOI] [PubMed] [Google Scholar]
- 60.Herman GE, Butter E, Enrile B, Pastore M, Prior TW, Sommer A. Increasing knowledge of PTEN germline mutations: Two additional patients with autism and macrocephaly. Am J Med Genet A. 2007;143:589–93. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- 61.Kwon CH, Luikart BW, Powell CM, et al. PTEN regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–88. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asato MR, Hardan AY. Neuropsychiatric problems in tuberous sclerosis complex. J Child Neurol. 2004;19:241–49. doi: 10.1177/088307380401900401. [DOI] [PubMed] [Google Scholar]
- 63.Curatolo P, Porfirio MC, Manzi B, Seri S. Autism in tuberous sclerosis. Eur J Paediatr Neurol. 2004;8:327–32. doi: 10.1016/j.ejpn.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 64.Bolton PF, Griffiths PD. Association of tuberous sclerosis of temporal lobes with autism and atypical autism. Lancet. 1997;349:392–95. doi: 10.1016/S0140-6736(97)80012-8. [DOI] [PubMed] [Google Scholar]
- 65.Seri S, Cerquiglini A, Pisani F, Curatolo P. Autism in tuberous sclerosis: Evoked potential evidence for a deficit in auditory sensory processing. Clin Neurophysiol. 1999;110:1825–30. doi: 10.1016/s1388-2457(99)00137-6. [DOI] [PubMed] [Google Scholar]
- 66.Asano E, Chugani DC, Muzik O, et al. Autism in tuberous sclerosis complex is related to both cortical and subcortical dysfunction. Neurology. 2001;57:1269–77. doi: 10.1212/wnl.57.7.1269. [DOI] [PubMed] [Google Scholar]
- 67.Eluvathingal TJ, Behen ME, Chugani HT, et al. Cerebellar lesions in tuberous sclerosis complex: Neurobehavioral and neuroimaging correlates. J Child Neurol. 2006;21:846–51. doi: 10.1177/08830738060210100301. [DOI] [PubMed] [Google Scholar]
- 68.Baker P, Piven J, Sato Y. Autism and tuberous sclerosis complex: Prevalence and clinical features. J Autism Dev Disord. 1998;28:279–85. doi: 10.1023/a:1026004501631. [DOI] [PubMed] [Google Scholar]
- 69.Zoghbi HY. Postnatal neurodevelopmental disorders: Meeting at the synapse? Science. 2003;302:826–30. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 70.Jamain S, Betancur C, Giros B, Leboyer M, Bourgeron T. Genetics of autism: From genome scans to candidate genes. Med Sci (Paris) 2003;19:1081–90. doi: 10.1051/medsci/200319111081. [DOI] [PubMed] [Google Scholar]
- 71.Laumonnier F, Bonnet-Brilhault F, Gomot M, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–57. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan J, Oliveira G, Coutinho A, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–32. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 73.Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–18. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 74.Vincent JB, Kolozsvari D, Roberts WS, Bolton PF, Gurling HM, Scherer SW. Mutation screening of X-chromosomal neuroligin genes: No mutations in 196 autism probands. Am J Med Genet B Neuropsychiatr Genet. 2004;129:82–84. doi: 10.1002/ajmg.b.30069. [DOI] [PubMed] [Google Scholar]
- 75.Gauthier J, Bonnel A, St-Onge J, et al. NLGN3/NLGN4 gene mutations are not responsible for autism in the Quebec population. Am J Med Genet B Neuropsychiatr Genet. 2005;132:74–75. doi: 10.1002/ajmg.b.30066. [DOI] [PubMed] [Google Scholar]
- 76.Blasi F, Bacchelli E, Pesaresi G, Carone S, Bailey AJ, Maestrini E. Absence of coding mutations in the X-linked genes neuroligin 3 and neuroligin 4 in individuals with autism from the IMGSAC collection. Am J Med Genet B Neuropsychiatr Genet. 2006;141:220–21. doi: 10.1002/ajmg.b.30287. [DOI] [PubMed] [Google Scholar]
- 77.Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: Synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 78.Feng J, Schroer R, Yan J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 79.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poliak S, Salomon D, Elhanany H, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–60. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–77. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 82.Arking DE, Cutler DJ, Brune CW, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–64. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bakkaloglu B, O'Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alarcon M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–59. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–97. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–28. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 88.Shaw-Smith C, Redon R, Rickman L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–48. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 90.Moeschler JB, Mohandas TK, Hawk AB, Noll WW. Estimate of prevalence of proximal 15q duplication syndrome. Am J Med Genet. 2002;111:440–42. doi: 10.1002/ajmg.10419. [DOI] [PubMed] [Google Scholar]
- 91.Jacquemont ML, Sanlaville D, Redon R, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–49. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–49. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beaudet AL. Autism: Highly heritable but not inherited. Nat Med. 2007;13:534–36. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- 95.Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 96.Pickles A, Starr E, Kazak S, et al. Variable expression of the autism broader phenotype: Findings from extended pedigrees. J Child Psychol Psychiatry. 2000;41:491–502. [PubMed] [Google Scholar]
- 97.Piven J, Palmer P, Landa R, et al. Personality and language characteristics in parents from multiple-incidence autism families. Am J Med Genet. 1997;74:398–411. [PubMed] [Google Scholar]
- 98.Ronald A, Happe F, Plomin R. The genetic relationship between individual differences in social and non-social behaviours characteristic of autism. Dev Sci. 2005;8:444–58. doi: 10.1111/j.1467-7687.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 99.Ronald A, Happe F, Bolton P, et al. Genetic heterogeneity between the three components of the autism spectrum: A twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–99. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 100.Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 101.August GJ, Stewart MA, Tsai L. The incidence of cognitive disabilities in the siblings of autistic children. Br J Psychiatry. 1981;138:416–22. doi: 10.1192/bjp.138.5.416. [DOI] [PubMed] [Google Scholar]
- 102.Gamliel I, Yirmiya M, Sigman M. The development of young siblings of children with autism from 4 to 54 months. J Autism Dev Dis. 2007;37:171–83. doi: 10.1007/s10803-006-0341-5. [DOI] [PubMed] [Google Scholar]
- 103.Ruser T, Arin D, Dowd M, et al. Communicative competence in parents of children with autism and parents of children with specific language impairment. J Autism Dev Dis. 2007;37:1323–36. doi: 10.1007/s10803-006-0274-z. [DOI] [PubMed] [Google Scholar]
- 104.Bolton P, Macdonald H, Pickles A, et al. Case-control family history study of autism. J Child Psychol Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 105.Folstein SE, Santangelo SL, Gilman SE, et al. Predictors of cognitive test patterns in autism families. J Child Psychol Psychiatry. 1999;40:1117–28. [PubMed] [Google Scholar]
- 106.Fombonne E, Bolton P, Prior J, Jordan H, Rutter M. A family study of autism: Cognitive patterns and levels in parents and siblings. J Child Psychol Psychiatry. 1997;38:667–83. doi: 10.1111/j.1469-7610.1997.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 107.Piven J, Palmer P. Cognitive deficits in parents from multiple-incidence autism families. Journal of Child Psychology and Psychiatry. 1997;35:877–900. doi: 10.1111/j.1469-7610.1997.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 108.Piven J, Gayle J, Chase GA, et al. A family history study of neuropsychiatric disorders in adults siblings of autistic individuals. J Am Acad Child Adolesc Psychiatry. 1990;29:177–83. doi: 10.1097/00004583-199003000-00004. [DOI] [PubMed] [Google Scholar]
- 109.Landa R, Piven J, Wzorek MM, Gayle JO, Chase GA, Folstein SE. Social language use in parents of autistic individuals. Psychol Med. 1992;22:245–54. doi: 10.1017/s0033291700032918. [DOI] [PubMed] [Google Scholar]
- 110.Landa R, Folstein SE, Isaacs C. Spontaneous narrative-discourse performance of parents of autistic individuals. J Speech Hear Res. 1991;34:1339–45. doi: 10.1044/jshr.3406.1339. [DOI] [PubMed] [Google Scholar]
- 111.Landa R, Garrett-Mayer E. Development in infants with autism spectrum disorders: A prospective study. J Child Psychol Psychiatry. 2006;47:629–38. doi: 10.1111/j.1469-7610.2006.01531.x. [DOI] [PubMed] [Google Scholar]
- 112.Toth K, Dawson G, Meltzoff AN, Greenson J, Fein D. Early social, imitation, play, and language abilities of young non-autistic siblings of children with autism. J Autism Dev Disord. 2007;37:145–57. doi: 10.1007/s10803-006-0336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stone WL, McMahon CR, Yoder PJ, Walden TA. Early social-communicative and cognitive development of younger siblings of children with autism spectrum disorders. Arch Pediatr Adolesc Med. 2007;161:384–90. doi: 10.1001/archpedi.161.4.384. [DOI] [PubMed] [Google Scholar]
- 114.Bishop DV, Maybery M, Wong D, Maley A, Hallmayer J. Characteristics of the broader phenotype in autism: A study of siblings using the children's communication checklist-2. Am J Med Genet B Neuropsychiatr Genet. 2006;141:117–22. doi: 10.1002/ajmg.b.30267. [DOI] [PubMed] [Google Scholar]
- 115.Dawson G, Estes A, Munson J, Schellenberg G, Bernier R, Abbott R. Quantitative assessment of autism symptom-related traits in probands and parents: Broader Phenotype Autism Symptom Scale. J Autism Dev Disord. 2007;37:523–36. doi: 10.1007/s10803-006-0182-2. [DOI] [PubMed] [Google Scholar]
- 116.Briskman J, Happe F, Frith U. Exploring the cognitive phenotype of autism: Weak central coherence in parents and siblings of children with autism: II. Real-life skills and preferences. J Child Psychol Psychiatry. 2001;42:309–16. [PubMed] [Google Scholar]
- 117.Merin N, Young GS, Ozonoff S, Rogers SJ. Visual fixation patterns during reciprocal social interaction distinguish a subgroup of 6-month-old infants at-risk for autism from comparison infants. J Autism Dev Disord. 2007;37:108–21. doi: 10.1007/s10803-006-0342-4. [DOI] [PubMed] [Google Scholar]
- 118.Cassel TD, Messinger DS, Ibanez LV, Haltigan JD, Acosta SI, Buchman AC. Early social and emotional communication in the infant siblings of children with autism spectrum disorders: An examination of the broad phenotype. J Autism Dev Disord. 2007;37:122–32. doi: 10.1007/s10803-006-0337-1. [DOI] [PubMed] [Google Scholar]
- 119.Yirmiya N, Gamliel I, Pilowsky T, Baron-Cohen S, Feldman R, Sigman M. The development of siblings of children with autism at 4 and 14 months: Social engagement, communication and cognition. J Child Psychol Psychiatry. 2006;47:511–23. doi: 10.1111/j.1469-7610.2005.01528.x. [DOI] [PubMed] [Google Scholar]
- 120.Golberg W, Jarvis KL, Osann K, et al. Brief report: Early social communication behaviors in the younger siblings of children with autism. J Autism Dev Disord. 2005;35:657–64. doi: 10.1007/s10803-005-0009-6. [DOI] [PubMed] [Google Scholar]
- 121.Bailey A, Palferman S, Heavey L, Le Couteur A. Autism: The phenotype in relatives. J Autism Dev Disord. 1998;28:369–92. doi: 10.1023/a:1026048320785. [DOI] [PubMed] [Google Scholar]
- 122.Murphy M, Bolton P, Pickles A, Fombonne E, Piven J, Rutter M. Personality traits of the relatives of autistic probands. Psychol Med. 2000;30:1411–24. doi: 10.1017/s0033291799002949. [DOI] [PubMed] [Google Scholar]
- 123.Piana H, Fortin C, Noulhiane M, Golse B, Robel L. Investigation of the behavioural phenotype of parents of autistic children through the new FAQ self-report. Encephale. 2007;33:285–92. doi: 10.1016/s0013-7006(07)92041-2. [DOI] [PubMed] [Google Scholar]
- 124.Piven WM, Landa R, Lainhart J, Bolton P, Chase G, et al. Personality characteristics of the parents of individuals with autism. Psychol Med. 1994;24:783–95. doi: 10.1017/s0033291700027938. [DOI] [PubMed] [Google Scholar]
- 125.Spiker D, Lotspeich L, Kraemer HC, et al. Genetics of autism: Characteristics of affected and unaffected children from 37 multiplex families. Am J Med Genet (Neuropsychiatr Genet) 1994;54:27–35. doi: 10.1002/ajmg.1320540107. [DOI] [PubMed] [Google Scholar]
- 126.Hughes C, Leboyer M, Bouvard M. Executive function in parents of children with autism. Psychol Med. 1997;27:209–20. doi: 10.1017/s0033291796004308. [DOI] [PubMed] [Google Scholar]
- 127.Ozonoff S, Rogers SJ, Farnham JM, Pennington BF. Can standard measured identify subclinical markers of autism? J Autism Dev Disord. 1993;23:429–41. doi: 10.1007/BF01046049. [DOI] [PubMed] [Google Scholar]
- 128.Baron-Cohen S, Hammer J. Parents of children with Asperger syndrome: What is the cognitive phenotype? J Cog Neurosci. 1997;9:548–54. doi: 10.1162/jocn.1997.9.4.548. [DOI] [PubMed] [Google Scholar]
- 129.Baron-Cohen S, Wheelwright S, Hill J, Raste Y, Plumb I. The “Reading the Mind in the Eyes” Test revised version: A study with normal adults, and adults with Asperger syndrome or high-functioning autism. J Child Psychol Psychiatry. 2001;42:241–51. [PubMed] [Google Scholar]
- 130.Dorris L, Espie CA, Knott F, Salt J. Mind-reading difficulties in the siblings of people with Asperger's syndrome: Evidence for a genetic influence in the abnormal development of a specific cognitive domain. J Child Psychol Psychiatry. 2004;45:412–18. doi: 10.1111/j.1469-7610.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 131.Happe F, Briskman J, Frith U. Exploring the cognitive phenotype of autism: Weak central coherence in parents and siblings of children with autism: I. Experimental tests. J Child Psychol Psychiatry. 2001;42:299–307. [PubMed] [Google Scholar]
- 132.Losh M, Piven J. Social-cognition and the broad autism phenotype: Identifying genetically meaningful phenotypes. J Child Psychol Psychiatry. 2007;48:105–12. doi: 10.1111/j.1469-7610.2006.01594.x. [DOI] [PubMed] [Google Scholar]
- 133.Dalton KM, Nacewicz BM, Alexander AL, Davidson RJ. Gaze-fixation, brain activation, and amygdala volume in unaffected siblings of individuals with autism. Biol Psychiatry. 2007;61:512–20. doi: 10.1016/j.biopsych.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 134.Shao Y, Raiford KL, Wolpert CM, et al. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet. 2002;70:1058–61. doi: 10.1086/339765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bradford Y, Haines J, Hutcheson H, et al. Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet (Neuropsychiatr Genet) 2001;105:539–47. [PubMed] [Google Scholar]
- 136.Chen GK, Kono N, Geschwind DH, Cantor RM. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Mol Psychiatry. 2006;11:214–20. doi: 10.1038/sj.mp.4001753. [DOI] [PubMed] [Google Scholar]
- 137.Buxbaum JD, Silverman J, Keddache M, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: Evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry. 2004;9:144–50. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- 138.Sakurai T, Ramoz N, Reichert JG, et al. Association analysis of the NrCAM gene in autism and in subsets of families with severe obsessive-compulsive or self-stimulatory behaviors. Psychiatr Genet. 2006;16:251–57. doi: 10.1097/01.ypg.0000242196.81891.c9. [DOI] [PubMed] [Google Scholar]
- 139.Shao Y, Cuccaro ML, Hauser ER, et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet. 2003;72:539–48. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brune CW, Kim SJ, Salt J, Leventhal BL, Lord C, Cook EH., Jr. 5-HTTLPR genotype-specific phonotype in chidren and adolescents with autism. Am J Psychiatry. 2006;163:2148–56. doi: 10.1176/ajp.2006.163.12.2148. [DOI] [PubMed] [Google Scholar]
- 141.Nurmi EL, Dowd M, Tadevosyan-Leyfer O, Haines JL, Folstein SE, Sutcliffe JS. Exploratory subsetting of autism families based on savant skills improves evidence of genetic linkage to 15q11-q13. J Am Acad Child Adolesc Psychiatry. 2003;42:856–63. doi: 10.1097/01.CHI.0000046868.56865.0F. [DOI] [PubMed] [Google Scholar]
- 142.Molloy CA, Keddache M, Martin LJ. Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Mol Psychiatry. 2005;10:741–46. doi: 10.1038/sj.mp.4001691. [DOI] [PubMed] [Google Scholar]
- 143.Conciatori M, Stodgell CJ, Hyman SL, et al. Association between the HOXA1 A218G polymorphism and increased head circumference in patients with autism. Biol Psychiatry. 2004;55:413–19. doi: 10.1016/j.biopsych.2003.10.005. [DOI] [PubMed] [Google Scholar]