Abstract
Lipid droplets play a critical role in a variety of metabolic diseases. Numerous proteomic studies have provided detailed information about the protein composition of the droplet, which has revealed that they are functional organelles involved in many cellular processes, including lipid storage and metabolism, membrane traffic, and signal transduction. Thus, the droplet proteome indicates that lipid accumulation is only one of a constellation of organellar functions critical for normal lipid metabolism in the cell. As a result of this new understanding, we suggested the name adiposome for this organelle. The trafficking ability of the adiposome is likely to be very important for lipid uptake, retention, and distribution, as well as membrane biogenesis and lipid signaling. We have taken advantage of the ease of purifying lipid-filled adiposomes to develop a cell-free system for studying adiposome-mediated traffic. Using this approach, we have determined that the interaction between adiposomes and endosomes is dependent on Rab GTPases but is blocked by ATPase. These methods also allowed us to identify multiple proteins that dynamically associate with adiposomes in a nucleotide-dependent manner. An adiposome-endosome interaction in vitro occurs in the absence of cytosolic factors, which simplifies the assay dramatically. This assay will enable researchers to dissect the molecular mechanisms of interaction between these two organelles. This chapter provides a detailed account of the methods developed.
1. Introduction
Imbalances in lipid storage and metabolism in human cells have been linked to the progression of many metabolic diseases, such as morbid obesity, type 2 diabetes, cardiovascular disease, and nonalcoholic fatty liver disease. Generally, these health consequences are driven by the excessive storage of lipids in cells. For example, obesity is the excessive storage of lipids in adipocytes. A common consequence of obesity is the inappropriate sequestration of lipids in liver and skeletal muscle cells, which is thought to drive insulin resistance and the eventual development of overt type 2 diabetes. Lipid accumulation in hepatocytes is also the first stage in the development of fatty liver disease. The accumulation of cholesterol in macrophages transforms the cells into foam cells, which initiates the development of atherosclerosis. However, a lack of neutral lipid storage, especially in adipose tissue, can cause lipodystrophies that are associated with similar metabolic complications. Thus, understanding the regulation of cellular lipid storage is key to preventing and treating these metabolic diseases.
All cells, from bacteria to mammals, are able to store neutral lipids in a cellular structure that has been given many names, including lipid droplets, lipid bodies, oil bodies, and fat bodies. The term lipid droplet, which describes the structure of a lipid storage depot, is the most commonly used appellation (Martin and Parton, 2006). Recently, we and other groups have partially identified the droplet proteome from various cell types and tissues. Unexpectedly the proteome contains proteins involved in lipid metabolism, membrane traffic, and signal transduction. This information has extended our understanding of the droplet and suggests that rather than being a simple storage depot, it is a complex organelle involved in the synthesis, degradation, and transport of cellular lipids that participate in energy balance, membrane biogenesis, and cellular signaling. This suggests that the droplet is only one morphologic form of a complex functional organelle and, therefore, deserves a new term that encompasses all of its properties. We have proposed the name adiposome for this organelle, in keeping with the long-standing tradition in cell biology to name organelles with a unique prefix followed by a common suffix such as endosome, lysosome, and peroxisome (Liu et al., 2004).
Among the newly identified adiposome-associated proteins are ones known to be involved in regulating membrane traffic (e.g., Rab GTPases), which led to the hypothesis that adiposomes are involved in lipid and/or protein traffic (Fujimoto et al., 2004; Liu et al., 2004). The recruitment of Rab proteins to adiposomes when lipolysis is stimulated supports this view (Brasaemle et al., 2004). To test this hypothesis and further investigate the molecular mechanism of adiposome-mediated trafficking, we developed a cell-free system to study the interaction between purified lipid-filled adiposomes and isolated early endosomes. Using this approach, we demonstrated that Rab proteins interact with adiposomes and found that these Rabs exhibit biological properties similar to those when associated with membrane organelles such as endosomes (Liu et al., 2007). As detailed later, this reconstitution system also enables one to study the interaction between adiposomes and early endosomes and to study the role of Rab proteins in this process (Liu et al., 2007).
2. Rab-Mediated Adiposome-Endosome Interaction in vitro
Taking advantage of the dramatic density difference between lipid-filled adiposomes and other endomembranes, we developed a cell-free system to study the dynamic behavior of this organelle and its interaction with endosomes. To study the dynamics, we incubated purified adiposomes with isolated cytosol and then reisolated adiposomes by flotation, followed by analysis of its protein profile. Using this assay, we observed the recruitment of small G proteins such as Rabs and Arfs, as well as cytoskeleton proteins, including actin and tubulin (Bartz et al., 2007b; Liu et al., 2007). To study the interaction of adiposomes with endosomes, we incubated the isolated organelles under defined conditions and then reisolated the adiposomes by flotation. The physical interaction between endosomes and adiposomes is detected by immunoblotting the adiposome fraction for endosome marker proteins. The activity of Rab proteins is regulated using GTP and GDP. Rab proteins on adiposomes and endosomes can be removed by treating each with Rab-GDP dissociation inhibitor (RabGDI). Rabs are recruited back to Rab-free adiposomes by incubating in the presence of cytosol and GTP. Using these methods, we discovered that the adiposome interaction with early endosome is dependent on GTPγS. Moreover, removing Rabs from both organelles totally abolished this interaction, demonstrating that Rab proteins are essential for the interaction (Liu et al., 2007). The methods used are detailed next.
2.1. Sample Preparation
2.1.1. Purification of adiposome
The purification method developed can be used to purify lipid-filled adiposomes from virtually any type of animal cell or tissue, including Chinese hamster ovary fibroblasts (CHO K2) (Liu et al., 2004), human fibroblasts (Bartz et al., 2007a), HeLa cells (Bartz et al., 2007b; Liu et al., 2004), 3T3 L1 adipocytes (Bartz et al., 2007a), human B cells (Bartz et al., 2007a), LNCaP cells, and mouse liver. Figure 24.1A shows the signature Coomassie colloidal blue staining pattern of lipid-filled adiposomes isolated from various sources. While the pattern is highly reproducible for each cell type, it varies between different cell types and tissues, which indicate that there are tissue-specific differences in the protein composition of the organelle.
Figure 24.1.
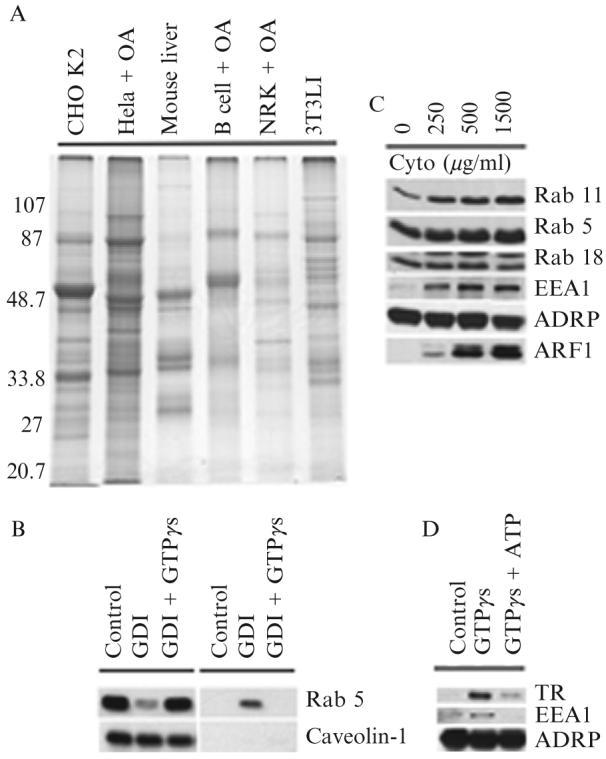
(A) Signature protein of adiposomes from different cells. Adiposomes were purified from the indicated cells grown in the presence or absence of oleic acid (OA). Proteins were precipitated with 100% acetone and dissolved in SDS sample buffer, and 10 μg of proteins was separated on 10% SDS-PAGE and stained with Coomassie colloidal blue. Adiposomes were isolated from 3T3 L1 cells after 8 days of differentiation. (B) Release of GDP-bound Rab 5 from adiposomes by RabGDI. Purified adiposomes were incubated in the presence of RabGDI (GDI) plus or minus GTPγs. Adiposomes (left) and reaction buffer (right) were separated and processed for immunoblotting to detect the indicated proteins. (C) Recruitment of Rabs from cytosol to adiposome. Purified adiposomes were processed to remove endogenous Rabs with RabGDI. Rab-depleted adiposomes were then incubated with the indicated concentrations of cytosol for 1 h. At the end of the reaction, adiposomes were reisolated, washed, and processed for immunoblotting to detect the indicated proteins. The adiposome-specific protein ADRP was used as a loading control. (D) Early endosomes bind adiposomes. Purified adiposomes were mixed with purified early endosomes in the presence or absence of GTPγs and ATP. Adiposomes were reisolated and unbound endosomes removed. Adiposomes were then processed for immunoblotting to detect the indicated proteins. ADRP was used as a loading control.TR, transferrin receptor.
Methods
Cells are cultured to 100% confluence in 150-mm plates containing 25 ml of standard tissue culture media. Cells are washed with 10 ml ice-cold phosphate-buffered saline (PBS) per plate, scraped into 5 ml ice-cold PBS plus 5 μl 100 mM phenylmethylsulfonyl fluoride (PMSF), pooled, and placed in a 50-ml tube. The cell suspension from nine plates is centrifuged at 500 g for 5 min at 4° and the supernatant is discarded. Cells are resuspended by vortexing in 8 ml buffer A (20 mM Tricine, pH 7.8, 250 mM sucrose) plus 8 μl of 100 mM PMSF and then incubated on ice for 20 min. The cells are homogenized with a nitrogen bomb (Parr Instrument Company, Moline, IL). The cells are placed in the bomb at 450 psi for 15 min on ice. The mixture is then released from the bomb chamber drop wise into a 50-ml tube and the lysate is centrifuged at 1000 g for 10 min at 4°. The slow release of the sample from the bomb under pressure is essential for proper homogenization. Seven milliliters of the supernatant fraction (PNS) is added to a SW41 tube, and then 3.5 ml of buffer B (20 mM HEPES, pH 7.4, 100 mM KCl, 2 mM MgCl2) is loaded on top of the PNS fraction using a gradient maker. It is important to be able to see a sharp interface. The gradient is centrifuged at 40,000 rpm for 1 h at 4°. Lipid-filled adiposomes will concentrate in a white band at the top of the gradient. Adiposomes are collected using a 1-ml pipette tip and transferred to a 1.5-ml microfuge tube, making sure to remove the most adiposomes with the least amount of buffer. Adiposomes are separated from buffer by centrifuging at 10,000 g for 5 min at 4°. The solution underlying the adiposome is removed using a gel-loading tip attached to a 1-ml tip. The tip is slid into solution on the tube surface to avoid disturbing the floating adiposomes. Enough buffer is removed until the adiposome fraction reaches the pipette tip opening. Removing the buffer can disturb the adioposme layer. Adiposomes are suspended in buffer form a cloudy solution. It is impossible to remove the buffer from this cloudy solution without losing material. To avoid loss the sample is centrifuged again and the buffer is removed. The centrifugation step is repeated at least three times to remove all the buffer. To separate out contaminating membranes, the adiposomes are resuspended in 1 ml buffer B by vortexing and centrifuged at 40,000 rpm for 5 min in a Beckman Optima Max ultracentrifuge using a TLA 100.3 rotor at 4°. After centrifugation, some of the adiposomes are retained on the side of the tube while the remainder float at the top of the solution. The floating adiposomes are collected and transferred into a new 1.5-ml Eppendorf tube. The rest of the solution is discarded. The pellet in the bottom of the tube is resuspended using 100 μl buffer B and is discarded. Another 100 μl of buffer B is used to wash the bottom of the tube. Do not touch the adiposomes on the side of the tube during these steps (carefully use a 200-μl tip). Adiposomes are transferred back into the original tube, which is vortexed to wash the adiposomes off the side of tube. When all of the adiposomes are resuspended, transfer them into a new 1.5-ml Eppendorf tube and adjust the volume to 500 μl with buffer B. Centrifuge the sample at 10,000 g for 5 min. Remove the solution using a gel-loading tip attached to a 1-ml tip, making sure not to disturb the floating adiposomes. Repeat the washing step two more times. After the final wash, the adiposomes are ready for biochemical, morphological, and functional analysis. Note: Two important clues are needed for good purification. (1) Handle the sample very gently because lipid-filled adiposomes are extremely fragile (especially large ones). Avoid harsh vortexing and use wide-end pipette tips (cut tip) for transferring. (2) Broken adiposomes tend to be retained on plastic surfaces and can be driven into the pellet by centrifugation. Therefore, make sure that all adiposomes are resuspended in buffer B and that any aggregates (broken adiposomes and contaminated membrane structures) are removed using new tubes and tips. Remove the pellet from the bottom of the tube using a gel-loading tip.
2.1.2. Purification of His-tagged RabGDI
The RabGDI is purified from the BL 21(DE3) Escherichia coli strain hosting the pRSETa-GDI construct as described previously (Bartz et al., 2003).
Methods
Briefly, 100 ml of cell lysate from a 10-liter culture is mixed and incubated with 10-ml preequilibrated Ni-NTA agarose beads (Qiagen) in 50-ml Falcon tubes. The samples are rotated on a wheel for 1 h at 4°. The beads are centrifuged to a pellet, washed, and then loaded into a column. RabGDI is eluted with 200 mM imidazole, and the sample is dialyzed overnight at 4° against buffer C (20 mM HEPES, pH 7.2, 10 mM 2-mercaptoethanol). Purified RabGDI is analyzed by SDS-PAGE to determine purity and protein concentration. The protein is further analyzed to measure its ability to remove Rab. The rest of the protein is snap frozen as aliquots and stored at -80°.
2.1.3. Isolation of cytosol
PNS (10 ml) from the adiposome purification step is loaded into a SW41 tube and centrifuged at 40,000 rpm for 1 h to remove any membranes. The cytosol is collected between adiposome (top) and membrane pellet (bottom) and is used fresh in each experiment.
2.1.4. Isolation of early endosomes
Early endosomes are purified by the method of Bartz et al. (2005) with minor modifications to isolate them from CHO K2 cells.
Methods
CHO K2 cells are cultured to 60% confluence, collected by scraping, and concentrated by pelleting through centrifugation. The cells are then resuspended in buffer B with proteinase inhibitor cocktail and homogenized firmly with a 1-ml syringe (slim) and 22-gauge × 1.5-in. needle (Aldrich) five times up and down. The homogenate is centrifuged in a 15-ml Falcon tube at 1000 g for 15 min at 4° to remove nuclei and unbroken cells. After centrifugation, the supernatant fraction is collected and adjusted to 40.6% sucrose using 62% sucrose in buffer B. The sample is transferred into a SW28 tube, overlaid with 12 ml of 35% sucrose in buffer B, 8 ml 20% sucrose in buffer B, and 2 ml of buffer B. The gradient is centrifuged at 28,000 rpm (103,745 g) for 3 h. The visible interface (2 ml) between 25 and 30% sucrose is collected in a 15-ml tube, mixed carefully two to three times with a disposable plastic pipette, placed in 200-μl aliquots in an Eppendorf tube (use cut tips), snap frozen in liquid N2, and stored at -80°.
2.2. Verification Proteins Associated with Adiposomes
It is important to verify by morphology that any protein of interest is associated with adiposomes and not a contaminating organelle. This can only be done by immunogold electron microscopy. For example, even though the proteome indicates that lipid-filled adiposomes are enriched in multiple Rabs, they could be in contaminating endoplasmic reticulum (ER) (Liu et al., 2007).
Methods
Purified adiposomes in 200 μl of buffer B are mixed with 200 μl of 6% paraformaldehyde in buffer B containing 0.15% crystallized BSA (buffer D) and incubated for 1 h at room temperature. The fixed sample is washed by flotation (centrifuge at 20,000 g for 3 min) with buffer B three times, 5 min each at room temperature, resuspended in buffer B plus 50 mM NH4Cl (buffer E) for 10 min, and washed again with buffer B two times, 5 min each. The sample is mixed with buffer D and incubated for 30 min at room temperature. Buffer D is removed and the sample is mixed with pAb or mAb, diluted 1:25 with buffer D, and incubated further for 15 h at 4°. The adiposomes are washed by flotation with buffer D and mixed with the appropriate α-IgG conjugated to 10 nm gold diluted 1:30 in buffer D for 2 h at room temperature. Adiposomes are washed again with buffer D three times, 5 min each, at 4°. Finally, samples are washed with 50 mM Na2HPO4, pH 8.0, 0.3 M NaCl, 10 mM 2-mercaptoethanol (buffer F), fixed with 1% GTA, and postfixed in 1% OsO4, all in buffer F. Samples are embedded in Epon.
2.3. Testing for Adiposome Interaction with Proteins
2.3.1. Release of Rab proteins from adiposomes by RabGDI
Rabs are small GTP-binding proteins that regulate membrane traffic by controlling targeting, tethering, docking, and fusion between various membrane systems. Previous studies demonstrate that Rab proteins are activated by GTP. When GTP is hydrolyzed to GDP, Rabs are released from the membrane into the cytosol by RabGDI (Holtta-Vuori et al., 2000). Cytosolic Rabs are recruited back to target membranes through the action of the RabGDI displacement factor, RabGDF (Dirac-Svejstrup et al., 1997). Therefore, a measure of whether an organelle-associated Rab is functional is if it can cycle off and on the organelle in a RabGDI- and RabGDF-dependent fashion. This assay can be carried out on isolated adiposomes.
Methods
Purified adiposomes (100 μl) are incubated in the presence of 2 mM GDP and 10 μM RabGDI in buffer B at 37° for 1 h. The reaction is vortexed briefly every 10 min to keep adiposomes in suspension because they tend to float to the top of solution. Adiposomes are repurified from the reaction buffer by flotation, and the reaction buffer and adiposome fraction are processed. The adiposome fraction is washed three times with 200 μl/time of buffer B using the same flotation method, and the protein is precipitated with 100% acetone. Proteins are precipitated from the reaction buffer with 7.2% trichloroacetic acid. The presence of Rabs or other proteins of interest in either fraction is measured by immunoblotting. The immunoblot is reprobed with an appropriate adiposome resident protein marker that is not released by washing, such as caveolin-1. Ral A, another adiposome-associated small G protein, is used as a negative control because it is not removed by RabGDI. Figure 24.1B (left) shows that RabGDI releases Rab 5 and that release is blocked by GTPγs. Therefore, as expected, RabGDI only removes Rabs from adiposomes that are bound to GDP. RabGDI-released Rab 5 appears in the incubation buffer (see Fig. 24.1, right).
2.3.2. Recruitment of cytosolic Rabs to adiposomes
Rab-depeleted adiposomes are used to study the recruitment of cytosolic proteins.
Methods
Rab-deficient adiposomes (50 μl) prepared as described earlier are incubated in the presence of 100 μl cytosol (250-1500 μg/ml) in buffer B plus 1 mM GTPγs for 1 h at 37°. The reaction is vortexed briefly every 10 min to keep adiposomes in suspension. Adiposomes are then reisolated, washed, and processed for immunoblotting to detect both the Rabs of interest and any other proteins. To avoid possible contamination from cytosol, adiposomes are washed five times with buffer B. Liposomes prepared from CHO K2 cell total lipids are used as a nonspecific binding control. To make liposomes, lipids are extracted by the method of Bligh and Dyer (1959) from the total membrane pellet of cultured cells. The solvent is removed from the lipids using a rotary evaporator followed by 20 min under low torr vacuum. Diethyl ether (5 ml) and buffer B (300 μl) are added to the lipid film. The ether is driven off under a stream of nitrogen with bath sonication. The resulting concentrated liposomes are diluted with 1.7 ml of buffer B and drawn through a 22-gauge needle three times.
Figure 24.1C shows an experiment demonstrating that isolated, RabGDI-treated adiposomes can recruit Rabs 5 and 11 plus the Rab 5 effector EEA1 as a function of cytosol concentration in the incubation media. Interestingly, Rab 18 is neither removed by RabGDI nor recruited from cytosol. Arf1 is also recruited to the adiposomes. The peripheral adiposome marker ADRP is not affected.
2.4. Rab-Dependent Interaction between Early Endosomes and Adiposomes
The GTP-dependent interaction of Rabs with adiposomes is similar to how Rabs function in intracellular membrane traffic, which suggests that adiposomes may interact with various endomembranes. Indeed, Rab 18 appears to mediate the close apposition of the ER to adiposomes in situ (Martin et al., 2005; Ozeki et al., 2005). Since we found that adiposomes bound cytosolic EEA1 (see Fig. 24.1C), we focused on the endosome interaction with adiposomes. Not only can the adiposome-endosome interaction be reconstituted, but the requirements for GTP, Rabs, and other proteins can be determined (Liu et al., 2007). Interestingly, the presence of cytosol in the assay does not affect the interaction, suggesting that the factors required for the interaction are completely contained in the interacting organelles. Early endosome binding to adiposomes is saturable, which suggests that there are a limited number of binding sites on each adiposome. Finally, in stark contrast to membrane-membrane interactions during membrane traffic, ATP inhibits the binding of endosome to adiposome (see Fig. 24.1D).
Methods
Purified adiposomes (50 μl) are mixed with 50 μl endosomes (100 μg/ml) in buffer B and incubated for 1 h at 37° in the presence or absence of 1 mM nucleotide. The reaction is vortexed briefly every 10 min to keep the adiposomes in suspension. Endosomes bound to adiposomes are separated from free endosomes by flotation using a centrifuge speed of 10,000 g for 4 min at 4°. The centrifuge speed is very critical because too high a speed can strip off specifically bound endosomes, whereas too low a speed will lead to nonspecific binding. Additional nonspecifically bound endosomes are removed by washing adiposomes three times with 200 μl buffer B as mentioned previously. With each spin the membrane pellet gets smaller. After three spins, the pellet is not readily visible unless the bottom of the tube is viewed with a bright light against a dark background. Proteins are precipitated and lipids are removed from the adiposomes using 1 ml/tube of 100% acetone followed by centrifugation at 20,000 g for 10 min. The bound endosomes are detected by immunoblotting using the pAb transferrin receptor and mAb EEA1 IgG. The lipid-filled adiposome marker ADRP is used as a loading control. To determine whether Rabs are required for adiposome-early endosome interaction, it is necessary to remove Rabs from both organelles using RabGDI. We used the method described earlier to remove Rabs from adiposomes. To remove them from endosomes, the isolated endosomes (200 μl) are incubated in the presence of 2 mM GDP and 10 μM RabGDI in buffer B at 37° for 1 h. Then the reaction is mixed with 600 μl of buffer B, loaded on the top of 200 μl of 2 M KCl in an Eppendorf tube, and centrifuged at 20,000 g for 30 min at 4°. Endosomes (400 μl) are collected from the interface between 2 M KCl and buffer B. The ability of Rab-depleted adiposomes and endosomes to interact is determined as described earlier.
3. Conclusion
The development of an in vitro method for reconstituting the interaction between lipid-filled adiposomes and either cytosolic or membrane proteins is a rich system for exploring the organellar properties of the adiposome. In conjunction with various genetic, molecular biology, and cell biology techniques, these methods should be very useful not only in understanding the function of adiposome-associated Rabs, but also the function of adiposomes in the synthesis, storage, and distribution of cellular lipids. While the assays are technically straightforward, they demand attention to details and fresh materials. The lipid-filled adiposome is fragile and cannot be frozen for any length of time.
ACKNOWLEDGMENTS
We thank Meifang Zhu for her valuable technical assistance and Brenda Pallares for administrative assistance. This work was supported by grants from the National Institutes of Health, HL 20948, GM 52016, the Cecil H. Green Distinguished Chair in Cellular and Molecular Biology, and the Perot Family Foundation to RGWA and GM070117 to John K. Zehmer.
REFERENCES
- Bartz R, Benzing C, Ullrich O. Reconstitution of vesicular transport to Rab11-positive recycling endosomes in vitro. Biochem. Biophys. Res. Commun. 2003;312:663–669. doi: 10.1016/j.bbrc.2003.10.172. [DOI] [PubMed] [Google Scholar]
- Bartz R, Benzing C, Ullrich O. Reconstitution of transport to recycling endosomes in vitro. Methods Enzymol. 2005;404:480–490. doi: 10.1016/S0076-6879(05)04042-5. [DOI] [PubMed] [Google Scholar]
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007a;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: Protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 2007b;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Holtta-Vuori M, Maatta J, Ullrich O, Kuismanen E, Ikonen E. Mobilization of late-endosomal cholesterol is inhibited by Rab guanine nucleotide dissociation inhibitor. Curr. Biol. 2000;10:95–98. [PubMed] [Google Scholar]
- Liu P, Bartz R, Zehmer JK, Ying YS, Zhu M, Serrero G, Anderson RG. Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta. 2007;1773:784–793. doi: 10.1016/j.bbamcr.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: Effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: A unified view of a dynamic organelle. Nat. Rev. Mol. Cell. Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]


