Abstract
Cytochrome P450 (P450) 2B6 metabolizes a number of clinically relevant drugs and is one of the most highly polymorphic human P450 enzymes, with the Lys262→Arg substitution being especially common in several genetic variants. Therefore, K262R (2B6*4) was created in the CYP2B6dH background (N-terminal-modified and C-terminal His-tagged) and expressed in Escherichia coli. The recombinant CYP2B6dH and K262R were purified and studied to investigate the effect of the Lys262→Arg substitution with six of the most potent drug inhibitors of CYP2B6, namely, clopidogrel, clotrimazole, itraconazole, raloxifene, sertraline, and ticlopidine. K262R showed a >3-fold increase in the Ki values with clopidogrel, itraconazole, and raloxifene and ∼6-fold increase in Ki with sertraline compared with CYP2B6dH. Likewise, K262R showed 2-, 4-, and >20-fold higher Ks values than CYP2B6dH with clopidogrel, sertraline, and itraconazole, respectively. In contrast, when tested with several known type II inhibitors of CYP2B enzymes, K262R showed a 10-fold lower IC50 with 4-(phenyl)pyridine and ∼2-fold lower IC50 with 4-(4-nitrobenzyl)pyridine or 1-(4-phenyl)benzylimidazole than CYP2B6dH. Subsequent analysis predicted possible in vivo drug-drug interactions between the CYP2B6 substrate efavirenz and drug inhibitors clopidogrel, clotrimazole, itraconazole, sertraline, and ticlopidine. Furthermore, Q172H/K262R (2B6*6), which is the most common genetic variant of CYP2B6 harboring K262R, was created in CYP2B6dH, expressed, purified, and characterized for inhibition. Q172H/K262R showed a >6-fold increase in Ki with sertraline and clopidogrel compared with CYP2B6dH. The results suggest that individuals, especially homozygotes, with the 2B6*4 or 2B6*6 allele might be less susceptible to drug interactions resulting from P450 inhibition.
Although cytochrome P450 (P450) 2B6 (CYP2B6) is expressed at relatively low levels in the liver (Guengerich, 2005), the enzyme metabolizes important pharmaceuticals including cyclophosphamide, propofol, promazine, methadone, S-mephenytoin, efavirenz, bupropion, imipramine, midazolam, artemisinin, and tamoxifen (Rendic, 2002; Lewis et al., 2004; Zanger et al., 2007). In addition, CYP2B6 possesses several important genetic variants; among them the most common are K262R (2B6*4), Q172H/K262R (2B6*6), and R487C (2B6*5). Frequencies of the three most common single nucleotide polymorphisms range from 14 to 49% for Q172H, 17 to 63% for K262R, and 0 to 14% for R487C depending on the ethnicity of the population studied (Lang et al., 2004). For example, studies in German males have found a K262R allele frequency of approximately 5% and a single nucleotide polymorphism frequency of 30% (Lang et al., 2001; Kirchheiner et al., 2003). Single-dose bupropion pharmacokinetic data obtained from 121 individuals showed 1.3-fold increased clearance by individuals with the 2B6*1/*4 genotype (Kirchheiner at al., 2003). A similar study of 169 individuals with efavirenz showed a 17% reduced area under plasma concentration in *1/*4 heterozygotes (Rotger et al., 2007). In vitro, K262R has been incorporated into the engineered CYP2B6dH (N-terminal-deleted and C-terminal Histag) by Hollenberg's group to study structure function. Compared with CYP2B6dH, K262R shows a >2-fold increased kcat for the metabolism of bupropion to hydroxybupropion (Bumpus et al., 2005) and ∼2-fold increased catalytic efficiency for the metabolism of efavirenz to 8-hydroxyefavirenz (Bumpus et al., 2006). It is interesting to note that, in contrast to CYP2B6dH, K262R is refractory to mechanism-based inactivation by 17α-ethynylestradiol or efavirenz, whereas susceptibility of the variant to inactivation is preserved with bergamottin, N,N′,N″-triethylenethiophosphoramide, and 8-hydroxyefavirenz (Bumpus et al., 2005, 2006).
Drug-drug interactions (DDIs), especially through inhibition of P450-mediated drug metabolism by a coadministered drug, are one of the primary causes of serious adverse events occurring in clinical practice (Dambro and Kallgren, 1988). The presence of polymorphic variants of P450 further complicates the prediction of in vivo DDIs. To address the issues of genotype-dependent DDIs as a result of enzyme inhibition, Kumar et al. (2006) have used five substrates and a battery of inhibitors with CYP2C9.1 and CYP2C9.3 variants. In a recent study, several clinically relevant drugs, such as clopidogrel, clotrimazole, itraconazole, ticlopidine, sertraline, and raloxifene, have been found to be potent inhibitors of CYP2B6 (Walsky et al., 2006). Therefore, in the present study we have studied the effect of the Lys262→ Arg substitution in K262R (2B6*4) and Q172H/K262R (2B6*6) genetic variants on the susceptibility of CYP2B6dH to inhibition by important clinical drugs, and predicted the effect on metabolism of the marker CYP2B6 substrate efavirenz in vivo.
Materials and Methods
Materials. 7-Methoxy-4-(trifluoromethyl) coumarin (7-MFC) and 7-hydroxy-4-trifluoromethylcoumarin were purchased from Invitrogen (Carlsbad, CA). NADPH, drug compounds, and most of the pyridine and imidazole inhibitors were bought from Sigma-Aldrich (St. Louis, MO). 5-Cyclohexylpentyl-β-d-maltoside (CYMAL-5) was from Anatrace (Maumee, OH). Recombinant NADPH P450 reductase and cytochrome b5 from rat liver were prepared as described previously (Harlow et al., 1997). Oligonucleotide primers for polymerase chain reaction were obtained from Sigma-Genosys (The Woodlands, TX). The molecular chaperone plasmid pGro7, which expresses GroES/EL (Nakajima et al., 1994), was obtained from TAKARA BIO (Shiba, Japan). The QuikChange Site-Directed Mutagenesis kit was obtained from Stratagene (La Jolla, CA). Ni-NTA affinity resin was purchased from QIA-GEN (Valencia, CA). All of the other chemicals were of the highest grade available and were obtained from standard commercial sources.
Mutagenesis, Expression, and Purification. To create K262R, CYP2B6dH was used as the template, and the forward and reverse primers were 5′-CCCAGCGCCCCCAGGGACCTCATCGAC-3′ and 5′-GTCGATGAGGTCCCTGGGGGCGCTGGG-3′, respectively. For Q172H/K262R, K262R was used as the template, and the forward and reverse primers were 5′-ACCTTCCTCTTCCATTCCATTACCGCC-3′ and 5′-GGCGGTAATGGAATGGAAGAGGAAGGT-3′, respectively. The resulting constructs were sequenced to verify the desired mutations and absence of unintended mutations (K262R and Q172H/K262R were analyzed at Protein Chemistry Laboratory, University of Texas Medical Branch, Galveston, TX and Retrogen, Inc., San Diego, CA, respectively). CYP2B6dH, K262R, and Q172H/K262R were coexpressed with GroES/EL in JM109 cells (Stratagene) as described previously (Kumar et al., 2007). The proteins were then extracted and purified by modifying the recently described procedure (Kumar et al., 2007). In brief, the cell extract was loaded onto Ni-NTA resin in the presence of the detergent CYMAL-5. Protein was eluted with 10 mM KPi, pH 7.4, containing 100 mM NaCl, 20% glycerol, 10 mM β-mercaptoethanol, 0.5 mM phenylmethylsulphonyl fluoride, and 40 mM histidine. CYMAL-5 was added to 4.8 mM, and the sample was subsequently loaded onto a CM-Sepharose column. After washing the CM-Sepharose column using 10 mM KPi buffer containing 0.2 mM DTT, 1 mM EDTA, 20% glycerol, and 100 mM NaCl, the protein was eluted using 500 mM NaCl in the above buffer. Eluted protein was dialyzed against 10 mM KPi buffer containing 10% glycerol and 1 mM EDTA with three changes. The P450 content was measured by reduced CO-difference spectra. Protein concentrations were determined using the Bradford protein assay kit (Bio-Rad, Hercules, CA).
Enzyme Inhibition. 7-MFC O-deethylation was measured in a final reaction volume of 100 μl as described earlier (Oezguen et al., 2008). In brief, the reaction mixture contained 150 μM 7-MFC in the standard reconstitution system (P450/NADPH P450 reductase/cytochrome b5, 1:4:2) at 5 pmol of P450 in 50 mM Hepes, pH 7.4, 15 mM MgCl2, and 2% MeOH. The reaction was performed at 37°C for 5 min using 1 mM NADPH. Nonlinear regression analysis was performed to fit the data using a four-parameter logistic function to derive the IC50 values for all the imidazole and pyridine derivatives. The Ki values were determined using the 7-MFC O-deethylation assay in a final reaction volume of 100 μl at 0.5 to 5.0 μM drug concentrations and 0 to 50 μM substrate concentrations. For all the inhibition studies, 10 pmol of P450 was used. The Ki was determined using global fit for competitive inhibition from SpectraLab (Davydov et al., 1995).
Spectral Binding. For binding studies, difference spectra were recorded using 1 μM P450 on a Shimadzu (Kyoto, Japan) 2401 PC spectrophotometer at 25°C as described earlier (Muralidhara et al., 2006). In brief, the difference in absorbance between the maxima and minima (ΔA) was recorded after the addition of a series of inhibitor concentrations in methanol to the sample cuvette and the same amount of methanol to the reference cuvette. The spectral dissociation constants (Ks) were obtained by fitting the data to the equation for “tight binding” 2ΔA = (ΔAmax/[E0]) [(KD + [I0] + [E0]) + (KD + [I0] + [E0])]2 – 4[E0][I0])1/2. All the data treatment and fitting of the inhibition and titration curves were performed with our SpectraLab software (Kumar et al., 2007).
Analysis of DDIs. The analysis for DDIs was carried out essentially as described by Kumar et al. (2006). In brief, the values for plasma concentrations (Cmax) of efavirenz as [S] and of drug inhibitors as [I] in a normal population were taken from the literature. The Ki values were from this investigation, and kcat and Km values for efavirenz hydroxylation were obtained from a previous study as follows: kcat = 4.3 and 7.9/min and Km = 14.3 and 15.9 μM for CYP2B6dH and K262R, respectively (Bumpus et al., 2006). These values for CYP2B6dH or K262R were fit to the equation for competitive inhibition to determine the predicted in vivo activity of the enzyme. The plasma concentrations of efavirenz and drug inhibitors are not known in the population harboring K262R; therefore, we have used the concentrations as reported in the normal population.
Results
Expression and Thermal Stability of CYP2B6dH and K262R. Heterologous expression of CYP2B6 and genetic variants such as M46V, G99E, K139E, Q172H, K262R, R140Q, and I391N in COS-1 cells yielded lower P450 expression, suggesting decreased P450 stability (Lang et al., 2004). Therefore, we investigated P450 expression of K262R (2B6*4) as described previously (Kumar et al., 2007). The expression of K262R in Escherichia coli under our standard conditions was ∼1.5-fold higher than CYP2B6dH. However, the thermal stability (Tm) of K262R was 2°C lower than CYP2B6dH (Supplemental Fig. 1). The results suggest no major difference in the expression or stability of K262R compared with the wild-type. In this study, we used the dH construct because it shows much higher bacterial expression and solubility and more facile purification than the full-length wild-type.
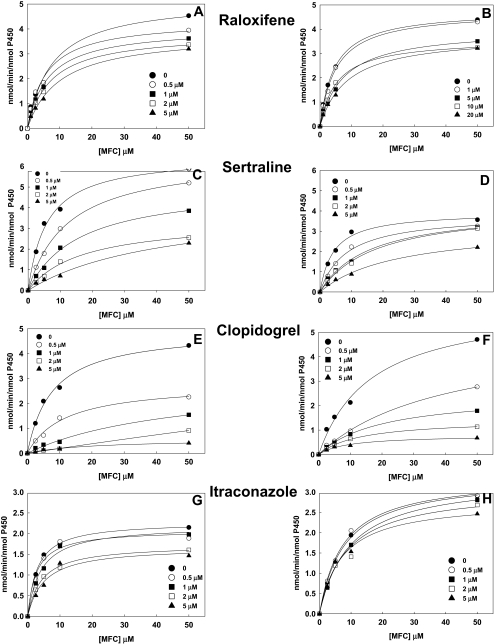
Inhibition and Spectral Binding of CYP2B6dH and K262R by Drugs. To investigate the effect of the Lys262→ Arg substitution on the interaction with the most potent drug inhibitors of CYP2B6 (Walsky et al., 2006), we selected clopidogrel, ticlopidine, clotrimazole, itraconazole, sertraline, and raloxifene (Supplemental Fig. 2). The results are presented in Fig. 1 and Table 1. The Ki values were determined for competitive inhibition of CYP2B6dH and K262R by the six drugs using 7-MFC at concentrations up to 50 μM and 0 to 20 μM inhibitor concentrations. The kcat and Km values for 7-MFC oxidation were 3.8/min and 5.1 μM for CYP2B6dH and 4.9/min and 5.0 μM for K262R, respectively. CYP2B6dH and K262R showed similar Ki values with clotrimazole and ticlopidine (Fig. 1; Table 1). However, K262R showed a >3-fold increase in the Ki values with clopidogrel, itraconazole, and raloxifene compared with CYP2B6dH. In addition, K262R showed ∼6-fold increase in Ki with sertraline compared with CYPB6dH.
Fig. 1.
Determination of Ki for inhibition of 7-MFC O-deethylation by CYP2B6dH and K262R in the presence of inhibitors (A–H). 7-MFC concentrations included in the assay were 2.5, 5, 10, and 50 μM, and the concentrations of the inhibitors used are provided in the plot. Global fitting of all the data from each experiment was used to obtain Ki. The fitting was done using SpectraLab as described under Materials and Methods.
TABLE 1.
Determination of Ki and Ks for CYP2B6dH and K262R with clinically important drugs
The inhibition was performed using a 7-MFC O-deethylation assay in a standard reconstitution system as described under Materials and Methods.
|
Drugs
|
Ki
|
Ks
|
ΔAmax
|
|||
|---|---|---|---|---|---|---|
| CYP2B6dH | K262R | CYP2B6dH | K262R | CYP2B6dH | K262R | |
| μM | μM | |||||
| Clopidogrel | 0.07, 0.12a | 0.36, 0.47 | 0.16 ± 0.11b | 0.33 ± 0.14 | 0.026 ± 0.002 | 0.030 ± 0.002 |
| Clotrimazole | 0.15, 0.11 | 0.11, 0.17 | N.D. | N.D. | N.D. | N.D. |
| Itraconazole | 1.42, 1.34, 1.14 | 4.40, 3.84, 4.34 | 0.07 ± 0.13 | 1.73 ± 0.44 | 0.022 ± 0.003 | 0.021 ± 0.002 |
| Raloxifene | 5.59, 2.60 | 15.8, 17.8 | N.D. | N.D. | N.D. | N.D. |
| Sertraline | 0.22, 0.38 | 1.70, 1.76 | 0.51 ± 0.16 | 2.06 ± 0.61 | 0.008 ± 0.0005 | 0.02 ± 0.002 |
| Ticlopidine | 0.11, 0.16 | 0.14, 0.12 | 0.28 ± 0.13 | 0.32 ± 0.11 | 0.023 ± 0.001 | 0.025 ± 0.001 |
N.D., not determined.
Ki values are shown from each independent determination.
Standard errors for fit to the tight ligand binding equation. The data are representative of at least two independent determinations. The variations between the experimentsare ≤20%.
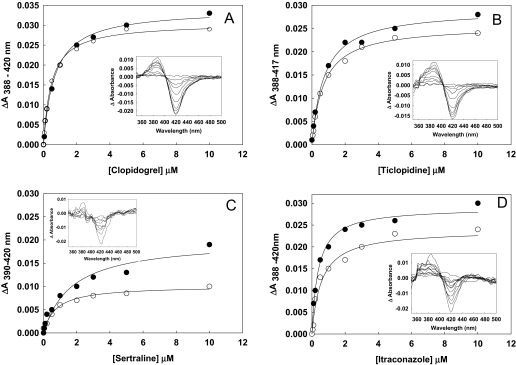
Clopidogrel, ticlopidine, itraconazole, and sertraline induced type I difference spectra with a peak at ∼388 nm and a trough at ∼420 nm (Fig. 2, inset). CYP2B6dH and K262R showed similar ΔAmax values with clopidogrel, itraconazole, and ticlopidine, whereas the ΔAmax with sertraline was 3-fold higher in K262R than the wild-type. Compared with CYP2B6dH, K262R showed approximately 2-, 4-, and 20-fold higher Ks values with clopidogrel, sertraline, and itraconazole, respectively (Fig. 2; Table 1). No significant changes were observed in the Ks values of ticlopidine between CYP2B6dH and K262R. However, the experiment could not be performed with raloxifene because it interfered with the measurement of the type I spectral change. At lower concentration of clotrimazole, the majority of CYP2B6dH and K262R P450 was converted into P420 (data not shown).
Fig. 2.
Representative type I difference spectra of ticlopidine, clopidogrel, sertraline, and itraconazole binding (A–D inset). The data were fit to the tight-binding equation, as described under Materials and Methods, to derive the Ks values as listed in Table 1.
Analysis of DDIs in CYP2B6dH and K262R. Clopidogrel, sertraline, and raloxifene are among the top 100 prescribed drugs in seniors (http://www.marylandspdap.com), who often use multiple drugs simultaneously, suggesting possible DDIs. Therefore, we analyzed possible DDIs between the established marker drug substrate of CYP2B6, efavirenz, and drug inhibitors of CYP2B6—clopidogrel, ticlopidine, clotrimazole, itraconazole, sertraline, and raloxifene. The analysis was carried out as described under Materials and Methods. The results predicted that whereas raloxifene would not alter efavirenz metabolism in vivo, clopidogrel or clotrimazole would almost completely abolish the metabolism of efavirenz (remaining activity = 2 and 6%, respectively) by CYP2B6 (Table 2). In addition, the metabolism of efavirenz would be reduced to 15, 48, and 57% in the presence of ticlopidine, sertraline, and itraconazole, respectively (Table 2). Although the Lys262→ Arg substitution would not alter the inhibition significantly in most cases, the substitution is predicted to reduce the metabolism of efavirenz to a lesser extent in the presence of itraconazole (20 versus 43% inhibition) and sertraline (16 versus 52% inhibition).
TABLE 2.
Potential in vivo DDIs between efavirenz with drug inhibitors
|
Inhibitors
|
Estimated in Vivo
Cmaxa
|
Predicted in Vivo
Activityb
|
|
|---|---|---|---|
| CYP2B6dH | K262R | ||
| /min | /min | ||
| NIL | 2.05 (100)c | 3.55 (100) | |
| Clopidogrel | 9.3 | 0.04 (1.9) | 0.27 (7.6) |
| Clotrimazole | 3.7 | 0.13 (6.3) | 0.23 (6.5) |
| Itraconazole | 1.9 | 1.16 (57) | 2.84 (80) |
| Raloxifene | 0.003 | 2.06 (100) | 3.55 (100) |
| Sertraline | 0.62 | 0.99 (48) | 3.00 (84) |
| Ticlopidine | 1.6 | 0.30 (15) | 0.47 (13) |
Cmax of the inhibitors and substrate were obtained from the following literature sources: www.mentalhealth.com/drug/p30-z02.html (sertraline); www.medscape.com/ (raloxifene); www.pharmgkb.org/ (ticlopidine, clopidogrel, and efavirenz); Burgess and Bodey (1972) (clotrimazole); Goodwin and Drew (2008) (itraconazole).
Predicted in vivo activity was determined using the equation for competitive inhibition. The values for [S] and [I] correspond to the estimated in vivo Cmax of efavirenz (13.0 μM) and drug inhibitor, respectively. The Ki values were taken from Table 1, whereas kcat and Km values were taken from a previous study (Bumpus et al., 2006). Predicted activities for K262R assume that both CYP2B6 alleles are the variant.
The values in parenthesis indicate the percentage activity.
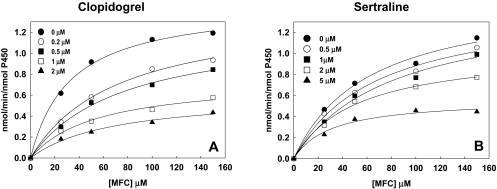
Inhibition of Q172H/K262R by Clopidogrel and Sertraline. Because of the low allele frequency of 2B6*4 and few if any homozygotes, we performed selected inhibition studies with the most common variant harboring K262R, namely, Q172H/K262R (2B6*6). This haplotype is found with high frequency (14–62%) as summarized by Zanger et al. (2007) and 21 to 60% (Rotger et al., 2007). The latter study reported 10% *6/*6 homozygotes. Clopidogrel and sertraline showed 4- and 6-fold higher Ki values, respectively, with K262R than CYP2B6dH; therefore, the drugs were tested with Q172H/K262R. The results are presented in Fig. 3. Q172H/K262R showed a 6-fold increase in the Ki value with clopidogrel (0.6 versus 0.1 μM) and >7-fold increase in the Ki value with sertraline (2.2 versus 0.3 μM) compared with CYP2B6dH. The results suggest that the binding affinity of Q172H/K262R with clopidogrel (Fig. 3A) and sertraline (Fig. 3B) was further reduced compared with K262R. For reference, the kcat and Km values for 7-MFC oxidation by Q172H/K262R were 1.3/min and 49 μM, which are >2-fold lower and ∼10-fold higher, respectively, than the corresponding values for CYP2B6dH or K262R.
Fig. 3.
Determination of Ki for inhibition of 7-MFC O-deethylation by Q172H/K262R in the presence of clopidogrel (A) or sertraline (B). 7-MFC concentrations included in the assay were 25, 50, 100, and 150 μM, and the concentrations of the inhibitors used are provided in the plot. Global fitting of all the data from each experiment was used to obtain Ki. Experiments were done in duplicate. The individual Ki values were clopidogrel (0.59, 0.63 μM) and sertraline (2.20, 2.16 μM).
Inhibition of CYP2B6dH and K262R by Pyridine and Imidazole Derivatives. We also investigated whether K262R shows altered inhibition by the known CYP2B4dH imidazole (Muralidhara et al., 2006) and CYP2B6 pyridine (Korhonen et al., 2007) type II inhibitors. Four compounds from each imidazole and pyridine groups were selected. Compared with CYP2B6dH, K262R showed 10-fold lower IC50 values with 4-(phenyl)pyridine and 2-fold lower IC50 values with 4-(4-nitrobenzyl)pyridine and 1-(4-phenyl)benzylimidazole (Supplemental Table 1). It is intriguing that with these two model type II inhibitors, the Lys262→ Arg substitution increased affinity unlike with the drugs.
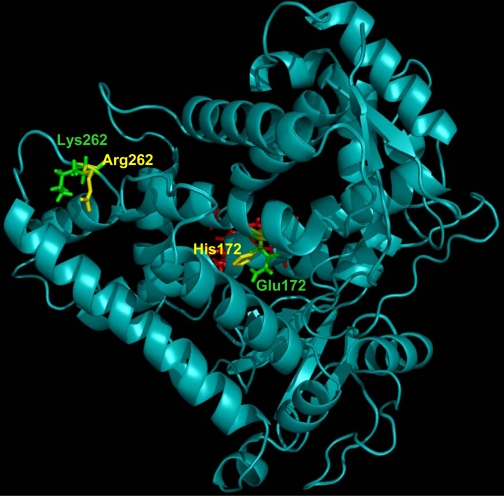
Molecular Modeling of CYP2B6dH. A CYP2B6dH homology model was generated using the 4-(4-chlorophenyl) imidazole-bound CYP2B4dH structure (1SUO) as a template by energy minimization using Insight II (Accelrys, San Diego, CA), and was represented using Pymol graphics (http://pymol.sourceforge.net/) (Fig. 4). Residues 172 and 262 are located in helix E and the G-H loop, respectively. It is interesting to note that CYP2B4 and CYP2B11 have a His residue at position 172, whereas CYP2B1, CYP2B4, and CYP2B11 enzymes have an Arg residue at 262, suggesting a possible specific role for Gln-172 or Lys-262 in CYP2B6. A molecular model of CYP2B6dH does not predict a role of residue 262 in substrate binding, except that the side chains of Lys and Arg have different orientations. Further in silico analysis suggests that Arg-262 interacts with His-252 (G-helix), Thr-255 (G-helix), and Asp-266 (H-helix) through H-bonds. These H-bonds are not found in Lys-262, suggesting that the additional interactions in K262R contribute to altered drug binding. We have recently shown the importance of H-bonds in CYP2B4dH among the nonactive site residues Glu-149, Asn-177, Arg-187, and Tyr-190 in substrate specificity, inhibitor selectivity, and protein stability (Oezguen et al., 2008).
Fig. 4.
Schematic representation of a three-dimensional homology model of CYP2B6dH. The heme is shown in red, whereas the wild-type Gln-172 and Lys-262 are in green and the variant His-172 and Arg-262 are in yellow.
Discussion
In this study the recombinant CYP2B6dH, K262R, and Q172H/K262R provided new insights into the interactions of the enzyme with clinically relevant drugs. First, by determining Ki values, we verified and extended previous findings of potent inhibition of CYP2B6 derived from IC50 values in liver microsomes or Sf9 cells. Second, the competitive nature of the inhibition shown in our experiments was substantiated by spectral binding assays with four of the drugs, which showed typical type I spectra. Third, decreased inhibitor potency of clopidogrel, itraconazole, raloxifene, and sertraline for K262R is in contrast to increased potency of three small type II inhibitors. Fourth, the role of the nonactive site residue at position 262 in CYP2B6 is consistent with our recent conserved sequence motif analysis of P450 family 2 enzymes. Finally, the lower inhibitor potency of sertraline and clopidogrel for K262R (CYP2B6*4) alone and especially in combination with Q172H (CYP2B6*6) suggests the real possibility of a diminished genotype-dependent drug interaction in vivo.
An earlier report showed that itraconazole yields a type II spectrum with CYP3A4 and can adopt multiple orientations within the CYP3A4 active site, including a catalytically productive mode (type I) and a slowly dissociating inhibitory mode (type II) (Pearson et al., 2006). However, a type I spectrum is observed with CYP2B6dH and K262R, suggesting that the orientation of itraconazole is such that the nitrogen of the ligand is not able to coordinate with the heme iron of the protein. The difference in the mode of binding of itraconazole in CYP3A4 and CYP2B6dH may contribute to the differences in their Ks values (0.019 versus 0.07 μM, respectively). A relatively large and flexible active site of CYP3A4 may facilitate the multiple modes of interactions with itraconazole.
Although molecular modeling suggests that Lys-262 in CYP2B6 does not contact ligands directly, this residue is found within a conserved sequence motif (CSM) in P450 family 2 enzymes (261PRDFIDVY268). This motif (CSM 11) is only present in the CYP2B and CYP2C subfamilies, where it has a very high rank order of conservation (Oezguen et al., 2008). In addition, analysis of the individual residues showed that Arg-262 is among the most conserved residues within CSM 11, further suggesting its functional and/or structural importance. It is interesting to note that Leu-264 is also among the most conserved residues within the motif (Oezguen et al., 2008), and a Leu264→ Phe substitution in CYP2B6dH enhances P450 expression and thermal stability (Kumar et al., 2007).
Because ticlopidine and clopidogrel have been shown to be mechanism-based inactivators of CYP2B6-catalyzed efavirenz hydroxylation (Richter et al., 2004; Walsky and Obach, 2007), an in vivo DDI between efavirenz and the inhibitors might be even more pronounced than indicated based on competitive inhibition alone. The area under plasma concentration ratio of hydroxybupropion to bupropion was reduced by 68 and 90% in the presence of clopidogrel and ticlopidine, respectively, compared with the control, which suggests that both clopidogrel and ticlopidine inhibit the activity of CYP2B6 significantly in vivo and in vitro (Turpeinen et al., 2005). We were surprised to find that despite the clear inhibition of CYP2B6 by clopidogrel, clotrimazole, itraconazole, sertraline, or ticlopidine, there is no medical documentation on combination therapy using these drugs that may lead to possible side effects. As an exception, there is a report on DDI between efavirenz and itraconazole in a patient with disseminated histoplasmosis and AIDS. The drug combination resulted in persistently elevated urinary histoplasma antigen levels and subtherapeutic plasma itraconazole concentrations (Koo et al., 2007). Our results and those of Walsky and Obach (2006) suggest that an extensive survey among patients who use CYP2B6-metabolized drugs in combination with the potent drug inhibitors is highly desirable. Furthermore, individuals, especially homozygotes, with the 2B6*4 or 2B6*6 allele might be less susceptible to drug interactions resulting from P450 inhibition.
Supplementary Material
Acknowledgments
We thank Dr. Surendra Negi from the University of Texas Medical Branch (UTMB) for providing the model of CYP2B6dH. We also thank Ling Sun (Pharmacology and Toxicology, UTMB) for creating K262R.
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants ES03619, ES06676].
Parts of this work were previously presented as follows: Talakad JC, Sun L, Kumar S, and Halpert JR (2008) Characterization of the P450 2B6 genetic variant K262R for temperature stability and drug binding. Experimental Biology-2008; 2008 Apr 2–9; San Diego, CA. American Society for Pharmacology and Experimental Therapeutics, Bethesda, MD.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.023655.
ABBREVIATIONS: P450, cytochrome P450; DDI, drug-drug interaction; 7-MFC, 7-methoxy-4-(trifluoromethyl)coumarin; CYMAL-5, 5-cyclohexylpentyl-β-d-maltoside; CSM, conserved sequence motif.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
References
- Bumpus NN, Kent UM, and Hollenberg PF (2006) Metabolism of efavirenz and 8-hydroxyefavirenz by P450 2B6 leads to inactivation by two distinct mechanisms. J Pharmacol Exp Ther 318 345–351. [DOI] [PubMed] [Google Scholar]
- Bumpus NN, Sridar C, Kent UM, and Hollenberg PF (2005) The naturally occurring cytochrome P450 (P450) 2B6 K262R mutant of P450 2B6 exhibits alterations in substrate metabolism and inactivation. Drug Metab Dispos 33 795–802. [DOI] [PubMed] [Google Scholar]
- Burgess MA and Bodey GP (1972) Clotrimazole (Bay b 5097): in vitro and clinical pharmacological studies. Antimicrob Agents Chemother 2 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambro MR and Kallgren MA (1988) Drug interactions in a clinic using COSTAR. Comput Biol Med 18 31–38. [DOI] [PubMed] [Google Scholar]
- Davydov DR, Deprez E, Hoa GH, Knyushko TV, Kuznetsova GP, Koen YM, and Archakov AI (1995) High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: evidence for conformational inhomogeneity in the oligomers. Arch Biochem Biophys 320 330–344. [DOI] [PubMed] [Google Scholar]
- Goodwin ML and Drew RH (2008) Antifungal serum concentration monitoring: an update. J Antimicrob Chemother 61 17–25. [DOI] [PubMed] [Google Scholar]
- Guengerich FP (2005) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd ed (PR Ortiz de Montellano ed) pp 377–530, Plenum Press, New York.
- Harlow GR, He YA, and Halpert JR (1997) Functional interaction between amino-acid residues 242 and 290 in cytochromes P-450 2B1 and 2B11. Biochem Biophys Acta 1338 259–266. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Klein C, Meineke I, Sasse J, Zanger UM, Mürdter TE, Roots I, and Brockmöller J (2003) Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13 619–626. [DOI] [PubMed] [Google Scholar]
- Koo HK, Hamill RJ, and Andrade RA (2007) Drug-drug interaction between itraconazole and efavirenz in a patient with AIDS and disseminated histoplasmosis. Clin Infect Dis 45 77–79. [DOI] [PubMed] [Google Scholar]
- Korhonen LE, Turpeinen M, Rahnasto M, Wittekindt C, Poso A, Pelkonen O, Raunio H, and Juvonen RO (2007) New potent and selective cytochrome P450 2B6 (CYP2B6) inhibitors based on three-dimensional quantitative structure-activity relationship (3D-QSAR) analysis. Br J Pharmacol 150 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Zhao Y, Sun L, Negi SS, Halpert JR, and Muralidhara BK (2007) Rational engineering of human cytochrome P450 2B6 for enhanced expression and stability: importance of a Leu264→ Phe substitution. Mol Pharmacol 72 1191–1199. [DOI] [PubMed] [Google Scholar]
- Kumar V, Wahlstrom JL, Rock DA, Warren CJ, Gorman LA, and Tracy TS (2006) CYP2C9 inhibition: impact of probe selection and pharmacogenetics on in vitro inhibition profiles. Drug Metab Dispos 34 1966–1975. [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, and Zanger UM (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11 399–415. [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Richter T, Zibat A, Kerb R, Eichelbaum M, Schwab M, and Zanger UM (2004) Multiple novel nonsynonymous CYP2B6 gene polymorphisms in Caucasians: demonstration of phenotypic null alleles. J Pharmacol Exp Ther 311 34–43. [DOI] [PubMed] [Google Scholar]
- Lewis DF, Lake BG, and Dickins M (2004) Substrates of human cytochromes P450 from families CYP1 and CYP2: analysis of enzyme selectivity and metabolism. Drug Metabol Drug Interact 20 111–142. [DOI] [PubMed] [Google Scholar]
- Muralidhara BK, Negi S, Chin CC, Braun W, and Halpert JR (2006) Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J Biol Chem 281 8051–8061. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Elovaara E, Gonzalez FJ, Gelboin HV, Raunio H, Pelkonen O, Vainio H, and Aoyama T (1994) Styrene metabolism by cDNA-expressed human hepatic and pulmonary cytochromes P450. Chem Res Toxicol 7 891–896. [DOI] [PubMed] [Google Scholar]
- Oezguen N, Kumar S, Hindupur A, Braun W, Muralidhara BK, and Halpert JR (2008) Identification and analysis of conserved sequence motifs in cytochrome P450 family 2: functional and structural role of a motif 187RFDYKD192 in CYP2B enzymes. J Biol Chem 283 21808–21816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JT, Hill JJ, Swank J, Isoherranen N, Kunze KL, and Atkins WM (2006) Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry 45 6341–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S (2002) Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev 34 83–448. [DOI] [PubMed] [Google Scholar]
- Richter T, Mürdter TE, Heinkele G, Pleiss J, Tatzel S, Schwab M, Eichelbaum M, and Zanger UM (2004) Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther 308 189–197. [DOI] [PubMed] [Google Scholar]
- Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Décosterd L, Blievernicht J, Saussele T, Günthard HF, Schwab M, et al. (2007) Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther 81 557–566. [DOI] [PubMed] [Google Scholar]
- Turpeinen M, Tolonen A, Uusitalo J, Jalonen J, Pelkonen O, and Laine K (2005) Effect of clopidogrel and ticlopidine on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Clin Pharmacol Ther 77 553–559. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Astuccio AV, and Obach RS (2006) Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J Clin Pharmacol 46 1426–1438. [DOI] [PubMed] [Google Scholar]
- Walsky RL and Obach RS (2007) A comparison of 2-phenyl-2-(1-piperidinyl)propane (ppp), 1,1′,1″-phosphinothioylidynetrisaziridine (thioTEPA), clopidogrel, and ticlopidine as selective inactivators of human cytochrome P450 2B6. Drug Metab Dispos 35 2053–2059. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, and Schwab M (2007) Polymorphic CYP2B6: molecular mechanism and emerging clinical significance. Pharmacogenomics 8 743–759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.