Abstract
We present results from a novel strategy that enables concurrent identification of protein-protein interactions and topologies in living cells without specific antibodies or genetic manipulations for immuno-/affinity purifications. The strategy consists of (i) a chemical cross-linking reaction: intact cell labeling with a novel class of chemical cross-linkers, protein interaction reporters (PIRs); (ii) two-stage mass spectrometric analysis: stage 1 identification of PIR-labeled proteins and construction of a restricted database by two-dimensional LC/MSMS and stage 2 analysis of PIR-labeled peptides by multiplexed LC/FTICR-MS; and (iii) data analysis: identification of cross-linked peptides and proteins of origin using accurate mass and other constraints. The primary advantage of the PIR approach and distinction from current technology is that protein interactions together with topologies are detected in native biological systems by stabilizing protein complexes with new covalent bonds while the proteins are present in the original cellular environment. Thus, weak or transient interactions or interactions that require properly folded, localized, or membrane-bound proteins can be labeled and identified through the PIR approach. This strategy was applied to Shewanella oneidensis bacterial cells, and initial studies resulted in identification of a set of protein-protein interactions and their contact/binding regions. Furthermore most identified interactions involved membrane proteins, suggesting that the PIR approach is particularly suited for studies of membrane protein-protein interactions, an area under-represented with current widely used approaches.
An essential component of the goal to elucidate global biological function is the determination of protein interaction networks. Current approaches for mapping protein-protein interactions include yeast two-hybrid system (1), affinity purification procedures based on immunoprecipitation (IP)1 or a single (e.g. FLAG tag (2)) or double affinity tag (e.g. TAP tag (3, 4)) followed by protein identification with mass spectrometry, protein microarray technology (5, 6), and computational prediction methods (7, 8). Although all these approaches demonstrate great promise in mapping protein-protein interactions on a proteome wide level, the resulting large scale data sets are often associated with high rates of false negatives and false positives (>50%), and poor overlap of data sets among different approaches used for the same system are often observed (9–11). Such observations suggest that no single method is flawless and comprehensive. The strengths and weaknesses of each method have been thoroughly reviewed (12–15). For example, traditional IP-based affinity purification methods require a specific antibody for every protein of interest that is a hindrance for widespread, large scale application. Tag-based methods overcome this limitation by fusing the bait protein genetically with an affinity tag that is applicable to all proteins. One of the most successful tag-based methods is TAP technology, which fuses two affinity tags to the bait protein, and nonspecific binding is significantly reduced with two sequential purification steps (3, 4). Although tag-based methods allow bait proteins to be expressed in vivo and interact with native physiological partners, recent studies showed that tagging can also cause overexpression of the bait protein that can result in association with chaperones and improper intercellular localization (16, 17). In addition, tagging one bait protein at a time for large scale studies can be tedious and costly. Another issue worth noting is that all affinity-based methods require cell lysis prior to purification of the associated complex of the bait protein. During cell lysis, the native cellular system is disturbed, and the bait protein is present in the lysis buffer, which is very different from the intracellular milieu. As described recently by Berggard et al. (13), the fact that the affinity between interacting proteins may be substantially different in vivo as compared with in vitro has not been carefully considered in the literature. We reported the first such comparison of mapping targeted protein interactions using both intact cells and cell lysates, and our results illustrated significantly different protein interaction data, highlighting the importance of identification of protein-protein interactions under native conditions (18).
Another challenge that affinity-based methods face is related to the inherent difficulty involved in maintaining the integrity of native protein complexes while removing the nonspecific bindings during washing steps. Most transient and weak protein-protein interactions may not survive through harsh washing steps; this is particularly true for interactions involving membrane proteins. For example, a high level of detergent normally required for maintaining the solubility of membrane proteins can also disturb non-covalent associations (15, 19). Chemical cross-linking can be used to stabilize and freeze protein-protein interactions by forming covalent bonds with proteins while proteins are present in the native cellular environment (15, 20, 21). The cross-linked protein complexes can remain intact during cell lysis and stringent washes. Therefore, cross-linking strategies have been successfully combined with affinity-based methods for studies in protein-protein interactions. In vivo cross-linking applications coupled with IP (22–27) and TAP tag (28, 29) techniques have been extensively reported and reviewed (15, 20, 21, 30–32).
Another important feature of chemical cross-linking methods is the potential for mapping topology of proteins and protein complexes (for reviews, see Refs. 15, 20, 21, and 30). If cross-linked residues/peptides can be identified, this information can yield clues about the contact/binding interfaces among protein complexes. Although in vivo cross-linking coupled with affinity purification can readily allow identification of interacting protein partners for a particular protein of interest with the detection of higher bands in gels or Western blot images, identification of cross-linked peptides/residues is not trivial even for purified protein complexes available in large quantity. Improved cross-linkers such as chemically cleavable cross-linkers (such as dithiobis(succinimidyl propionate)) (33), isotope-encoded cross-linkers (34), and cross-linkers with affinity tags (35, 36) have been used to aid in assignment of cross-linked residues in mapping topological structures of purified proteins and protein complexes. Factors that have slowed or inhibited the advancement of cross-linking strategies for large scale in vivo applications stem from the complex nature inherent with the cross-linking approach. Cross-linking reactions are often carried out with a large excessive amount of cross-linkers to increase reaction rate and product yield, thus resulting in highly complex mixtures. Proteolysis performed prior to MS analysis further increases the degree of complexity, and the desired cross-linked peptides are only a tiny fraction (<0.1%) of the total theoretical peptide combinations (37). In addition, interpretation of MSMS fragmentation spectra of the cross-linked peptides is challenging because simultaneous fragmentation of the cross-linker itself with one or more peptides can result in spectral complexity that precludes peptide/protein identification. The Aebersold group (37) recently reported breakthrough results illustrating identification of cross-linked peptides directly from complex Escherichia coli cell lysates with advanced informatics software development. Their efforts resulted in identification of three interactions from homo-oligomers and two interactions from hetero-oligomers, which suggests strongly growing interest in cross-linking methods for protein interaction identification and as yet untapped potential in this challenging research area.
To help overcome the limitations of current cross-linking methods, we have pursued a chemistry-based strategy by developing a novel class of cross-linkers called protein interaction reporters (PIRs). The spacer chain of a traditional cross-linker is modified in our PIR compounds with the incorporation of two specific mass spectrometry-cleavable bonds, a mass-encoded reporter tag, and an affinity tag (see Fig. 1a). Affinity-based purification is used to enrich PIR-labeled native protein complexes; however, no genetic manipulation is required as used in FLAG tag and TAP tag methods nor is any antibody required for each protein of interest because the affinity moiety is part of the PIR cross-linkers. This feature conceivably allows further extension of the PIR strategy to native biological systems other than cells, such as tissues and biological fluids. When combined with a novel two-stage mass spectrometric analysis strategy, the PIR approach allows large scale mapping of protein-protein interactions and their sites of interaction in native biological systems. Here we report the initial application of the PIR strategy to the microbial system Shewanella oneidensis MR-1 cells and the results of identification of a set of protein-protein interactions including information on contact/binding interfaces from living cells. These results represent the first such successful application of a chemical cross-linking strategy to map interactions and interaction sites from native living cells in a large scale, non-targeted way. The tunable chemistry of the PIR strategy will ultimately allow multiplexing an array of PIR cross-linkers with different structures and reporters for more comprehensive mapping of protein-protein interactions from native biological systems.
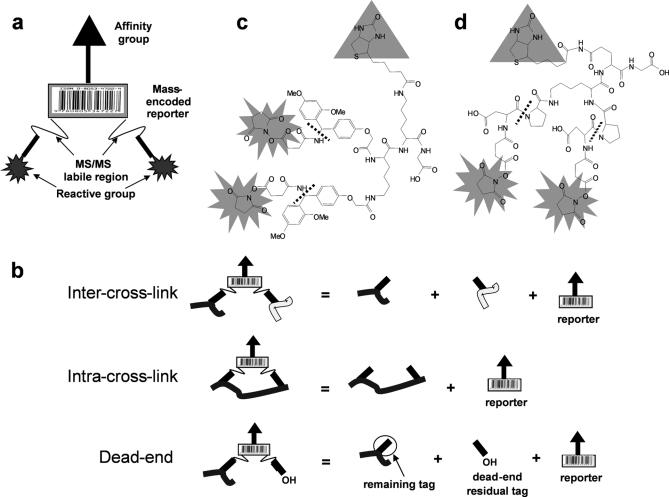
Fig. 1.
PIR structure. a, conceptual modular design of novel cross-linkers, PIRs. b, the specific fragmentation pattern of PIR-labeled peptide distinguishes dead-end, intra-, and intercross-linked peptides. The neutral mass of the precursor ion equals the sum of the neutral masses of its product ions. c, structure of a biotinylated Rink-based PIR. The singly and doubly charged reporter ions are 1122.5 and 561.7, respectively. d, structure of a biotinylated DP-based PIR. The MSMS labile bonds are indicated by the dashed lines, the reactive groups are NHS esters, and the affinity group is biotin. The singly charged reporter ion is 752.4.
EXPERIMENTAL PROCEDURES
Materials—
All chemicals were purchased from Sigma unless otherwise noted. Fmoc-protected amino acids and HMPB-MBHA resins used for PIR synthesis were purchased from Novabiochem. Water used for preparing buffers and solutions was 18-megaohm deionized water produced with a Barnstead Nanopure Water System.
PIR Synthesis—
The PIR cross-linkers were synthesized using a 431A peptide synthesizer (Applied Biosystems, Foster City, CA). Glycine was coupled to HMPB-MBHA resin, and then the biotin group was introduced in the form of N-α-Fmoc-N-ɛ-biotinyl-l-lysine. The second lysine in the form of N-α,ɛ-di-Fmoc-l-lysine was coupled to the biotinylated lysine and was then used as the branch point for the cross-linker. The Rink groups (for Rink-based PIR) or aspartic acid and proline (for DP-based PIR) were coupled to lysine in forms of Fmoc-amino acid. Carboxyl groups were then introduced by coupling the primary amines of the Rink groups or aspartic acid with succinyl anhydride. Subsequently the two carboxyl groups were activated by forming the esters with N-hydroxysuccinimide (NHS). The crude product was cleaved using either 0.5 or 1.0% TFA in chloroform and then neutralized with pyridine. The chloroform and TFA pyridine salts were removed under vacuum. The crude product was purified using reversed phase HPLC, and the final product had a purity of about 90%.
Two-stage Analysis—
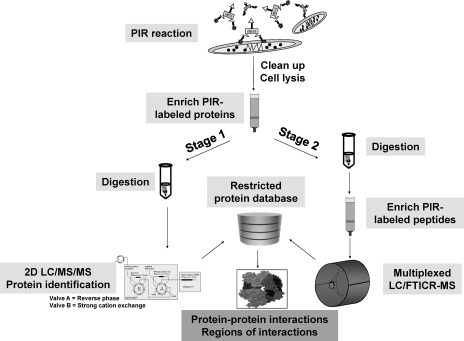
To fully utilize the advanced features of PIRs, a two-stage mass spectrometric approach was developed (see Fig. 2). Stage 1 analysis was carried out with avidin capture of in vivo labeled proteins followed by digestion and shotgun two-dimensional LC/MSMS for protein identification as described previously (38).
Fig. 2.
Diagram of two-stage mass spectrometric strategy. After in vivo labeling, protein extraction, and affinity capture of labeled proteins, enriched PIR-labeled proteins are divided into two parts. Stage 1 involves digestion and shotgun LC/MSMS for protein identification to constitute a restricted protein database. Stage 2 is performed by digesting PIR-labeled proteins first followed by another affinity enrichment of PIR-labeled peptides. Analysis of the labeled peptides is carried out with multiplexed LC/FTICR-MS. Accurate masses of the labeled peptides are measured and used for protein identification by searching against the restricted database compiled from stage 1. 2D, two-dimensional.
Stage 2 analysis was carried out by labeling 0.2-g cell pellets suspended in 4 ml of PBS buffer (150 mm sodium phosphate and 100 mm NaCl (pH 7.5)), and the cross-linkers were added to the suspended cell pellets to produce a final concentration of 1 mm. The cross-linking reaction was carried out at 4 °C for 1 h and quenched by 1 m ammonium bicarbonate. Extensive washing steps with PBS after cross-linking reactions were used to eliminate most nonspecific contamination as reported previously (38). Then cells were lysed in 2% Nonidet P-40, PBS solution by sonication for 2 min. The cell lysates were centrifuged at 15,000 × g at 4 °C for 45 min. The pellets were discarded, and supernatants were collected in four clean tubes with a total amount of ∼4 ml. Protein concentration of the supernatants was determined to be ∼5 mg/ml by a Bradford assay (Bio-Rad). The PIR-labeled proteins were enriched by adding a 50-μl slurry of monomeric avidin beads (Pierce) to each tube, and the reaction was incubated for 2 h with gentle mixing at room temperature. Then the avidin beads were washed three times with 500 μl of 100 mm NH4HCO3 (pH 7.8). After the last wash, the avidin beads were suspended in a 100-μl solution of 8 m urea and 100 mm NH4HCO3 (pH 7.8). Both enriched PIR-labeled proteins and avidin were reduced directly on the beads with 10 mm DTT at 56 °C for 1 h and alkylated with 20 mm iodoacetamide for 1 h at room temperature. After reduction and alkylation, additional 10 mm DTT was added to neutralize the unreacted iodoacetamide for another 1 h at room temperature. The bead suspension was then diluted 4 times with 100 mm NH4HCO3 (pH 7.8) buffer, and 10 μg of trypsin (Promega, Madison, WI) was added for overnight digestion at 37 °C. After digestion, 1 mm PMSF was added to quench the trypsin activity. A 50-μl slurry of monomeric avidin beads was added to the digestion mixture and gently mixed for 2 h at room temperature. After beads were washed three times with 100 mm NH4HCO3 (pH 7.8), PIR-labeled peptides were eluted from avidin beads with 200 μl of 70% acetonitrile and 0.5% TFA solution four times. Finally the volume of the combined eluent was reduced to ∼30 μl using a speed vacuum, and ∼10 μl was used for multiplexed LC/FTICR-MS analysis.
Multiplexed LC/FTICR-MS—
A Bruker Daltonics Apex-Q 7-tesla FTICR mass spectrometer (Billerica, MA) was used for multiplexed LC/FTICR-MS experiments. The nano-ESI source, nano-LC systems, reversed phase nanocolumn, and LC gradient methods were the same as those used with the ion trap mass spectrometer as described previously (39). Ions formed by the nano-ESI source entered the instrument through a glass capillary and then passed through a hexapole followed by a quadrupole and then a second hexapole, which was used for ion accumulation and PIR activation. After the second hexapole, ions were guided through a series of ion transfer optics and entered the ICR cell where ions were detected after excitation of cyclotron motion, resulting in accurate mass measurements. Xmass 7.0.6 and HyStar 3.0 software programs were used to acquire LC/FTICR-MS data. All data sets were acquired with 131,072 points, and each spectrum was resultant from averaging four scans. The optimized collision energy, −22 V, which efficiently fragmented the labile bonds in PIR molecules but not peptide bonds, was applied during alternating scans throughout the LC elution. External calibration of the instrument was performed to a mass accuracy of less than 1 ppm with direct infusion of BSA tryptic peptides immediately prior to the LC/FTICR-MS experiments.
Data Analysis—
FTICR-MS data were analyzed using the program ICR-2LS. The time domain ICR signals were apodized with a Welch function, zero-filled once, and then Fourier transformed before calibration. The spectrum was calibrated with six or more peaks to about 1 ppm or less, and then the calibration equation was applied to the instrument. All masses were reported in neutral mass forms in a .PEK text file that was further used by the program X-links (40). Lock mass calibration using reporter ions was performed to correct all the neutral masses produced in the .PEK file to further improve mass measurement accuracy (MMA). All possible tryptic peptides of the restricted protein database were produced by X-links with the constraint of at least one internal lysine in the sequence (40). 10-ppm error tolerance was used in X-links searches for both cross-link type identifications (using the relationship between the precursor ion in the MS scan and the released peptide and reporter ions in the subsequent multiplexed MSMS scan) and cross-linked peptide sequence identifications (using the measured accurate mass of the released peptide ions in the multiplexed MSMS scans). MS2 and MS3 validation experiments were performed using a quadrupole ion trap mass spectrometer (Esquire HCT, Bruker Daltonics) as described previously (39). GPMAW (General Protein/Mass Analysis for Windows; version 6.21) was used to calculate MSMS fragmentation. The whole genome sequence of S. oneidensis MR-1 was completed in 2001, and the database containing the entire genome (4,854 ORFs) was downloaded in July 2006 from The Institute for Genomic Research.
RESULTS
PIR Structure—
The PIR strategy is enabled by incorporating two labile bonds in the spacer chain of the cross-linker that can be cleaved with high specificity in the mass spectrometer resulting in release of a mass-encoded reporter ion (Fig. 1a). This allows the detection of the cross-linked peptide or peptide complex during a low energy precursor scan and then detection of the intact peptide masses released in the subsequent high energy scan with accurate mass and/or additional MSMS analysis. The conceptual mathematical relationships that exist between PIR-labeled precursors, released peptides, and reporters are illustrated in Fig. 1b, and these relationships can be used to facilitate differentiation of cross-link types and identification of cross-linked peptides with informatics software tools. The tunable chemistry of PIR structure allows cross-linking a variety of different proteins by incorporation of different labile bonds, affinity tags, and reactive groups. For the current study, we used a biotin group as the affinity tag, NHS esters as the reactive group, and the Rink group (Fig. 1c) and Asp-Pro (DP) bond (Fig. 1d) as low energy MSMS-cleavable bonds.
The relatively large size of PIR cross-linkers has been a concern for obtaining useful structural constraint information (21). However, our previous studies using the model RNase S noncovalent complex showed that the distance (14 Å) between two cross-linked residues was much smaller than the maximum length (43 Å) of PIR (39). Furthermore Rinner et al. (37) recently reported identification of the same intercross-link in RNase A using the smaller cross-linker disuccinimidyl suberate. These results suggest that in solution the PIR is constrained to give rise to a shorter distance between the two reactive groups than the fully extended length; this is in agreement with the simulated estimates of the realistic lengths of 32 commercial cross-linkers in solution reported by Green et al. (41). Finally it is also worth considering that within the cellular environment many potential labeling sites that would allow identification of interactions span a wide range of distances. Therefore, flexibility in the cross-linker structure that can allow labeling sites over a range of distances is most critical to enable identification of larger numbers of interactions. Once identified as an interaction that exists in cells, these complexes can be studied for more detailed structural information using a wide variety of conventional molecular biology, analytical techniques, and computational methods.
The NHS was used as the reactive group in PIR cross-linkers. The NHS is the most frequently used group in protein labeling reagents (21) such as iTRAQ (isobaric tags for relative and absolute quantitation) (42), one of the most widely used protein quantitation reagents, and chemical cross-linkers such as disuccinimidyl suberate, dithiobis(succinimidyl propionate), bis(sulfosuccinimidyl) suberate, etc. (Pierce). The reason for this is multifold. First, the most common targets for labeling proteins are the primary amine groups, which are present on the large majority of proteins due to the high occurrence of lysine side chain ɛ-amines. Second, NHS forms a stable amide linkage to the primary amines in proteins at physiological condition (pH 7.0–7.5), which is important for in vivo labeling. Finally the NHS ester reaction has very rapid reaction rates with primary amines (43). Our studies (38) and those of others (23, 25) showed that a 5-min reaction time was sufficient for completion of labeling. The fast reaction rate is critical for in vivo experiments because it can quickly “snapshot” or “freeze” the protein complexes with stable covalent bonds before living cells are extensively perturbed.
Two-stage Analysis—
To fully utilize the advanced features of the PIR strategy, a novel two-stage mass spectrometric approach was developed (Fig. 2). Intact cells were labeled with PIR compounds under near physiological conditions. After labeling, proteins were extracted, and PIR-labeled proteins were enriched by avidin capture. Then the enriched PIR-labeled proteins were divided into two parts for the subsequent stage 1 and stage 2 analyses. Stage 1 analysis was carried out with digestion of PIR-labeled proteins followed by protein identification with shotgun two-dimensional LC/MSMS (44). The goal of stage 1 analysis is to construct a candidate protein pool as complete as possible that provides a “lookup” table for stage 2 analyses. We previously used the Rink-based PIR compound as a general biotinylation reagent for labeling intact cells (38), and the ∼400 identified labeled proteins were used to constitute the restricted database for the current study. Stage 2 analysis was performed by further digestion of enriched PIR-labeled proteins on avidin beads followed by another affinity capture of PIR-labeled peptides. These peptides were then subjected to multiplexed LC/FTICR-MS experiments to measure both the intact PIR-labeled peptide masses in the low energy precursor scan and the masses of the intact peptides in the following PIR activation scan. Finally we used an informatics software tool, X-links (40), to identify cross-links in two steps: step 1 involved characterization of the precursor-product relationships and cross-link types based on the equations depicted in Fig. 1b; and step 2 involved assignment of cross-linked peptide sequences/proteins of origin with accurate peptide mass-based protein identification strategy.
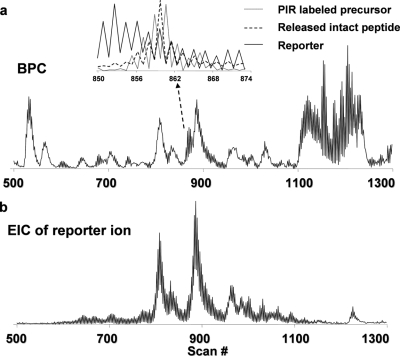
Fig. 3 shows an example data set from a multiplexed LC/FTICR-MS analysis of stage 2 sample, including the base peak chromatogram and the extracted ion chromatogram (EIC) for the expected reporter ion. Fig. 3a, inset, shows the overlaid EICs for an identified PIR-labeled precursor ion, the released intact peptide ion, and the reporter ion. The observed complementarities of the peak intensity patterns and the high degree of overlap of the LC elution profiles between these traces strongly suggest a precursor-product relationship. This information can supplement multiplexed LC/MS analysis to help define precursor-product relationships and cross-link types and can be used to further constrain our search results.
Fig. 3.
LC/FTICR-MS chromatograms of PIR-labeled peptides with alternating low energy (−4 V) and high energy (−22 V) applied in the collision cell. a, base peak chromatogram (BPC). b, EIC of reporter ion. The inset in a is the overlaid EICs of a PIR-labeled precursor ion (dotted line) and its released peptide ion (dashed line) and reporter ion (solid line).
Because MMA is the most important factor in identification of both cross-link types and cross-linked peptide sequences, we routinely calibrated the FTICR-MS instrument externally to ∼1-ppm MMA with BSA tryptic peptide mixture immediately prior to LC/FTICR-MS analysis. However, space charge effects and variation of ion populations (45) can severely reduce MMA for LC/FTICR-MS experiments. Introduction of a standard mass used as an internal lock mass calibrant via dual spray has been used to improve MMA for LC/FTICR-MS analysis (46). The PIR strategy provides an inherent lock mass, i.e. the known mass of reporter ion. Thus, we calibrated the LC/FTICR-MS data set using reporter ion as lock mass, and MMA for reporter ions was corrected to ∼3 ppm (supplemental Fig. 1). However, the observed MMA for detected peptide ions can be worse than the lock mass because our approach works with all detected ions irrespective of their detected S/N. Low S/N peaks are more prone to error because of the effect of noise on the extraction of the detected peak centroid. Thus, to be conservative, 10 ppm was chosen as the mass tolerance limit for all X-links searches in the current report. Future work will implement automatic gain control as used by the LTQ-FT instrument (47) to further improve MMA.
Identification of Relationships/Cross-link Types—
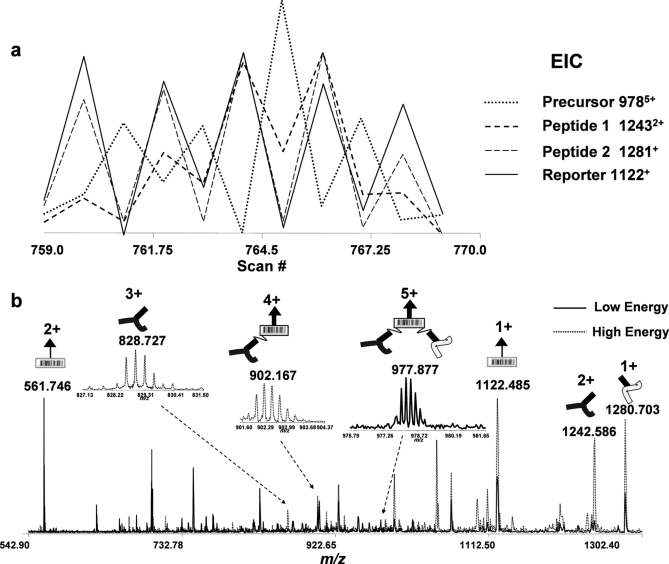
The description of the program X-links and the estimated false discovery rate are reported elsewhere (40). Briefly the program first searches the observed neutral masses for relationships between precursors and released peptide and reporter species to assign dead-end, intra-, and intercross-links using the equations defined in Fig. 1b. Second the PIR-labeled peptide sequences and proteins of origin are identified using the measured accurate peptide masses to search against the restricted protein database. Fig. 4 shows an example relationship identification resultant from X-links searches. Fig. 4a illustrates a plot of overlaid EICs of an intercross-linked precursor (m/z 9785+), released peptide 1 (m/z 12432+), released peptide 2 (m/z 1281+), and reporter (m/z 1122+) ions, respectively. The precursor ion was observed with an alternating intensity pattern opposite to that of the reporter and released peptide ions. Fig. 4b shows superimposed spectra extracted from a low energy precursor ion scan and subsequent high energy product ion scan with corresponding precursor and product ions labeled schematically. To confirm the putative X-links assignment, we repeatedly performed LC/MSMS with a quadrupole ion trap mass spectrometer, isolated the precursor ions (m/z 9785+), and fragmented them with CID. All intense fragment peaks observed in the MSMS spectrum were accounted for by the fragmentation of one or two PIR labile bonds (Fig. 5a), validating the relationship identified with X-links and further illustrating the high cleavage specificity of PIR bonds. More validation examples of relationship identifications are provided in supplemental Figs. 2, 3, and 4.
Fig. 4.
An example of intercross-link determination from multiplexed spectra based on the mathematical relationship (precursor = peptide 1 + peptide 2 + reporter). a, overlaid EICs of m/z 9785+ (precursor 9785+; dotted line), m/z 12432+ (peptide 1 12432+; dashed bold line), m/z 1281+ (peptide 2 1281+; dashed line) and m/z 1122+ (reporter 1122+; solid line) ions, respectively. b, overlaid FTICR mass spectra extracted from scan 765 (Low Energy; solid line) and scan 766 (High Energy; dotted line). All major products resultant from cleavage of one and two labile bonds in PIR are assigned and labeled schematically.
Fig. 5.
An example of validation of intercross-link determination and peptide sequence identification for the precursor ion with neutral mass of 4884.3531. a, isolation and MS2 of precursor ion (m/z 9785+). All peaks produced from cleavage of one and two PIR labile bonds are labeled schematically. b, isolation and MS3 of the released peptide ion with m/z 12432+ and 1280+, respectively.
Identification of Cross-linked Peptide Sequences/Proteins of Origin—
Peptide sequences and proteins of origin were identified using accurate peptide masses by searching against the restricted protein database. Proteins identified in the restricted database represent only that fraction of PIR-labeled proteins with highest abundance and PIR reactivity/accessibility because of the limited dynamic range of detection. However, it should be noted that stage 2 samples were similarly constrained, and thus the detected peptides were likely from the most abundant and PIR-reactive proteins present in the stage 1 database. Statistically stage 1 analysis expected a much greater probability of identifying proteins because any of the many tryptic peptides of a given protein can be used for identification. Stage 2 identification required analysis of the specific PIR-labeled peptides. Thus, although the stage 1 database was likely not comprehensive, it was much more so than the stage 2 data sets, and therefore most PIR-labeled peptides were expected to originate from proteins identified in stage 1 studies.
Table I shows the summary of identified intercross-linked peptides using Rink-based PIR. A selected list of dead-end labeled peptides and intracross-linked peptides is shown in supplemental Tables 1 and 2, respectively. It is worth noting that most identified proteins are membrane proteins; this is consistent with our previous reports that PIR molecules preferentially label membrane proteins (38). Our results also show that several identified proteins were labeled with multiple peptide sequences indicating their high abundance or accessibility. For example, SO1778 (MtrC), SO0404 (hypothetical protein), and SO1779 (OmcA) were identified by four, four, and seven different peptide sequences, respectively. On the other hand, the same peptide sequence can be labeled multiple times; this was observed previously in our efforts (39) and those of others (35, 36) to cross-link purified protein complexes. For example, FNPAKSELTYYLSNNFYDAK (The bold and underlined K was used both in the text and Table 1 to indicate the labeled lysine residue.), a peptide from SO0404, was detected in two different intercross-links and one dead-end labeled peptide, suggesting that this residue/region is highly exposed and accessible. In addition, several identified intercross-linked peptides in Table I were resultant from the same protein, e.g. OmcA, MtrC, SO0404, and SO4410 (glutamine synthetase, GlnA), which may be produced by intracross-link within the same protein sequence or intercross-link from subunits of homo-oligomers. It is worth noting that the current mass tolerance (10 ppm) used in this report is not sufficient to uniquely identify all the cross-linked peptides as demonstrated in the previous simulation study (40). Many PIR-labeled peptide masses resulted in more than one hit (supplemental Table 3). Further improvement of MMA or performing another MSMS scan on released peptides will help increase the ability to produce unambiguous protein interaction identification.
Table I.
A summary of intercross-linked peptides and proteins identified using Rink-based PIR
| Precursor mass | Peptide mass | Identified peptide | Error | Protein of origin | Typea | Molecular mass |
|---|---|---|---|---|---|---|
| Da | Da | ppm | kDa | |||
| 5047.3774 | 1911.9354 | TDNTYPGTKVPTSFAWK | 0.67 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 1815.8954 | GFALSNSKVSTYNEATK | 2.69 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 | |
| 4916.3157 | 2271.0398 | LMSEDQIKTTFADVAGCDEAK | 1.35 | SO1197, cell division protein (FtsH) | IM | 71 |
| 1325.7302 | NTPVEKGGIGQVK | 3.38 | SO2756, antioxidant | C | 22 | |
| 4906.2116 | 2641.1930 | ASFSGNSDIMVLEPDSEFFKYMK | 5.68 | SO0606, nf protein (HflC) | IM | 33 |
| 945.4501 | DKNTPSER | 1.57 | SO3343, conserved hypothetical protein | OM | 21 | |
| 4884.3531 | 2384.1255 | FNPAKSELTYYLSNNFYDAK | 2.92 | SO0404, hypothetical protein | OM | 130 |
| 1180.6630 | LVYTTTKDLK | 6.56 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 | |
| 4800.2140 | 2384.1324 | FNPAKSELTYYLSNNFYDAK | 3.61 | SO0404, hypothetical protein | OM | 130 |
| 1096.5665 | GFKGFSQAQK | 1.68 | SO1897, isodehydrogenase (Ivd) | C | 42 | |
| 4717.2362 | 2580.3263 | KFGTFTVAETGWPALDPLFIENK | 2.32 | SO4680, conserved hypothetical protein | C | 41 |
| 817.4073 | AWGKDNK | 1.17 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 | |
| 4429.1018 | 1815.8948 | GFALSNSKVSTYNEATK | 3.02 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 1293.6500 | GLKTDNTYPGTK | 5.01 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 | |
| 4353.0545 | 2222.1268 | SQINPKFQDLLTADVSFSGR | 2.85 | SO0404, hypothetical protein | OM | 130 |
| 811.3956 | FKSGDMK | 7.10 | SO0404, hypothetical protein | OM | 130 | |
| 4256.0728 | 2119.1187 | GKPHVAFSHLIHNVHNANK | 0.02 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 |
| 817.4082 | AWGKDNK | 1.90 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 | |
| 4228.0116 | 524.3454 | LHKK | 2.52 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 |
| 2384.1452 | FNPAKSELTYYLSNNFYDAK | 4.66 | SO0404, hypothetical protein | OM | 130 | |
| 3920.8081 | 1468.7459 | GYQWQAYINAKK | 0.27 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 1132.535 | EKVGETEADR | 0.99 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 | |
| 3793.8837 | 1293.6629 | GLKTDNTYPGTK | 5.21 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 1180.6690 | LVYTTTKDLK | 0.88 | SO1778, decaheme cytochrome c (MtrC) | OM | 71 | |
| 3744.775 | 1448.6644 | WNGTDTNSAAEKR | 1.81 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 976.5662 | AKFSNIGIK | 7.54 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 | |
| 3702.7157 | 1293.6484 | GLKTDNTYPGTK | 6.25 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 |
| 1089.5310 | EVTDEAGNKK | 0.80 | SO2001, 5-nucleotidase (UshA) | OM | 61 | |
| 3440.6819 | 729.3697 | KEYYK | 0.33 | SO3545, OmpA family protein | OM | 40 |
| 1391.7772 | YQQLVKTNAISK | 0.80 | SO4693, type I secretion membrane fusion protein | OM | 40 | |
| 3355.6241 | 1015.5170 | MENPEIKR | 4.94 | SO3064, amidophosphoribosyltransferase (PurF) | C | 56 |
| 1020.5345 | GLSAKAFDGR | 0.62 | SO0243, ribosomal protein L5 | C | 20 | |
| 3300.5526 | 848.4773 | MKSKLDK | 1.98 | SO1854, hypothetical protein | OM | 66 |
| 1132.5320 | EKVGETEADR | 3.29 | SO1779, decaheme cytochrome c (OmcA) | OM | 79 | |
| 3291.5894 | 982.5242 | SKEAANHVK | 4.78 | SO2366, response regulator | C | 59 |
| 989.5303 | EISMNIKR | 2.54 | SO0253, ribosomal protein S13 | C | 13 | |
| 3272.5790 | 937.5061 | IAGSMGFKK | 0.71 | SO0228, translation elongation factor G (FusA-1) | C | 77 |
| 1015.5167 | MENPEIKR | 4.65 | SO3064, amidophosphoribosyltransferase (PurF) | C | 56 | |
| 3246.6089 | 987.5094 | AEFAKTHGK | 4.43 | SO1521, iron-sulfur cluster-binding protein | C | 102 |
| 939.5683 | GKPVWVVR | 3.06 | SO0608, ubiquinol-cytochrome c | IM | 21 | |
| 3218.6457 | 850.4869 | FNTLTKK | 5.01 | SO4410, glutamine synthetase, type I (GlnA) | C | 52 |
| 1048.6404 | IPVVPSPKGR | 1.13 | SO4410, glutamine synthetase, type I (GlnA) | C | 52 | |
| 3179.4984 | 920.4166 | WADKMDR | 0.92 | SO0719, TonB-dependent receptor, putative | OM | 84 |
| 939.5683 | GKPVWVVR | 3.06 | SO0608, ubiquinol-cytochrome c | IM | 21 | |
| 3021.4109 | 811.3898 | FKSGDMK | 0.04 | SO0404, hypothetical protein | OM | 130 |
| 890.4986 | LGVFGKDR | 1.35 | SOA0110, lipoprotein, putative | OM | 150 | |
| 2194.0687 | 437.2634 | KYK | 0.91 | SO4211, preprotein translocase (SecA) | C | 103 |
| 437.2634 | YKK | 0.91 | SO1295, major outer membrane lipoprotein | OM | 9 |
Type of identified proteins: OM, IM, and C denote outer membrane, inner membrane, and cytoplasmic proteins, respectively.
All the intercross-linked peptide masses listed in Table I were also used to search against the unrestricted database with a 10-ppm constraint (supplemental Table 4). In agreement with the previous simulation, most peptide masses generated multiple protein hits at 10-ppm MMA. At this mass tolerance, a restricted database and/or MS3 analyses are required to further differentiate between multiple possibilities.
Although the accurate mass-based strategy for protein identification is highly reliable (40, 48), validation of a few examples was necessary for the initial application of PIR technology. This required MSMS of isolated PIR-labeled precursor and another isolation and fragmentation of the released peptide, thus MS3. Fig. 5b illustrates an example of MS3 validation of intercross-linked peptides found in X-links analysis and identified by accurate mass to be peptides from the proteins MtrC and SO0404. These data were obtained by isolating the intercross-linked precursor ion (m/z 9785+), activating the PIR labile bonds, releasing the peptides, isolating the intact peptide ions at m/z 12432+ and 1280+, respectively, and then fragmenting each. It is worth noting that two MS3 mass spectra in Fig. 5b did not generate comprehensive fragment ions with decent S/N because of the dominant neutral loss of the precursor ions and sensitivity limitations of MS3 analysis. To further validate the peptide sequence identification by accurate mass and MS3, a medium sized peptide, LVYTTTKDLK, with the expected tag on internal lysine was synthesized, and the singly charged precursor ion was subjected to tandem mass spectrometry (supplemental experimental material and supplemental Fig. 8). These experiments resulted in spectra that showed the same neutral loss and backbone fragmentation peaks (MH+ − H2O, y7, y4, and y4 − NH3) as were observed with the endogenous PIR-labeled peptide shown in Fig. 5b, supporting the fragment assignments. These results confirmed the accurate mass-based identification and MS3 validation analysis. Furthermore this approach with synthetic peptides represents an additional, general, and useful approach that can be used to help confirm all identified PIR-labeled proteins and peptides. Additional MS3 sequence validation examples are included in supplemental Figs. 2, 4, and 5. It should be noted that MS3 analysis is manually obtained during repeated LC separation of stage 2 samples, and thus both throughput and sensitivity of current validation experiments are low. Only those precursors with relatively high peak intensities in MS and MS2 scans can generate sufficient signals in MS3. On the other hand, these results illustrate the benefits of using high throughput multiplexed LC/MS and accurate mass-based protein identification because none of the identified products need ever be among the most abundant species to be identified by accurate mass so long as they are detected.
DISCUSSION
The primary advantage of PIR technology that distinguishes it from other available methods is the capability of concurrently mapping protein-protein interactions and their binding interfaces in native biological systems. Two essential components enable PIR applications at the systems level: novel PIR cross-linker structures and novel two-stage mass spectrometric analysis. The first application of Rink-based PIR with the use of S. oneidensis cells at proteome wide scale resulted in identification of over 30 different intercross-linked peptides; 24 of them with unambiguous identification are included in Table I. 17 peptide pairs were derived from interprotein cross-links, whereas seven were resultant from either intraprotein cross-links or intersubunit cross-links of homodimers. Importantly although our initial PIR applications do not show the hundreds to thousands of interactions one might detect with other leading methods, such as yeast two-hybrid and TAP tag technology, these results are distinguished by the fact that the obtained information is relevant to interactions detected in the native cellular environment and by the fact that the regions of close proximity of the interacting proteins can be determined. This constitutes information not readily achieved by any of the leading strategies. Furthermore it is important to consider that such chemistry-based strategies are eminently extendable to include an array of chemical structures that can label and identify a wider range of protein interactions in cells. For example, the initial application of the more hydrophilic DP-based PIR molecules generated more than 10 intercross-linked peptides, most of which were different from those identified with Rink-based PIR. An example of an identified intercross-link between ribosomal protein L10 (SO0222, RplJ) and ribosomal protein L7/L12 (SO0223, RplL) using DP-based PIR is shown in supplemental Fig. 6. PIR cross-linkers with different structures and different physical properties can be used to simultaneously label and identify interactions among many different classes of proteins. As a consequence, the PIR strategy is scalable to enable multiplexed application of an array of PIR compounds with variable physical properties, e.g. hydrophobic character, charges, reactivity, etc., and independent reporter masses to enable even larger numbers of protein interactions to be identified from cells in a single experiment.
Among the identified cross-linked interactions, most involved outer membrane proteins. These results suggest that the current Rink-based PIR is particularly suitable for studies of membrane protein interactions, which is challenging with currently available techniques. In addition, the PIR approach allowed us to recapitulate some previously known interactions. For example, both in vitro (49) and in vivo (18, 50) studies showed that outer membrane cytochromes OmcA and MtrC form a complex to facilitate electron transport in S. oneidensis. The identification of intercross-linked peptides (precursor mass 3793.8837 in Table I) between OmcA and MtrC further confirmed the direct interaction of these two proteins (supplemental Fig. 7) and provides insight into the regions of protein sequences that are in close proximity in cells. Furthermore outer membrane proteins SO0404, SO2001 (5-nucleotidase UshA), and SOA0110 (lipoprotein, putative) identified in the previous targeted OmcA-MtrC interaction network study using IP methods (18) were also identified in the present results in the intercross-links (two from MtrC-SO0404, one from OmcA-SO2001, and one from SO0404-SOA0110). In the previous report, the OmcA-MtrC interaction network was mapped from protein identification of a gel band that appeared above 250 kDa. This high molecular mass gel band was the result of cross-linking protein complexes of three or more components and did not directly identify connectivity among interacting proteins. Identification of intercross-linked residues/peptides produced direct evidence of pairwise protein-protein interactions and enables one to pinpoint contact regions of the interactions.
Taken together, our initial results using PIR technology on living cells of S. oneidensis have demonstrated the capability for probing protein-protein interactions and protein topology concurrently without the need of specific antibodies and genetic manipulations of the system. This strategy can be further used as an in vivo tool to investigate, snapshot, and compare potential binding sites of proteins and protein complexes in cells at different growth phases, culture conditions, cell cycles, external stresses, etc. Future work will include further improvement of the throughput, efficiency, and MMA for stage 2 analysis and application of a library of PIRs with different reporter ions and isotopic labeling that will ultimately allow large scale quantitative profiling of protein-protein interactions and protein topologies in living cells.
Footnotes
Published, MCP Papers in Press, October 20, 2008, DOI 10.1074/mcp.M800232-MCP200
The abbreviations used are: IP, immunoprecipitation; TAP, tandem affinity purification; PIR, protein interaction reporter; Fmoc, N-(9-fluorenyl)methoxycarbonyl; HMPB, 4-hydroxymethyl-3-methoxyphenoxybutyric acid; MBHA, 4-methylbenzhydrylamine hydrochloride; NHS, N-hydroxysuccinimide; MMA, mass measurement accuracy; EIC, extracted ion chromatogram; S/N, signal to noise.
This work was supported, in whole or in part, by National Institutes of Health Grants 1 R01 RR023334-01A1, 1S10RR017805-01, and 1S10RR022538-01 from the National Center for Research Resources. This work was also supported by Office of Science (Biological and Environmental Research), United States Department of Energy Grant DE-FG02-04ER63924.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Fields, S., and Song, O. ( 1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 2.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 3.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. ( 1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 4.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. ( 2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 5.Zhu, H., and Snyder, M. ( 2003) Protein chip technology. Curr. Opin. Chem. Biol. 7, 55–63 [DOI] [PubMed] [Google Scholar]
- 6.Ramachandran, N., Hainsworth, E., Bhullar, B., Eisenstein, S., Rosen, B., Lau, A. Y., Walter, J. C., and LaBaer, J. ( 2004) Self-assembling protein microarrays. Science 305, 86–90 [DOI] [PubMed] [Google Scholar]
- 7.Schachter, V. ( 2002) Bioinformatics of large-scale protein interaction networks. BioTechniques (suppl.) 16–18, 20–24, 26–27 [PubMed]
- 8.Aloy, P., and Russell, R. B. ( 2006) Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 7, 188–197 [DOI] [PubMed] [Google Scholar]
- 9.von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S., and Bork, P. ( 2002) Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 [DOI] [PubMed] [Google Scholar]
- 10.Lu, L., Arakaki, A. K., Lu, H., and Skolnick, J. ( 2003) Multimeric threading-based prediction of protein-protein interactions on a genomic scale: application to the Saccharomyces cerevisiae proteome. Genome Res. 13, 1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aloy, P., and Russell, R. B. ( 2004) Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 22, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 12.Ramachandran, N., Larson, D. N., Stark, P. R., Hainsworth, E., and LaBaer, J. ( 2005) Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 272, 5412–5425 [DOI] [PubMed] [Google Scholar]
- 13.Berggard, T., Linse, S., and James, P. ( 2007) Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833–2842 [DOI] [PubMed] [Google Scholar]
- 14.Kocher, T., and Superti-Furga, G. ( 2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 15.Gingras, A. C., Gstaiger, M., Raught, B., and Aebersold, R. ( 2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 16.Bouwmeester, T., Bauch, A., Ruffner, H., Angrand, P. O., Bergamini, G., Croughton, K., Cruciat, C., Eberhard, D., Gagneur, J., Ghidelli, S., Hopf, C., Huhse, B., Mangano, R., Michon, A. M., Schirle, M., Schlegl, J., Schwab, M., Stein, M. A., Bauer, A., Casari, G., Drewes, G., Gavin, A. C., Jackson, D. B., Joberty, G., Neubauer, G., Rick, J., Kuster, B., and Superti-Furga, G. ( 2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 17.Gingras, A. C., Aebersold, R., and Raught, B. ( 2005) Advances in protein complex analysis using mass spectrometry. J. Physiol. 563, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, H., Tang, X., Munske, G. R., Zakharova, N., Yang, L., Zheng, C., Wolff, M. A., Tolic, N., Anderson, G. A., Shi, L., Marshall, M. J., Fredrickson, J. K., and Bruce, J. E. ( 2008) In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical cross-linkers. J. Proteome Res. 7, 1712–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid, K. A., Hevi, S., Chen, Y., Le Caherec, F., and Chuck, S. L. ( 2002) A proteomic approach identifies proteins in hepatocytes that bind nascent apolipoprotein B. J. Biol. Chem. 277, 22010–22017 [DOI] [PubMed] [Google Scholar]
- 20.Back, J. W., de Jong, L., Muijsers, A. O., and de Koster, C. G. ( 2003) Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 331, 303–313 [DOI] [PubMed] [Google Scholar]
- 21.Sinz, A. ( 2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 22.Orlando, V., and Paro, R. ( 1993) Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75, 1187–1198 [DOI] [PubMed] [Google Scholar]
- 23.Orlando, V., Strutt, H., and Paro, R. ( 1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214 [DOI] [PubMed] [Google Scholar]
- 24.Schmitt-Ulms, G., Hansen, K., Liu, J., Cowdrey, C., Yang, J., DeArmond, S. J., Cohen, F. E., Prusiner, S. B., and Baldwin, M. A. ( 2004) Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat. Biotechnol. 22, 724–731 [DOI] [PubMed] [Google Scholar]
- 25.Vasilescu, J., Guo, X., and Kast, J. ( 2004) Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 26.Kim, S., Wong, P., and Coulombe, P. A. ( 2006) A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441, 362–365 [DOI] [PubMed] [Google Scholar]
- 27.Tardiff, D. F., Abruzzi, K. C., and Rosbash, M. ( 2007) Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl. Acad. Sci. U. S. A. 104, 19948–19953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerrero, C., Tagwerker, C., Kaiser, P., and Huang, L. ( 2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 29.Tagwerker, C., Flick, K., Cui, M., Guerrero, C., Dou, Y., Auer, B., Baldi, P., Huang, L., and Kaiser, P. ( 2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 30.Trakselis, M. A., Alley, S. C., and Ishmael, F. T. ( 2005) Identification and mapping of protein-protein interactions by a combination of cross-linking, cleavage, and proteomics. Bioconjug. Chem. 16, 741–750 [DOI] [PubMed] [Google Scholar]
- 31.Vasilescu, J., and Figeys, D. ( 2006) Mapping protein-protein interactions by mass spectrometry. Curr. Opin. Biotechnol. 17, 394–399 [DOI] [PubMed] [Google Scholar]
- 32.Novak, P., and Giannakopulos, A. E. ( 2007) Chemical cross-linking and mass spectrometry as structure determination tools. Eur. J. Mass Spectrom. (Chichester, Eng.) 13, 105–113 [DOI] [PubMed] [Google Scholar]
- 33.Bennett, K. L., Kussmann, M., Bjork, P., Godzwon, M., Mikkelsen, M., Sorensen, P., and Roepstorff, P. ( 2000) Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping—a novel approach to assess intermolecular protein contacts. Protein Sci. 9, 1503–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seebacher, J., Mallick, P., Zhang, N., Eddes, J. S., Aebersold, R., and Gelb, M. H. ( 2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 [DOI] [PubMed] [Google Scholar]
- 35.Trester-Zedlitz, M., Kamada, K., Burley, S. K., Fenyo, D., Chait, B. T., and Muir, T. W. ( 2003) A modular cross-linking approach for exploring protein interactions. J. Am. Chem. Soc. 125, 2416–2425 [DOI] [PubMed] [Google Scholar]
- 36.Sinz, A., Kalkhof, S., and Ihling, C. ( 2005) Mapping protein interfaces by a trifunctional cross-linker combined with MALDI-TOF and ESI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 1921–1931 [DOI] [PubMed] [Google Scholar]
- 37.Rinner, O., Seebacher, J., Walzthoeni, T., Mueller, L., Beck, M., Schmidt, A., Mueller, M., and Aebersold, R. ( 2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, X., Yi, W., Munske, G. R., Adhikari, D. P., Zakharova, N. L., and Bruce, J. E. ( 2007) Profiling the membrane proteome of Shewanella oneidensis MR-1 with new affinity labeling probes. J. Proteome Res. 6, 724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, X., Munske, G. R., Siems, W. F., and Bruce, J. E. ( 2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 40.Anderson, G. A., Tolic, N., Tang, X., Zheng, C., and Bruce, J. E. ( 2007) Informatics strategies for large-scale novel cross-linking analysis. J. Proteome Res. 6, 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green, N. S., Reisler, E., and Houk, K. N. ( 2001) Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 10, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. ( 2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 43.Lomant, A. J., and Fairbanks, G. ( 1976) Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J. Mol. Biol. 104, 243–261 [DOI] [PubMed] [Google Scholar]
- 44.Washburn, M. P., Wolters, D., and Yates, J. R., III ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 45.Ledford, E. B., Jr., Rempel, D. L., and Gross, M. L. ( 1984) Space charge effects in Fourier transform mass spectrometry. Mass calibration. Anal. Chem. 56, 2744–2748 [DOI] [PubMed] [Google Scholar]
- 46.Nepomuceno, A. I., Muddiman, D. C., Bergen, H. R., III, Craighead, J. R., Burke, M. J., Caskey, P. E., and Allan, J. A. ( 2003) Dual electrospray ionization source for confident generation of accurate mass tags using liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 75, 3411–3418 [DOI] [PubMed] [Google Scholar]
- 47.Peterman, S. M., Dufresne, C. P., and Horning, S. ( 2005) The use of a hybrid linear trap/FT-ICR mass spectrometer for on-line high resolution/high mass accuracy bottom-up sequencing. J. Biomol. Tech. 16, 112–124 [PMC free article] [PubMed] [Google Scholar]
- 48.Conrads, T. P., Anderson, G. A., Veenstra, T. D., Pasa-Tolic, L., and Smith, R. D. ( 2000) Utility of accurate mass tags for proteome-wide protein identification. Anal. Chem. 72, 3349–3354 [DOI] [PubMed] [Google Scholar]
- 49.Shi, L., Chen, B., Wang, Z., Elias, D. A., Mayer, M. U., Gorby, Y. A., Ni, S., Lower, B. H., Kennedy, D. W., Wunschel, D. S., Mottaz, H. M., Marshall, M. J., Hill, E. A., Beliaev, A. S., Zachara, J. M., Fredrickson, J. K., and Squier, T. C. ( 2006) Isolation of high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross, D. E., Ruebush, S. S., Brantley, S. L., Hartshorne, R. S., Clarke, T. A., Richardson, D. J., and Tien, M. ( 2007) Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73, 5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]